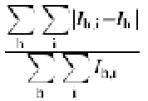Abstract
InlB, a surface-localized protein of Listeria monocytogenes, induces phagocytosis in non-phagocytic mammalian cells by activating Met, a receptor tyrosine kinase. InlB also binds glycosaminoglycans and the protein gC1q-R, two additional host ligands implicated in invasion. We present the structure of InlB, revealing a highly elongated molecule with leucine-rich repeats that bind Met at one end, and GW domains that dissociably bind the bacterial surface at the other. Surprisingly, the GW domains are seen to resemble SH3 domains. Despite this, GW domains are unlikely to act as functional mimics of SH3 domains since their potential proline-binding sites are blocked or destroyed. However, we do show that the GW domains, in addition to binding glycosaminoglycans, bind gC1q-R specifically, and that this binding requires release of InlB from the bacterial surface. Dissociable attachment to the bacterial surface via the GW domains may be responsible for restricting Met activation to a small, localized area of the host cell and for coupling InlB-induced host membrane dynamics with bacterial proximity during invasion.
Keywords: cell invasion/gC1q-R/GW domain/internalin/SH3 domain
Introduction
Many infectious microbes enter host cells that normally are not phagocytic by inducing phagocytosis. Among these is the Gram-positive, facultative intracellular pathogen Listeria monocytogenes, which is responsible for meningitis, abortions, gastroenteritis and septicemia in humans (Lorber, 1997; Aureli et al., 2000). Listeria monocytogenes induces its own uptake into non-phagocytic host cells through the actions of InlA and InlB, two related virulence factors that localize to the bacterial surface. These two proteins activate signaling pathways through different membrane-bound receptors: InlA binds the cell adhesion protein E-cadherin (Mengaud et al., 1996), and InlB binds and activates the receptor tyrosine kinase Met (Shen et al., 2000). While InlA promotes invasion of enterocytes in crossing the intestinal barrier (Lecuit et al., 2001), InlB appears to be more important for subsequent dissemination and infection of other tissues (Gaillard et al., 1996). InlB promotes invasion of a broad variety of cell types, including hepatocyte, endothelial and epithelial cell lines (Bierne and Cossart, 2002; Cabanes et al., 2002), and causes activation of a number of signaling pathways, including phosphoinositide 3-kinase, Ras-MAPK and NF-κB (Ireton et al., 1996; Mansell et al., 2001). Signaling events elicited by InlA or InlB lead to actin-mediated zippering of the host membrane around the bacterium and internalization.
InlA and InlB belong to the internalin family of Listeria proteins. The ∼20 members of this family are characterized by N-terminal leucine-rich repeats (LRRs) (Glaser et al., 2001). These motifs form a curved, tube-like structure, whose concave face generally acts as a protein- or ligand-binding surface (Kobe and Deisenhofer, 1995; Marino et al., 2000). LRRs are found in evolutionarily widespread and functionally diverse proteins, and are prominent in the innate immune system of animals and in the disease resistance genes of plants. The InlA LRRs are required to bind E-cadherin (Lecuit et al., 1997) and the InlB LRRs are sufficient to bind Met (Shen et al., 2000). The structure of the InlB LRRs is known, and reveals potential Met-binding sites on its concave face (Marino et al., 1999; Schubert et al., 2001). Other internalins besides InlA and InlB also affect virulence, although their specific targets are unknown (Raffelsbauer et al., 1998; Schubert et al., 2001).
While InlA and InlB share several properties, a key difference suggests a unique mode of action for InlB. Rather than binding covalently to the peptidoglycan via an ‘LPXTG’ motif like InlA, InlB is attached non-covalently and reversibly. When added exogenously, InlB binds most but not all Gram-positive bacterial surfaces (Jonquières et al., 1999). This attachment occurs between lipoteichoic acid (LTA) on the bacterial cell wall and C-terminal GW domains, named for a conserved Gly–Trp (GW) dipeptide. These ∼80 residue domains are unique to InlB among the internalins but are present in other proteins of Gram-positive bacteria. The non-covalent attachment results in release of nearly half of surface-attached InlB into a soluble form (Jonquières et al., 1999). Unlike InlA, InlB binds multiple host components besides its primary receptor Met. Recently, InlB was demonstrated to bind the glycosaminoglycan (GAG) heparin through its GW domains, and GAGs were shown to enhance InlB- mediated invasion (Jonquières et al., 2001). This suggests a direct role for the GW domains in invasion.
InlB also binds a soluble, doughnut-shaped trimeric protein known as gC1q-R (Jiang et al., 1999; Braun et al., 2000). Evidence exists for InlB interaction with gC1q-R at the mammalian cell surface, although the cellular localization and function of gC1q-R are controversial (Peerschke and Ghebrehiwet, 2001; van Leeuwen and O’Hare, 2001). Still, three results support a role for gC1q-R in InlB-mediated invasion (Braun et al., 2000). First, an antibody against gC1q-R blocks invasion in a dose-dependent manner. Secondly, C1q also blocks invasion in a dose-dependent manner, presumably by competing with InlB for association with gC1q-R. Thirdly, a guinea pig cell line that is non-permissive to InlB- mediated invasion is made permissive by transfection with human gC1q-R. How gC1q-R binds to InlB and promotes invasion is not known.
To address the mode of InlB action, we have determined its crystal structure and carried out experiments to analyze its association with heparin and gC1q-R. The structure shows a highly extended molecule with the LRRs at one end and GW domains at the other. Surprisingly, we find that the GW domains are structurally and evolutionarily related to SH3 domains, which typically are found in eukaryotic or viral signal transduction proteins and bind proline-rich targets. The GW domains are also related to but distinct from prokaryotic SH3-like sequences called SH3b domains (Ponting et al., 1999; Whisstock and Lesk, 1999). We infer that the GW domains are unlikely to mimic SH3 domains functionally, as their potential peptide-binding sites are destroyed or blocked. However, we demonstrate that the GW domains do mediate specific binding to gC1q-R and, further, that this binding requires soluble rather than bacterial surface-attached InlB. These data provide evidence for InlB action on host cells following release from the bacterial surface. The dissociable mode of attachment to the bacterial surface via the GW domains may act to restrict Met activation to a small, local area of the host cell and to coordinate membrane dynamics with bacterial proximity during invasion.
Results
Structure determination
InlB crystals have a high solvent content (∼77%) and diffract anisotropically with a resolution limit of ∼3 Å along the c* unit cell edge and of 2.65 Å resolution along the others. Phases were determined by multiwavelength anomalous dispersion (MAD) using terbium or samarium derivatives (Table I), and the resulting electron density was improved by solvent flipping (Figure 1A). Terbium binds at a crystal contact and enhances resolution by ∼0.5 Å, and model refinement was carried out with data from a terbium-derivatized crystal [Table I, Tb(2)λ1]. The free R-factor of 30.2% is acceptable for a 2.65 Å resolution structure (Kleywegt and Brünger, 1996), although it is elevated due to the anisotropy, which is also reflected in the high average B-factor (Figure 1B; Table I). However, the Rfree/Rwork ratio is better than average and indicates a model that is not subject to systematic error (Tickle et al., 1998).
