Abstract
Phytochelatin synthase (PCS) is a key enzyme for heavy-metal detoxification in plants. PCS catalyzes the production of glutathione (GSH)-derived peptides (called phytochelatins or PCs) that bind heavy-metal ions before vacuolar sequestration. The enzyme can also hydrolyze GSH and GS-conjugated xenobiotics. In the cyanobacterium Nostoc, the enzyme (NsPCS) contains only the catalytic domain of the eukaryotic synthase and can act as a GSH hydrolase and weakly as a peptide ligase. The crystal structure of NsPCS in its native form solved at a 2.0-Å resolution shows that NsPCS is a dimer that belongs to the papain superfamily of cysteine proteases, with a conserved catalytic machinery. Moreover, the structure of the protein solved as a complex with GSH at a 1.4-Å resolution reveals a γ-glutamyl cysteine acyl–enzyme intermediate stabilized in a cavity of the protein adjacent to a second putative GSH binding site. GSH hydrolase and PCS activities of the enzyme are discussed in the light of both structures.
Keywords: cysteine protease, heavy-metal detoxification, phytochelatin synthase
Phytochelatins (PCs) are known to be the main heavy-metal-detoxifying peptides in the plant kingdom (1). PCs are glutathione (GSH)-derived peptides of the general formula (γ-Glu-Cys)nGly (with n = 2–11), which have been identified in a wide variety of plant species and in some microorganisms. They act as heavy metal (mainly Cd2+) chelators and favor their vacuolar sequestration (2–4). PCs are synthesized posttranslationally in the presence of heavy-metal ions by the PC synthase (PCS), a γ-glutamyl-cysteine transpeptidase (5).
Genes encoding PCS have been isolated and characterized in Arabidopsis thaliana (AtPCS1) (6), Schizosaccharomyces pombe (SpPCS) (4), and wheat (Triticum aestivum; TaPCS1) (7) but also in the model nematode Caenorhabditis elegans (CePCS1) (8, 9). Sequence alignment of PCS proteins reveals a high degree of similarity in the N-terminal domain whereas the C-terminal region turns out to be extremely variable. Limited proteolysis and mutant analyses confirmed that the N-terminal core domain is sufficient to confer a PCS activity and therefore can be referred to the catalytic domain (7, 10, 11). However, the C-terminal domain was shown to ensure higher PCS activity, improved protein stability, and response to a broader spectrum of heavy-metal ions (10).
Several groups have proposed that PCS is a bisubstrate enzyme, in which the donor molecule is the GSH and the acceptor is another GSH molecule or a PCn molecule (12). Recently, Rea and coworkers (13) have gone further and showed by in vitro radiolabeling experiments on AtPCS1 that this class of enzyme corresponds to a dipeptidyl transferase whose mechanism takes place in two distinct steps: (i) the formation of a γ-glutamyl-cysteine acyl–enzyme intermediate resulting from the GSH deglycination followed by (ii) the transfer of the γ-glutamylcysteine unit from the acyl–enzyme to the acceptor molecule (GSH or PCn).
After the pioneering in vivo experiments of Zenk and coworkers (1, 14), several groups have shown in vitro in various organisms that no significant activity was detectable in the absence of Cd2+ or other heavy-metal ions (4–6). However, the specific role of divalent metals in PC synthesis is still controversial. Cobbett (12) first proposed that the variable C-terminal domain would bind, via conserved cysteine residues, the heavy-metal ions and then transfer them to the catalytic N-terminal domain. Later, Maier et al. (15) performed Cd-binding assays using peptide libraries of PCS and found that five conserved cysteines in the N-terminal domain of AtPCS1 could bind Cd2+ and were also essential for the PCS activity. Rea and coworkers (13, 16) suggested another model, in which Cd2+ does not directly bind to the protein but rather forms a heavy-metal–GS2 (or PCn) thiolate complex, which would act as the acceptor molecule. According to those authors, three conserved residues in the N-terminal domain, Cys-56, His-162, and Asp-180, when substituted by site-directed mutagenesis, completely abolish the activity of AtPSC1 (17). Their actual assumption is that the first γ-Glu-Cys acyl intermediate of reaction is a thioester, resulting from the nucleophilic attack of Cys-56 on the GSH Cys—Gly peptidic bond. Also based on sequence homology, Rea et al. (17) proposed that the PCS mechanism could be similar to that of papain and, more generally, cysteine proteases. The PCS and cysteine protease mechanisms would only differ in their second step, i.e., the nucleophilic attack on the thioester intermediate: in the PCS case, the acceptor molecule is a second GSH molecule or a PCn molecule, and, in the cysteine protease case, the acceptor is a water molecule, resulting in a net hydrolysis. Such a mechanism is compatible with the ability of PCS to catalyze the cleavage of glycine from GS-conjugated xenobiotics, thus contributing to their degradation (18).
