Abstract
Background
L-threonine is an indispensable amino acid. One of the major L-threonine degradation pathways is the conversion of L-threonine via 2-amino-3-ketobutyrate to glycine. L-threonine dehydrogenase (EC 1.1.1.103) is the first enzyme in the pathway and catalyses the reaction: L-threonine + NAD+ = 2-amino-3-ketobutyrate + NADH. The murine and porcine L-threonine dehydrogenase genes (TDH) have been identified previously, but the human gene has not been identified.
Results
The human TDH gene is located at 8p23-22 and has 8 exons spanning 10 kb that would have been expected to encode a 369 residue ORF. However, 2 cDNA TDH transcripts encode truncated proteins of 157 and 230 residues. These truncated proteins are the result of 3 mutations within the gene. There is a SNP, A to G, present in the genomic DNA sequence of some individuals which results in the loss of the acceptor splice site preceding exon 4. The acceptor splice site preceding exon 6 was lost in all 23 individuals genotyped and there is an in-frame stop codon in exon 6 (CGA to TGA) resulting in arginine-214 being replaced by a stop codon. These truncated proteins would be non-functional since they have lost part of the NAD+ binding motif and the COOH terminal domain that is thought to be involved in binding L-threonine. TDH mRNA was present in all tissues examined.
Conclusions
The human L-threonine 3-dehydrogenase gene is an expressed pseudogene having lost the splice acceptor site preceding exon 6 and codon arginine-214 (CGA) is mutated to a stop codon (TGA).
Background
Liver failure is a cause of considerable mortality; therefore bioartificial livers may offer significant therapeutic benefit. Porcine-derived hepatocytes are being used in clinical studies of bioartificial livers [1,2]. There may, however, be significant differences in the activities of liver enzymes between species [3]. These differences are also important considerations when the pharmaceutical industry conducts new drug metabolism and pharmacokinetic studies on key mammal species [4].
The liver plays a critical role in regulating the circulating concentrations of amino acids. The regulation of amino acid supply to bioartificial organs and maintaining the activity of the amino acid-metabolising enzymes will be important in their development. Active maintenance of the optimal amino acid concentrations will offer the possibility of prolonging the differentiated function of hepatocytes in bioartificial livers [5].
L-threonine is one of three indispensable amino acids since mammals do not possess the necessary enzymes for the transamination of threonine [6]. However, the transient apoenzyme form of L-threonine dehydratase bound to pyridoxamine 5'-phosphate dehydratase can carry out a half-transamination of L-threonine [7]. Once L-threonine is oxidised through various catabolic pathways it is lost for the purposes of protein synthesis. Therefore, it is important for determining levels of human nutrition that factors regulating threonine oxidation are identified. The international recommended intake of L-threonine for adults is 7 mg.kg-1.d-1[8]. However, this has recently been challenged as being too low and a recommendation of 15 mg.kg-1.d-1 has been suggested [9].
There are two major L-threonine degradation pathways. L-threonine is either catabolised by L-threonine 3-dehydrogenase to 2-amino-3-ketobutyrate or by L-serine/threonine dehydratase (EC 4.2.1.16) to NH4+ and 2-ketobutyrate. In both eukaryotic and prokaryotic cells, the conversion of L-threonine via 2-amino-3-ketobutyrate to glycine takes place in a two-step process [10,11]. L-threonine dehydrogenase catalyses the reaction: L-threonine + NAD+ = 2-amino-3-ketobutyrate + NADH. The subsequent reaction between 2-amino-3-ketobutyrate and coenzyme A to form glycine and acetyl-CoA is catalysed by 2-amino-3-ketobutyrate coenzyme A ligase (EC 2.3.1.29 also called glycine acetyltransferase; gene names KBL and GCAT). L-threonine dehydrogenase and 2-amino-3-ketobutyrate coenzyme A ligase are associated physically on the inner membrane-matrix of mitochondria where the two enzymes form a complex with a stoichiometry of one threonine dehydrogenase tetramer to two 2-amino-3-ketobutyrate coenzyme A ligase dimers [12-14].
In mammals, the removal of the 1-carbon of threonine is thought to occur through threonine dehydrogenase, this carbon being incorporated into glycine and released as CO2 by the mitochondrial glycine cleavage system. In normally-fed pigs and rats 80 to 87% of L-threonine catabolism occurs via threonine dehydrogenase [15,16] and in isolated rat and cat hepatocytes 35% to 50% of threonine oxidation occurs through this pathway [17,18]. Zhao et al. measured threonine oxidation in adult humans by the production of labelled CO2 from L-[1-13C]threonine and concluded that the conversion of threonine to glycine through the threonine dehydrogenase pathway was not important if it exists at all [19]. However, the dehydrogenase pathway is thought to account for 44% of total threonine oxidation in infants [20]. More recently, the same group used a more sensitive method to measure the production of labelled glycine and CO2 in adults and suggested that there was some threonine catabolism via the threonine dehydrogenase pathway, but it accounted for only 7–11% of total [21]. This suggested that the L-serine/threonine dehydratase pathway is the dominant route in the catabolism of threonine in humans.
Recently, the sequences of the murine and porcine TDH cDNAs have been reported [22]. The amino-terminal regions of these proteins have characteristics of a mitochondrial targeting sequence and are related to the UDP-galactose 4-epimerases, with both enzyme families having an amino-terminal NAD+ binding domain. The sequence of the human KBL transcript, the second enzyme in the pathway, has also been described [23] and this study aimed to identify the human TDH gene.
