Abstract
Sixteen distinct sites distributed on all five Arabidopsis (Arabidopsis thaliana) chromosomes have been tagged using different fluorescent proteins and one of two different bacterial operator-repressor systems: (1) a yellow fluorescent protein-Tet repressor fusion protein bound to tet operator sequences, or (2) a green or red fluorescent protein-Lac repressor fusion protein bound to lac operator sequences. Individual homozygous lines and progeny of intercrosses between lines have been used to study various aspects of interphase chromosome organization in root cells of living, untreated seedlings. Features reported here include distances between transgene alleles, distances between transgene inserts on different chromosomes, distances between transgene inserts on the same chromatin fiber, alignment of homologous chromosomes, and chromatin movement. The overall findings are consistent with a random and largely static arrangement of interphase chromosomes in nuclei of root cells. These transgenic lines provide tools for in-depth analyses of interphase chromosome organization, expression, and dynamics in living plants.
Although the arrangement of interphase chromosomes is thought to be important for regulating nuclear function and gene expression (Misteli, 2004; Bolzer et al., 2005; Wegel and Shaw, 2005), little information is currently available about interphase chromosome organization in living plant cells (Lam et al., 2004; Tessadori et al., 2004). Most studies so far have used fluorescence in situ hybridization (FISH) to visualize interphase chromosomes in nonliving, fixed material (e.g. Fransz et al., 2002; Pecinka et al., 2004). As an alternate technique, bacterial operator-repressor systems combined with fluorescent proteins offer a unique opportunity to visualize fluorescence-tagged loci in nuclei of living, unfixed cells. The operator repeats are integrated into the genome as a transgene array, which then specifically binds the respective nuclear-localized repressor protein that is fused with a fluorescent protein such as green fluorescent protein (GFP). The tagged loci appear as bright fluorescent dots when viewed with appropriate filters under a fluorescence microscope. Using a fluorescence microscope equipped with a motorized z axis and image-processing software, it is possible to make optical sections through nuclei and reconstruct them in three dimensions to determine spatial relationships among fluorescence-tagged loci. This technique has been employed in yeast, Drosophila, and mammalian cells to analyze interphase chromosome organization and dynamics (Gasser, 2002; Spector, 2003), but so far has been used to study only a limited number of genomic insertion sites in plant cells (Kato and Lam, 2001, 2003; Esch et al., 2003; Matzke et al., 2003; Pecinka et al., 2005).
We report here on 16 transgenic Arabidopsis (Arabidopsis thaliana) lines in which distinct chromosomal sites carrying arrays of either tet or lac operators are tagged with the corresponding repressor proteins fused with either yellow fluorescent protein (YFP), red fluorescent protein (dsRed), or GFP. We have used these lines to analyze various aspects of interphase chromosome arrangement and movement in root cells of living, untreated seedlings.
RESULTS AND DISCUSSION
Constructs, Generation, and Screening of Transgenic Lines
Three transgene constructs comprising tet or lac operator repeats and genes encoding the respective nuclear-localized repressor protein-fluorescent protein (RP-FP) fusion proteins were used in this study. Two of these constructs have been reported previously: Tet repressor-enhanced YFP (EYFP), which binds to tet operators (Fig. 1, construct 5), and Lac repressor-enhanced GFP (EGFP), which binds to lac operators (Fig. 1, construct 25; Matzke et al., 2003). A third construct, Lac repressor-dsRed, which binds to lac operators, is reported here (Fig. 1, construct 16). In the three constructs, the genes encoding the RP-FP fusion proteins are under the control of the nominally constitutive 35S promoter of the Cauliflower mosaic virus. This allows visualization of fluorescent signals in nuclei of living plants that have not undergone any inducing treatments, which can potentially disturb interphase nuclear organization. The constructs were introduced into the Arabidopsis ecotype Columbia (Col-0) genome by the floral-dip method and transformants were selected on kanamycin-containing medium. Following prescreening of kanamycin-resistant T1 seedlings for nuclei with distinct signals appearing as bright, fluorescent dots (Matzke et al., 2003), 16 independent transgenic lines that contained T-DNAs segregating as single loci (nine YFP, six dsRed, and one GFP) were chosen for further analysis.
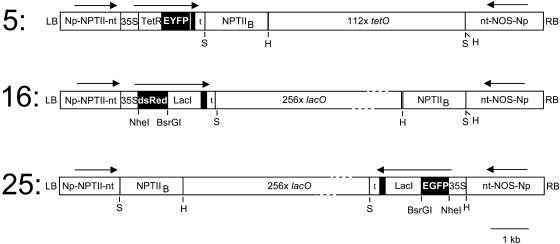
Figure 1.
Constructs. Each construct contains a neomycin phosphotransferase II (NPTII) gene under the control of the NOS promoter (Np) and terminator (nt) for selection of transformed plant cells on kanamycin, an NPTIIB gene for selection of bacteria on kanamycin, and an intact NOS gene. Construct 5 encodes a Tet-repressor (TetR)-EYFP fusion protein and contains 112 copies of the tet operator; 16 encodes a Lac-repressor (LacI)-dsRed fusion protein and contains 256 copies of the lac operator; 25 encodes a LacI-EGFP and contains 256 copies of the lac operator. The fusion proteins are under the control of the 35S promoter and terminator (t) and contain three tandem copies of the SV40 nuclear localization signal (narrow vertical black bar). Arrows show the direction of transcription. LB, Left T-DNA border; RB, right T-DNA border; S, SalI; H, HindIII.