To go beyond these first insights into the PCS mechanism, the three-dimensional structure of PCS or of its nearest homologues is greatly needed. No structural information on PCS is currently available. Recent sequencing has revealed a PCS-related gene in the genome of cyanobacteria (19, 20). The corresponding alr0975 gene derived from Nostoc encodes a protein (called NsPCS) sharing 36% identity at the amino acid level with AtPCS1. The sequence contains only the conserved N-terminal domain of the PCS eukaryotic family. NsPCS has been found to catalyze essentially the first step in PC synthesis, which converts GSH to γ-glutamyl cysteine, whereas only weakly net synthesis of PC2 occurs, i.e., the transfer of the γ-glutamylcysteine unit from the acyl–enzyme to another GSH molecule. No significant difference on the NsPCS activity was found in the presence or in the absence of heavy-metal ions (11, 19, 20). Here, we report crystal structures of (i) NsPCS at 2.0-Å resolution and (ii) the γ-glutamyl cysteine acyl–enzyme intermediate at 1.4-Å resolution. To our knowledge, this enzyme–substrate complex structure under its acyl form is the first one reported for a cysteine protease. The overall structure and a detailed analysis of the active site are described. Our results confirm the previous prediction that PCS belongs to the papain superfamily of cysteine proteases, with the structurally conserved “catalytic triad” and oxyanion hole in the active site. Both structures also cast new light on the putative role of the other eukaryotic conserved cysteines and on the origin of the differences between the cyanobacterial and the eukaryotic PCS catalytic activities. Overall, these structures contribute to a better understanding of the PCS mechanism and provide a structural framework for further functional studies.
Materials and Methods
Bacterial Expression and Protein Purification. NsPCS without the 24 N-terminal residues putatively coding for a signal peptide was cloned in pJC40 with a 10His Tag at the N terminus (20). The recombinant protein was overexpressed in Escherichia coli BL21 strain, and cells were first incubated at 37°C overnight on 10 ml of LB medium. The cells were pelleted, washed in sterile water, and then resuspended into 1 liter of methionine-minus medium from Molecular Dimensions, supplemented with 40 mg of l-selenomethionine (SeMet), l-lysine, l-threonine, and l-phenylalanine [these three last amino acids create an inhibition of the methionine biosynthesis pathway (21)]. Protein expression was induced at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside. After 3 h of growth, cells were pelleted, resuspended in buffer A [containing 20 mM phosphate (pH 8.0), 500 mM NaCl, 20 mM imidazole, and 1 mM Tris(2-carboxyethyl)phosphine], and disrupted with a French press at 7 MPa. The resulting soluble fraction was loaded on a nickel-charged column (HisTrap column, Amersham Pharmacia), and the protein was eluted by an imidazole step gradient (150 mM wash and 500 mM elution). Because NsPCS precipitated in buffer A after 2 h at room temperature or during the following concentration steps, the protein was then exchanged into buffer B [50 mM Mes (pH 5.5), 50 mM NaCl, and 1 mM Tris(2-carboxyethyl)phosphine] and concentrated up to 10 mg·ml–1 before crystallization trials. The yield of SeMet NsPCS was typically of 5 mg per liter of culture. The incorporation of SeMet was confirmed by MALDI mass spectrometry.
Crystallization. All of the crystallization experiments were carried out at 293 K by using the hanging drop vapor diffusion technique. Optimal crystallization conditions were obtained by a microseeding technique. The best crystals of the native protein were obtained by mixing 2 μl of SeMet-labeled NsPCS with 2 μl of a reservoir solution (1 ml) containing 100 mM Mes (pH 5.0), 15–20% (wt/vol) PEG 4K, and 100–400 mM NaCl. Thin plates (approximate dimensions: 10 × 100 × 100 μm3) grew in ≈2 days and belong to space group P21 with unit-cell parameters as follows: a = 47.86 Å, b = 62.47 Å, c = 76.55 Å, and β = 101.38° (see Table 1, which is published as supporting information on the PNAS web site). Crystals of the acyl–enzyme were obtained at acidic pH by mixing 2 μl of SeMet-labeled NsPCS with 0.2 μl of a 200 mM GSH solution and 2 μl of a reservoir solution containing 100 mM citric acid (pH 2.6–3.4), 100 mM NaCl, and 10–20% (wt/vol) PEG 4K. Although crystals have the same shape as the native crystals, they grow in ≈2 weeks and correspond to a different crystal form (Table 1).