Results
Human TDH gene
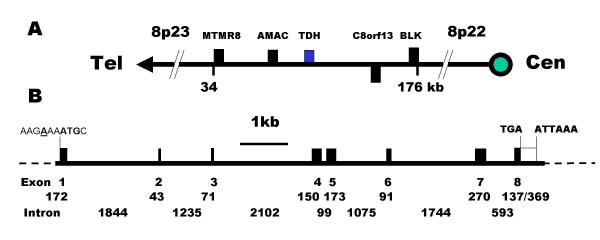
The murine L-threonine 3-dehydrogenase gene (TDH) has recently been identified [22]. The human genome was searched using this murine sequence and only a single putative human TDH gene was found to be present. The putative human TDH gene is located on clone RP11-110L10, chromosome 8p23-22 (GenBank accession No. AC011959, Whitehead Institute/MIT Center for Genome Research, USA), between the gene for acyl-malonyl condensing enzyme and hypothetical protein, C8orf13 gene (accession No. XP_088377) (Figure 1A). By comparison with the murine TDH gene and the porcine cDNA, 8 human exons were identified spanning 10 kb (Figure 1B). The predicted start of the ORF is in good sequence context for the initiation of translation [24].
Figure 1.
Chromosomal localisation and gene structure of the human TDH gene. (A) The human TDH gene is located at 8p23 and the neighbouring genes are: myotubularin related protein 8, MTMR8: acyl-malonyl condensing enzyme, AMAC: threonine dehydrogenase, TDH: hypothetical protein, C8orf13; B lymphoid tyrosine kinase, BLK. Genes encoded by + and - stands are shown above and below the line respectively. The telomeric and centromeric directions are indicated. (B) The human TDH gene spans 10 kb and consists of 8 exons. The putative initiation methionine codon, stop codon and poly-adenylation signal are shown. Closed boxes indicate the ORF and the sizes (in bp) of the exons and introns are indicated.
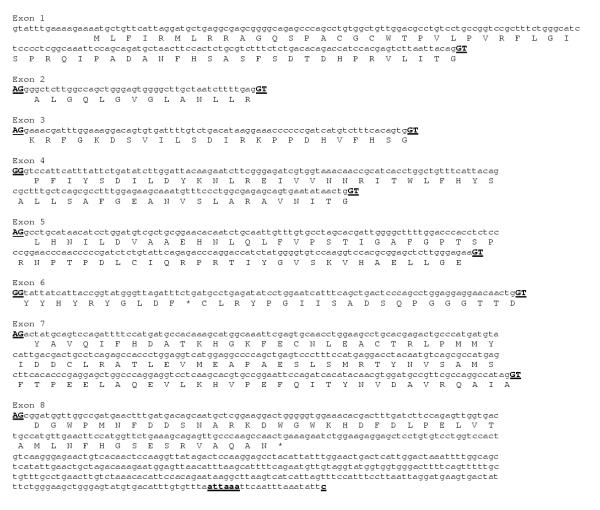
The sequence of the human TDH exons (Fig. 2) has 84% identity with the porcine cDNA at the nucleotide level in the ORF and the theoretical full-length human protein has 85% identity and 97% similarity with the porcine TDH protein [22], suggesting that this is the human TDH gene. The first potential polyadenylation signal site (ATTAAA) is also conserved between species.
Figure 2.
Sequence and translation of the human TDH exons. A comparison of the human genomic sequence with the murine TDH gene enabled the identification of 8 homologous exons on clone RP11-110L10. A translation of the exons is shown below the nucleotide sequence. The acceptor and donor splice sites (AG >) at the ends of each exon are shown in uppercase and underlined. The potential polyadenylation signal and site (attaaa and c) are shown in bold and underlined.
Human TDH protein
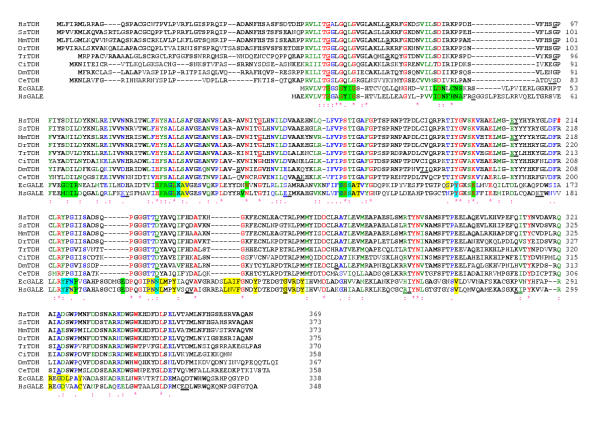
The predicted human TDH protein possesses very weak similarity to the human uridine diphosphogalactose-4-epimerase protein (GALE). GALE catalyses the interconversion of UDP-galactose and UDP-glucose in the metabolic pathway that converts galactose into glucose1-phosphate, and also catalyses the interconversion of UDP-GalNAc and UDP-GlcNAc. An alignment of the human TDH protein with other eukaryotic TDH protein sequences and GALE proteins from Homo sapiens and Escherichia coli identified conserved residues likely to contact the nicotinamide-adenine-dinucleotide cofactor (NAD+) (Fig. 3) [25]. They are Gly-58, Gly-61, Gly-64, Asp-84, Asp-102, Ile-103, Leu-104, His-123, Leu-127, Val-142, Asn-143, Ser-165, Tyr-191, Lys-195 and Tyr-218 [26-29]. In view of the different substrates utilised by the 2 enzymes, of those 22 residues that contact the sugar substrate in GALE only one, Asp-325, is conserved (excluding those residues involved in contact with both sugar and NAD+).
Figure 3.