Table I. X-ray data collection and refinement.
| Data collection | |||||||
|---|---|---|---|---|---|---|---|
| Data set | Smλ1 | Smλ2 | Smλ3 | Tb(1)λ1 | Tb(1)λ2 | Tb(1)λ3 | Tb(2)λ1 |
| Wavelength (Å) | 1.8455 | 1.8461 | 1.7220 | 1.6493 | 1.6499 | 1.6100 | 1.1500 |
| Resolution range (Å) | 50–3.00 | 50–3.00 | 50–2.95 | 50–2.75 | 50–3.00 | 50–3.32 | 50–2.65 |
| Highest shell (Å) | 3.11–3.00 | 3.11–3.00 | 3.06–2.95 | 2.85–2.75 | 3.12–3.00 | 3.50–3.32 | 2.74–2.65 |
| Completenessa | 89.6 (39.6) | 89.5 (44.8) | 95.0 (89.5) | 84.0 (57.4) | 83.9 (37.2) | 86.1 (83.6) | 86.3 (59.0) |
| Redundancy | 8.0 | 8.0 | 8.0 | 6.4 | 6.4 | 6.4 | 4.2 |
| I/σIa | 11.9 (1.3) | 12.4 (1.2) | 12.4 (1.5) | 16.4 (1.6) | 15.6 (1.2) | 14.8 (1.6) | 14.4 (1.5) |
|
Rmergea,b |
16.4 (54.0) |
15.1 (57.3) |
16.2 (68.0) |
13.0 (64.6) |
10.6 (74.2) |
11.6 (72.9) |
8.1 (56.1) |
| Phasing | |||||||
| Dispersive and anomalous differencesc | Sumarium |
Terbium |
|||||
| |
λ1 |
λ2 |
λ3 |
λ1 |
λ2 |
λ3 |
|
| λ1 | 11.2 | 9.5 | |||||
| λ2 | 5.3 | 8.0 | 7.3 | 6.6 | |||
| λ3 | 9.2 | 7.8 | 6.1 | 10.1 | 6.3 | 6.7 | |
| Phasing powerd | |||||||
| (isomorphous/anomalous) | |||||||
| Centric | 030/– | 4.9/– | –/– | 1.1/– | 5.1/– | –/– | |
| Acentric | 0.43/4.1 | 5.4/3.6 | –/2.7 | 1.7/3.3 | 5.7/2.7 | –/1.8 | |
| Figure of merit | |||||||
| Resolution rangea (Å) | 50–3.45 (3.69–3.45) | 50–3.45 (3.69–3.45) | |||||
| Centrica | 037 (0.20) | 0.47 (0.24) | |||||
| Acentrica |
0.62 (0.45) |
|
|
0.62 (0.45) |
|
|
|
| Refinement | |||||||
| Resolutiona (Å) | 50–2.65 (2.74–2.65) | ||||||
| Rcrysta,e | 27.3 (46.0) | ||||||
| Rfreea,e | 30.2 (48.8) | ||||||
| No. of reflections | |||||||
| Working set | 35 702 | ||||||
| Test set | 1706 | ||||||
| No. of atoms | |||||||
| Protein | 4172 | ||||||
| Solvent/ion | 65/12 | ||||||
| R.m.s. deviation | |||||||
| Bonded B-factor (Å2)f | 3.6 | ||||||
| From ideal geometry | |||||||
| Lengths (Å) | 0.0064 | ||||||
| Angles (°) | 1.344 | ||||||
| Average B-factors (Å2) | |||||||
| Protein | 93.6 | ||||||
| Non-protein | 88.4 | ||||||
| Ramachandran regions (%) | |||||||
| Most favored | 69.5 | ||||||
| Additional allowed | 28.0 | ||||||
| Generously allowed | 2.5 | ||||||
aHighest resolution shell in parentheses.
cAnomalous and dispersive differences = 100 × r.m.s. ΔF/r.m.s. F, where ΔF for anomalous differences is (F+h – F–h)/2 (diagonal element) and for dispersive differences is Fλi – Fλj (off-diagonal).
dIsomorphous and anomalous phasing power are FH/E and FH”/E, respectively. FH and FH” are the mean amplitude and anomalous component, respectively, of the heavy atom structure factor. E is the r.m.s. lack-of-closure error.
eR = 100 × 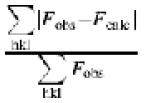 , where Rfree is calculated for a randomly chosen 5% of reflections (F > 0) omitted from refinement, and Rcryst is calculated for the remaining 95% of reflections (F > 0) included in refinement.
, where Rfree is calculated for a randomly chosen 5% of reflections (F > 0) omitted from refinement, and Rcryst is calculated for the remaining 95% of reflections (F > 0) included in refinement.
fR.m.s. deviation between B-factors of bonded atoms.
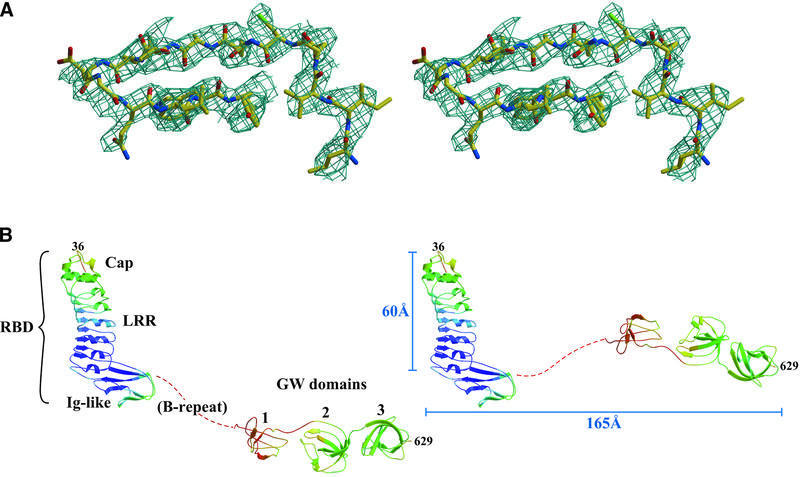
Fig. 1. Structure of InlB. (A) Stereo view of experimental electron density (contoured at 1σ) in the third GW domain. Phases were calculated from samarium MAD data and modified by solvent flipping. (B) InlB in ribbon representation, with a color gradient of main chain B-factors (blue, ≤50 Å2; red, ≥140 Å2). The red dotted line represents the B-repeat, which was not modeled. Two possible conformers, differing in the path of the B-repeat, are shown.
Overall structure of InlB
InlB is an elongated molecule that has an ‘L’ shape in the crystal (Figure 1B). The short arm of the ‘L’ spans ∼60 Å and the long arm ∼165 Å. The N-terminus is composed of three motifs that together form the Met receptor-binding domain (RBD), as previously described (Schubert et al., 2001). The first two of these structural motifs, an N-terminal cap (residues 36–76) and an LRR motif (residues 77–240), form the short arm of the ‘L’ and are sufficient to bind Met (Shen et al., 2000). An immunoglobulin-like (Ig-like) segment (residues 241–319) projects at nearly a right angle from the base of the LRR and forms the third part of the RBD. The RBD in intact InlB is structurally similar to the previously published RBD fragment (Cα r.m.s.d. 0.84 Å) (Schubert et al., 2001), except for a loop consisting of residues 288–291 (Cα r.m.s.d. 3.9 Å). This loop has higher than average B-factors in both structures, and may also be constrained differently in intact InlB as opposed to the RBD fragment. In addition, some small differences exist in secondary structure, most notably in residues 237–248, which form the first β-strand of the Ig-like domain in the 1.6 Å resolution RBD fragment structure (Schubert et al., 2001) but lack β-strand characteristics in this medium resolution structure.