The sites of T-DNA insertion in the 16 lines were determined initially by genetic mapping (Supplemental Fig. 1). The integration sites were confirmed and defined in more detail by using cosmid rescue cloning, lambda cloning, or thermal asymmetric interlaced (TAIL)-PCR to isolate the T-DNA inserts and then sequencing the flanking plant DNA. With the exception of the short arm of chromosome 2, each chromosome arm contains at least one T-DNA insertion site, and, with the exception of chromosome 4, each chromosome has at least one YFP and one dsRed insert (Fig. 2). The chromosome sequence coordinates of the T-DNA insertions and primers for detection of the inserts by PCR are listed for 14 lines in Supplemental Table I; for two inserts on chromosome 1, 5:56 and 16:52, only the identity of the bacterial artificial chromosome clone containing the T-DNA is currently available. Additional features of the genomic insertion sites are presented in Supplemental Table II. A Southern-blot analysis demonstrated that all lines contained the appropriate operator arrays, which were usually present in scrambled and/or multiple copies (Matzke et al., 2003; data not shown).
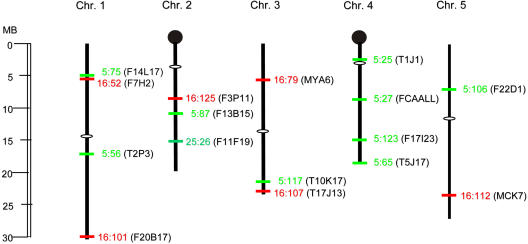
Figure 2.
Genomic insertion sites of fluorescence-tagged T-DNAs. Nine YFP-tagged sites are shown in green and numbered beginning with the prefix 5. Six dsRed-tagged sites are shown in red and numbered beginning with the prefix 16. One GFP-tagged site is shown in green and numbered beginning with the prefix 25. Bacterial artificial chromosome numbers containing the insertions are shown in parentheses. White ovals indicate centromeres. Black knobs represent nucleolar organizing regions. All lines display essentially a wild-type phenotype; the exceptions are lines 5:75 and 16:125, which have a somewhat bushy phenotype.
Root cells were chosen to investigate the fluorescent signals in each line because they are nonphotosynthetic and hence have a low background fluorescence. An initial screen at low magnification of living seedlings mounted on indented microscope slides was used to locate a region of the root that produces optimal signals, which are visualized as bright fluorescent dots in a nucleoplasm of low diffuse background fluorescence (Matzke et al., 2003). Often a suitable region is followed by a region where the RP-FP fusion protein is overexpressed, filling the nucleoplasm and often obscuring the fluorescent dots (Supplemental Fig. 2). Despite the use of the nominally constitutive 35S promoter, the gene encoding the RP-FP fusion protein does not seem to be expressed in every cell because signals are not observed in all nuclei. Moreover, the frequency of RP-FP fusion gene expression varies among seedlings within a line and among individual lines as do the intensities of the signals (Supplemental Table III). It is not known whether the lower-than-expected frequencies result from cell-to-cell variation in the activity of the 35S promoter, gene silencing, or other factors that remain to be identified.
Distances between Transgene Alleles
The 16 transgenic lines were bred to homozygosity. The relatively uniform distribution of the transgene inserts throughout the Arabidopsis genome (Fig. 2) provided an opportunity to investigate whether alleles at any of the insertion sites showed preferential associations, such as pairing or fixed interallelic distances (Fig. 3A). For this, three-dimensional (3D) reconstructions and allelic distance measurements were made for all lines on approximately 10 to 20 root nuclei in which two fluorescent dots could be observed (see Supplemental Fig. 3 for examples). The allelic distances were plotted against nuclear diameter (determined as described in “Materials and Methods”). Complicating the analysis to some extent was the fact that not all nuclei in root cells are round. Most round nuclei were between 3 to 7 μm in diameter, whereas larger nuclei were often irregularly or spindle shaped. In these cases, the estimated nuclear diameter was based on the longest nuclear end-to-end distance. The allelic distances in all lines vary in a similar manner, with a general trend toward larger interallelic distances as nuclear diameter increases, although low values in large nuclei can also be observed (Supplemental Fig. 4). This relationship applies even for insertion sites that are close to telomeres (16:101 on chromosome 1, 16:107 on chromosome 3, and 5:65 on chromosome 4) or in the vicinity of heterochromatic regions, in particular 5:25, which is between the centromere and the heterochromatic knob of chromosome 4 (Fig. 2).
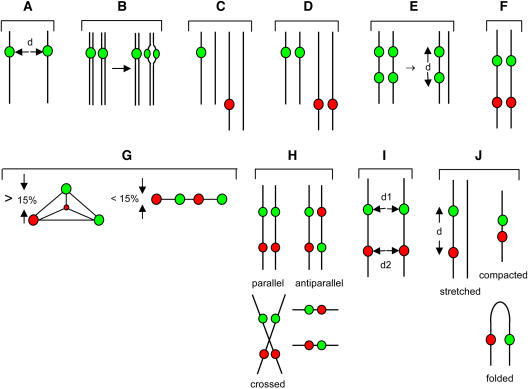
Figure 3.
