Abstract
The Human Brain Project consortium continues to struggle with effective sharing of tools. To facilitate reuse of its tools, the Stanford Psychiatry Neuroimaging Laboratory (SPNL) has developed BrainImageJ, a new software framework in Java. The framework consists of two components—a set of four programming interfaces and an application front end. The four interfaces define extension pathways for new data models, file loaders and savers, algorithms, and visualization tools. Any Java class that implements one of these interfaces qualifies as a BrainImageJ plug-in—a self-contained tool. After automatically detecting and incorporating new plug-ins, the application front end transparently generates graphical user interfaces that provide access to plug-in functionality. New plug-ins interoperate with existing ones immediately through the front end. BrainImageJ is used at the Stanford Psychiatry Neuroimaging Laboratory to develop image-analysis algorithms and three-dimensional visualization tools. It is the goal of our development group that, once the framework is placed in the public domain, it will serve as an interlaboratory platform for designing, distributing, and using interoperable tools.
In the last decade, neuroscience researchers have begun to exploit computer-based infrastructures and tools to produce, maintain, and share a growing body of knowledge about human brain structure and function. Limited interoperability of computer-based tools poses a risk for the continued growth of knowledge about the human brain and for effective use of that knowledge by the medical research and clinical communities. In its program announcement, the Human Brain Project (HBP) of the National Institutes of Health acknowledged the difficulty of integrating diverse scientific data and encouraged the development of frameworks for integrating and synthesizing data from different sources. The HBP sought to promote effective use of neuroscience resources by requiring its participants to concentrate on approaches and technologies that are generalizable, extensible, and interoperable.1
In this paper we present BrainImageJ, a Java-based framework that supports the goals outd by the HBP. BrainImageJ facilitates efficient creation of computational tools for neuroscience and immediate sharing of those tools across different operating systems. Features of the framework help developers in the Stanford Psychiatry Neuroimaging Laboratory (SPNL) prototype, refine, and distribute neuroimaging tools that are modular, cross-platform, and easily shared. In this paper we present and discuss the design considerations, architecture, and implementation of BrainImageJ. In addition, we demonstrate its scope with respect to a specific application in the SPNL—that is, expert-based segmentation of cortical gyri on three-dimensional reconstructions of the cortex.
Background
Sharing of knowledge and tools is an ongoing goal of informatics research,2 and academic and industry communities have worked toward this objective in several areas. Researchers in knowledge-based systems have constructed controlled vocabularies and thesauri3 that allow software systems to incorporate terminologies from multiple sources into heterogeneous databases and to integrate diverse databases.4,5 In addition, special interest groups have contributed standards for archiving and communicating textual and visual medical information.6,7 Finally, distribution of software packages in the public domain has made it possible for research laboratories to benefit from tools developed in other laboratories.8–10
An important, emerging area of informatics research is concerned with the development of unifying data models.11,12 On a conceptual level, these models aid in the integration and synthesis of data from different sources and serve as a mechanism for precise communication of specific theories between scientists.1 When formally expressed in a programming language, they become computational data models, which serve as the foundations for software tools that communicate in a model-specific manner. Public computational data models have the potential to accelerate interoperability efforts by providing a common language for research groups to create and share tools that work together. The ongoing development and refinement of computational data models is an important mechanism for furthering interoperability.13
In spite of these efforts toward sharing knowledge and tools, interoperability in the research community is still limited by various factors. One is the existence of data in multiple formats, which complicates the sharing of files between different laboratories. Data-format standards that have been developed, such as DICOM for picture archiving and communication7 and Health Level 7 for communication between hospital information systems,14 are targeted at interoperability in clinical and industry settings. These standards tend to be overly cumbersome for use in research groups. Because fully supporting a comprehensive standard is a significant task,15 researchers often prefer simpler formats for laboratory-specific communication and archival and tool development. Furthermore, the emergence of new data models will generate heterogeneous data sets in domains that have no standard formats.
Another barrier to interoperability is the platform-specific nature of many existing computational frameworks. Even though public-domain software packages are excellent examples of tool sharing, their applicability is limited by the set of supported platforms (operating systems)—only laboratories that possess the necessary platform can benefit. Supporting multiple platforms is a re-engineering or porting task that is typically beyond the scope of the original package developers, particularly if they work in small research laboratories. As a result, the most popular public-domain tool kits tend to be commercial,16,17 and the tool kits of research laboratories tend to be shared in smaller circles. Allocation of resources to supporting multiple platforms slows both the creation of new research tools and the evolution of existing ones.
Even when a package can be distributed to another laboratory, successful sharing often requires customization. The cost of tailoring a tool kit to fit the specific needs of a laboratory is another factor that inhibits the interoperability of public-domain software. However, this problem can be minimized through the application of modern software engineering principles to the design of the package. Object-oriented programming enables easy incorporation of independent modules into existing frameworks and supports a development model in which functionality is added incrementally as systems are used and tested.18 On a higher level of abstraction, software design patterns document solutions to recurring problems, to facilitate development of modular software that is easy to maintain and extend.19
The Java programming language is an attractive solution to problems related to supporting multiple platforms and enabling easy customization on those platforms. The Java Virtual Machine (JVM) is a platform-specific program that interprets platform-independent Java software and allows Java code to run on different platforms without being recompiled. The number of platforms with a JVM implementation continues to grow,20 and testing in the SPNL has demonstrated smooth portability across Windows, Linux, and Solaris operating systems. Java supports rapid development and easy customization of software with features such as automatic memory management, object-oriented syntax, strong type-checking, and a protected runtime environment.21 Performance is an important issue, because computationally intensive portions of Java code often execute several times slower than natively compiled code. However, just-in-time compilers, which are present in many modern JVMs, have the potential to almost completely eliminate this performance hit. These compilers automatically translate interpreted Java programs into native instructions at runtime, without compromising portability.22,23
We now describe the influence of interoperability issues on the design considerations for our system, and the role of modern software engineering techniques in its implementation.