The long arm of the ‘L’ continues from the Ig-like domain through a poorly ordered segment of 72 residues known as the B-repeat region into three GW domains (Figure 1B). InlB has a single B-repeat, while other internalins have more (up to nine in Listeria innocua LIN2724) (Glaser et al., 2001). No function has been attributed to the InlB B-repeat region or other B-repeat regions. Electron density for the B-repeat, which spans ∼30 Å and is more closely associated with the Ig-like section than the GW domains, is weak and could not be modeled reliably. Additionally, B-repeats from two InlB molecules meet at a crystal contact, making it difficult to distinguish between two alternative conformations of InlB in the crystal (Figure 1B, left and right). The existence of these two possibilities, which differ in orientation between the RBD and GW domains, does not affect functional interpretation. The flexibility of the B-repeat domain raises the possibility that the ‘L’ shape observed for InlB may be driven by crystal contacts rather than representing its shape on the bacterial surface or in solution.
Biochemical evidence for domain arrangement
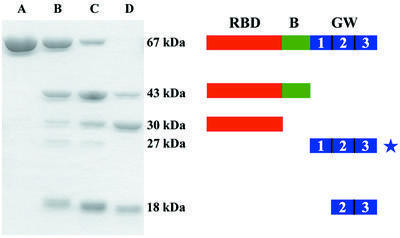
Proteolytic mapping verifies the flexibility of the B-repeat region and the domain arrangement seen in the crystal. Digestion of InlB using thermolysin (Figure 2), chymotrypsin or papain yields similar patterns, allowing general conclusions to be drawn. Within 1 h, InlB (67 kDa, residues 36–630) is cleaved into two major fragments: an ∼43 kDa polypeptide containing the RBD and B-repeat (InlB-RBD + B, residues 36–393) and an ∼18 kDa polypeptide containing the last two GW domains (InlB- GW[2–3], residues 464–630). A faint product at ∼27 kDa is also observed and probably contains all three GW domains. The first GW repeat is the most proteolytically susceptible domain in InlB, consistent with it having the highest average B-factor. The B-repeat is more stable than the first GW domain, but is removed over the course of 20 h, resulting in trimming of InlB-43 to an ∼30 kDa fragment (InlB-RBD, residues 36–320) that contains only the RBD.
Fig. 2. Proteolysis of InlB. Time course of InlB proteolysis. InlB was digested with thermolysin for 0, 1, 3 and 20 h (lanes A, B, C and D, respectively) and analyzed by 12% SDS–PAGE. A schematic of proteolytic products is shown to the right of corresponding fragments. Fragments were identified by N-terminal sequencing and mass spectrometry (MALDI), except for the 27 kDa fragment (starred) whose identification is tentative.
GW domains resemble SH3 domains
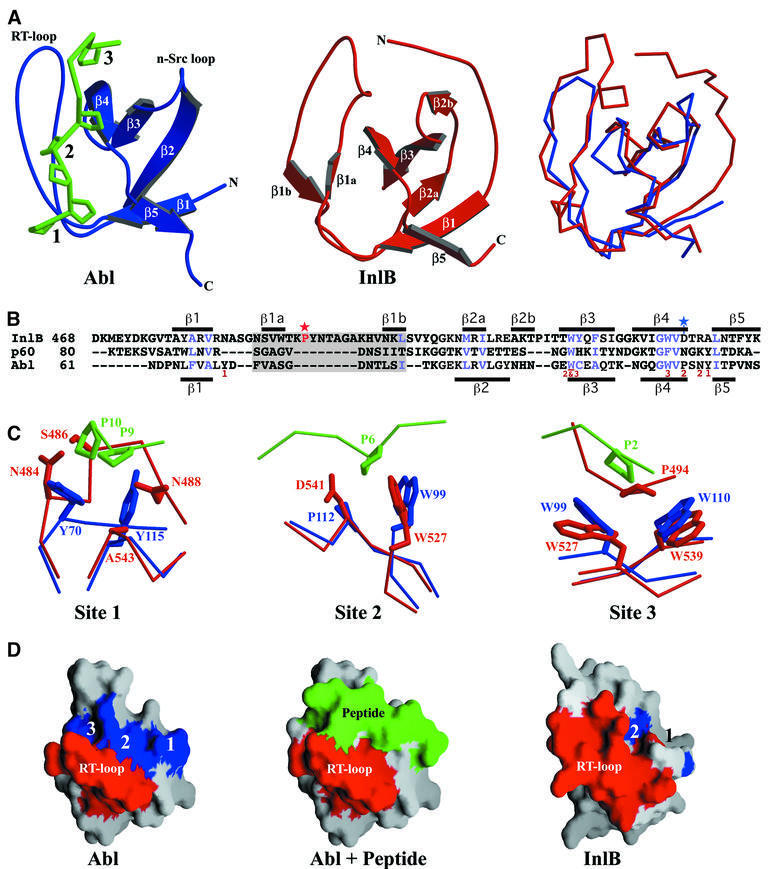
Strikingly, the InlB structure reveals that GW domains are related to SH3 domains (Figure 3A). A common evolutionary origin for these domains is suggested by conservation of hydrophobic core-forming residues (Figure 3B, blue). GW domains are also related to but distinct from recently described SH3-like prokaryotic sequences called SH3b domains (Figure 3B, p60) (Ponting et al., 1999; Whisstock and Lesk, 1999); the structure of an SH3b domain has not yet been determined. SH3 domains are found in signal transduction proteins and function as adaptors that bind proline-rich target sequences (Kuriyan and Cowburn, 1997). The GW domain is composed of seven β-strands, five of which are organized into an open barrel conformation like that of SH3 domains (Figure 3A). The eponymous GW dipeptide, located in the fourth β-strand (Figure 3B), is more conserved in GW domains than in SH3 domains (Larson and Davidson, 2000). The tryptophan is strictly conserved in GW proteins, while the glycine, which appears to be conserved for steric rather than conformational reasons, is found in all but two GW proteins (LMO1076 and Leuconostoc mesenteroides alternansucrase). Both the glycine and tryptophan are buried in GW proteins, while the equivalent residues in SH3 proteins are surface accessible, perhaps explaining the greater conservation in GW proteins.
Fig. 3. GW domains resemble SH3 domains. (A) Ribbon representation of GW and SH3 domains. Left: the Abl SH3 domain (blue), with bound peptide (green, backbone representation with prolines shown). The three peptide-binding pockets are numbered. Middle: InlB GW domain 2. Right: superposition of Abl SH3 (blue) and InlB GW (red), in Cα representation. (B) Structure-based sequence alignment of InlB GW domain 2, the L.monocytogenes p60 SH3b domain and the Abl SH3 domain. Residues responsible for peptide binding in the Abl SH3 domain are marked with numbers corresponding to binding pockets. Core residues conserved in GW and Abl are in blue, and secondary structure is indicated for GW domain 2 (top) and Abl (bottom). Gray shading marks the RT-loop, a red star indicates the intramolecular proline contact in InlB site 3, and a blue star indicates the substituted residue at InlB site 2. (C) Peptide-binding sites of Abl SH3 (blue, and bound peptide in green) and equivalent locations in InlB GW domains (red). (D) Molecular surface representations of the Abl SH3 domain and InlB GW domain 2. Numbers correspond to proline-binding sites (blue) in Abl and potential sites in the GW domain (blue). The RT-loop is colored red, and peptide bound to the SH3 domain is in green.
Although structural mimics of SH3 domains, GW domains are unlikely to be functional mimics. SH3 domains have three distinct proline-binding sites formed in part by the RT-loop, which connects the β1 and β2 strands (Figure 3A and B) (Musacchio et al., 1994). In GW domains, the RT-loop is longer, forming additional β-strands (β1a and β1b), and occupies two of the three potential proline-binding sites. Interestingly, GW site 3 has a contact that mimics an SH3–ligand interaction, binding a proline from the longer RT-loop (Figure 3C). However, this intramolecular contact blocks the site from intermolecular ligand interaction. Site 3 is formed by a pair of tryptophans that are highly conserved in SH3 and GW domains; in GW domains, one of the tryptophans is part of the GW dipeptide. The RT-loop residue contacting site 3 is conserved as a proline or valine (Figure 3B, red star), implying that this site is blocked in all GW domains.