Aspects of interphase chromosomes studied here. For illustrative purposes, interphase chromosomes are depicted as black lines. A, Distance between transgene alleles in homozygous lines (d) was determined for all 16 lines (YFP, dsRed, GFP; one color shown for simplicity). B, Splitting of signal might be due to separation of sister chromatids in polytene nuclei. C, Double hemizygous transgene insertions (dsRed and YFP/GFP) on nonhomologous chromosomes. D, Double homozygous transgene insertions (dsRed and YFP/GFP) on nonhomologous chromosomes. E, Double homozygous YFP insertions on homologous chromosomes (left). A backcross to untransformed plants produces double hemizygous YFP insertions on the same homolog (right). F, Double homozygous transgene insertions (dsRed and YFP/GFP) on homologous chromosomes. G, Transgene insertion sites depicted in F were used to study 3D arrangements. When connected by lines, the four signals can be arranged as a tetrahedron. This was defined here as a 3D space in which the smallest height (observed by rotating the connected dots 360° vertically and horizontally) exceeded 15% of the nuclear diameter. The four signals from the lines depicted in F can also be flat and lie in a plane. This was defined here as any arrangement in which the smallest height was less than 15% of the nuclear diameter. H, When transgene inserts are in the same plane (flat), chromosomes containing the signals can be in parallel/crossed or antiparallel orientations. These are extremes of a continuum that can also include perpendicular orientations (Supplemental Table IV). I, For flat parallel signals in the same plane, the distances between the YFP and dsRed alleles (d1 and d2, respectively) can provide information about alignment of homologs in the region between the YFP and dsRed inserts (d1 = d2). J, Lines containing transgene insertion sites depicted in F were backcrossed to untransformed plants to produce double hemizygous insertions on the same homolog (see also 3E, right). These lines can be used to measure cis-distances between insertions on the same chromatin fiber, which provide clues about chromosome folding and/or compaction.
The shortest interallelic distance measured in this study was approximately 0.5 μm (Supplemental Fig. 4). This approaches the resolution limit of this technique in budding yeast (Saccharomyces cerevisiae), where two GFP spots closer than 300 nm cannot be resolved (Bystricky et al., 2004). Whether 0.5 μm can be considered to represent allelic pairing is unclear. Indeed, the analysis highlighted the uncertainty about how pairing should be assessed with this method, which allows visualization of transgene inserts from all angles in three dimensions. For example, would pairing be visualized from all viewing angles as a single large dot, approximately double the size of two smaller dots, or as two smaller dots side by side with little or no empty space between them? Conceivably, pairing could potentially lead to loss of signal if close proximity of alleles results in silencing of genes encoding RP-FP fusion proteins. Resolving these issues will require further analysis and an agreement on what constitutes pairing as assessed by this technique. Despite the uncertainties, the observations that most interallelic distances are greater than 1 μm and that they tend to increase as nuclear diameter increases suggest that if pairing of active (i.e. visible) loci occurs, it is not a constant feature of all nuclei within the region of the root examined. Furthermore, alleles throughout the genome do not appear to be at fixed distances relative to each other; instead, the interallelic distances can accommodate changes in nuclear size. Whether these conclusions extend to nuclei in other cell types and to other insertion sites in the genome remains to be investigated. A recent FISH study examining two tagged T-DNAs on the top arm of chromosome 3 in leaf nuclei suggested that pairing, which was thought to be mediated by common lac operator repeats, occurred more often at these sites than expected from a random arrangement (Pecinka et al., 2005).
Signal Splitting
In a number of homozygous lines (16:52, 5:87, 25:26, 5:117, 16:107, 5:25, 5:27, 5:65; Supplemental Table III), signals from individual alleles were often observed to split into two (or more) smaller signals. Splitting could affect only one allele or both alleles simultaneously (Fig. 4). Splitting reduced the strength of the signal and hence was often detected only after deconvolution, which removes out-of-focus haze and blur from stacks of optical sections and enhances the sharpness and clarity of the image. Understanding the basis of the splitting phenomenon will require more detailed investigation. One possibility is that splitting reflects stochastic separation of sister chromatids in polytene chromosomes of some root nuclei (Fig. 3B). Alternatively, cells could be in the G2 stage of the cell cycle, when sister chromatids split visibly in some chromosomal regions as judged by in situ labeling (Schubert et al., 2005).
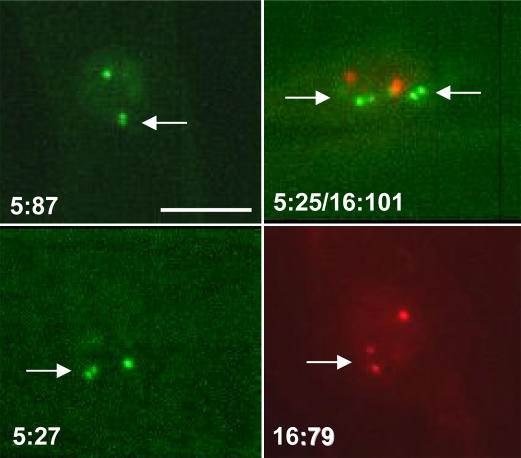
Figure 4.
Splitting of signals. Arrows point to split signals affecting one allele only in homozygous lines 5:87, 5:27, and 16:79, and simultaneous splitting of both YFP alleles in the double homozygous line 5:25/16:101. White bar in 5:87 indicates 5 μm and is the same for all images shown.
Distances between Transgene Inserts on Different Chromosomes (Ectopic Sites)
To determine whether there is any preferential association between unlinked transgene inserts, intercrosses were made to produce lines containing dsRed and YFP signals on different chromosomes. For this, the homozygous dsRed line 16:101 (chromosome 1) was intercrossed with six homozygous YFP lines: 5:75 (chromosome 1), 5:87 (chromosome 2), 5:117 (chromosome 3), 5:25 (chromosome 4), 5:123 (chromosome 4), and 5:106 (chromosome 5; Fig. 2). F1 progeny of these crosses are hemizygous for the two transgene inserts (Fig. 3C, which depicts the two inserts on nonhomologous chromosomes; for the 5:75/16:101 combination, the two inserts are on homologs of chromosome 1). The double hemizygous progeny were analyzed with respect to the distance between the dsRed and YFP signals. As with the interallelic distances, the ectopic distances were variable, with a trend toward increased values as the nuclear diameter increased (Supplemental Fig. 5). There was no evidence for preferential associations of any ectopic T-DNA insertions. Note that, in these cases, the YFP and dsRed inserts contain heterologous operator arrays (tet or lac, respectively) and hence would not be prone to ectopic pairing of operator elements.