System Design
Design Considerations
Two interrelated goals directed the design of BrainImageJ. Our first goal was to develop a framework that would stream the evolution and maintenance of neuroimaging tools in the SPNL. Second, we aimed to facilitate interoperability of tools and files through public computational data models.
The first goal arose out of the user requirements of two target groups in the SPNL—end users of tools for analyzing neuroimaging data, and developers of those tools. Each group presented different functional requirements that shaped the final architecture of the framework. End users required an extensible paradigm that would allow support for new data formats, data-processing algorithms, and visualization tools to be easily added in the form of plug-ins, or self-contained modules. They also emphasized the importance of a sophisticated application front end for efficient interaction with plug-in tools. Modern application features were expected, including an intuitive graphical user interface (GUI), multi-step undo/redo options, and multi-threaded processing.
In contrast, developers wanted to avoid the implementation of these modern application features, preferring to focus on the research-oriented functionality of new tools. As a result, they requested that the framework provide transparent support for application-level features. In addition, they required support for rapid prototyping and incremental testing, which would allow them to maintain a short development cycle. Both end users and developers required portability of the framework, so that it could be used on the various computer platforms in the SPNL. These platforms included Windows, Linux, MacOS, and Unix. Thus, the requirements of end users and developers dictated much of the structure and functionality of the framework.
The second set of goals emerged as we attempted to generalize the applicability of the framework. To accelerate interoperability in new research domains, we sought to foster the ongoing development of computational data models by requiring seamless integration of new data models through plug-ins. To facilitate the ongoing development of tools in each domain, we also required instant interoperability of file loaders, algorithms, and visualization tools that would be developed for a specific data model.
Architecture and Implementation
The BrainImageJ framework consists of two major components. The first component comprises four extension pathways, which are sets of programming interfaces. These interfaces guide the development of plug-in extensions in the four areas identified in the design considerations—data models, file loaders and savers, algorithms, and visualization tools. The second component of the framework is an application front end. The front end detects plug-ins, dynamically builds user interfaces to activate their functionality, and transparently enables the modern application services requested by end users (Figure 1▶). In this section, we detail the four extension pathways and elaborate on their relationship to the application front end.
Figure 1.
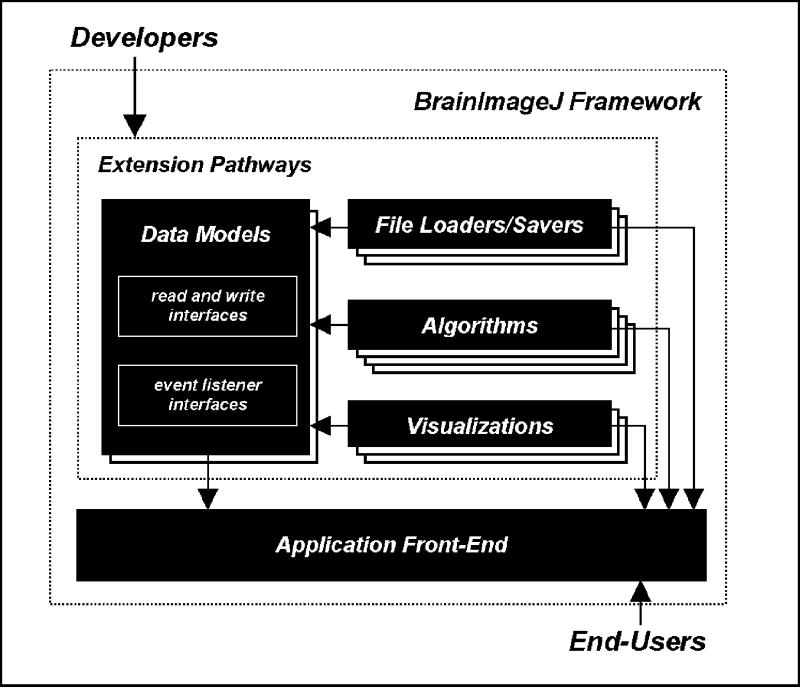
The BrainImageJ architecture, and its relationship to developers and end-users. Plug-ins in the file loader and saver, algorithm and visualization extension pathways communicate in terms of a common data-model plug-in. The front end automatically incorporates new plug-ins, and provides them with transparent support for modern application features.
Data Models
The data-model extension pathway allows an arbitrary number of computational data models to be developed and plugged into the framework. A data-model plug-in provides a unifying language for communication between collections of interoperable file loaders, algorithms, and visualization tools. The characteristics of a data-model plug-in are captured in two sets of programming interfaces—its read and write interfaces, which define the properties of the model that can be accessed and modified, and its event-listener interfaces, which define objects that the framework notifies when data is modified. We will illustrate the properties of these interfaces with DataCells3D—a data-model plug-in that represents volumetric image data.
The read interface for a data-model plug-in is an abstract class with methods for accessing its current state; the write interface extends the read interface with additional methods for modifying the current state. For example, the read interface for a DataCells3D includes a method for obtaining the intensities of voxels, and the write interface allows the intensities to be changed. This separation of read and write capabilities allows the application front end to transparently optimize processing operations. The front end uses a well-known protocol for concurrent reader-writer access, which guarantees the safety of the data while maximizing performance24: processes that only read data are granted simultaneous, multi-threaded access to the data, but processes that make modifications are executed in sequence.
The event-listener interfaces employ the Observer design pattern,19 so that the framework can synchronize the display of visualization tools with the current state of the data. Distinctions between different kinds of data-change events allow the system to provide a specific response to each type of change. For example, the DataCells3D event-listener contains different call-back methods for changes in voxel intensity and changes in the dimensions of the voxel grid. The front end accepts responsibility for broadcasting these changes to visualization tools.