Data Collection and Phasing. Native and acyl–enzyme crystals were soaked 1 min in mother liquor containing 30% glycerol and 50% ethylene glycol, respectively, and then directly flash-frozen in liquid nitrogen before data collection. Diffraction data on the native crystal were collected at three wavelengths at beamline BM30 (European Synchrotron Radiation Facility, Grenoble, France) by using a 180-mm MAR CCD detector, and diffraction data on the acyl–enzyme crystal were collected at beamline ID29 on an ADSC Q210 2D detector. Data sets were processed with mosflm (22) and scaled by using scala from the ccp4 suite (23, 24).
Structure Determination and Refinement. Ten selenium positions were found from the MAD data by the program solve (25). From these positions the NCS operators for a dimer could be derived, and, subsequently, 70% of the molecule was automatically built by the program. Several rounds of iterative model building and refinement were performed by using the programs turbo (26) and refmac (27). The final native model contains two molecules (consisting of amino acids 29–239), 259 solvent molecules, and two ions assigned as calcium. Electron density is missing for the His-tag and the four N-terminal residues as well as the three C-terminal residues.
The acyl–enzyme structure was solved by molecular replacement by using the program molrep (28) with the native dimeric structure as a search model. After several cycles of model building and refinement with refmac, the final model displays the same disordered regions as the native model, with additional disorder for residues 87–94, 106–108 in molecule A, and 29–31 and 88–91 in molecule B. Additionally, the main chain and side chain of Arg-180 (a Ramachandran outlier in the native structure) display a large disorder in both molecules and have been modeled as a glycine. The final model contains two molecules, 391 solvent molecules, and four ions (assigned as two calcium and two chlorides). For both structures, all of the residues fall in the favored region of the Ramachandran plot except for the above-mentioned Arg-180. procheck (29) was used to assign secondary structure and revealed that structural parameters were consistently better than or within the average for structures at comparable resolution. Graphics presented in this article were generated by using the program pymol (www.pymol.org).
Results
Overall Structure. We determined the crystal structures of NsPCS in its native form and as a complex with GSH, its natural substrate. The structure of the selenomethionine-labeled NsPCS in its native form was first solved by MAD and then refined to 2-Å resolution with an R-factor of 20.%0 (Rfree = 25.7%; see Table 1). The protein–GSH complex was trapped and crystallized at noncatalytically active pH values (≈3). The resulting structure, solved by molecular replacement and refined to 1.4-Å resolution (R = 17.4%; Rfree = 18.7%), corresponds to an acyl–enzyme complex. In both the native and the acyl–enzyme crystal forms, the asymmetric unit contains two monomers. With an rms deviation <0.5 Å, these monomers are almost identical; the few structural differences will be detailed below. Each monomer has an overall “crescent” shape (with approximate dimensions of 50 × 30 × 30 Å3) with α/β fold containing eight α-helices and six β-strands (Fig. 1A). A structural homology search with dali (30) revealed that the NsPCS monomer is structurally most similar to staphopain [z = 11.5 (31)], IdeS [z = 10.8 (32)], and bleomycin hydrolase [z = 10.4 (33)]. As a result, and despite very low sequence similarity (sequence identity is in all cases <10%), NsPCS fold definitely shows that it belongs to the papain superfamily of cysteine proteases. Moreover, the main catalytic residues responsible for the protease activity are structurally conserved.
Fig. 1.
Overall structure of the NsPCS dimer and its relationship with the papain family. (A) Stereo view of the overall NsPCS dimer. The ribbon is colored in gray for one monomer and in green for the other one. A comparison with the papain fold shows additional secondary structures drawn here in yellow or red depending on the monomer to which they belong. (B) Stereo view of the overlap of the NsPCS (green) and papain (white) active-site residues.
In the two different crystal forms that we solved, two NsPCS molecules are clamped as a dimer (Fig. 1 A). The homodimerization is mediated mainly by the two N-terminal helices, which are absent from the papain fold and therefore represent a specificity of the PCS family. Upon dimerization, ≈1,130 Å2 are buried, corresponding to 12% of the solvent-accessible surface of each monomer. Although 13 intermolecular hydrogen bonds are observed at the dimer interface, 71% of the interacting surface is nonpolar with numerous Van der Waals contacts.