Site 1 is also blocked by the longer RT-loop (Asn484 and Ser486) and, additionally, the two tyrosines that form this site in SH3 domains are not conserved in GW domains. Lastly, site 2, while not blocked by the RT-loop, is destroyed by various large polar residues that substitute for a highly conserved proline in SH3 domains (Figure 3B, blue star).
The RT-loop in GW domains is unlikely to shift away and expose the potential proline-binding site. The RT-loop buries 1260 Å2 of surface area in packing against the site through hydrophobic interactions and hydrogen bonding between main chain atoms of the loop and those of the site. In addition, the loop has B-factors representative of the rest of the domain. These observations are consistent with the RT-loop forming an integral and stable part of the GW domain.
InlB GW domains bind gC1q-R
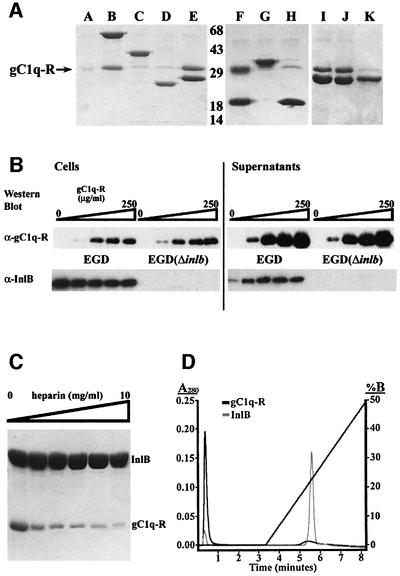
The InlB GW domains have been shown to bind the non-protein ligands LTA, a Gram-positive cell wall component, and heparin, a mammalian GAG (Jonquières et al., 1999, 2001). We now show that the GW domains also bind a protein ligand, gC1q-R. His-tagged InlB or InlB fragments were incubated with purified, recombinant gC1q-R, and association was assayed using Ni-NTA– agarose pull-down. Intact InlB associates with gC1q-R, as does a fragment composed of the three GW domains or just the second and third GW domains, while fragments lacking the GW domains fail to bind gC1q-R (Figure 4A). Binding between the InlB GW domains and gC1q-R is specific, as seen by the lack of association between gC1q-R and GW domains from the L.monocytogenes protein Ami. Although 40% identical in sequence to InlB GW domains 2 and 3, Ami GW domains 5 and 6 fail to bind gC1q-R (Figure 4A, lane H). Likewise, a construct containing the third to sixth Ami GW domains also fails to bind gC1q-R (Figure 4A, lane G). We also examined whether InlB association with gC1q-R is dependent on divalent cations, as EDTA was shown previously to elute gC1q-R from an InlB affinity column (Braun et al., 2000). We, however, find no dependence on divalent cations, as gC1q-R binds to the GW domains in the presence of 1 mM EGTA or EDTA (Figure 4A, lanes I and J). This indicates the potential existence of other components that stabilize association between InlB and gC1q-R in a divalent cation-dependent manner.
Fig. 4. InlB GW domains associate with gC1q-R. (A) Ni-NTA–agarose bead pull-down assay, visualized using 12% SDS–PAGE, of gC1q-R incubated with (A) no added protein, (B) InlB (residues 36–630), (C) InlB-RBD + B (36–393), (D) InlB-LRR (36–248), (E) InlB- GW[1–3] (399–630), (F) InlB-GW[2–3] (464–630), (G) Ami-GW [3–6] (436–755), (H) Ami-GW[5–6] (593–755), (I) InlB-GW[1–3] plus 1 mM EDTA, (J) InlB-GW[1–3] plus 1 mM EGTA, and (K) InlB-GW[1–3] plus 500 mM NaCl. The arrow indicates the position of gC1q-R, and molecular weights (kDa) are indicated. (B) gC1q-R binds soluble but not bacterial surface-attached InlB. Listeria monocytogenes EGD and EGD(ΔinlB) were incubated with increasing gC1q-R concentrations. Western blot of cell pellets (left) and supernatants (right) using an anti-gC1q-R monoclonal antibody (top) or anti-InlB polyclonal antibodies (bottom). Lanes correspond to 0, 10, 50, 100 and 250 µg/ml gC1q-R. (C) Dose-dependent competition of heparin with gC1q-R for InlB binding (left). Ni-NTA–agarose bead pull-down of His-tagged InlB incubated with gC1q-R and increasing concentrations of heparin, visualized using 12% SDS–PAGE. Lanes correspond to 0, 0.5, 1, 2.5, 5 and 10 mg/ml heparin. (D) Heparin affinity of InlB and gC1q-R. InlB (gray) or gC1q-R (black) were applied to a heparin column and eluted using a salt gradient (sloping line).
To determine whether association with gC1q-R requires release from the bacterial surface, we asked whether gC1q-R binds InlB while it is still attached to the bacterial surface. gC1q-R was incubated with L.monocytogenes EGD or EGD(ΔinlB) (Dramsi et al., 1995), and the amount of bacterially associated gC1q-R was assessed by western blot using a monoclonal antibody (Figure 4B, top). While a small amount of gC1q-R does associate with these bacteria, this association is not specific to InlB, as seen by the equal gC1q-R association with both EGD and EGD(ΔinlB). Significantly, gC1q-R competes with the bacterial surface for InlB binding. Increasing concentrations of gC1q-R result in increasing amounts of released, soluble InlB from EGD, as detected by western blot using a polyclonal antibody (Figure 4B, bottom). The amount of released, soluble InlB appears to saturate at 50 µg/ml gC1q-R. These results demonstrate that gC1q-R binds soluble rather than bacterial surface-attached InlB, and support a model in which soluble InlB interacts functionally with the mammalian cell surface. A similar result has been observed for heparin, in which heparin competes with the bacterial surface for InlB (Jonquières et al., 2001).
We next asked whether association of InlB with heparin and gC1q-R could occur simultaneously. Complexes of gC1q-R and InlB, immobilized on Ni-NTA–agarose beads through a His tag on InlB, were incubated with increasing concentrations of heparin. A competitive, dose-dependent release by heparin of gC1q-R from InlB is observed (Figure 4C). gC1q-R does not itself bind heparin, as detemined by heparin affinity chromatography. gC1q-R flows through a heparin column, while, as previously reported (Jonquières et al., 2001), InlB binds and is eluted by high salt (975 mM) (Figure 4D). These data indicate that InlB forms only binary complexes with gC1q-R or heparin, and that these ligands must act sequentially rather than simultaneously.