Selfing of double hemizygous plants produced some F2 progeny that were doubly homozygous for both transgene inserts (Fig. 3D). Distance measurements between the two YFP alleles and the two dsRed alleles in the double homozygotes were similar to those obtained with the single homozygous lines and continued to indicate no preferential allelic associations (data not shown). Overall, the results on distances between ectopic insertions are similar to those observed here for alleles and are compatible with a random arrangement of interphase chromosomes.
3D Arrangements
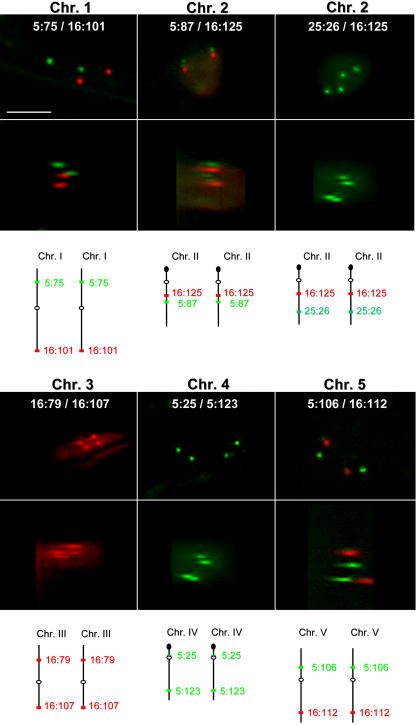
Some of the most interesting material came from lines that were doubly homozygous for either two YFP (or two dsRed) inserts or one dsRed and one YFP (or GFP) insert on the same chromosome (Fig. 3E, left, and F, respectively). The four signals define a space and hence give information about 3D spatial relationships among loci. To obtain these lines, intercrosses were made to generate lines with the following combination of dsRed and YFP (or GFP) signals: 5:75/16:101 (chromosome 1), 5:87/16:125 (chromosome 2), 25:26/16:125 (chromosome 2), and 5:106/16:112 (chromosome 5). In the case of chromosome 4, which did not have any dsRed inserts, two YFP signals were combined (5:25/5:123). Four signals could be observed in nuclei of these five double homozygous lines (Fig. 5), although the frequency was usually lower than the frequencies of observing two signals in each of the homozygous parental lines (Supplemental Table III; Supplemental Fig. 6). With the 25:26/16:125 combination on chromosome 2, in which both T-DNA inserts contain lac operator arrays but encode either a GFP- or dsRed-Lac repressor fusion protein, only the GFP-Lac repressor fusion protein was expressed such that four green signals (and no red signals) were usually observed (Fig. 5). For chromosome 3, the two possible YFP-dsRed combinations (5:117/16:79 and 5:117/16:107) resulted in signals that were too weak for proper imaging. Therefore, we used a double combination comprising two dsRed inserts, 16:79/16:107, in which four dots were visible (although only in a few nuclei per seedling root; Fig. 5; Supplemental Table III). The lower-than-expected frequencies in the double homozygous lines are currently unexplained, but they might be due to interactions between T-DNA inserts that lead to gene silencing.
Figure 5.
Double homozygous lines containing two transgene inserts on the same chromosome. The combinations of transgene inserts are shown in white letters and their positions on a given chromosome are depicted below the images. The top images are top views of the maximal projection (all deconvoluted planes from the stack collapsed into a single plane); the bottom images are rotated 90° to give a side view. White bar in 5:75/16:101 indicates 5 μm and is the same for all images shown.
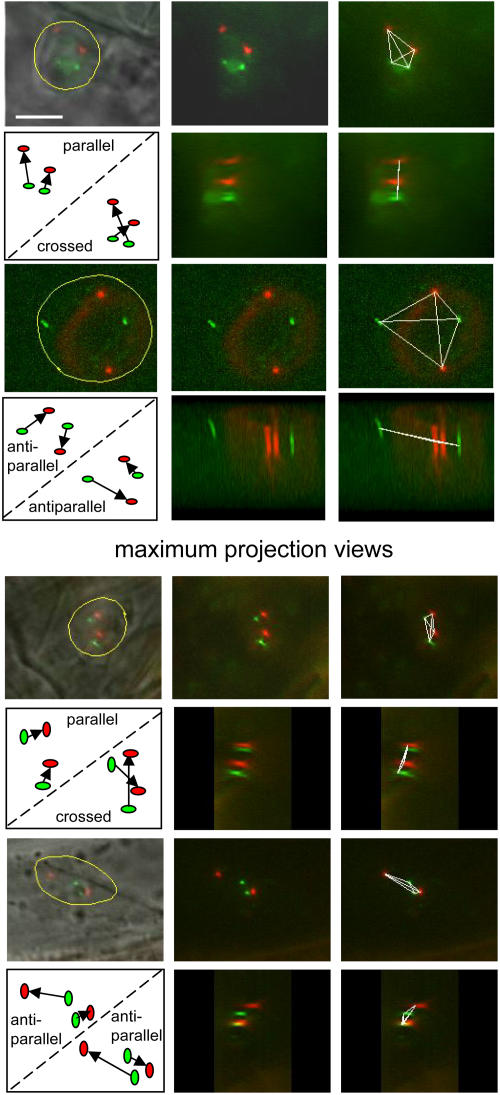
The analysis of 3D relationships is facilitated in double homozygous lines containing dsRed and YFP signals on the same chromosome because the two loci can be distinguished by color. Three such lines are available in the present collection: 5:75/16:101 (chromosome 1), 5:87/16:125 (chromosome 2), and 5:106/16:112 (chromosome 5; Fig. 5). Several features of the 3D arrangement could be discerned with these lines. First, the use of image-processing software to connect the four dots and rotate them 360° in all directions revealed that they either formed the corners of a tetrahedron or were more-or-less flat in a single plane (Fig. 3G). Second, in the case of a plane, the signals could be arranged in a way that was consistent with either a more-or-less parallel/crossed, perpendicular, or antiparallel orientation of homologs (Fig. 3H). Examples of parallel/crossed and antiparallel for 5:75/16:101 (chromosome 1) and 5:106/16:112 (chromosome 5) are shown in Figure 6; the complete set of data for all three lines is shown in Supplemental Table IV. For a parallel orientation, the distances between the YFP and dsRed alleles should be similar, essentially reflecting alignment of homologs in the region between the two T-DNA insertion sites (d1 ≅ d2; Fig. 3I). This was indeed the case for line 5:87/16:125 (Supplemental Fig. 7, middle), in which the YFP and dsRed inserts are separated by only 2.4 Mb on chromosome 2 (Fig. 2). The equivalence of d1 and d2 was independent of their absolute values, such that d1 and d2 could range from approximately 1.5 to 8 μm. These findings serve as a control to validate the distance measurements, since one would expect two adjacent loci separated by a relatively short physical distance to have similar interallelic distances in a given nucleus with a flat, parallel arrangement of homologs.