File Loaders and Savers
The file loader and saver extension pathway allow BrainImageJ to accommodate plug-ins for loading and saving multiple data formats. Load and save plug-ins convert data from a file format into an instance of a particular data-model. For example, the DataCells3D TIFF Loader plug-in constructs a DataCells3D object from a TIFF-formatted file. For laboratories that agree on a common data model, file interoperability requires only that each laboratory provide a plug-in to load their preferred format.
To simplify the introduction of new file formats, the BrainImageJ front end transparently supports application services, such as obtaining configuration parameters, monitoring progress and canceling execution. Configuration parameters are often needed before a loader or saver can operate. For example, a plug-in that loads raw-image data needs to be told the dimensions of the volume, and a plug-in that saves a DataCells3D in DICOM format would need to know whether to compress the stored data. These parameters could be obtained in different ways—from the user through dialog boxes or from a script file. Plug-ins simply declare configuration parameters with a name, description, and data type. The DICOM-format saver declares a parameter with the following name, description, and data type—Compressed, Save the file compressed, boolean. Using these parameter descriptions the front end dynamically generates dialog boxes that solicit parameter values from the user or reads the values from a script.
The ability to monitor and cancel time-consuming operations is necessary to maintain a responsive user interface. Unfortunately, implementing these features in a multithreaded environment is difficult and error-prone, because of implementation complexities concerned with efficient monitoring, concurrent data access, and avoiding thread deadlock.25 Consequently, BrainImageJ contains code support in its base classes to simplify the implementation of plug-ins that pause, resume, or terminate execution in response to user intervention and provide visual feedback about the percentage completion of their tasks. Furthermore, the front end assumes all responsibility for creating and managing threads that parallelize computation. Thus, the framework provides the efficiency and responsiveness of a multithreaded environment with minimal burden on plug-in developers.
Algorithms
The algorithm extension pathway allows a diverse set of analysis and modification algorithms to be developed as plug-ins. BrainImageJ integrates these plug-ins seamlessly into the application GUI, providing the same underlying support for configuration, progress monitoring, and cancellation as for loaders and savers.
BrainImageJ groups algorithm plug-ins into algorithm families, which are abstract classes defined by the types of input and output data models. For example, the DataCells3D Modification family defines plug-ins that accept a single DataCells3D object for modification. Plug-ins in this family include image filters and segmentation algorithms. The DataCells3D Comparison family accepts two DataCells3D models for cross-analysis. Developers add new algorithm families by defining an abstract class with new input and output pairs. Algorithm plug-ins are concrete subclasses of algorithm families and provide implementations that operate on data inputs.
The algorithm extension pathway packages units of processing as individual objects in an expression of the Command design pattern.19 The BrainImageJ front end exploits the self-contained packaging of each algorithm plug-in to provide three services. First, it detects algorithm plug-ins, organizes them in menus, and executes algorithms that are selected from a menu. Second, it parallelizes execution of multiple algorithms, subject to the concurrent reader-writer protocols described in the Data Models section. Finally, the front end logs all algorithms that are applied and provides a complete undo/redo history for each data model.
To provide a user-friendly application GUI and increase user efficiency, the front end menus are context-sensitive. For example, the menu item for each algorithm is enabled and disabled to reflect its applicability with respect to the currently selected data model. To provide this functionality the front end tracks the selected data set by monitoring various GUI events, such as the selection of windows by the user. If the data type of the selected data set does not match the data type of an algorithm's input parameter, that algorithm's menu item is disabled. The file save, close, undo and redo menus are also context-sensitive, synchronized to operate on the selected data set.
Visualization
The visualization extension pathway allows users to develop custom GUI components and incorporate them into the application front end. Although the framework eases the development of simple plug-ins, it does not exclude the possibility of developing arbitrarily sophisticated visualizations. Java supports graphics that work consistently across different machine architectures, with its cross-platform GUI components (AWT and Swing libraries)26 and low-level graphics (Java 2D and the optional Java 3D packages).27,28 Figures 2 and 3▶▶ show SPNL visualization tools that were developed with these tool kits.
Figure 2.
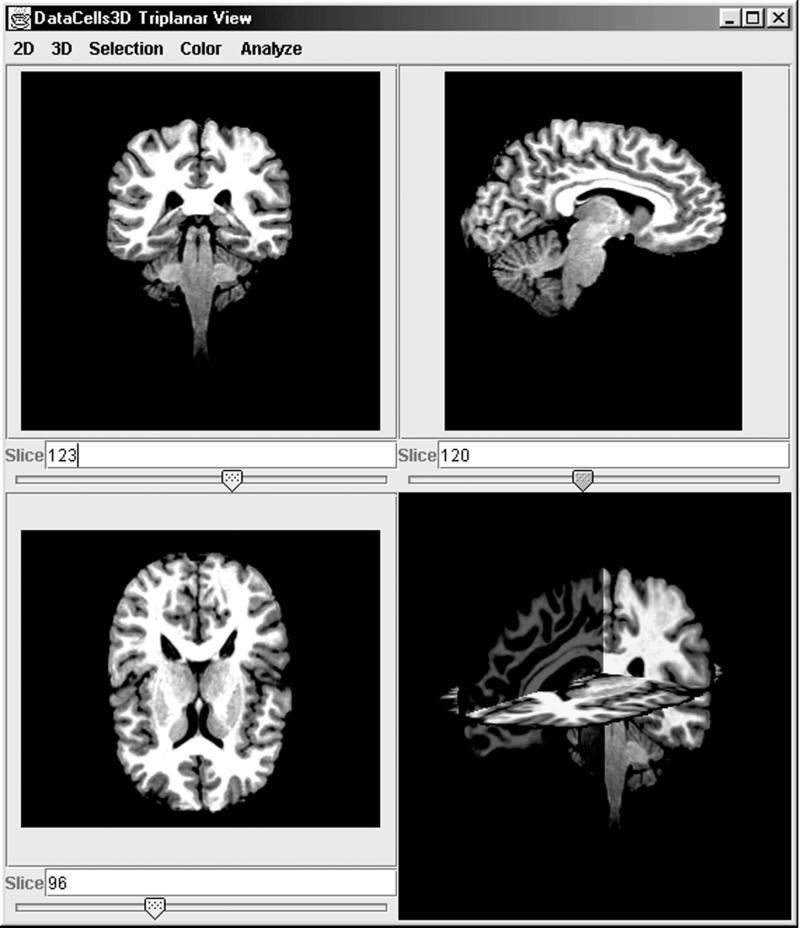
A visualization plug-in that displays axial slices of a DataCells3D model. The bottom right pane shows an interactive three-dimensional composition of the three slices.