Clustal alignment of the predicted human protein with other eukaryotic L-threonine dehydrogenase (TDH) protein sequences and 2 UDP-galactose 4-epimerase sequences (GALE). The species and the derivation of the sequences are as follows: Homo sapiens, HsTDH, genomic DNA (accession No. AC011959); HsGALE, cDNA (NP_000394); Sus scrofa, (pig), Ss, cDNA (AY095535); Mus musculus, mouse, Mm, cDNA (AY116662); Danio rerio (zebrafish), Dr, ESTs (electronic contigous sequence); Takifugu rubripes (puffer fish), Tr, genomic DNA, (AF411956); Ciona intestinalis (a tunicate), Ci, ESTs; Drosophila melanogaster, Dm, genomic DNA gene CG5955 and ESTs; Caenorhabditis elegans, Ce, genomic DNA (U64847) and ESTs; Escherichia coli, EcTDH (P07913) and EcGALE (AAC73846). The locations of the exon/exon boundaries are shown on the translated protein as underlined residues. Conserved residues are indicated by a (*), strongly similar residues by a (:) and weakly similar residues by a (.). Residues that are common to at least two protein sequences are shown in bold. The in-frame stop codon in the human sequence is indicated by a red (#). Gale protein residues in contact with the ligand nicotinamide-adenine-dinucleotide are highlighted in green and those residues in contact with the sugar ligands, uridine-5'-diphosphate-mannose and uridine-5'-diphosphate-4-deoxy-4-fluoro-alpha-d-galactose are highlighted in yellow. Those residues that have been shown to contact both NAD and sugar are highlighted in light blue [26-29].
Human TDH predicted secondary structure
Using the translation of the human genomic sequence (with the stop codon replaced with the conserved arginine residue found in other TDH proteins) the predicted secondary structure of the likely "ancestral" TDH protein was determined using Psi-Pred [30] and aligned with the crystal structure of GALE from E. coli[29,31] (Fig. 4). The human ancestral TDH protein would have been a mixed alpha-helices/beta-sheet protein with a NAD+ binding Rossmann-fold and belongs to the tyrosine-dependent oxidoreductase protein family (also known as short-chain dehydrogenases). The characteristic Tyr-x-x-x-Lys couple (residues 191 and 195) found in all family members is important for catalysis with the conserved tyrosine serving as the active-site base [33]. Two domains were identified, with the larger amino-terminus domain (residues 53–234) having a NAD+ binding motif and the smaller carboxy-terminus domain (residues 218–329) likely to be involved in L-threonine binding. It is likely that the substrate is located in the cleft between the two domains.
Figure 4.
Predicted secondary structure of human "ancestral" TDH protein was determined using the Psi-Pred program and was aligned with that of the crystal structure of uridine diphosphogalactose-4-epimerase protein (GALE) from Escherichia coli[29,31] using 3D-PSSM. The labels are: human "ancestral" TDH protein predicted secondary structure, HsTDH_PSSM; human "ancestral" TDH protein sequence, HsTDH_seq; E. coli GALE protein sequence, EcGale_seq; E. coli GALE protein secondary structure, 1udc_SS; alpha-helix, H, highlighted in light blue; beta-sheet, E, highlighted in yellow; c = turn, coil or loop. Identical residues are shown and additionally, a ":" indicates positive equivalence.
Mutations in the human TDH gene
However, there are 3 mutations that disrupt the gene transcription and translation on the sequence of genomic clone RP11-110L10 (Fig. 2). There is a loss of the acceptor splice sites in exon 4 and 6 and an in-frame stop codon in exon 6 (from the expected CGA to TGA) resulting in arginine-214 being replaced by a stop codon. Together, these mutations suggested that the human TDH gene is a pseudogene.
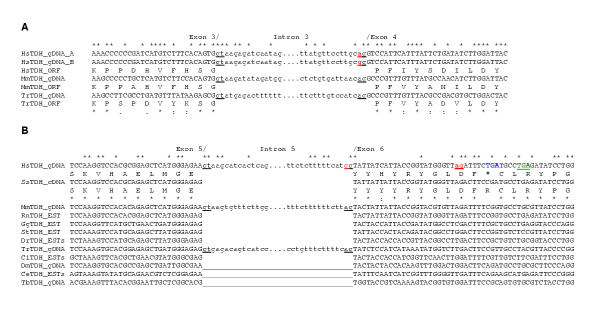
To determine whether these mutations were present in other species, sequence alignments around the acceptor splice sites in exons 4 and 6 were constructed (Fig. 5). The genomic sequences from the mouse and puffer fish (Takifugu rubripes or Fugu) and another human genomic clone (AF131216, Institute of Molecular Biotechnology, Germany) have intact acceptor splice sites in exon 4 suggesting that the loss of this splice site is a single nucleotide polymorphism (SNP) (Fig. 5A). In both human genomic DNA sequences the acceptor splice site in exon 6 is lost, but it is intact in both the mouse and puffer fish sequences (Fig. 5B). Similarly, the stop codon is present in both human genomic DNA sequences, but it is not present in genomic DNA, cDNA and EST sequences from other species. In human genomic DNA sequence (AC011959) there is also an A to T mutation in codon 201 (ATG to TTG, in exon 5) resulting a methionine to leucine substitution.
Figure 5.
Comparison of the human TDH gene sequences in the region of exon 4 and 6 acceptor sites with those of other species. (A) The splice donor and acceptor sites are indicated (gt and ag respectively). There is a splice site mutation in exon 4 in one of the human genomic sequences (gg, coloured red). The translations of the genes are shown below the DNA sequences (ORF). The species and accession numbers are: Homo sapiens, HsTDH_gDNA_A (AF131216); HsTDH_gDNA_B (AC011959); Mus musculus, MmTDHgDNA (14.54000001-55000000 supercontig, Sanger Centre, UK) and puffer fish, Takifugu rubripes, TrTDH_gDNA (AF411956). (B) There is an in-frame stop codon (TGA, coloured blue) and a splice site mutation in exon 6 in the human genomic sequence (AF131216). A cryptic splice site (ag, coloured red) was utilised in cDNA clone 1 that altered the reading frame, resulting in another stop codon (TGA, coloured green). The species and accession numbers are: pig, Sus scrofa, SsTDH_cDNA (AY095535); MmTDH_cDNA (AY116662); rat, Rattus norvegicus, RmTDH_EST (BF557448); chicken, Gallus gallus, GgTDH_EST (BM491791); western clawed frog, Silurana tropicalis, StTDH_EST (AL638064); zebrafish, Danio rerio, DrTDH_ESTs; the tunicate, Ciona intestinalis, CiTDH_ESTs; fruit fly, Drosophila melanogaster, DmTDH_gDNA (AAF51607); the nematode, Caenorhabditis elegans, CeTDH_ESTs and the euglenoid, Trypanosoma brucei, TbTDH_gDNA (AC084047). There are no introns in this region of the fly, nematode and trypanosome genes (__). Identical nucleotides and residues are shown by *, strongly similar residues by: and weakly similar residues by (.).