Figure 6.
Parallel/crossed and antiparallel orientations. Examples are shown for the YFP and dsRed inserts on chromosome 1 (5:75/16:101) and on chromosome 5 (5:106/16:112). In the four cases shown (six boxes each), the signals are shown in the nucleus (bounded in yellow), then from the top and side views. The dots are connected by white lines, then turned in three dimensions to see whether they fall into a line or a very shallow tetrahedron (<15% of nuclear diameter) and are thus considered flat arrangements (Fig. 3G; Supplemental Table IV). The drawings show that it is only possible to connect the dots in either a parallel/crossed or antiparallel orientation. White bar in 5:75/16:101 indicates 5 μm and is the same for all images shown.
There was less frequent equivalence of d1 and d2 as the YFP and dsRed inserts became separated by much larger distances in lines 5:75/16:101 (chromosome 1) and 5:106/16:112 (chromosome 5; Supplemental Fig. 7, top and bottom, respectively). Thus, when considering interallelic distances at two widely spaced loci, d1 and d2 could be quite different. These data suggest that alignment of homologs (defined here as a flat, parallel arrangement in which d1 = d2, regardless of the absolute value of d1 and d2; Fig. 3I) is most often detected over relatively short regions (e.g. 2.4 Mb in line 5:87/16:125).
The models shown in Figure 3 depict the chromosomes as linear entities, which is an oversimplification. However, the models provide a basis for interpreting the 3D results, which demonstrate that the relative orientation of the dsRed and YFP signals on homologous chromosomes is not fixed. Indeed, evidence for all possible variations (Fig. 3, G–I) was obtained in root nuclei of a given line, supporting a random arrangement of interphase chromosomes in root cells of living, untreated seedlings.
cis-Distances between Transgene Inserts on the Same Chromatin Fiber
Although the distances between YFP alleles or dsRed alleles can be measured in plants that are doubly homozygous for the respective inserts (previous section), it was not possible in those cases to discern which YFP signal and which dsRed signal were together on the same chromatin fiber (e.g., parallel could not be distinguished from crossed; Fig. 3H). To address the question of distances between transgene inserts on the same chromatin fiber, we backcrossed all six lines that are doubly homozygous for two transgene inserts on the same chromosome (Fig. 5) to nontransgenic plants. The progeny of these crosses are double hemizygous on the same homolog for either two YFP inserts (Fig. 3E, right) or one YFP (or GFP) and one dsRed insert (Fig. 3J). With these plants, it is possible to measure the distance in cis between two transgene inserts that are known to be present on the same chromatin chain. This can potentially provide information on chromosome folding or compaction (Fig. 3J) since the approximate linear distance (in megabase pairs) between the two signals is known (Table I).
Table I.
Possible fold compaction of DNA between fluorescence-tagged transgene inserts on the same chromosome based on maximal interlocus distance measurements in interphase nuclei of root cellsa
Conversions: 1 bp double helical DNA (B form) = 0.34 nm (Lehninger, 1975); 1,000 bp (1 kb) = 340 nm; 3 kb equals approximately 1 μm; and 3 Mb equals approximately 1 mm.