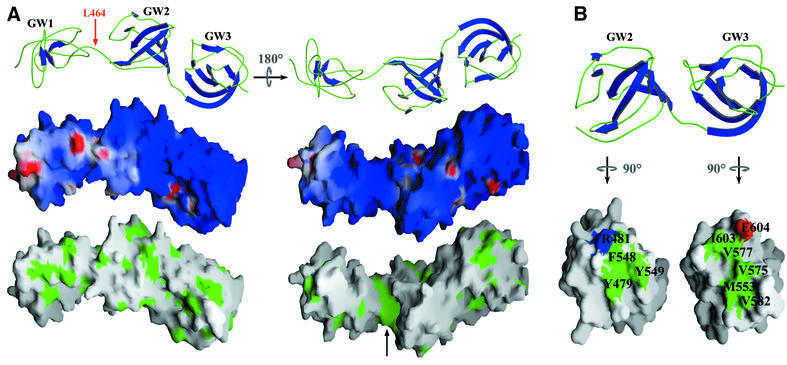
Surface features of InlB GW domains
The surface features of the InlB GW domains explain some of their ability to bind multiple ligands (Figure 5A). All three ligands of the InlB GW domains, i.e. LTA, heparin and gC1q-R, are acidic molecules. The surfaces of the GW domains are entirely basic, except for a few small, isolated areas of negative charge. The InlB GW domains have a predicted isoelectric point of ∼10 and, although almost all known GW domains are basic, the InlB GW domains present the most extreme case. Consistent with electrostatic interactions, high ionic strength is found to disrupt association between the GW domains and gC1q-R (Figure 4A, lane K) or heparin (Figure 4D) (Jonquières et al., 2001). It should be noted that gC1q-R lacks proline-rich regions and, as explained above, the GW domains are unlikely to engage in SH3-like proline binding. Although electrostatic forces are important for binding, they do not alone explain specificity, as evidenced by the highly basic GW domains of Ami failing to interact with gC1q-R (Figure 4A). Rather, specificity must arise from other interactions, possibly conferred by a small hydrophobic groove located between the first and second GW domains (Figure 5A).
Fig. 5. Surface features of InlB GW domains. (A) Top: ribbon representation of the three InlB GW domains. The first GW domain is proteolytically sensitive and cleaved from the second and third protease-resistant GW domains at Leu464. Middle: electrostatic surface potential of the GW domains (red = –10 kT, blue = +10 kT). Bottom: exposed hydrophobic residues (green) mapped to the molecular surface of the GW domains. The black arrow indicates the hydrophobic groove between domains 1 and 2. (B) Basis for GWA–GWB pairwise association. Top: ribbon representations of GWA domains (left) and GWB domains (right). Bottom: molecular surface representation (green, hydrophobic; red, acidic; blue, basic), with GWA and GWB rotated to show interface residues (numbered).
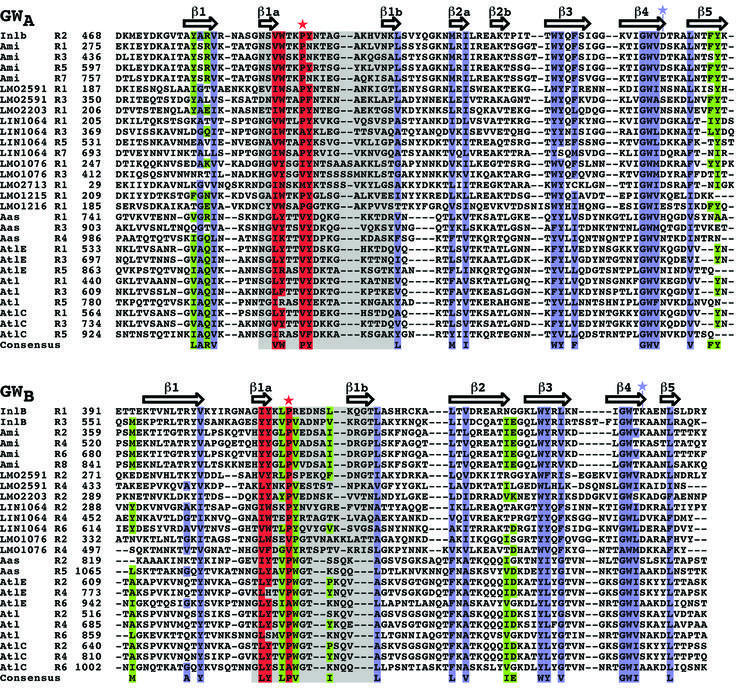
Pairwise association in GW domains: GWA and GWB
More than 90% of known GW domains are found in proteins with multiple, tandem GW domains, and sequence identity between alternate domains is greater than between adjacent ones (average of 58 versus 27%, respectively). This is best understood as GW domains assorting into two subtypes, GWA and GWB (Figure 6), that alternate in sequence but share a common SH3-like structural core (Cα r.m.s.d. GW[2] versus GW[3]: 2.4 Å). The InlB structure reveals the basis for this alternation: GWA–GWB pairs form stable structural units, with an interface made up of three hydrophobic residues conserved in GWA domains and five hydrophobic residues conserved in GWB domains (Figure 5B, and Figure 6, green). The second and third InlB GW domains comprise one such GWA–GWB pair. The pairwise interaction is also aided by a conserved hydrogen bond between Arg481 in the second GW domain (GWA) and Glu604 in the third domain (GWB).
Fig. 6. Sequence alignment of GWA and GWB domains. Sequences were aligned with CLUSTAL W (Thompson et al., 1994). Residues conserved with SH3 domains are blue, and those conserved uniquely in GW domains are red; the RT-loop is gray. Conserved residues involved in GWA–GWB pairwise association are in green. Secondary structure assignments for InlB (GW domain 2 for GWA and domain 3 for GWB) are shown above the sequences. The red star indicates the intramolecular proline in InlB site 3, and the blue star the substituted residue at InlB site 2. (DDBJ/EMBL/GenBank accession Nos: InlB, NP_463963; Ami, NP_466081; LMO2591, NP_466114; LMO2203, NP_465727; LIN1064, NP_470401; LMO1076, NP_464601; LMO2713, NP_466235; LMO1215, NP_464740; LMO1216, NP_464741; Aas, T30290; AtlE, AAB63571; Atl, NP_371577; AtlC, AAK17065.)
These interface residues promote pairing between N-terminal GWA domains and C-terminal GWB domains, but are not found between N-terminal GWB and C-terminal GWA domains. Indeed, variable sequences of 3–36 residues often separate N-terminal GWB domains from C-terminal GWA domains. However, no N-terminal GWA is disrupted by a linker sequence from a C-terminal GWB. Proteins with large numbers of GW repeats, such as the staphylococcal autolysins, may then contain long chains of GWA–GWB pairs tethered by flexible linkers.
The first GW domain of InlB is of the GWB subtype and does not have a GWA pairing partner preceding it. The lack of pairing explains its proteolytic susceptibility, its higher than average B-factors and its relative lack of regular secondary structure. This domain does not pack tightly against the succeeding GW domain (GWA), burying only 649 Å2 of surface area in contrast to the 1072Å2 buried in the GWA–GWB packing of the second and third domains. The third GW domain, which like the first is of the GWB subtype but is paired, is not susceptible to proteolysis and contains significantly more regular secondary structure. Only 13% of identified GW domains are unpaired (mostly of the GWA type). Each of these unpaired GW domains is missing some or all of the residues involved in GWA–GWB pairing (Figure 6), indicating the loss of selection pressure on pair-forming residues in isolated GW domains.
Discussion
A number of proteins are known to promote entry of microbial pathogens into host cells that normally are not phagocytic. Among these are L.monocytogenes InlA and InlB, Yersinia pseudotuberculosis invasin (Hamburger et al., 1999), Shigella flexneri Ipa proteins (Tran Van Nhieu et al., 2000) and Streptococcus pyogenes F1 protein (Ozeri et al., 1998). The structure of invasin has been determined, showing a highly elongated rod-like protein (Hamburger et al., 1999), in some ways reminiscent of the elongated structure of InlB. The elongation of invasin, which is tethered to the outer membrane, serves to project a C-terminal domain ∼180 Å from the bacterial surface for interaction with integrins on host cells. However, the elongation of InlB appears to serve a different purpose.