Figure 3.
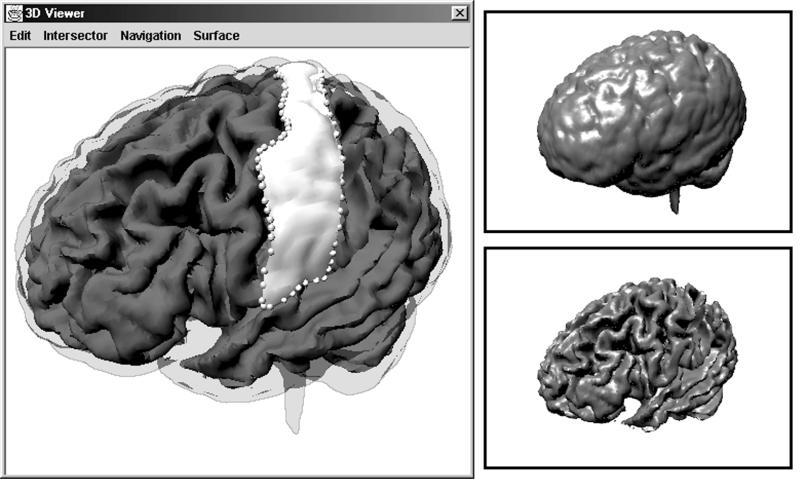
Surface Editor, a visualization plug-in that renders Surface objects. The inner and outer cortical surfaces (top right and bottom right) extracted with the Isosurface Extraction Algorithm. A composite, interactive view (left), which also depicts magnified surface points corresponding to mouse clicks (spheres), and a gyrus segmented from the outer cortical surface (opaque white).
BrainImageJ eases the task of creating visualization tools by providing services for automated GUI management. Instead of requiring visualization plug-ins to generate their own menus and toolbars, the visualization extension pathway allows plug-ins to package their executable commands in hierarchic sets. The front end accepts these hierarchic sets and dynamically organizes them in menus and toolbars. This paradigm allows the front end to standardize the appearance of visualization plug-ins by providing the user with global GUI customization options. Another service provided by the front end is transparent updating of visualization tools when data changes. Views of the data are synchronized by monitoring the state of all data models and maintaining a list of active visualizations for each model. When a data-change event occurs in a model, the front end broadcasts that event to all its active visualizations.
Plug-in Creation and Installation
To create a BrainImageJ plug-in, a developer constructs a subclass of an appropriate extension-pathway base class. The subclass inherits functionality from the base class. Successful compilation of the new subclass thus guarantees its implementation of the programming interface required by the front end to provide transparent application services.
Installing plug-ins into BrainImageJ is straightforward. After compiling a plug-in into a class file, a developer incorporates the plug-in into the framework by copying its class file into the user-configurable plug-in search path. A plug-in will typically be shared as a precompiled class file, which is directly incorporated into the framework by moving the class file into the search path. The framework utilizes the Java reflection capability, which provides runtime-type information,29 to load plug-ins, analyze their class hierarchies, and incorporate them into the appropriate extension pathway.
Application to Neuroimaging: Segmentation of Gyri
In this section we demonstrate the utility and scope of the BrainImageJ framework with a specific application designed by the SPNL development team to enable more efficient segmentation of cortical gyri. Our motivation for developing these tools was increasing interest in segmenting cortical substructures, as seen in the numerous studies that correlate schizophrenia with altered morphology of the cortex.30,31 Although tracking morphologic changes in more specific structures is desirable, fewer studies examine morphology at the level of gyri. Those that do tend to have small sample sizes, because of the cost and difficulty of segmenting gyri on two-dimensional brain sections.32
Segmenting gyri with traditional techniques is difficult for two reasons. First, gyri have highly complex and variable morphology.33 Second, it is often difficult to identify cortical protrusions that correspond to a particular gyrus on two-dimensional sections. The tools described in this section take advantage of the fact that experts identify gyri more easily in three-dimensional views of the data. Our surface drawing application allows expert-based segmentation by outlining regions-of-interest on three-dimensional reconstructions of the cortical surface.
Surface Drawing Plug-ins
The application makes use of two types of data—magnetic resonance (MR) volumetric data and geometric surface data in the form of a polygon mesh. The former is represented by a DataCells3D data model, as described above. The second is represented by a Surface data-model plug-in. The read interface for Surface objects includes an important method for obtaining its intersection point with a given ray. In the SPNL, MR data are stored in TIFF format and loaded with a DataCells3D TIFF Loader plug-in. Surface data are stored in a simple, SPNL-specific format using a Polygon-Mesh Saver plug-in.
Two algorithm plug-ins were implemented to create and manipulate Surface objects. The first, Isosurface Extraction Algorithm, derives a polygon-mesh approximation for an isosurface in volumetric data using the Marching Cubes algorithm.34 Isosurface Extraction Algorithm accepts the volumetric data input as a DataCells3D, and outputs the polygon-mesh as a Surface object. The second algorithm plug-in, Subsurface Extraction Algorithm, accepts two inputs—a Surface and a list of real-valued points defining a loop on the surface. It outputs a new Surface representing the portion of the input Surface that is contained in the loop. The algorithm consists of three main steps. First, a boundary for the subsurface is established by joining the given points with segments that lie on the mesh. Second, polygons are split along the boundary if they intersect it. Finally, a new polygon-mesh is created containing only the subset of polygons that lie within the boundary.