The Active Site. Detection and analysis of the residues involved in the catalysis benefit from previous mutagenesis studies (17) as well as from the considerable work done on other cysteine proteases and notably on papain-like proteins (34). The active site of NsPCS is formed by the so-called catalytic triad composed of residues Cys-70 (equivalent to Cys-25 in papain), His-183 (His-159 in papain), and Asp-201 (Asn-175 in papain) (Fig. 1B). Cys-70 is the catalytic cysteine, and Asp-201, with its side chain involved in a hydrogen bond with His-183 Nδ1, is responsible for both the correct orientation and polarization of the imidazolium group of His-183. Overall, the rms deviation between all atoms of the catalytic triad in NsPCS and papain is 0.65 Å. Furthermore, Gln-64 in NsPCS occupies an analogous position to Gln-19 in papain, whose side-chain amide group, together with the main-chain amide group of Cys-25, define the so-called “oxyanion hole.” This oxyanion hole is known to polarize the carbonyl group of the substrate scissile bond. The large structural similarities observed between the active site of both papain-like proteins and NsPCS have functional consequences and suggest a catalytic mechanism of the same kind. The main differences between the two families around the active site concern the other residues in interaction with Asp-201. In typical cysteine proteases, Asp-201 is shielded by an aromatic residue (Trp-177 in papain). In NsPCS, a short residue, Ser-203, takes this position, which results in an open space filled by the side chain of Arg-173 that is hydrogen bonded via its guanidine group to Ser-203 (OH) and Asp-201 (Oδ1). This unusual geometry might be important for NsPCS catalysis, as will be discussed below.
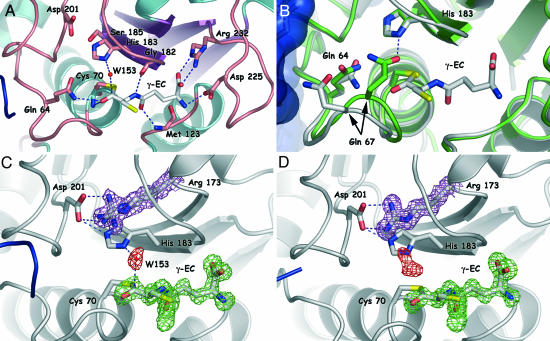
The Acyl–Enzyme Complex. The structure of the acyl–enzyme complex shows that, as expected in papain-like proteins, the carbonyl oxygen of the thioester points toward the oxyanion hole formed by Gln-64 Nε2 and Cys-70 N (Fig. 2A). The γ-EC is also held in place by other numerous hydrogen bonds involving the main chain of Gly-182 and Met-123 and the side chain of Arg-232 and Asp-225. The γ-EC orientation is such that the sulfur atom of the substrate points toward the solvent medium. A significant residual density is linked to this sulfur atom and could be due to its partial oxidation (Fig. 2 C and D). The carbonyl oxygen of the thioester linkage is distorted from planarity by 37° in molecule A and 30° in molecule B. These angles are intermediate between the planarity observed in the elastase/β-casomorphine-7 acyl–enzyme complex and the tetrahedral intermediate resulting from a nucleophilic attack on the thioester bond (35, 36). The catalytic water molecule W153, found in only one molecule of the dimer (molecule A), is ideally placed for this nucleophilic attack toward the synthesis of free γ-EC. W153, which is 2.70 Å away from the Nε2 of the imidazole ring of His-183 and 3.15 Å from the thioester carbon, forms indeed an angle of 103° relative to the carbonyl bond, a value close to the expected Bürgi's angle of 105° (37).
Fig. 2.
Residues involved in the binding of γ-EC and structural changes upon acylation. (A) View of selected residues in molecule A around the γ-EC binding site. Hydrogen bonds between the protein and the γ-EC moiety are depicted as blue broken lines. (B) Superimposition of the native NsPCS structure (green) and the acyl–enzyme complex (white). Note that Oε1 of Gln-64 in the native structure occupies the position of W153 (not shown for clarity) in the acyl–enzyme intermediate. (C and D) Comparison of the electron-density map around the γ-EC moiety in molecule A (C) and molecule B (D). In both cases, the electron-density omit map of γ-EC (contoured at +2.5σ and colored in green) is superimposed with the 2m Fo – Fc map around the position of the catalytic water and of the residue Arg-173 (contoured at +1σ and drawn in red and magenta, respectively). In molecule B of the acyl–enzyme intermediate, the electron density depicted in red is clearly not compatible with the presence of a water molecule (see text for details). In all of the figures, the second monomer is colored in blue.
Structural Changes Accompanying Acylation Reaction. Apart from a small reorientation of the two monomers relative to each other, two main structural rearrangements accompany the acylation reaction. First, in the native form, the B-loop 1 is in a “closed” conformation, with the side chain of Gln-67 hydrogen bonded to the side chain of the catalytic histidine (His-183). Instead, in the acyl–enzyme complex, the B-loop 1 is in an “open” configuration and reveals an open space close to the active site (Fig. 2B). Interestingly, in the native protein the Oε1 of Gln-67 takes the same position as the catalytic water molecule in the acyl–enzyme complex. The second main structural change is the destabilization of the protruding loop in the acyl–enzyme complex. Indeed, two segments and a total of 11 residues in this loop are not resolved in the electron-density map calculated at a 1.4-Å resolution. Because part of this loop is in the vicinity of the above-mentioned B-loop 1, the two structural changes may be connected.