Genotyping of individuals by restriction enzyme digests and DNA sequencing
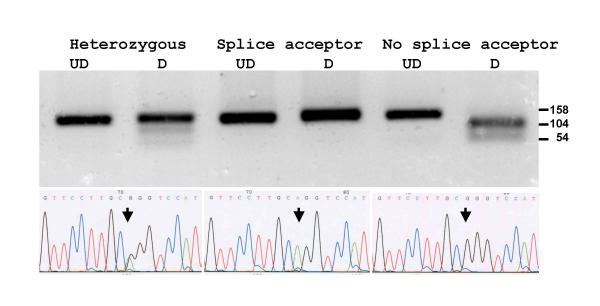
To determine whether these mutations in the 2 human genomic sequences were sequencing errors or just present in those individuals utilised for determining the human genome, the region encompassing the mutations was amplified by PCR from a number of individuals, sequenced and digested with restriction enzymes. Amplicons from the exon 4 splice acceptor site were digested with the restriction enzyme AciI. If the acceptor splice site was not present (G^CGG rather than GCAG, acceptor splice site underlined) the 158 bp amplicon was digested to produce restriction fragments of 104 and 54 bp. The restriction enzyme digest pattern and DNA sequence of 3 individuals are illustrated (Fig. 6) in which one individual has the splice site intact, another has lost the splice site and the third has the heterozygous condition. Digestion is not as complete as would be expected in the heterozygous condition, presumably due to heteroduplex formation during the PCR. This SNP was found in 20 out of 40 chromosomes examined.
Figure 6.
Amplicons from individuals with different genotypes for the exon 4 splice acceptor site SNP were digested with the restriction enzyme AciI (upper panel) and DNA sequenced (lower panel). Undigested samples, UD and digested samples, D. Arrows indicate the polymorphic nucleotide. In an individual homozygous for the splice acceptor site (AG, middle panel) no AciI digest site is present. In an individual homozygous for loss of the splice acceptor site (GG, right panel) the 158 bp amplicon is cut into fragments of 104 and 54 bp. In an individual heterozygous for the AG splice acceptor site (RG, left panel) both alleles are present and the amplicon is only partially digested.
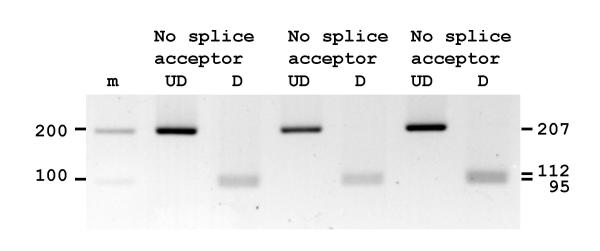
Amplicons from the exon 6 splice acceptor site from 3 individuals were digested with the restriction enzyme Hsp92II. If the acceptor splice site was not present (CATG^g rather than CATAg) the 207 bp amplicon was digested to produce restriction fragments of 112 and 95 bp. Three individuals examined had lost the splice site (Fig. 7). DNA sequencing verified the loss of the splice site and the presence of the in-frame stop codon in exon 6 in all 20 individuals examined (data not shown).
Figure 7.
Amplicons from 3 individuals for the exon 6 splice acceptor site were digested with the restriction enzyme Hsp92II. All amplicons were digested showing loss of the splice site. Undigested samples, UD and digested samples, D.
Copy DNA sequences of the human TDH gene
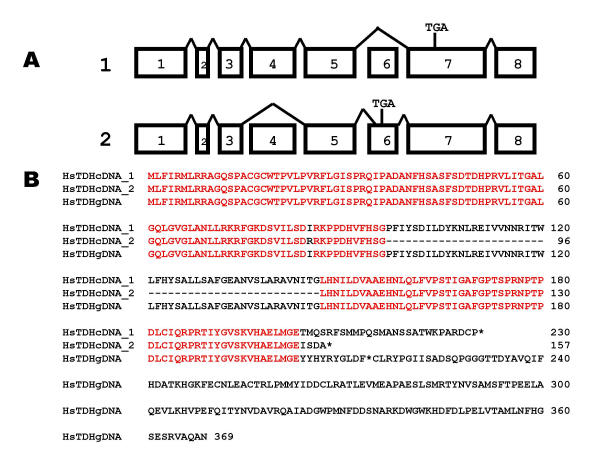
The human TDH gene is expressed. PCR amplification from cDNA libraries using primers designed to amplify the TDH ORF was carried out and amplicons of approximately 1000 bp were obtained from cDNAs from 2 individuals, cloned and sequenced. The 1019 bp sequence of clone 1 (AY101186) skipped exon 6, resulting in a premature stop codon in exon 7, (Fig. 8A) and would encode a 230 residue ORF (Fig. 8B). The 935 bp sequence of clone 2 (AY101187) skipped exon 4 and utilised a cryptic splice site in exon 6, 24 bp downstream of the expected site (Fig 5B), resulting in a premature stop codon in exon 6 (Fig. 8A) and would encode a 157 residue ORF (Fig. 8B). The human TDH gene is therefore a pseudogene since all individuals examined contain at least 2 mutations that on translation would generate truncated proteins that would be non-functional since they would be unable to make appropriate contacts with the substrates, L-threonine and NAD+.
Figure 8.