Visualization plug-ins were developed for each of the two data models. The Triplanar Viewer plug-in displays axial-slice views of MR data. Slices perpendicular to each axis are presented separately as well as combined in an interactive three-dimensional view (Figure 2▶). Contrast and false-color enhancement aid in the selection of intensity values at the inner and outer boundaries of the cortex. The three-dimensional view, rendered with the Java 3D library, incorporates perspective, local lighting, and texture mapping for depth cueing.
Java 3D was also used in the implementation of the Surface Editor component, which renders Surface objects (Figure 3▶). These renderings can be interactively rotated, translated, and zoomed to be explored in six degrees of freedom; frame rates are presented below in the section on Performance Testing. Gouraud shading and specular highlighting are used to increase the realism of the rendering. The tool allows users to draw on the displayed surface by clicking and dragging. Mouse events are translated into three-dimensional points on the rendered surface in a two-step process. The two-dimensional location of each mouse event is projected as a ray into the rendered scene; this ray is then intersected with the Surface to yield a three-dimensional intersection point.
Applying Plug-ins to Segment a Gyrus
The following processing pipe enumerates the steps taken by an expert in segmenting a gyrus (Figure 4▶).
Figure 4.
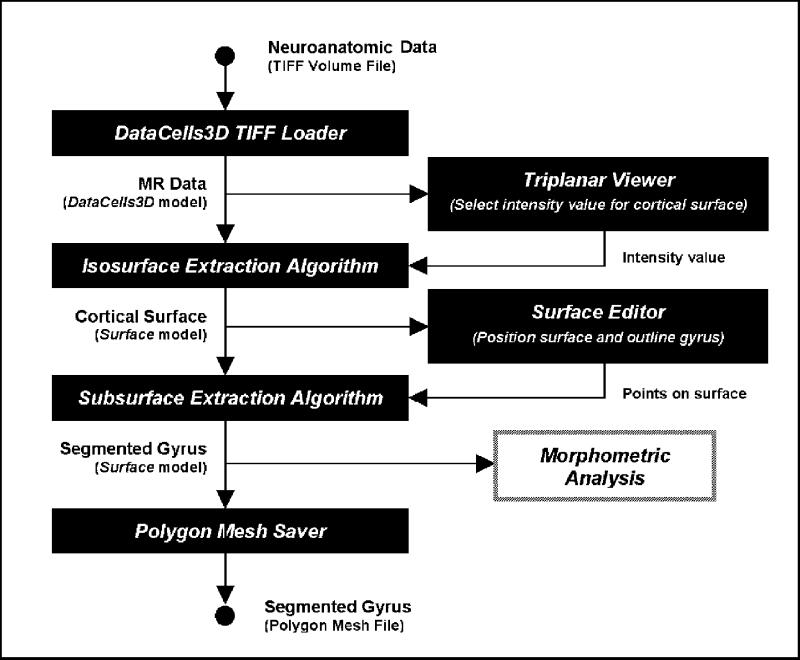
The processing pipe for expert-based segmentation of a gyrus from MR data using the surface-drawing BrainImageJ plug-ins.
Load MR data from disk using DataCells3D TIFF Loader.
Visualize MR data with Triplanar Viewer to select intensity values for isosurfaces approximating the cortex.
Activate Isosurface Extraction Algorithm to derive isosurfaces based on the intensities provided by step 2.
Visualize the cortical surface with Surface Editor, and out the gyrus of interest by clicking and dragging with the mouse.
Activate Subsurface Extraction Algorithm on the surface points obtained in step 4 to extract the gyrus as a subsurface.
Save the segmented gyrus Surface using Polygon-Mesh Saver for further morphometric analysis.
For these experiments, we use 256 × 256 × 212 volume data that have been manually “skull-stripped” to remove non-brain data, and “resliced” to ensure that voxels are cubic. These preprocessing steps were done in BrainImage, an earlier imaging framework developed at the SPNL for MacOS. The final results of the six-step processing pipe can be seen in Figure 3▶, a screenshot of Surface Editor that shows the surfaces of the cortex, the outline of the gyrus, and the extracted gyrus Surface.
Life Cycle of Plug-ins
One of the benefits of the BrainImageJ design is that individual plug-in tools are modular and easily reusable. The separation of file loading and saving, data analysis and transformation, and visualization into discrete plug-in tools allows developers to reuse portions of existing applications in developing new ones. Repositories of reusable plug-ins provide building blocks for assembling new application skeletons quickly, allowing developers to more quickly flesh out new functionality. Existing plug-ins need not be modified, or even recompiled, to be incorporated into new applications.
A simple example of plug-in reuse is illustrated in Figure 5▶, which depicts another application that is being developed in the SPNL. The application is a processing pipe that supports the development of new algorithms for correcting bias in MR data due to field inhomogeneities. The skeleton of this application is built of the Surface-drawing plug-ins described in the previous section. DataCells3D TIFF Loader and Saver plug-ins are reused for file manipulation. The Triplanar Viewer plug-in is reused to visualize application output, helping in debugging and experimentation. Developers create only one new plug-in—an inhomogeneity correction algorithm plug-in that accepts a DataCells3D and modifies it to correct bias. BrainImageJ glues these plug-ins together into a usable application, freeing developers from much of the application-level coding.
Figure 5.