There are some subtle but significant changes when comparing the active site of molecules A and B of the dimer in the acyl–enzyme structure (Fig. 2 C and D). First, as already mentioned, the catalytic water molecule W153 is present only in molecule A. Instead, a small but contiguous electron density is present in molecule B. This density could tentatively be ascribed to a glycine (a product of the reaction) or an ethylene glycol (used for the cryoprotection of the crystals). However, in both cases the refinement of these compounds was not satisfactory and we therefore left this density unassigned. Second, the electron-density map around Arg-173 clearly shows that its side chain adopts two conformations in molecule A. Of these two orientations, one is similar to what is found in the native structure and the other is similar to that found in molecule B. Another disparity visible from Fig. 2 C and D is the dynamics of the protruding loop, which appears to be different in molecule A, where residues 87–94 are disordered, as compared with molecule B, where the disorder occurs from residue 88 to residue 91. This difference is located relatively close to the active site and may thus influence the reactivity of the enzyme.
Discussion
As highlighted by the similarity of their fold and the superimposition of their active site, we have shown that NsPCS belongs to the papain superfamily of cysteine proteases. It is therefore expected that the peptide hydrolase activity of NsPCS, i.e., the deglycination of GSH, resembles the general and well known mechanism of papain-like cysteine proteases. By analogy with such a mechanism, Cys-70, His-183, and Asp-201 (equivalent to Cys-56, His-162, and Asp-180 in AtPCS1) correspond to the catalytic triad in NsPCS. Moreover, it is also highly probable that Cys-70 and His-183 form a thiolate–imidazolium ion pair, stabilized by Asp-201, and that the nucleophilic attack of Cys-70 on the Cys—Gly peptide bond is favored by the oxyanion hole, made up of Cys-70 and Gln-64. In a second step, the water molecule W153, whose nucleophilicity is enhanced by the proximity of His-183 and Asp-201, is then ideally placed to attack the thioester bond and liberate the γ-glutamylcysteine. We could trap the acyl–enzyme complex at very acidic pH (≈3). The pKa values characteristic of the formation of the thiolate–imidazolium ion pair (Cys-S–/(His)-ImH+) of papain-like proteins are pKa1 ≈ 3 and pKa2 ≈ 8. pKa1 was found as low as 2.5 in some papains (38), which proves that the formation of the thiolate–imidazolium ion pair and therefore the acylation step can occur even at a pH of ≈3. On the other hand, at this pH, the imidazolium group remains protonated. As a result, His-183 cannot act as a base, which makes the catalytic water molecule W153 unreactive and the acyl–enzyme stable. In a similar way, Wilmouth and colleagues (35, 36) succeeded in stabilizing at pH 5 an acyl–enzyme intermediate between a serine protease, the porcine pancreatic elastase, and the heptapeptide human β-casomorphin-7. We have also determined that the carbonyl oxygen of the thioester linkage is distorted from planarity and that the catalytic water molecule W153 forms an angle close to the expected Bürgi's angle for a nucleophilic attack on a carbonyl bond. The resulting geometry suggests that, as in the serine and cysteine protease mechanism, the hydrolysis reaction in NsPCS occurs through a tetrahedral intermediate. Consequently, because the amino acids of the catalytic triad and oxyanion hole are strictly conserved (Fig. 3), the crystal structures of NsPCS and of its acyl–enzyme conjugate enable us to conclude that the deglycination step in all of the PCS must occur through a very similar mechanism to that of a cysteine protease. The cysteine protease catalytic machinery is independent of the addition of any specific ion, which may explain why the cleavage of glycine from GSH in PCS-like proteins is not activated by Cd2+ or other heavy-metal ions (17). The NsPCS acyl–enzyme structure reveals also that the cysteine sulfur atom of the γ-EC moiety is exposed toward the solvent. It proves that the active site can easily accommodate a GSH-S-conjugate and catalyze its deglycination. The degradation of GSH-S-conjugate is currently proposed to be another function of PCS (18).
Fig. 3.