| Chromosome | Inserts | Approximate Distance between Inserts | Approximate DNA Length | Maximal Observed Distance | Fold Compaction |
|---|---|---|---|---|---|
| Mb | mm | μm | |||
| 1 | 5:75/16:101 | 25 | 8.3 | 8.7 | 960× |
| 2 | 5:87/16:125 | 2.4 | 0.8 | 2.1 | 380× |
| 2 | 25:26/16:125 | 6.7 | 2.2 | 5.1 | 430× |
| 3 | 16:79/16:107 | 17.5 | 5.8 | 6.7 | 865× |
| 4 | 5:25/5:123 | 12.3 | 4.1 | 7.9 | 520× |
| 5 | 5:106/16:112 | 16.4 | 5.5 | 7.4 | 740× |
| Average 650× |
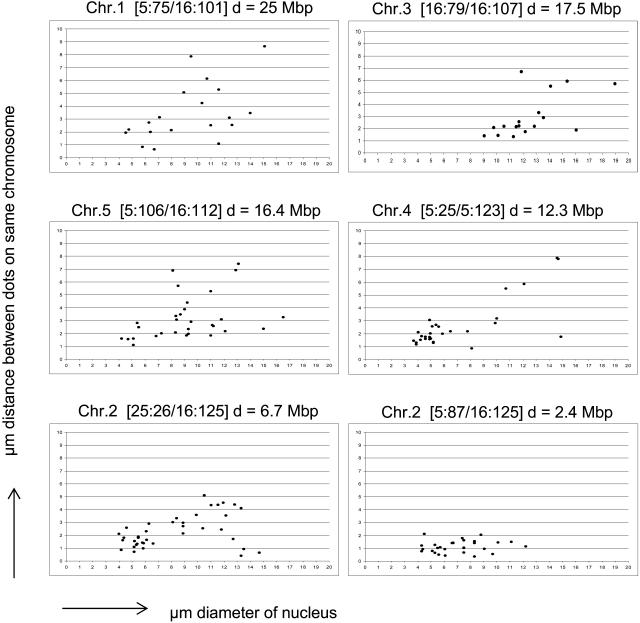
Of the six lines tested, four of them have the signals on opposite sides of the centromere (5:75/16:101 on chromosome 1, 16:79/16:107 on chromosome 3, 5:25/16:123 on chromosome 4, and 5:106/16:112 on chromosome 5; Fig. 5). In these lines, the cis-distances were quite variable (Fig. 7, top and middle), indicating considerable nucleus-to-nucleus variability in chromosome folding and/or compaction. The cis-distances also appear to be proportional to some extent on nuclear diameter, as indicated by a tendency to increase as nuclear diameter increased, but lower values can also be seen in some larger nuclei, possibly as a consequence of folding (Fig. 3J). These results suggest that individual chromosome arms can be quite flexible when adopting their positions in interphase nuclei.
Figure 7.
cis-Distances between hemizygous transgene inserts on the same chromatin fiber. The identity of the lines and the approximate distance (d) separating the transgene inserts on the same chromatin chain in megabase pairs are shown at the top of each graph and in Table I. Each point represents the cis-distance in a single nucleus determined after optical sectioning, deconvolution, and 3D reconstruction.
A more constrained pattern was obtained with line 5:87/16:125 (chromosome 2), in which the YFP and dsRed signals are on the same side of the centromere and separated by only 2.4 Mb (Fig. 5). Here the cis-distance remained in most cases below 2 μm (Fig. 7, bottom right). The YFP and dsRed signals in this line are distinguishable in the fluorescence microscope but always close together (Fig. 5; Supplemental Fig. 8), indicating that 2.4 Mb is still above the minimum resolvable cis-distance between two differently colored inserts on the same chromatin fiber. A second chromosome 2 line that has the same dsRed insert (16:125), but a more distantly spaced GFP insert (25:26; distance 6.7 Mb) on the same chromosome arm (Fig. 5), shows increasing cis-distances up to a maximum measurement of 5.1 μm but also low values (Fig. 7, bottom left). Whether some low values might represent enhanced pairing in cis of lac operator repeats associated with both the GFP and dsRed inserts in this line is not known. cis-Pairing of common operator repeats is also potentially possible in line 16:79/16:107 on chromosome 3, in which both inserts have lac operators, and line 5:25/5:123 on chromosome 4, in which both inserts have tet operators. However, unlike the T-DNA inserts in line 25:26/16:125, the inserts in these lines are on opposite sides of the centromere. Whether this might influence the chances of cis-pairing is not known.
It should be emphasized that interpretations of the cis-distance measurements assume that no chromosome rearrangements, such as inversions (Pecinka et al., 2005), have taken place during T-DNA integration. Under this assumption, the fold compaction associated with the maximum measured distances ranged from 380- to 960-fold (Table I). These values are only very rough estimates, since it is likely that the degree of compaction and/or folding vary considerably along a chromatin fiber. A larger collection of more closely spaced inserts would be valuable for studying chromatin compaction in plant interphase nuclei. Based on a similar approach of using fluorescence-tagged tet and lac operators on the same chromosome, the average compaction ratio of interphase chromatin in budding yeast was determined to be approximately 40-fold (Bystricky et al., 2004).
Movement of Chromosomes
Fluorescence-tagging of transgenes has permitted analysis of interphase chromosome dynamics in cells of yeast, Drosophila, and mammals (Gasser, 2002). A study on Arabidopsis using fluorescence-tagged T-DNAs on chromosome 3 in diploid nuclei of guard cells indicated that the maximal radius of the confinement area was 0.21 μm (Kato and Lam, 2003). In the absence of a nuclear envelope marker, which can be used to control for movement of the nucleus itself, we studied chromosome dynamics with respect to whether the distance between alleles varies over a period of 80 min. A strong signal that is stable for the duration of the experiment is required. Homozygous line 16:112, in which the dsRed insert is present toward the end of the long arm of chromosome 5 (Fig. 2), fulfilled this requirement. Consistent with the previous study (Kato and Lam, 2003), we observed a maximal change in the allelic distance of 0.2 μm over the observation period (Supplemental Fig. 9). Although we cannot rule out movements over very short time frames (e.g. seconds), the allelic arrangement appears overall to be quite stable. Additional studies are needed to determine whether similar results will be obtained with transgenes inserted at other chromosomal locations and in nuclei from other cell types. Interphase chromosomes might also change their positions during development or under various inducing or environmental stress conditions (Tumbar et al., 1999; Dietzel et al., 2004). The ability to view the fluorescence-tagged loci in living cells provides a means to study these and other problems of interphase chromosome organization.