Unlike invasin, in InlB, the bacterial surface attachment domains are also involved in binding host ligands which are important to invasion of mammalian cells. This is possible because while invasin has a putative β-barrel domain that integrates non-dissociably into the bacterial outer membrane, the InlB GW domains are attached dissociably to LTA. Interactions between the GW domains and host components require detachment from the bacterial surface, as shown by competition between host ligands and bacterial surface components. InlB also appears to be buried in the Gram-positive cell wall (Jonquières et al., 1999), hindering the accessibility of its N-terminal LRR portion for interaction with Met. These data argue that elongation in InlB is not important for projection from the bacterium but rather for proper conformation in associations with host ligands—Met through the LRRs, and GAGs or gC1q-R through the GW domains—as a soluble, released molecule.
The GW domains are related to SH3 (Kuriyan and Cowburn, 1997) and prokaryotic SH3b domains (Ponting et al., 1999; Whisstock and Lesk, 1999). Interestingly, searches that identified SH3b domains failed to identify GW domains as part of the SH3 family. However, our structural evidence along with sequence considerations establish that GW domains are indeed divergent members of the SH3 family. The pattern of conservation of hydrophobic core-forming residues is similar among SH3, SH3b and GW domains (Figure 3B), and distinguishes them from other bacterial proteins, such as the diphtheria toxin repressor, which have structural but not sequence homology to SH3 domains.
GW domains are notably different from SH3 and SH3b domains in having a longer RT-loop. This has functional consequence in that the longer loop blocks two of the three SH3-like peptide-binding sites. This and substitutions at some of the SH3-like peptide-binding sites indicate that GW domains are unlikely to bind ligands using the same motifs as SH3 domains. By these criteria, it remains possible that SH3b domain proteins, such as the L.monocytogenes p60, are able to bind proline-rich targets. Additionally, GW domains are seen to assort into two subtypes, which associate as GWA–GWB structural pairs, an arrangement not observed in SH3 domains. The pairing appears to stabilize these small protein domains, as evidenced by the unpaired first GW domain of InlB being highly susceptible to proteolysis.
While SH3b domains are found in both Gram-negative and Gram-positive bacteria and form a family of ∼100 non-redundant proteins (Schultz et al., 2000), GW domains have only been identified in Gram-positive bacteria and form a small protein family (Cabanes et al., 2002). Based on structure-based sequence alignment, we are able to identify L.mesenteroides alternansucrase and Bacillus subtilis YubE as new members of the GW protein family. Furthermore, searches of partial genome sequences and nucleotide databases indicate the potential existence of GW domains in a diverse group of bacteria, including Clostridia spp., Geobacillus, Cytophaga hutchin sonii, Mesorhizobium loti and Nostoc punctiforme. This would extend their occurrence beyond Gram-positive bacteria.
The GW and SH3b families, however, do share some similarities. All known GW proteins have putative signal sequences targeting them for export to the bacterial surface, and a predominant number of SH3b domain proteins examined also localize to the extracellular space. Furthermore, a number of SH3b proteins have bacterial peptidoglycan lytic domains, as do almost all of the GW proteins, with InlB being one of the exceptions. The function of SH3b domains is not completely understood but, in the case of the Staphylococcus simulans protein lysostaphin, the SH3b domain is responsible for targeting this lytic enzyme to the cell surface of competing bacterial strains (Baba and Schneewind, 1996).
The function of GW domains has been ascribed to bacterial surface attachment, but these domains appear to have functions beyond attachment, and not all GW proteins are expected to be surface attached. This latter prediction comes from the observation that the strength of bacterial surface attachment is modulated by the number of GW domains. InlB, with three domains, partitions nearly evenly between bacterial surface-attached and released forms (Jonquières et al., 2001). However, an InlB variant carrying only one GW domain is completely released, and one carrying eight is completely retained (Braun et al., 1997; Jonquières et al., 1999). A number of GW proteins have a single GW domain, and would therefore be expected to be released rather than attached. Additionally, a number of GW proteins have putative transmembrane regions that render the GW domain redundant for attachment. Taken together, these observations indicate that GW domains could have functions besides attachment. For InlB, this encompasses binding to host ligands, as has been observed for other GW proteins. GW domains in Staphylococcus saprophyticus Aas and Staphylococus caprae AtlC confer binding to fibronectin, and in Staphylococcus epidermidis AtlE to vitronectin (Heilmann et al., 1997; Hell et al., 1998; Allignet et al., 2001). Like InlB, these GW domain-containing proteins are also virulence factors. Additionally, the GW domains of L.monocytogenes Ami have been shown to confer adhesion to mammalian cells (Milohanic et al., 2001).
What could be the purpose of dissociable attachment of InB to the bacterial surface? An intriguing idea is that this method of attachment permits localized release of GW domain-containing proteins. In the case of InlB, localized release may be important for activating Met in the vicinity of the bacterium. Interestingly, host membrane ruffling is elicited in the absence of bacteria by soluble InlB, while ruffles are not evident during internalization of L.monocytogenes, which occurs through membrane zippering. The reason for this may be the following. Membrane ruffles elicited by released, soluble InlB may be stabilized and prevented from retracting by a proximate bacterium, thereby promoting expansion of transient ruffles into concerted membrane zippering and productive internalization. On the other hand, membrane ruffling or other membrane changes without a nearby bacterium to engulf would, of course, be unproductive. Therefore, it may be important for invasion to couple Met activation spatially and temporally with bacterial proximity. The dissociable attachment of InlB to the bacterial surface seems perfectly suited to allow such control, highlighting how pathogens may be adapted to exploit host ligands maximally for their own profit.
Materials and methods
Protein cloning and purification
InlB was expressed in Escherichia coli as previously described (Braun et al., 1997), and purified by nickel chelation chromatography (Poros MC) from bacteria lysed by sonication (in 600 mM NaCl, 100 mM sodium phosphate buffer pH 8.0, 15 mM imidazole, 5 mM β-mercaptoethanol). Nucleic acids were removed by 0.5% polyethyleneimine precipitation; the supernatant was precipitated with 80% saturated ammonium sulfate, and resuspended and dialyzed in buffer A [500 mM NaCl, 75 mM Tris pH 8.0, 5 mM EDTA, 1 mM dithiothreitol (DTT)]. InlB was purified further by size exclusion chromatography (Superdex 200), and concentrated to ∼28 mg/ml (ε280 = 97 510/M/cm) by dialysis against 30% polyethylene glycol (PEG) 20 000 in buffer A. Concentrated InlB was dialyzed in buffer A (except containing 10 mM Tris pH 8.0 and 1 mM EDTA), and stored as flash-frozen aliquots at –80°C.
InlB-LRR (residues 36–248) was expressed and purified as previously described (Marino et al., 1999). InlB-RBD + B (residues 36–398) was expressed in E.coli as previously described (Braun et al., 1998), and purified using the InlB-LRR protocol. InlB-GW[1–3] (residues 399–630) was expressed in E.coli as previously described (Braun et al., 1999) and purified using the InlB protocol.
gC1q-R (residues 75–282) was expressed in E.coli as previously described (Krainer et al., 1991), and purified as follows. Bacteria were lysed by sonication (in 50 mM HEPES pH 8.0, 5% glycerol, 100 mM KCl, 2 mM EDTA and 2 mM DTT) and subjected to ammonium sulfate cuts of 65 and 80% saturation. The pellet from the 80% cut was resuspended, dialyzed in 20 mM HEPES, 5% glycerol, 50 mM NaCl, 2 mM EDTA, 2 mM DTT, and purified by anion exchange chromatography (Poros HQ/M). gC1q-R was purified further by size exclusion chromatography (Superdex 200) in 25 mM HEPES pH 8.0, 150 mM NaCl, 2 mM EDTA, 2 mM DTT. Purified gC1q-R was dialyzed against 10 mM HEPES, pH 8.0, 1 mM DTT and 0.5 mM EDTA, concentrated to 20 mg/ml [ε280(calc) = 22 190/M/cm], and stored as flash-frozen aliquots at –80°C.