Sequence alignment of NsPCS with selected eukaryotic PCS sequences. Red stars denote the position of the catalytic triad. Red squares and triangles correspond to the residues whose main chain or side chain, respectively, is hydrogen-bonded to the γ-EC in the acyl–enzyme complex. Shading along the secondary structure elements represents the residues at the dimeric interface. In the consensus sequence, red boxes are drawn when the residues are strictly conserved along the alignment, whereas a simple letter is indicated when the sequences are conserved in all eukaryotic PCSs but Nostoc. Nos, Nostoc; Sp, S. pombe; Ce, C. elegans; At, A. thaliana PCS; Ta, T. aestivum; Nt, Nicotiana tabacum; Cons, consensus sequence.
NsPCS, in its native form or as a complex with GSH, was found dimeric in both types of crystal. Because the dimerization (i) is observed in two different crystal forms, (ii) results in an extended buried area, and (iii) is mediated mainly by hydrophobic contacts, it is very likely that the dimer is also the active form in solution. Many residues are conserved at the dimeric interface in the PCS family (Fig. 3), suggesting that the active form of all PCS-like proteins is also a dimer. It is noteworthy that Grill et al. (5) found out that PCS activity from Silene cucubalus elutes on gel filtration at a volume corresponding to a molecular mass of 95 kDa. Knowing that the molecular mass of PCS molecules falls around 50 kDa, it fits with a dimer, which is in agreement with our result. In the crystallographic dimer, the two monomers differ at several places. First, the catalytic water molecule W153 is visible in only one of them. Second, the observable conformations of Arg-173 and the number of water molecules in close interaction with this residue are different in the two monomers. Finally, the protruding loop, which is rather close to the active site, does not show the same disorder in both monomers. The two monomers are therefore structurally and mechanistically different, which suggests that some cooperativity could exist between them.
The two NsPCS structures are not sufficient to elucidate the mechanism of PC synthesis, i.e., the reaction of another GSH molecule or of a PCn molecule on the acyl–enzyme. NsPCS, unlike all other PCS-like proteins, can hardly synthesize PCn ≥ 2 (11, 19, 20); furthermore, we did not find in the acyl–enzyme structure any other fixed GSH or closely related molecule. The fact that the C-terminal part is absent in NsPCS is not crucial for the PCS activity because the N-terminal domain of AtPCS1 alone is active (10), which suggests that the acceptor binding site is in the N-terminal domain. Furthermore, if we suppose that the PC synthesis mechanism from the acyl–enzyme is similar to the deglycination step, the amino group of the incoming GSH is expected to take the place of the nucleophilic water molecule W153. As a consequence, the second substrate binding site should be close enough to the first GSH binding site. An empty zone, in connection with the cavity occupied by GSH, is actually observed in the acyl–enzyme structure and could accommodate another GSH molecule (Fig. 4). The incoming GSH molecule could be stabilized by conserved residues such as Arg-173, Asp-201, Gln-64, Lys-206, or Tyr-207, all of them involved in the formation of the cavity. Remarkably, two regions around this zone, the B-loops 1 and 3, are well conserved among all of the members of the PCS family, except NsPCS. Notably, Gln-67 (in the B-loop 1) and Arg-180 (in the B-loop 2) are replaced in the other PCS-like proteins by much less volumic residues, a proline and a glycine, respectively. We are therefore tempted to propose that the NsPCS difficulties to synthesize PCs is related to the nature and the orientation of these two loops, which could hinder the binding of the second substrate. On the other hand, the protruding loop, which is highly disordered in the acyl–enzyme complex and close to the active site, could also play a role, because the NsPCS sequence differs also from the consensus sequence in this region.
Fig. 4.
Analysis of the surface conservation among the PCS family. (A and B) Mapping of the sequence conservation in the native (A) and acyl–enzyme (B) structures. In each case the surface is colored according to the alignment found in Fig. 3. Red patches correspond to identical and homologous sequence conservation (red and gray shading in the alignment). Residues conserved in all PCSs but Nostoc are colored in yellow, except for cysteine residues falling in this category, which are colored in blue.
The role of the conserved cysteines for the PCS activity is currently debated. Four cysteines (referred to as Cys-90, Cys-91, Cys-109, and Cys-113 in PCS from A. thaliana) are conserved in all PCS-like proteins except NsPCS. It has been thought that these cysteines could participate in the fixation of heavy atom ions known to activate the synthesis of PCs (12, 15). Recently, Vatamaniuk et al. (13) showed by site-directed mutagenesis that none of these four cysteines decreased the ability of AtPCS1 to confer Cd2+ tolerance or mediate PC synthesis. Tsuji et al. (11) confirmed these results by showing that the activity of the AtPSC1 N-terminal domain remains unchanged when the four cysteines are replaced by the NsPCS corresponding residues Ala, Val, Arg, and Ser. We found that these four residues are very close to each other in the NsPCS structure. Based on the sequence alignment of Fig. 3, we can suppose that the corresponding cysteines in eukaryotic PCS form a “cysteine patch” stabilized by disulfide bonds. As illustrated in Fig. 4, this cysteine patch would be far from the active site and also from the putative acceptor binding site. Some care remains required because the region around Ala-111 and Val-112 is strongly unconserved, and a misalignment of the NsPCS sequence is conceivable. Nevertheless, the analysis of the NsPCS structures tends to confirm that these four cysteines residues are not determining for the PC synthesis, as pointed out by Vatamaniuk et al. (13) and Tsuji et al. (11).