CONCLUSION
We have developed 16 transgenic lines that have fluorescence-tagged T-DNAs at different locations in the Arabidopsis genome. The precise insertion sites have been determined in most cases so that the transgene inserts can be detected by PCR analysis. The lines have been used to study various features of interphase chromosome disposition and 3D spatial relationships. The results obtained so far indicate considerable variation in interphase chromosome arrangement in root cells of living, intact seedlings and generally support previous findings from FISH analyses of interphase chromosomes in Arabidopsis (Fransz et al., 2002; Pecinka et al., 2004). The types of analyses performed in this study can be extended to other cell types, developmental stages, or environmental conditions.
There are several advantageous features of the transgenic lines reported here. The combination of two easily distinguishable fluorescent proteins (YFP and dsRed) that bind to heterologous operator repeats (tet and lac, respectively) eliminates the possibility that unlinked arrays of operator repeats might pair (Pecinka et al., 2005). Thus, these combinations allow 3D analyses of chromosomal sites under conditions in which interphase nuclear architecture is largely unperturbed, although local distortions induced by the operator repeat arrays cannot be ruled out. Minimal disruption of nuclear organization is also ensured by the fact that no treatments are required to induce expression of the RP-FP proteins in these lines. Living seedlings can simply be removed from growth medium, mounted on microscope slides, immediately viewed and optically sectioned under the fluorescence microscope, and then replaced on medium to resume growth. In all of the lines reported here, the signals are visible in the fluorescence microscope before deconvolution (Supplemental Table III). Indeed, signals in dsRed lines 16:101 and 16:112 are strong enough to be seen under low magnification (160×; data not shown).
The primary drawback of these lines is the lower-than-expected frequency of expression of the RP-FP fusion protein. This problem affects all individual lines to varying extents and can be exacerbated in the progeny of some intercrosses (Supplemental Table III). Whether the low frequencies of RP-FP fusion protein expression are more pronounced in root cells than in other cell types remains to be investigated. The contribution of various types of gene silencing to the low signal frequencies is being examined by crossing the tagged transgenic lines with mutants defective in either transcriptional or posttranscriptional gene silencing. In addition, we are examining whether supplying the RP-FP fusion proteins (under the control of different promoters) in trans will alleviate this problem. Even with the frequencies currently attainable, however, these lines provide useful material for analyzing plant interphase chromosomes in their natural state.
MATERIALS AND METHODS
Constructs and Production of Transgenic Plants
Constructs 5 (tetY) and 25 (lacG) and the production of transgenic Arabidopsis (Arabidopsis thaliana) plants (Col-0) using these constructs have been described previously (Matzke et al., 2003). The construct 16 (lacR) was created by replacing EGFP with DsRed2 (purchased from CLONTECH-BD Biosciences) at the pBC stage of the fusion construct using NheI and BsrGI (Fig. 1). At the pBC stage of the fusion construct, the 35S promoter can easily be replaced with another promoter using an XhoI/NheI cleavage (Matzke et al., 2003).
Genetic Mapping and Recovery of Flanking DNA
Homozygous transgenic Arabidopsis plants (Col-0) were crossed to ecotype Landsberg erecta (Ler) to obtain mapping populations. T-DNA insertions were roughly mapped using a mapping population of around 30 F2 plants and sets of simple sequence length polymorphism (Bell and Ecker, 1994; Lukowitz et al., 2000) and cleaved amplified polymorphic sequence markers (Konieczny and Ausubel, 1993), which detect polymorphisms between the Col-0 and Ler ecotypes (Supplemental Fig. 1). The primer sequence for the simple sequence length polymorphism marker MAC9 on the lower arm of chromosome 5 (developed by W. Aufsatz, personal communication) is 5′-GTC ATG TCAC TGG GGA TAA G-3′; 5′-ATG TGT AAC ACC CAT TGG AC-3′.
To recover flanking DNA of the T-DNA inserts, a combination of approaches was used: TAIL-PCR (Weigel and Glazebrook, 2002), lambda cloning using a Lambda FIX II/XhoI partial fill-in vector kit and Gigapack III gold packaging extract (both from Stratagene), and cosmid rescue cloning using the cosmid pWEB::TNC cloning kit (from Epicentre, purchased from Biozym Diagnostics GmbH), which allows direct selection of T-DNA inserts carrying the NPTIIB gene for bacterial selection. Inserts of cosmids were end sequenced using M13 forward and T7 standard primers. In cases where Arabidopsis DNA was detected, we further sequenced using a nopaline synthase (NOS) promoter primer oriented toward the T-DNA border (5′-TTC TGT CAG TTC CAA ACG-3′) and could usually recover the T-DNA-plant DNA junction. If Arabidopsis DNA sequences were not detected by using the NOS promoter primer, we performed primer walking (starting at the Arabidopsis DNA detected by end sequencing) until the T-DNA-plant DNA junction was reached. Primers were designed to allow these T-DNA inserts to be detected in progeny by simple PCR assays (Supplemental Table I).
Seedling Growth Conditions
Seeds were sterilized and germinated on sterile solid Murashige and Skoog medium in petri dishes. The dishes were placed at 4°C for 2 to 3 d, then moved to a Percival incubator and grown at 23°C in a 16-h-light/8-h-dark cycle. Seedlings used in all experiments were approximately 10 to 20 d old.
Fluorescence Microscopy and Image Analysis
Whole seedlings were mounted in tap water on indented slides. Leaves were placed in the indented region and the root was stretched out on the regular slide surface. A coverslip placed over the entire seedling was sealed with rubber cement. Photographs and stacks (40 images at 0.2-μm z-axis steps; exposure time for each image, 1 s; total elapsed time for one stack, approximately 2 min) were taken using a Zeiss Axioplan 2 fluorescence microscope (Zeiss) equipped with appropriate filter cubes for YFP, dsRed, and GFP, and with a Quantix CCD camera from Photometrics (purchased from Zeiss) run with MetaView software (Visitron Systems GmbH).