Ami GW[3–6] (residues 436–755) and Ami GW[5–6] (residues 593–755) were cloned by PCR from L.monocytogenes EGD genomic DNA. InlB-GW[2–3] (residues 464–630) was cloned by PCR from plasmid pET28b-1 (Braun et al., 1997). These constructs contain artifactual sequences at the N-terminus (MG) for expression and cloning purposes and at the C-terminus (LEHHHHHH) for purification. These proteins were purified using the InlB protocol.
InlB crystallization and data collection
Crystals were obtained in 1.9 M Li2SO4, 10% glycerol, 1 mM DTT, 100 mM MES pH 6.5 by the hanging drop method. Crystals grew in space group C2221 with unit cell dimensions of a=48.5Å, b = 330.9 Å and c = 182.4 Å, and one protein molecule per asymmetric unit. Crystals were cryoprotected by soaking for ∼30 min in 2 M Li2SO4, 500 mM NaCl, 100 mM MES pH 6.5 and 11% (w/v) i-erythritol (Sigma) supplemented with either 50 mM SmCl3 or 65 mM TbCl3, and flash cooled in liquid N2. Samarium MAD data were collected at the Advanced Photon Source (Argonne, IL,), beamline ID19, and terbium MAD data were collected at the National Synchrotron Light Source (Brookhaven, NY), beamline X25A. Data were indexed, integrated and scaled using the HKL2000 program suite (Otwinowski and Minor, 1997).
Phase determination
Samarium atom positions were determined with SOLVE (Terwilliger and Berendzen, 1996), and later adjusted using dispersive and anomalous difference Fourier maps calculated with partial model (containing the RBD and GW[3]) phases. Terbium atom positions were determined using the same partial model phases. The final samarium heavy atom model has six heavy atom sites and the terbium model has seven, with five in common. Heavy atom positions were refined in SHARP (de La Fortelle and Bricogne, 1997), and solvent flattening (77% solvent) was carried out with Solomon (CCP4, 1994).
Model building and refinement
The structure of the InlB-LRR (residues 36–242, PDB 1D0B) was placed by inspection into an experimental electron density map calculated with phases derived from samarium MAD data, and residues 243–319 were built in O (Jones et al., 1991). Other portions of InlB were modeled through iterative rounds of model building and refinement, using CNS (Brünger et al., 1998), against samarium or terbium MAD data. Refinement in these rounds was performed using cycles of rigid body and domain B-factor refinement, followed by cycles of conjugate gradient minimization and per-residue B-factor refinement. Phases calculated from the model (residues 36–319, or RBD) were combined by σa weighting (CCP4, 1994) with phases calculated from the samarium MAD experiment, and GW[3] (residues 551–629) was built and refined in four further rounds. Phases calculated from the resulting model (containing RBD and GW[3]) were combined with phases calculated from the terbium MAD experiment, and GW[2] (residues 468–550) was built and refined in three further rounds. A model for GW[1] (residues 392–467) was constructed based on homology to GW[3] and used as a starting point for building. GW[1] was built and refined in five additional rounds.
The resulting model, containing residues 36–319 and 391–629, was refined, using alternating cycles of conjugate gradient minimization and restrained atomic B-factor refinement, against data collected at 1.15 Å wavelength from a terbium-derivatized crystal. Fifty independent runs of Cartesian-simulated annealing were carried out, with the best run yielding decreases of 0.58 and 0.78% in Rfree and Rcryst, respectively. A conservative set of 65 waters (>5σ Fo – Fc peak size, and within 2.5–3.4 Å of a hydrogen bond donor or acceptor), a single sulfate ion and seven terbiums were added to the model before the final rounds of refinement.
gC1q-R binding assays
Binding assays were performed using Ni-NTA–agarose beads. Beads were washed four times with binding buffer (50 mM Tris pH 8.0, 50 mM NaCl, 10 mM imidazole, 1 mM DTT), then incubated with 75 µl of His-tagged constructs of InlB, InlB-LRR, InlB-GW[1–3], InlB-GW[2–3], InlB-RBD, Ami-GW[3–6] or Ami-GW[5–6], each at 65 µM. Beads were washed three times with binding buffer, mixed with 65 µl of gC1q-R (100 µM) in binding buffer, and incubated at room temperature for 15 min. Beads were washed four times with binding buffer, boiled in 2× SDS–PAGE sample buffer, and analyzed by 12% SDS–PAGE. Binding assays were also carried out as above in 1 mM EGTA, 1 mM EDTA or 500 mM NaCl. Heparin competition assays were carried out as above, except that once gC1q-R was bound, beads were rinsed three times with binding buffer and incubated for 30 min at 25°C in 150 µl of binding buffer supplemented with 0, 0.5, 1, 2.5, 5 or 10 mg/ml heparin, then washed and analyzed.
Listeria monocytogenes EGD and EGD(ΔinlB) were grown overnight at 37°C with shaking. Bacteria were washed three times with phosphate-buffered saline (PBS; 15 mM sodium phosphate buffer pH 7.4, 150 mM NaCl) and 1.5 ml (A600 = 1.0) aliquots were centrifuged (16 000 g, 2 min), resuspended in 300 µl of PBS containing varying concentrations (0, 10, 50, 100 or 250 µg/ml) of gC1q-R, and incubated at 25°C for 30 min. Cell suspensions were then centrifuged and supernatants were separated from cell pellets. Cell pellets were rinsed three times in PBS, and both supernatants and cell pellets were analyzed by western blot. Briefly, samples were resolved by 10% SDS–PAGE, transferred to PVDF membranes, blocked with 5% bovine serum albumin (BSA) and blotted with either a mouse monoclonal antibody to gC1q-R (Covance) or rabbit polyclonal antibodies to InlB, which were raised with purified InlB as antigen at the UCSD animal facility. Detection was carried out using horseradish peroxidase-conjugated anti-mouse- or anti-rabbit-Fc antibodies and the ECL-plus (Amersham) detection reagent.
InlB and gC1q-R heparin binding experiments were carried out using heparin affinity chromatography (Poros HE). InlB or gC1q-R (0.25 mg) was applied to a 1.6 ml column in buffer HA (300 mM NaCl, 15 mM HEPES pH 7.4); the column was washed with buffer HA and eluted with a gradient of 0–50% buffer HB (3000 mM NaCl, 15 mM HEPES pH 7.4), and absorbance at 280 nM was monitored.
Proteolysis
Proteolysis experiments were carried out with thermolysin at a 50:1 InlB:thermolysin mass ratio at 37°C in 100 mM MES pH 6.5, 500 mM NaCl, 2 mM CaCl2, 0.15 mM ZnSO4 and 1 mM DTT. The final concentration of InlB was 1 mg/ml and the total reaction volume was 100 µl. Aliquots of 5 µl were taken at varying times, mixed with 5 µl of thermolysin stop buffer (2× SDS–PAGE sample buffer supplemented with 50 mM EDTA), boiled and analyzed by 12% SDS–PAGE.
Molecular figures
Figures were prepared using Molscript and Raster3d (Kraulis, 1991; Merrit and Bacon, 1997), GRASP (Nicholls et al., 1991) or Bobscript (Esnouf, 1997).
Coordinates
Coordinates and structure factors have been deposited in the Protein Data Bank (1M9S).