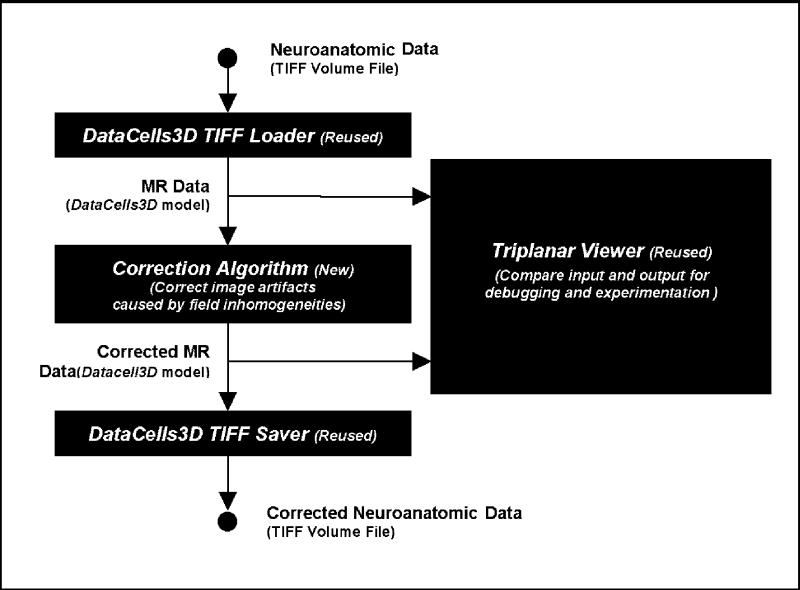
An example of plug-in reuse in the development cycle: an application to correct image artifacts in MR data caused by field inhomogeneity, in which developers reuse three plug-ins and implement one new algorithm plug-in.
Portability and Performance Testing
In this section we discuss our experiments on the portability of the BrainImageJ framework and SPNL-developed plug-ins. We tested portability on three platforms with implementations of Java 2—Windows (2000), Linux (Redhat 6.1), and Unix (Solaris 7). We separated our portability tests into two categories—tests on correctness of execution, and tests on execution performance.
Correct execution is essentially guaranteed by the nature of the JVM, so our tests in this area can be regarded as a check of the quality of the JVM implementation for the three test platforms. Testing involved normal use of the framework and plug-ins for image visualization and analysis, exercising each of JVM's capabilities in basic computation, file input/output, and two-dimensional/three-dimensional graphics. On Windows, no execution errors were detected. On Unix, no runtime errors were detected; however, the source code had to be recompiled, but no modifications were necessary. On early versions of Java 2 for Linux, minor problems occurred with the GUI. In some cases, menus were not drawn correctly or windows were not sized as requested. In our experiments, however, trivial changes to the code fixed these discrepancies.
While correct execution across different platforms is necessary for portability, interactive programs must run at comparable speeds on different platforms to remain usable. Our performance tests addressed this practical aspect of portability by measuring the speed of Java on the three platforms. These tests were not designed to exhaustively measure all aspects of JVM performance. Instead, they aimed to provide basic performance comparisons. Results of the tests are shown in Figure 6▶. Test 1 targeted basic computational speed, measuring the average number of voxels processed per second while repeatedly iterating through and modifying a 256 × 256 × 212 volume. Test 2 targeted the speed at which JVM accessed the file system (IO performance), by measuring the number of bytes that were read per second while loading several 256 × 256 × 212 volumes. Test 3 targeted three-dimensional rendering speed by measuring the number of frames drawn per second for two complex three-dimensional scenes. The scenes used were the left and top right images of Figure 3▶, which contain, respectively, 205,000 and 67,000 triangles. These scenes were rendered at a resolution of 512 × 512 pixels.
Figure 6.
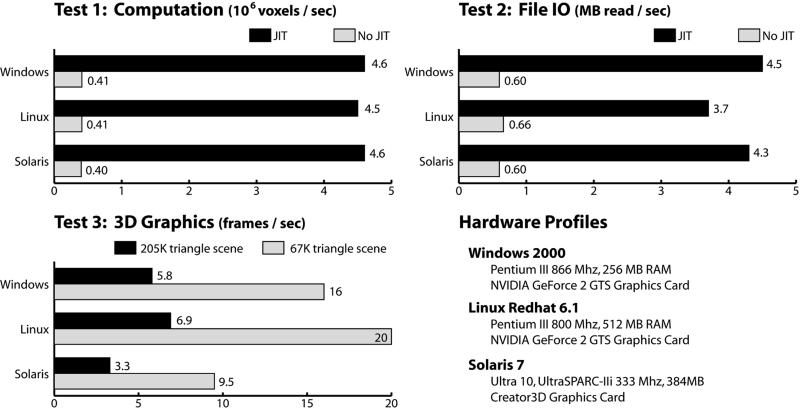
Comparative performance of basic computation, file IO and 3D graphics rendering for SunTM JVMs on Windows, Linux and Solaris.
Performance on computation and IO tests was even across all three platforms. Just-in-time compilation improved performance on these tests dramatically (Figure 6▶), in some cases by an order of magnitude. In contrast, the three-dimensional graphics test was not affected by just-in-time compilation. This lack of just-in-time acceleration indicates that the CPU was relatively idle in the tests, demonstrating that Java 3D makes use of graphics hardware acceleration. The Linux and Windows machines were equipped with GeForce 2 GTS graphics chips, which are relatively common and inexpensive in new x86-processor computers. Frame rates of 16 to 20 Hz for the 67,000 triangle scene, and 5 to 7 Hz for the 205,000 scene were achieved. The Solaris machine, equipped with a Creator3D graphics card, rendered about half as quickly.
Applicability and Limitations
Our portability and performance tests indicate that it is currently feasible to use BrainImageJ, on at least three platforms, for visualization of complex three-dimensional data sets at multi-frame per second rates. On the platform with the best graphics performance (Linux), we achieved “real-time” rendering rates for scenes with 67,000 triangles. Performance was still interactive for research purposes when rendering scenes with 205,000 triangles. Even though these visualization tools will run on any platform with Java 3D, they are limited in practice to platforms with hardware that can accelerate three-dimensional rendering to interactive speeds.