The human TDH pseudogene is expressed. (A) Clones from 2 individuals were sequenced and mapped to the TDH gene. Clone 1 skipped exon 6, and the resulting frame-shift generated a premature stop codon in exon 7 (TGA). Clone 2 skipped exon 4 and utilised a cryptic splice site in exon 6, and the resulting frame-shift generated a premature stop codon in exon 6. (B) The translation of clone 1 (exon 6 skipped) encoded a truncated 230 residue ORF. The skipped exon 4 in clone 2 does not alter the reading frame. However, the use of a cryptic splice site in exon 6, results in a premature stop codon in exon 6. Stop codons are indicated by *.
Expression of the TDH pseudogene in human tissue and cell types
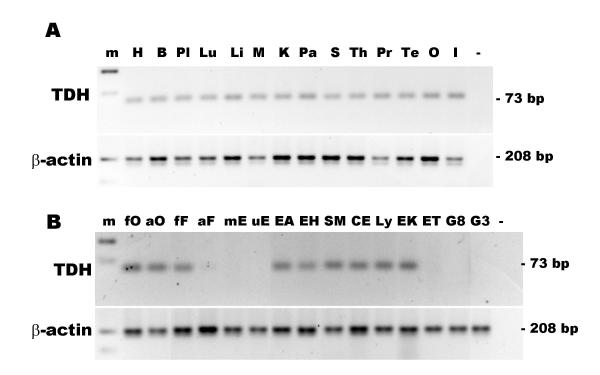
Reverse-transcriptase PCR was used to determine the presence of the TDH pseudogene mRNA in different tissues and cell types. TDH expression was found in all tissues examined (Fig. 9A). TDH mRNA was present in most cell types examined, but was below the level of detection in endothelial cells, glioma cell lines and some leukaemia cell lines (Fig. 9B). Together, this suggests that the TDH promoter is still active and is regulated in different cell types.
Figure 9.
Expression of the TDH pseudogene in human tissues and cell types. (A) Expression of the TDH pseudogene in human tissues. The lanes are: 100 bp marker, m; heart, H; brain, B; placenta, Pl; lung, Lu; liver, L; skeletal muscle, M; kidney, K; pancreas, Pa; spleen, S; thymus; Th; prostate, Pr; testis, Te; ovary, O; small intestine, I; negative control, (-). (B) Expression of the TDH pseudogene in human cells. RT-PCR for TDH (top panel) and β-actin (bottom panel). The cell types examined were: foetal osteoblast, fO; adult osteoblasts, aO; pulmonary foetal fibroblast, HFL-1, fF; adult fibroblasts, aF; placental microvascular endothelial, mE; umbilical vein endothelial, uE; alveolar epithelial adenocarcinoma, A549, EA; bronchial epithelial adenocarcinoma, H322, EH; bronchial smooth muscle, SM; colorectal adenocarcinoma, CaCo2, CE; Epstein-Barr-transformed lymphocyte Ly; erythroleukaemia, K562, EK; erythroleukaemia TF1, ET; glioma, U87, G8; glioma, U373, G3; and negative control (-).
Discussion
A search of the human genomic sequences for the TDH gene identified only one candidate location on chromosome 8p23-22. This is the expected site of the human TDH gene since there are a number of genes that neighbour the TDH genes in the genomes of human, mouse and puffer fish that are found in common in this locus. They are the myotubularin related protein 8, the B-lymphoid tyrosine kinase and the hypothetical protein C8orf13. There are differences in the gene order and orientation in the human and puffer fish TDH loci indicating that there have been re-arrangements in this locus since the divergence of the human and puffer fish lineages [34].
The human TDH gene contains 3 mutations that disrupt the gene transcription and translation. There is a loss of the acceptor splice sites preceding exons 4 and 6 and an in-frame stop codon in exon 6. A database search of expressed human sequences identified other TDH cDNAs and ESTs similar to the 2 cDNA sequences described. The sequence of the human cerebellum full-length cDNA clone FLJ25033 (AK057762, NEDO human cDNA sequencing project University of Tokyo, Japan) has 2 5'UTR exons, the first of which corresponds to exon -1 on the mouse genomic DNA sequence [22]. This clone skips both exons 4 and 6, and includes the polyadenylation site. The ORF would encode a truncated 180 residue protein. Three very similar human ESTs (AI005002, AI243637 and AI809781) have exon 4 skipped and use the cryptic acceptor splice site in exon 6 that was found in clone 2 and possesses the in-frame stop codon.
The human TDH gene is a pseudogene because in all individuals examined the gene contains 1 or more mutations that disrupt RNA splicing which give rise to a variety of mRNAs in different individuals. In both pig and mouse TDH cDNAs there was no evidence of alternatively splicing [22]. In humans, arginine-214 is replaced by a stop codon. On translation all the human mRNAs would generate a variety of truncated proteins ranging from 157 to 230 residues, some of which would also contain deletions of residues 96–146; whereas the size of the core of TDH enzyme (residues 48–364) differs by only 2 residues in other species, including some bacterial TDH proteins [22]. The human truncated proteins would be non-functional since they would be unable to make appropriate contacts with the substrates, L-threonine and NAD+. They have lost most of the carboxy-terminal domain, which by homology with GALE [26-29], would be expected to bind L-threonine and also have lost various regions of the NAD+ binding motif that extends from exon 1 to exon 6. In comparison, even single point missense mutations in the human GALE gene disrupt protein function resulting in epimerase-deficiency galactosemia [35].