In conclusion, we have proved that NsPCS belongs structurally and mechanistically to the papain superfamily of cysteine proteases. A catalytic triad and an oxyanion hole are responsible for the deglycination of a GSH donor molecule with a mechanism similar to that described for cysteine proteases. The attack of the acceptor molecule on the acyl–enzyme intermediate remains uncertain. However, the analysis of the NsPCS structures enables us to propose a location for the acceptor binding site and also to give some structural clue as to why, unlike the other PCS-like proteins, NsPCS synthesizes only very weakly PCs. The orientation of specific loops close to the active site could indeed obstruct the binding of the acceptor GSH molecule. Both structures also bring a new element about the role of conserved cysteine residues. In the NsPCS structure, these cysteines are far from the active sites, which supports the view that they are not crucial for PC synthesis. Finally, we propose that, as in the crystal, NsPCS is dimeric in solution. Some subtle but significant differences between these two dimers also suggest some cooperative mechanism in PCs synthesis.
Supplementary Material
Acknowledgments
We thank Dr. S. Clemens (Institute of Plant Biochemistry, Halle, Germany) for kindly providing us the pJC40-Alr0975 plasmid and for fruitful discussions; Dr. G. Peltier, Dr. S. Sauge-Merle, and S. Cuiné [Commissariatá l'Energie Atomique (CEA)/Cadarache, France] for their initial and continuous support in the project; and Dr. J. Lavergne (CEA/Cadarache, France) for critical reading of the manuscript. We are also grateful to the ID-29 and BM-30 staff (European Synchrotron Radiation Facility) for technical assistance in synchrotron data collection. This work was supported by the CEA.
Author contributions: P.A. and D.P. designed research; D.V., P.A., and D.P. performed research; and D.V., P.A., and D.P. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PC, phytochelatin; GSH, glutathione; PCS, PC synthase; NsPCS, PCS from Nostoc; AtPCS1, PCS from Arabidopsis thaliana; SeMet, l-selenomethionine.
Data deposition: The coordinates and structure factors for the native and acyl–enzyme structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2BTW and 2BU3, respectively).
References
- 1.Grill, E., Winnacker, E. L. & Zenk, M. H. (1985) Science 230, 674–676. [DOI] [PubMed] [Google Scholar]
- 2.Cobbett, C. S. (1999) Trends Plant Sci. 4, 335–337. [DOI] [PubMed] [Google Scholar]
- 3.Cobbett, C. & Goldsbrough, P. (2002) Annu. Rev. Plant Biol. 53, 159–182. [DOI] [PubMed] [Google Scholar]
- 4.Clemens, S., Kim, E. J., Neumann, D. & Schroeder, J. I. (1999) EMBO J. 18, 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grill, E., Loffler, S., Winnacker, E.-L. & Zenk, M. H. (1989) Proc. Natl. Acad. Sci. USA 86, 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vatamaniuk, O. K., Mari, S., Lu, Y. P. & Rea, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 7110–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha, S. B., Smith, A. P., Howden, R., Dietrich, W. M., Bugg, S., O'Connell, M. J., Goldsbrough, P. B. & Cobbett, C. S. (1999) Plant Cell 11, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, S., Schroeder, J. I. & Degenkolb, T. (2001) Eur. J. Biochem. 268, 3640–3643. [DOI] [PubMed] [Google Scholar]
- 9.Vatamaniuk, O. K., Bucher, E. A., Ward, J. T. & Rea, P. A. (2001) J. Biol. Chem. 276, 20817–20820. [DOI] [PubMed] [Google Scholar]
- 10.Ruotolo, R., Peracchi, A., Bolchi, A., Infusini, G., Amoresano, A. & Ottonello, S. (2004) J. Biol. Chem. 279, 14686–14693. [DOI] [PubMed] [Google Scholar]