A different kind of real-world limitation to BrainImageJ portability is the absence of Java 2 implementation for some target platforms. For example, even though MacOS supports Java 1.1, it is not expected to support Java 2 and Java 3D before mid- to late 2001. In contrast, the first Java 2 implementations appeared on Windows and Solaris in late 1998. This discrepancy indicates the relatively long period that may be required for new Java technology to spread to various platforms. In addition, our correctness-of-execution tests show that it takes time for a JVM to stabilize: early Java 2 implementations for Linux did not behave in exactly the same way as Java 2 for Windows or Solaris. Some testing across platforms may be necessary for practical portability.
Finally, realistic use of BrainImageJ may be limited by the goals of the developer and end user. Although the performance tests show that just-in-time compilation and Java 3D already enable the use of BrainImageJ for interactive visualization of large three-dimensional data sets, Java is unlikely to match the performance of three-dimensional visualization applications that are optimized for a specific platform. Such applications can utilize platform-specific interfaces that cannot be exposed in Java's portable application program interfaces. As a result, BrainImageJ may not be suitable for developers who depend on the special capabilities of their particular hardware. Choosing BrainImageJ as a working environment trades some platform-specific power for cross-platform interoperability and ease of development.
Discussion
BrainImageJ addresses and attempts to alleviate several barriers to research-related interoperability. The most fundamental barrier is the lack of tools supporting the creation of computational data models. Through the data model extension pathway, BrainImageJ perpetuates dedicated development and refinement of such models. This paradigm is based on two principles—that computational data models are necessary for effective development and sharing of interoperable tools13 and that new models will be required to support integration and synthesis of diverse brain data.1
The second barrier to interoperability addressed by BrainImageJ is the existence of files in different data formats. By promoting communication of data in terms of a computational data model rather than a particular file format, BrainImageJ turns file sharing into a vehicle for archiving and transferring a data model. We designed the file loader and saver extension pathway to be a simple and natural mechanism for sharing data in different formats. It is simple because files in any format are shared by providing the relevant loader plug-in along with the files. It is natural because any laboratory that wants to share its data would necessarily possess the plug-ins to load and save their format. This paradigm will be especially helpful for data interoperability in emerging domains that have no standard formats.
BrainImageJ also addresses the interoperability barrier posed by the existence of multiple target operating systems. Guaranteeing that an application executes correctly and efficiently across different operating systems is a difficult, ongoing task. The choice of Java as a programming language transfers this burden from developers in the research community to JVM developers in the Java industry. Thus, the portability, efficiency, and reliability of BrainImageJ tools improve as the industry continues to implement new JVMs and refine existing ones, at no cost to researchers. Meanwhile, the BrainImageJ extension pathways allow plug-in developers to focus on experimentation with research-oriented functionality without limiting the future portability and interoperability of their efforts.
Aside from leveraging the inherent portability of the Java language, BrainImageJ supports interoperability by leveraging design patterns that simplify the customization of research tools. As discussed in the Background section, customization is essential for tailoring tools to the specific needs of each laboratory. BrainImageJ supports customization through both components of the framework. First, the four extension pathways encourage the development of small, self-contained modules, thus simplifying the creation of extensions. Second, the front end makes it easy for extensions to become part of a sophisticated application, because it automatically integrates new plug-ins and provides transparent support for modern application services. As a result, customization through extension is free from the responsibility for maintaining complex features such as undo/redo histories, user-customizable GUIs, and multi-threading of processing tasks.
This support for extension and customization helps developers create tools that are more user-oriented. Development is complicated by the common inability of end users to formally define their needs and the lack of domain-specific knowledge on the part of developers.35 BrainImageJ minimizes these problems in two ways. First, by directing growth in terms of small, self-contained plug-ins, the four extension pathways promote a shorter development cycle that enables closer communication between developers and end users. Second, by incorporating even rapid prototypes into sophisticated user interfaces, the application front end provides a platform for efficient end-user testing that provides developers with more rapid feedback. Experience at the SPNL has shown that BrainImageJ supports a development paradigm of iterative design, assessment, and refinement.1 This paradigm has helped SPNL end users define their needs precisely and has kept SPNL developers focused on user requirements.
Conclusion and Further Work
The BrainImageJ framework arose as an abstraction for the common factors in many of the neuroimaging research projects at the SPNL. For developers, the framework enables a software development cycle that concentrates on research-related functionality. For end users, it provides a customizable collection of plug-ins that can be easily adapted and extended to support new research directions. The framework is currently being used for neuroscience research only in the SPNL, and is used primarily for visualization and surface characterization. We hope to release the software in the public domain by early 2002, after the user interfaces have been further refined for real-world usability.
These refinements are the main thrust of our current work on the framework. In addition, we are working to add application front-end services that incorporate distributed computing capabilities. These services will allow the system to parallelize concurrent algorithm tasks across multiple networked machines to exploit the total computing environment more effectively. Finally, we will enable the application front end to communicate directly with databases, using file loader and saver plug-ins as decoders for data stored in object-oriented databases.
The BrainImageJ framework is a relatively simple set of interfaces and generalized services designed to facilitate effective tool development and distribution. Just as it supports communication between developers and end users in the SPNL, the framework can be used to facilitate communication between different research laboratories. Once it has been released in the public domain, we hope that BrainImageJ will become a vehicle for collaboration and interoperability in the neuroscience research community.
Acknowledgments
The authors thank Dr. Stephan Eliez and the SPNL staff for their feedback and help.
This work was supported by Human Brain Project grant HD 31715-08 from the National Institutes of Health and the National Institute of Child Health and Human Development, by grant MH01142 and MJ50047 from the National Institute of Mental Health, and by a grant from the Packard Foundation.
References
- 1.The Human Brain Project: Phase I Feasibility Studies. NIH Guide. Bethesda, Md.: National Institutes of Health, Oct 6, 1999;24(35).