At present, no known human genetic disease has been associated with defects in the threonine dehydrogenase and 2-amino-3-ketobutyrate coenzyme A ligase biochemical pathway. This may be because the enzymatic activity of L-serine/threonine dehydratase is sufficient to metabolise L-threonine in humans. The mitochondrial threonine dehydrogenase enzyme is thought to act in the maintenance of free somatic threonine concentration derived from dietary threonine [15] suggesting that humans may not be able to regulate the level of circulating threonine as well as other mammals. In vertebrates, L-threonine is degraded by two major enzymatic pathways and attempts to determine their relative contribution in humans has been by indirect methods. Zhao et al. measured threonine oxidation by the production of labelled CO2 in adult humans from labelled threonine and concluded that the conversion of threonine to glycine through the threonine dehydrogenase pathway was not important if it exists at all [19]. More recently, Darling et al. used a more sensitive method to measure the production of labelled glycine and CO2 and suggested that there was some threonine catabolism via the threonine dehydrogenase pathway, but it accounted for only 7–11% of total [21]. Direct demonstration of the absence of TDH enzymatic activity in human tissues such as liver remains to be demonstrated. In contrast, in normally-fed pigs and rats 80 to 87% of L-threonine catabolism occurs via threonine dehydrogenase [15,16]. Since the TDH gene is not functioning in humans, what enzyme(s) could be responsible for the small percentage of threonine catabolism to glycine? In bacteria, threonine aldolase also yields glycine, but mammals are thought to lack the "genuine" threonine aldolase [36]. However, serine hydroxymethyltransferase exhibits a low threonine aldolase activity in mammalian liver [36].
Whether the first dehabilitating mutation was the loss of the splice acceptor site preceding exon 6 or the arginine-214 to stop codon mutation is unknown, nor is it known when in human evolution that a functional TDH gene was lost. However, the arginine codon CGA is used in only 11.1% of arginine residues in humans, being prone to mutate to a stop codon. The TDH gene has been conserved throughout evolution, being found in bacteria and other mammals [22], so why has it not been conserved in man? How could such a mutation establish its self in the ancestral population? It is likely that the mutation must have conferred some selectable advantage, under the prevailing environmental conditions, on those individuals who carried it, and arose at a time when the ancestral population was small. Under conditions of protein starvation, threonine dietary intake could have been a limiting factor on growth, survival and successful reproduction and a reduction in the rate of threonine catabolism would have conferred a selective advantage on those individuals with defective TDH gene. Additionally, if we consider that the threonine catabolism pathway is also a glycine synthesis pathway, then a reduction in the production of glycine in inhibitory glycinergic neurons [37] may have contributed to human neural evolution.
Since humans lack a functioning threonine dehydrogenase enzyme and human parasites such as trypanosomes do possess one (Edgar and Horn, unpublished results, GenBank AF529241), the parasite threonine dehydrogenase enzyme is a potential target for therapeutic intervention. Indeed, in trypanosomes such as Trypanosoma brucei, which causes sleeping sickness, L-threonine dehydrogenase is an important metabolic enzyme and inhibition of this enzyme by a wide range of sulphydryl reagents, such as tetraethylthiuram disulphide, leads to a loss of trypanosome viability [38-40].
Loss of other metabolically important genes in man results in disease susceptibility. Man and primates are scurvy-prone, having lost the L-gulono-gamma-lactone oxidase gene that is found in most other eukaryotes, and are unable to synthesize L-ascorbic acid [41]. Man and primates have also lost the urate oxidase enzyme that catalyses the conversion of uric acid to allantoin. This results in a high concentration of uric acid in the blood, predisposing man to hyperuricemia that can lead to gouty arthritis and renal stones [42]. Humans have recently lost 2 functional genes involved in sialic acid function. They are the CMP-N-acetylneuraminic acid hydroxylase gene (CMAH) [43] and the siglec-like molecule (Siglec-L1) [44]. However, no diseases have yet been associated with the loss of these genes. The CMAH enzyme converts the sialic acid, N-acetylneuraminic acid to N-glycolylneuraminic acid, potentially affecting recognition by a variety of endogenous and exogenous sialic acid-binding lectins. Siglecs are immunoglobulin superfamily member lectins that selectively recognize different sialic acids types and siglec-L1 preferentially recognizes N-glycolylneuraminic acid. To date, no disease has been associated with the loss of the TDH gene, but humans may not be able to metabolise high protein diets as efficiently as other mammals.
Conclusions
The human L-threonine 3-dehydrogenase gene is an expressed pseudogene, which accounts for the very low levels of threonine oxidation measured in humans by the production of labelled CO2 from labelled threonine. This suggests that the L-serine/threonine dehydratase pathway may be the only route in the catabolism of threonine in humans. The presence of a functional TDH gene in key mammal species such as pigs and mice and its loss in man should be taken into consideration when utilising hepatocytes in bioartificial livers and pharmacokinetic studies.
Methods
Genotyping of genomic DNA from individuals by restriction enzyme digests and DNA sequencing
Human genomic DNA was isolated from peripheral blood [45]. Regions of genomic DNA surrounding exon 4 and 6 splice acceptor sites were amplified by PCR using primers ATTGCGTGGCTACCAGTGA and CGATCTCCCGAAGATTCTTGT for exon 4 and GAATGAGATTTCAGAAAAAGGCAGG and AACCAGTTGTTCCTCCTCCA for exon 6, using the Advantage 2 cDNA polymerase mix (BD-Clontech, UK). Amplification conditions were: 30 cycles of 94°C for 10 sec and 57°C for 10 sec. PCR products were desalted using the QIAquick PCR purification kit (Qiagen, UK). Amplicons from the exon 4 splice acceptor site were digested in appropriate buffer with the restriction enzyme AciI. Amplicons from the exon 6 splice acceptor site were digested with the restriction enzyme Hsp92II. The amplicons were also sequenced directly using the big dye terminator cycle sequencing ready reaction kit and AmpliTaq DNA polymerase FS and run on an ABI 377 automated DNA sequencers (both from PE Applied Biosystems, UK).
Molecular cloning of human L-threonine dehydrogenase cDNAs
Clones encoding the human L-threonine dehydrogenase cDNA sequence were obtained by touchdown PCR amplification from liver and lung cDNA libraries using primers derived from regions of the sequence of the human genomic DNA clone RP11-110L10 which had homology to the 5' and 3' ends of the ORF of the porcine TDH cDNA sequence. The primers were: ATGCTGTTCATTAGGATGCTGA and GTTGGCTTGGGCAACTCTG and the cycling conditions were 94°C for 10 sec, 64°C for 10 sec and 72°C for 1 min less -1.0°C per cycle for 8 cycles followed by 24 cycles with an annealing temperature of 56°C. PCR products were examined by agarose gel electrophoresis and stained with ethidium bromide. For cloning, PCR products were excised from low-melting point agarose gels and the agarose digested with agarase (Promega, UK). The PCR products were cloned into the T-A vector pCR4-TOPO (Invitrogen, The Netherlands) and sequenced in both directions.