- 11.Tsuji, N., Nishikori, S., Iwabe, O., Matsumoto, S., Shiraki, K., Miyasaka, H., Takagi, M., Miyamoto, K. & Hirata, K. (2005) Planta 222, 181–191. [DOI] [PubMed] [Google Scholar]
- 12.Cobbett, C. S. (2000) Plant Physiol. 123, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vatamaniuk, O. K., Mari, S., Lang, A., Chalasani, S., Demkiv, L. O. & Rea, P. A. (2004) J. Biol. Chem. 279, 22449–22460. [DOI] [PubMed] [Google Scholar]
- 14.Grill, E., Winnacker, E. L. & Zenk, M. H. (1987) Proc. Natl. Acad. Sci. USA 84, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier, T., Yu, C., Kullertz, G. & Clemens, S. (2003) Planta 218, 300–308. [DOI] [PubMed] [Google Scholar]
- 16.Vatamaniuk, O. K., Mari, S., Lu, Y. P. & Rea, P. A. (2000) J. Biol. Chem. 275, 31451–31459. [DOI] [PubMed] [Google Scholar]
- 17.Rea, P. A., Vatamaniuk, O. K. & Rigden, D. J. (2004) Plant Physiol. 136, 2463–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck, A., Lendzian, K., Oven, M., Christmann, A. & Grill, E. (2003) Phytochemistry 62, 423–431. [DOI] [PubMed] [Google Scholar]
- 19.Tsuji, N., Nishikori, S., Iwabe, O., Shiraki, K., Miyasaka, H., Takagi, M., Hirata, K. & Miyamoto, K. (2004) Biochem. Biophys. Res. Commun. 315, 751–755. [DOI] [PubMed] [Google Scholar]
- 20.Harada, E., von Roepenack-Lahaye, E. & Clemens, S. (2004) Phytochemistry 65, 3179–3185. [DOI] [PubMed] [Google Scholar]
- 21.Doublié, S. & Carter, C. W. J. (1992) in Crystallization of Nucleic Acids and Proteins: A Practical Approach, ed. Giégé, R. D. R. (Oxford Univ. Press, Oxford), pp. 311–317.
- 22.Leslie, A. G. W. (1993) in Proceedings of the CCP4 Study Weekend: Data Collection and Processing, eds. Sawyer, L., Isaacs, N. & Bailey, S. (Science and Engineering Research Council Daresbury Laboratory, Daresbury, U.K.), pp. 44–51.
- 23.Evans, P. R. (1997) Joint CCP4 and ESF-EACBM Newsletter 33, 22–24. [Google Scholar]
- 24.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763.15299374 [Google Scholar]
- 25.Terwilliger, T. C. (2003) Methods Enzymol. 374, 22–37. [DOI] [PubMed] [Google Scholar]
- 26.Roussel, A. & Cambillau, C. (1991) turbo (Silicon Graphics, Mountain View, CA).
- 27.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 28.Vagin, A. & Teplyakov, A. (2000) Acta Crystallogr. D. Biol. Crystallogr. 56, 1622–1624. [DOI] [PubMed] [Google Scholar]
- 29.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 30.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 31.Filipek, R., Szczepanowski, R., Sabat, A., Potempa, J. & Bochtler, M. (2004) Biochemistry 43, 14306–14315. [DOI] [PubMed] [Google Scholar]
- 32.Wenig, K., Chatwell, L., von Pawel-Rammingen, U., Bjorck, L., Huber, R. & Sondermann, P. (2004) Proc. Natl. Acad. Sci. USA 101, 17371–17376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, W., Johnston, S. A. & Joshua-Tor, L. (1998) Cell 93, 103–109. [DOI] [PubMed] [Google Scholar]
- 34.Menard, R. & Storer, A. C. (1994) Methods Enzymol. 244, 486–500. [DOI] [PubMed] [Google Scholar]
- 35.Wilmouth, R. C., Clifton, I. J., Robinson, C. V., Roach, P. L., Aplin, R. T., Westwood, N. J., Hajdu, J. & Schofield, C. J. (1997) Nat. Struct. Biol. 4, 456–462. [DOI] [PubMed] [Google Scholar]
- 36.Wilmouth, R. C., Edman, K., Neutze, R., Wright, P. A., Clifton, I. J., Schneider, T. R., Schofield, C. J. & Hajdu, J. (2001) Nat. Struct. Biol. 8, 689–694. [DOI] [PubMed] [Google Scholar]
- 37.Bürgi, H. B., Dunitz, J. D. & Shefter, E. (1974) J. Am. Chem. Soc. 95, 5065–5067. [Google Scholar]
- 38.Pinitglang, S., Watts, A. B., Patel, M., Reid, J. D., Noble, M. A., Gul, S., Bokth, A., Naeem, A., Patel, H., Thomas, E. W., et al. (1997) Biochemistry 36, 9968–9982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.