- 2.Musen MA, Schreiber AT. Architectures for intelligent systems based on reusable components. Artif Intell Med. 1995;7:189–99. [DOI] [PubMed] [Google Scholar]
- 3.Gu H, Perl Y, Geller J, Halper M, Liu L-M, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000;7(1):66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohane IS, Greenspun P, Fackler J, Cimino C, Szolovits P. Building national electronic medical record systems via the World Wide Web. J Am Med Inform Assoc. 1996;3(3):191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nadkarni PM, Marenco L, Chen R, Skoufos E, Shepherd G, Miller P. Organization of heterogeneous scientific data using the EAV/CR representation. J Am Med Inform Assoc. 1999;6(6):478–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creighton C. A literature review on communication between picture archiving and communication systems and radiology information systems and/or hospital information systems. J Digit Imag. 1999;12(3):138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidgood WD Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997;4(3):199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu SW, Eriksson H, Gennari JH, Shahar Y, Musen MA. Ontology-based configuration of problem-solving methods and generation of knowledge-acquisition tools: application of PROTEGE-II to protocol-based decision support. Artif Intell Med. 1995;7(3):257–89 [DOI] [PubMed] [Google Scholar]
- 9.NIH Image Web site. 2000. Available at: http://rsb.info.nih.gov/nih-image/. Accessed Oct 12, 2000.
- 10.Stanford Psychiatry Neuroimaging Laboratory. BrainImage software. Stanford University Child and Adolescent Psychiatry Web site. 2000. Available at: http://www-cap.stanford.edu/research/neuroimaging/imageanalysis/brainimage.html. Accessed Oct 12, 2000.
- 11.Poliakov AV, Hinshaw KP, Rosse C, Brinkley JF. Integration and visualization of multimodality brain data for language mapping. Proc AMIA Annu Symp. 1999:349–53. [PMC free article] [PubMed]
- 12.Gardner D. Cortical Neuron Net Data Model. Cornell University Medical College Web site. Sep 1999. Available at: http://cortex.med.cornell.edu/dataModel/cdm2000.html. Accessed Sep 11, 1999.
- 13.Annual Spring Meeting of Human Brain Project Research. National Institutes of Health, Bethesda, MD; Jun 30, 1999.
- 14.Kohane IS, Greenspun P, Fackler J, Cimino C, Szolovits P. Building national electronic medical record systems via the World Wide Web. J Am Med Inform Assoc. 1996;3(3):191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riesmeier J, Eichelberg M, Jensch P. An approach to DICOM image display handling the full flexibility of the standard's specification. Proc SPIE 1999;3658:363–9. [Google Scholar]
- 16.Advanced Visual Systems Web site. Available at: http:// www.avs.com/. Accessed Oct 1, 2000.
- 17.Interactive Data Language. Research Systems, Inc. Web site. Available at: http://www.rsinc.com/idl/index.cfm. Accessed Oct 1, 2000.
- 18.Brooks FP. No silver bullet. In: Brooks FP. The Mythical Man-Month. Reading, Mass.: Addison-Wesley, 1995.
- 19.Gamma E, Helm R, Johnson R, Vlissides J. Design Patterns: Elements of Reusable Object-Oriented Software. Reading, Mass.: Addison-Wesley, 1995.
- 20.Java Platform Ports. Sun Microsystems Web site. 2000. Available at: http://java.sun.com/cgi-bin/java-ports.cgi. Accessed Oct 12, 2000.
- 21.Gosling J, Joy B, Steele G. The Java Language Specification. Reading, Mass.: Addison-Wesley, 1996.
- 22.Plezber MP, Cytron RK. Does “Just in Time” Equal “Better Late than Never”? Proceedings of the 24th Annual ACM SIGPLAN-SIGACT Symposium on Principles of Programming Languages; Paris, France; Jan 15–17, 1997.
- 23.Java HotSpot Technology. Sun Microsystems Web site. 1999. Available at: http://java.sun.com/products/hotspot/. Accessed Nov 6, 1999.
- 24.Lee D. Concurrent Programming in Java: Design Principles and Patterns. 2nd ed. Reading, Mass.: Addison-Wesley, 1999.
- 25.Lee D. Cancellation, Concurrent Programming in Java [on supplement to Lee24]. 1999. Available at: http://gee.cs.oswego.edu/dl/cpj/cancel.html, 1999. Accessed Dec 10, 1999.
- 26.Eckstein R, Loy M, Wood D. Java Swing. Sebastopol, Calif.: O'Reilly & Associates, 1998.
- 27.Hardy VJ. Java 2D Graphics. Palo Alto, Calif.: Sun Microsystems Press, 2000.
- 28.Sowizral H, Rushforth K, Deering M. The Java 3D API Specification. Reading, Mass.: Addison-Wesley, 1998.
- 29.Van der Linden P. Just Java 2. 4th ed. Upper Saddle River, NJ: Prentice Hall, 1999.
- 30.Lawrie SM, Abukmeil SS. Brain abnormality in schizophrenia: a systematic and quantitative review of volumetric magnetic resonance imaging studies. Br J Psychiatry. 1998;172:110–20. [DOI] [PubMed] [Google Scholar]
- 31.McCarley RW, Wible CG, Frumin M, Hirayasu Y, Levitt JJ, Fischer A, Shenton ME. MRI anatomy of schizophrenia. Biol Psychiatry. 1999;45(9):1099–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchanan RW, Vladar K, Barta PE, Pearlson GD. Structural evaluation of the prefrontal cortex in schizophrenia. Am J Psychiatry. 1998;155(8):1049–55. [DOI] [PubMed] [Google Scholar]
- 33.Ono M, Kubik S Abernathey CD. Atlas of the Cerebral Sulci. New York: Thieme Medical, 1990.
- 34.Lorensen WE, C HE. Marching cubes: a high-resolution 3D surface reconstruction algorithm. Comput Graphics. 1987; 21(4):163–9. [Google Scholar]
- 35.Booch G. Object-oriented analysis and design, with applications. 2nd ed. San Francisco, Calif.: Benjamin Cummings, 1994.