Tissue and cellular distribution of L-threonine dehydrogenase mRNA by RT-PCR
Human cDNA from 14 tissues (BD-Clontech) was analysed for the relative expression of TDH and the housekeeping gene, β-actin. Approximately, 0.2 ng of oligo-dT primed cDNA (derived from poly A+ RNA) from each tissue was amplified by PCR using Taq Gold polymerase (AB Applied Biosystems). For the analysis of TDH expression in 15 cultured cell isolates and cell lines, total RNA was extracted using guanidine thiocyanate and treated with DNase-I to remove any contaminating genomic DNA (total RNA isolation system, Promega). Total RNA was reverse transcribed with random hexamer primers using an AMV RNase H- reverse transcriptase, ThermoScript (Invitrogen). Approximately, cDNA derived from 50 ng of total RNA was used in each PCR. Tissue and cell master mixes were divided into gene specific mixes with the addition of PCR primers to a final concentration of 200 μM. The L-threonine dehydrogenase primers were: exon 7 and 8 boundary, GCCAGGCCATAGCGGAT and exon 8, TGTTTCCACCCCCAGTCCTT and produced a 73 bp amplicon and the β-actin primers were as previously described [46] and produced a 208 bp amplicon. The amplification conditions were; a 10 min hot start to activate the polymerase followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The amplicons were examined by ethidium bromide stained agarose gel electrophoresis.
Acknowledgments
Acknowledgements
I thank Athina Milona for assistance with the genotyping and the Advanced Biotechnology Centre for DNA sequencing (Charing Cross Campus, Imperial College).
References
- Strain AJ, Neuberger JM. A bioartificial liver – state of the art. Science. 2002;295:1005–1009. doi: 10.1126/science.1068660. [DOI] [PubMed] [Google Scholar]
- Tsiaoussis J, Newsome PN, Nelson LJ, Hayes PC, Plevris JN. Which hepatocyte will it be? Hepatocyte choice for bioartificial liver support systems. Liver Transpl. 2001;7:2–10. doi: 10.1053/jlts.2001.20845. [DOI] [PubMed] [Google Scholar]
- Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Cross DM, Bayliss MK. A commentary on the use of hepatocytes in drug metabolism studies during drug discovery and development. Drug Metab Rev. 2000;32:219–240. doi: 10.1081/DMR-100100574. [DOI] [PubMed] [Google Scholar]
- Dabos KJ, Nelson LJ, Bradnock TJ, Parkinson JA, Sadler IH, Hayes PC, Plevris JN. The simulated microgravity environment maintains key metabolic functions and promotes aggregation of primary porcine hepatocytes. Biochim Biophys Acta. 2001;1526:119–130. doi: 10.1016/S0304-4165(01)00097-6. [DOI] [PubMed] [Google Scholar]
- Hoskin SO, Gavet S, Milne E, Lobley GE. Does glutamine act as a substrate for transamination reactions in the liver of fed and fasted sheep? Br J Nutr. 2001;85:591–597. doi: 10.1079/bjn2001332. [DOI] [PubMed] [Google Scholar]
- Leoncini R, Vannoni D, Di Pietro MC, Guerranti R, Rosi F, Pagani R, Marinello E. Restoration of rat liver L-threonine dehydratase activity by pyridoxamine 5'-phosphate: the half-transaminating activity of L-threonine dehydratase and its regulatory role. Biochim Biophys Acta. 1998;1425:411–418. doi: 10.1016/S0304-4165(98)00094-4. [DOI] [PubMed] [Google Scholar]
- FAO/WHO/UNU Energy and protein requirements. Report of a Joint FAO/WHO/UNU Expert Consultation. World Health Organ Tech Rep Ser. 1985;724:1–206. [PubMed] [Google Scholar]
- Borgonha S, Regan MM, Oh SH, Condon M, Young VR. Threonine requirement of healthy adults, derived with a 24-h indicator amino acid balance technique. Am J Clin Nutr. 2002;75:698–704. doi: 10.1093/ajcn/75.4.698. [DOI] [PubMed] [Google Scholar]
- Dale RA. Catabolism of threonine in mammals by coupling of L-threonine 3-dehydrogenase with 2-amino-3-oxobutyrate-CoA ligase. Biochim Biophys Acta. 1978;544:496–503. doi: 10.1016/0304-4165(78)90324-0. [DOI] [PubMed] [Google Scholar]
- Boylan SA, Dekker EE. L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12. J Biol Chem. 1981;256:1809–1815. [PubMed] [Google Scholar]
- Aoyama Y, Motokawa Y. L-Threonine dehydrogenase of chicken liver. Purification, characterization, and physiological significance. J Biol Chem. 1981;256:12367–12373. [PubMed] [Google Scholar]
- Pagani R, Leoncini R, Righi S, Guerranti R, Lazzeretti L, Marinello E. Mitochondrial L-threonine dehydrogenase. Biochem Soc Trans. 1992;20:379S. doi: 10.1042/bst020379s. [DOI] [PubMed] [Google Scholar]
- Tressel T, Thompson R, Zieske LR, Menendez MI, Davis L. Interaction between L-threonine dehydrogenase and aminoacetone synthetase and mechanism of aminoacetone production. J Biol Chem. 1986;261:16428–16437. [PubMed] [Google Scholar]
- Bird MI, Nunn PB. Metabolic homoeostasis of L-threonine in the normally-fed rat. Importance of liver threonine dehydrogenase activity. Biochem J. 1983;214:687–694. doi: 10.1042/bj2140687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballevre O, Cadenhead A, Calder AG, Rees WD, Lobley GE, Fuller MF, Garlick PJ. Quantitative partition of threonine oxidation in pigs: effect of dietary threonine. Am J Physiol. 1990;259:E483–491. doi: 10.1152/ajpendo.1990.259.4.E483. [DOI] [PubMed] [Google Scholar]