Frequency Plots
Frequency plots allow visualization of the variability of the signal frequency in interphase nuclei of root cells of a given line: Starting at the root tip, nuclei with signals (fluorescent dots) are counted in consecutive areas that are visible in the microscope using the 63× objective, which equals approximately 0.4-mm distance × 50 sections or about 20 mm (=2 cm) of root. After the root tip area, one can count per section (using DAPI staining of nuclei) about 100 elongated root cells on average (Dolan et al., 1993). Typical frequencies for a given line can be reduced drastically when combined with another locus (Supplemental Table III; see Supplemental Fig. 6 for an example).
Distance Measurements
Distance measurements were made in three dimensions after deconvolution of the image stacks using AutoDeblure 9.2, configuration WF (wide field), edition gold (AutoQuant Imaging, purchased from Bitplane AG) in MetaMorph (Meta Imaging Series 5.0; Universal Imaging, purchased from Visitron) and/or Imaris 4.1.3 (Bitplane AG). Using the region measurement tool of MetaMorph, the perimeter of each nucleus was manually drawn on the maximal projections following the background fluorescence that fills the nuclei (nuclei were turned in three dimensions to find the maximal area; if odd shaped, the most reasonable estimates were made), light micrographs, or overlays (whatever allowed the most reliable way to follow the nuclear rim), and the nuclear area A was determined. Nuclear diameters were calculated using the equation D = 2(A/pi)″0.5 (Parada et al., 2004).
Determination of Height of Tetrahedra in YFP/dsRed Double Homozygous Lines
Fluorescent signals (dots) were connected in MetaMorph with white lines using the “Measure XYZ Distance” function. After applying the 3D reconstruction function of MetaMorph, the resulting tetrahedra were turned in three dimensions until three spots formed one line (spanning a triangular plane). Using the caliper function of MetaMorph, the distance of the fourth spot relative to that plane was determined. The minimum height of all possibilities was determined. If this value was less than 15% of the nuclear diameter, the arrangement was considered flat.
Monitoring Allelic Distance over Time
The distance between dsRed alleles in homozygous line 16:112 was measured as a function of time. Every 5 min a stack (21 images at 0.2-μm z-axis steps) was collected for 80 min. Distance measurements were carried out after deconvolution (AutoDeblur) using the point detection function of Imaris (Supplemental Fig. 9). Fixed material was prepared according to a published procedure (Kato and Lam, 2003).
Acknowledgments
We thank Marie-Therese Hauser for suggesting that splitting might be due to separation of sister chromatids in polytene chromosomes.
This work was supported by the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung (grant no. 16545–B12).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Antonius J.M. Matzke (antonius.matzke@gmi.oeaw.ac.at).
The online version of this article contains Web-only data.
References
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, et al (2005) Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM (2004) Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci USA 101: 16495–16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel S, Zolghadr K, Hepperger C, Belmont AS (2004) Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing β-globin regulatory sequences. J Cell Sci 117: 4603–4614 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD (2003) A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894 [DOI] [PubMed] [Google Scholar]
- Fransz P, de Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatic loops emanate. Proc Natl Acad Sci USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S (2002) Visualizing chromatin dynamics in interphase nuclei. Science 296: 1412–1416 [DOI] [PubMed] [Google Scholar]
- Kato N, Lam E (2001) Detection of chromosomes tagged with green fluorescent protein in live Arabidopsis plants. Genome Biol 2: RESEARCH0045 [DOI] [PMC free article] [PubMed]
- Kato N, Lam E (2003) Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J Cell Sci 116: 2195–2201 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel F (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Watanabe K (2004) Visualizing chromosome structure/organization. Annu Rev Plant Biol 55: 537–554 [DOI] [PubMed] [Google Scholar]
- Lehninger A (1975) Biochemistry, Ed 2. Worth Publishers, New York, p 863
- Lukowitz W, Gillmor CS, Scheible W-R (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJM, van der Winden J, Matzke M (2003) Tetracycline operator/repressor system to visualize fluorescence-tagged T-DNAs in interphase nuclei of Arabidopsis. Plant Mol Biol Rep 21: 9–19 [Google Scholar]
- Misteli T (2004) Spatial positioning: a new dimension in genome function. Cell 119: 153–156 [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T (2004) Tissue-specific spatial organization of genomes. Genome Biol 5: R44 [DOI] [PMC free article] [PubMed]
- Pecinka A, Kato N, Meister A, Probst AV, Schubert I, Lam E (2005) Tandem repetitive transgenes and fluorescent chromatin tags alter local interphase chromosome arrangement in Arabidopsis thaliana. J Cell Sci 118: 3751–3758 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak M, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113: 258–269 [DOI] [PubMed] [Google Scholar]
- Schubert V, Klatte M, Pecinka A, Meister A, Jasencakova Z, Schubert I (2005) Sister chromatids are often incompletely cohesed in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics doi/10.1534/genetics.105.048363 [DOI] [PMC free article] [PubMed]
- Spector DL (2003) The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72: 573–608 [DOI] [PubMed] [Google Scholar]
- Tessadori F, van Driel R, Fransz P (2004) Cytogenetics as a tool to study gene regulation. Trends Plant Sci 9: 147–153 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS (1999) Large-scale chromatin unfolding and remodelling induced by VP16 acidic activation domain. J Cell Biol 145: 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Shaw P (2005) Gene activation and deactivation related changes in the three-dimensional structure of chromatin. Chromosoma doi/10.1007/s00412-005-0015-7 [DOI] [PubMed]
- Weigel D, Glazebrook J (2002) How to isolate a gene defined by a mutation. In Arabidopsis. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 143–170