Acknowledgments
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) grant R01 AI47163 (P.G.) and a grant from the American Heart Association (P.G.). P.G. is a W.M.Keck Distinguished Young Scholar; M.M. was supported in part by NIH training grant GM07240.
References
- Allignet J., Aubert,S., Dyke,K. and Solh,N. (2001) Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun., 69, 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aureli P., Fiorucci,G.C., Caroli,D., Marchiaro,G., Novara,O., Leone,L. and Salmaso,S. (2000) An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med., 342, 1236–1241. [DOI] [PubMed] [Google Scholar]
- Baba T. and Schneewind,O. (1996) Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J., 15, 4789–4797. [PMC free article] [PubMed] [Google Scholar]
- Bierne H. and Cossart,P. (2002) InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J. Cell Sci., 115, 3357–3367. [DOI] [PubMed] [Google Scholar]
- Braun L., Dramsi,S., Dehoux,P., Bierne,H., Lindahl,G. and Cossart,P. (1997) InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol., 25, 285–294. [DOI] [PubMed] [Google Scholar]
- Braun L., Nato,F., Payrastre,B., Mazie,J. and Cossart,P. (1999) The 213-amino-acid leucine-rich repeat region of the Listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol., 34, 10–23. [DOI] [PubMed] [Google Scholar]
- Braun L., Ohayon,H. and Cossart,P. (1998) The InlB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol. Microbiol., 27, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Braun L., Ghebrehiwet,B. and Cossart,P. (2000) gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J., 19, 1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cabanes D., Dehoux,P., Dussurget,O., Frangeul,L. and Cossart,P. (2002) Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol., 10, 238–245. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dramsi S., Biswas,I., Maguin,E., Braun,L., Mastroeni,P. and Cossart,P. (1995) Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol. Microbiol., 16, 251–261. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graphics, 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Gaillard J.L., Jaubert,F. and Berche,P. (1996) The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J. Exp. Med., 183, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P. et al. (2001) Comparative genomics of Listeria species. Science, 294, 849–852. [DOI] [PubMed] [Google Scholar]
- Hamburger Z.A., Brown,M.S., Isberg,R.R. and Bjorkman,P.J. (1999) Crystal structure of invasin: a bacterial integrin-binding protein. Science, 286, 291–295. [DOI] [PubMed] [Google Scholar]
- Heilmann C., Hussain,M., Peters,G. and Gotz,F. (1997) Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol., 24, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Hell W., Meyer,H.W. and Gatermann,S.G. (1998) Cloning of aas, a gene encoding a Staphylococcus saprophyticus surface protein with adhesive and autolytic properties. Mol. Microbiol., 29, 871–881. [DOI] [PubMed] [Google Scholar]
- Ireton K., Payrastre,B., Chap,H., Ogawa,W., Sakaue,H., Kasuga,M. and Cossart,P. (1996) A role for phosphoinositide 3-kinase in bacterial invasion. Science, 274, 780–782. [DOI] [PubMed] [Google Scholar]
- Jiang J., Zhang,Y., Krainer,A. and Xu,R. (1999) Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl Acad. Sci. USA, 96, 3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jonquières R., Bierne,H., Fiedler,F., Gounon,P. and Cossart,P. (1999) Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol., 34, 902–914. [DOI] [PubMed] [Google Scholar]
- Jonquières R., Pizarro-Cerda,J. and Cossart,P. (2001) Synergy between the N- and C-terminal domains of InlB for efficient invasion of non-phagocytic cells by Listeria monocytogenes. Mol. Microbiol., 42, 955–965. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Brünger,A.T. (1996) Checking your imagination: applications of the free R value. Structure, 4, 897–904. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature, 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Krainer A.R., Mayeda,A., Kozak,D. and Binns,G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- Kraulis P. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of proteins. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kuriyan J. and Cowburn,D. (1997) Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct., 26, 259–288. [DOI] [PubMed] [Google Scholar]
- Larson S. and Davidson,A. (2000) The identification of conserved interactions within the SH3 domain by alignment of sequences and structures. Protein Sci., 9, 2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M., Ohayon,H., Braun,L., Mengaud,J. and Cossart,P. (1997) Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun., 65, 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit M., Vandormael-Pournin,S., Lefort,J., Huerre,M., Gounon,P., Dupuy,C., Babinet,C. and Cossart,P. (2001) A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science, 292, 1722–1725. [DOI] [PubMed] [Google Scholar]
- Lorber B. (1997) Listeriosis. Clin. Infect. Dis., 24, 1–9. [DOI] [PubMed] [Google Scholar]
- Mansell A., Khelef,N., Cossart,P. and O’Neill,L.A. (2001) Internalin B activates nuclear factor-κB via Ras, phosphoinositide 3-kinase and Akt. J. Biol. Chem., 276, 43597–43603. [DOI] [PubMed] [Google Scholar]
- Marino M., Braun,L., Cossart,P. and Ghosh,P. (1999) Structure of the InlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L.monocytogenes. Mol. Cell, 4, 1063–1072. [DOI] [PubMed] [Google Scholar]
- Marino M., Braun,L., Cossart,P. and Ghosh,P. (2000) A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl Acad. Sci. USA, 97, 8784–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengaud J., Ohayon,H., Gounon,P., Mege,R.M. and Cossart,P. (1996) E-cadherin is the receptor for internalin, a surface protein required for entry of L.monocytogenes into epithelial cells. Cell, 84, 923–932. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Milohanic E., Jonquieres,R., Cossart,P., Berche,P. and Gaillard,J.L. (2001) The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol., 39, 1212–1224. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Saraste,M. and Wilmanns,M. (1994) High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat. Struct. Biol., 1, 546–551. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Ozeri V., Rosenshine,I., Mosher,D.F., Fassler,R. and Hanski,E. (1998) Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol., 30, 625–637. [DOI] [PubMed] [Google Scholar]
- Peerschke E.I. and Ghebrehiwet,B. (2001) Human blood platelet gC1qR/p33. Immunol. Rev., 180, 56–64. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Avarind,L., Schultz,J., Bork,P. and Koonin,E.V. (1999) Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol., 289, 729–745. [DOI] [PubMed] [Google Scholar]
- Raffelsbauer D., Bubert,A., Engelbrecht,F., Scheinpflug,J., Simm,A., Hess,J., Kaufmann,S.H. and Goebel,W. (1998) The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Gen. Genet., 260, 144–158. [DOI] [PubMed] [Google Scholar]
- Schubert W. et al. (2001) Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. J. Mol. Biol., 312, 783–794. [DOI] [PubMed] [Google Scholar]
- Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Naujokas,M., Park,M. and Ireton,K. (2000) InlB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell, 103, 501–510. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1996) Correlated phasing of multiple isomorphous replacement data. Acta Crystallogr. D, 52, 749–757. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle I., Laskowski,R. and Moss,D. (1998) Rfree and the Rfree ratio. I. Derivation of expected values of cross-validation residuals used in macromolecular least-squares refinement. Acta Crystallogr. D, 54, 547–557. [DOI] [PubMed] [Google Scholar]
- Tran Van Nhieu G., Bourdet-Sicard,R., Dumenil,G., Blocker,A. and Sansonetti,P.J. (2000) Bacterial signals and cell responses during Shigella entry into epithelial cells. Cell Microbiol., 2, 187–193. [DOI] [PubMed] [Google Scholar]
- van Leeuwen H.C. and O’Hare,P. (2001) Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J. Cell Sci., 114, 2115–2123. [DOI] [PubMed] [Google Scholar]
- Whisstock J.C. and Lesk,A.M. (1999) SH3 domains in prokaryotes. Trends Biochem. Sci., 24, 132–133. [DOI] [PubMed] [Google Scholar]