- House JD, Hall BN, Brosnan JT. Threonine metabolism in isolated rat hepatocytes. Am J Physiol Endocrinol Metab. 2001;281:E1300–1307. doi: 10.1152/ajpendo.2001.281.6.E1300. [DOI] [PubMed] [Google Scholar]
- Hammer VA, Rogers QR, Freedland RA. Threonine is catabolized by L-threonine 3-dehydrogenase and threonine dehydratase in hepatocytes from domestic cats (Felis domestica). J Nutr. 1996;126:2218–2226. doi: 10.1093/jn/126.9.2218. [DOI] [PubMed] [Google Scholar]
- Zhao XH, Wen ZM, Meredith CN, Matthews DE, Bier DM, Young VR. Threonine kinetics at graded threonine intakes in young men. Am J Clin Nutr. 1986;43:795–802. doi: 10.1093/ajcn/43.5.795. [DOI] [PubMed] [Google Scholar]
- Darling PB, Dunn M, Sarwar G, Brookes S, Ball RO, Pencharz PB. Threonine kinetics in preterm infants fed their mothers' milk or formula with various ratios of whey to casein. Am J Clin Nutr. 1999;69:105–114. doi: 10.1093/ajcn/69.1.105. [DOI] [PubMed] [Google Scholar]
- Darling PB, Grunow J, Rafii M, Brookes S, Ball RO, Pencharz PB. Threonine dehydrogenase is a minor degradative pathway of threonine catabolism in adult humans. Am J Physiol Endocrinol Metab. 2000;278:E877–884. doi: 10.1152/ajpendo.2000.278.5.E877. [DOI] [PubMed] [Google Scholar]
- Edgar AJ. Molecular cloning and tissue distribution of mammalian L-threonine 3-dehydrogenases. BMC Biochem. 2002;3:19. doi: 10.1186/1472-2091-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar AJ, Polak JM. Molecular cloning of the human and murine 2-amino-3-ketobutyrate coenzyme A ligase cDNAs. Eur J Biochem. 2000;267:1805–1812. doi: 10.1046/j.1432-1327.2000.01175.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Hegeman AD, Wesenberg G, Chapeau MC, Frey PA, Holden HM. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:6294–6304. doi: 10.1021/bi970025j. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Holden HM. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1998;37:11469–11477. doi: 10.1021/bi9808969. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem. 2001;276:15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Henderson JM, Fridovich-Keil JL, Holden HM. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J Biol Chem. 2002;277:27528–27534. doi: 10.1074/jbc.M204413200. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Bauer AJ, Rayment I, Frey PA, Holden HM. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 A resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJE. Enhanced Genome Annotation using Structural Profiles in the Program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- Brenner S, Venkatesh B, Yap WH, Chou CF, Tay A, Ponniah S, Wang Y, Tan YH. Conserved regulation of the lymphocyte-specific expression of lck in the Fugu and mammals. Proc Natl Acad Sci USA. 2002;99:2936–2941. doi: 10.1073/pnas.032680599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlers TM, Christacos NC, Harreman MT, Fridovich JL-Keil. Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia. Am J Hum Genet. 1999;64:462–470. doi: 10.1086/302263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Gomi T, Fujioka M. Serine hydroxymethyltransferase and threonine aldolase: are they identical? Int J Biochem Cell Biol. 2000;32:289–301. doi: 10.1016/S1357-2725(99)00113-2. [DOI] [PubMed] [Google Scholar]
- Legendre P. The glycinergic inhibitory synapse. Cell Mol Life Sci. 2001;58:760–793. doi: 10.1007/PL00000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross GA, Klein RA, Linstead DJ. Utilization of amino acids by Trypanosoma brucei in culture: L-threonine as a precursor for acetate. Parasitology. 1975;71:311–326. doi: 10.1017/s0031182000046758. [DOI] [PubMed] [Google Scholar]
- Linstead DJ, Klein RA, Cross GA. Threonine catabolism in Trypanosoma brucei . J Gen Microbiol. 1977;101:243–251. doi: 10.1099/00221287-101-2-243. [DOI] [PubMed] [Google Scholar]
- Baker JR, Selden LF. Trypanosoma (Schizotrypanum) dionisii as a model for chemotherapeutic studies related to Chagas's disease. Trans R Soc Trop Med Hyg. 1981;75:80–85. doi: 10.1016/0035-9203(81)90015-8. [DOI] [PubMed] [Google Scholar]
- Nishikimi M, Fukuyama R, Minoshima S, Shimizu N, Yagi K. Cloning and chromosomal mapping of the human nonfunctional gene for L-gulono-gamma-lactone oxidase, the enzyme for L-ascorbic acid biosynthesis missing in man. J Biol Chem. 1994;269:13685–13688. [PubMed] [Google Scholar]
- Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: primary structure and evolutionary implications. Proc Natl Acad Sci USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T, Varki NM, Varki A. A second uniquely human mutation affecting sialic acid biology. J Biol Chem. 2001;276:40282–40287. doi: 10.1074/jbc.M105926200. [DOI] [PubMed] [Google Scholar]
- John SW, Weitzner G, Rozen R, Scriver CR. A rapid procedure for extracting genomic DNA from leukocytes. Nucleic Acids Res. 1991;19:408. doi: 10.1093/nar/19.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerwald N, Cohen J, Herrera RJ, Brenner CA. Analysis of gene expression in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT-PCR. Mol Hum Reprod. 1999;5:1034–1039. doi: 10.1093/molehr/5.11.1034. [DOI] [PubMed] [Google Scholar]