Abstract
The macrolide rapamycin specifically binds the 12-kD FK506-binding protein (FKBP12), and this complex potently inhibits the target of rapamycin (TOR) kinase. The identification of TOR in Arabidopsis (Arabidopsis thaliana) revealed that TOR is conserved in photosynthetic eukaryotes. However, research on TOR signaling in plants has been hampered by the natural resistance of plants to rapamycin. Here, we report TOR inactivation by rapamycin treatment in a photosynthetic organism. We identified and characterized TOR and FKBP12 homologs in the unicellular green alga Chlamydomonas reinhardtii. Whereas growth of wild-type Chlamydomonas cells is sensitive to rapamycin, cells lacking FKBP12 are fully resistant to the drug, indicating that this protein mediates rapamycin action to inhibit cell growth. Unlike its plant homolog, Chlamydomonas FKBP12 exhibits high affinity to rapamycin in vivo, which was increased by mutation of conserved residues in the drug-binding pocket. Furthermore, pull-down assays demonstrated that TOR binds FKBP12 in the presence of rapamycin. Finally, rapamycin treatment resulted in a pronounced increase of vacuole size that resembled autophagic-like processes. Thus, our findings suggest that Chlamydomonas cell growth is positively controlled by a conserved TOR kinase and establish this unicellular alga as a useful model system for studying TOR signaling in photosynthetic eukaryotes.
The macrolide antibiotic rapamycin is a product of the bacterium Streptomyces hygroscopicus. Rapamycin was originally identified as a potent antifungal agent (Vezina et al., 1975) and much later was found to exhibit immunosuppressive activity due to its capacity to block the growth and proliferation of T cells (Schreiber, 1992; Sigal and Dumont, 1992). More recently, rapamycin has been found to display anticancer properties (Bjornsti and Houghton, 2004).
Rapamycin inhibits cell growth in many types of cells, including the budding yeast Saccharomyces cerevisiae. Studies performed in yeasts uncovered the unique mechanism of action of rapamycin. Both receptor and functional target were initially identified in S. cerevisiae (Heitman et al., 1991). Rapamycin first binds the 12-kD FK506-binding protein (FKBP12) and this complex inhibits the target of rapamycin (TOR) Ser/Thr kinase. FKBP12 is a member of the FK506- and rapamycin-binding protein (FKBP) family. This group of proteins, together with the cyclosporin A receptors, is collectively referred to as immunophilins because of their ability to tightly bind the immunosuppressive drugs rapamycin, FK506, or cyclosporin A (Schreiber, 1991; Fruman et al., 1994). Similar to other immunophilins, FKBP12 has peptidyl prolyl cis/trans isomerase activity that is involved in protein-folding processes. Studies have identified FKBP12 in bacteria, fungi, animals, plants (for review, see Schreiber, 1991; Fruman et al., 1994; He et al., 2004), and more recently in the green alga Chlamydomonas reinhardtii (Vallon, 2005). However, the physiological function of this protein is still poorly understood. FKBP12 is the only immunophilin that interacts with TOR in the presence of rapamycin and with calcineurin, a Ca2+ and calmodulin-dependent protein phosphatase, in the presence of FK506. FKBP12 has also been shown to interact with other important signaling molecules in the absence of its drug ligands. In mammals, FKBP12 associates with and modifies the activity of the transforming growth factor-β receptor (Wang et al., 1994, 1996) and the Ca2+-releasing ryanodine receptor (Brillantes et al., 1994). In Arabidopsis (Arabidopsis thaliana), FKBP12 interacts with AtFIP37, a phosphatidyl-inositol kinase essential for development (Vespa et al., 2004). FKBP12 is essential in mammals since a knockout mouse dies during embryonic development (Shou et al., 1998), while loss of FKBP12 function in yeasts has no effect on cell viability (Heitman et al., 1991; Dolinski et al., 1997).
When bound to rapamycin, FKBP12 associates with and inactivates TOR. The findings that dominant mutations in either TOR1 or TOR2 confer complete resistance to the growth-inhibitory properties of rapamycin allowed the identification of TOR in S. cerevisiae (Heitman et al., 1991). After the original identification of TOR in yeasts, TOR was identified in fungi, mammals, flies, worms, and plants, suggesting that TOR is conserved in all eukaryotic life forms (for review, see Crespo and Hall, 2002; Inoki et al., 2005). The TOR kinases are large (approximately 280 kD) proteins with a C-terminal region with strong sequence similarity to the catalytic domain of phosphatidylinositol 3-kinase. The FKBP12-rapamycin complex interacts with the FKBP12-rapamycin-binding (FRB) domain in TOR, adjacent to the catalytic kinase domain. Despite extensive studies on rapamycin action, the mechanism by which FKBP12-rapamycin inhibits TOR function remains unknown. TOR inactivation by rapamycin treatment results in a nutrient starvation response, suggesting that TOR responds to nutrient availability (Barbet et al., 1996). In S. cerevisiae, TOR signaling has been proposed to respond to nitrogen and possibly carbon sources (Beck and Hall, 1999; Shamji et al., 2000; Crespo et al., 2002), while in mammals and flies, TOR function is controlled by amino acid availability and growth factors (Inoki et al., 2005). Studies of TOR signaling in yeasts and mammals have demonstrated that TOR is a central controller of cell growth (Schmelzle and Hall, 2000).
The recent identification and functional analysis by reverse genetics of the Arabidopsis TOR (AtTOR) gene has revealed that AtTOR plays an important role in controlling plant cell growth (Menand et al., 2002). Disruption of the AtTOR gene leads to the premature arrest of endosperm and embryo development (Menand et al., 2002), demonstrating that similar to other eukaryotes, TOR is essential for cell growth in Arabidopsis. In contrast to yeasts, mammals, or flies, the vegetative growth of Arabidopsis and other plants such as Oryza sativa, Nicotiana tabacum, or Brassica napus is not sensitive to rapamycin (Menand et al., 2002). A feasible explanation to rapamycin resistance of land plants might be the inability of plant FKBP12 to bind this drug. Indeed, Luan and colleagues previously reported that plant FKBP12 has evolved structural changes that hamper this protein to mediate the action of its drug ligands against the functional targets (Xu et al., 1998). Interestingly, a yeast three-hybrid analysis performed with the FRB domain of AtTOR suggests that this domain is still functional for the formation of the FKBP12-rapamycin-FRB ternary complex (Menand et al., 2002). Recently, it has been indicated that growth of the photosynthetic unicellular alga C. reinhardtii is sensitive to rapamycin (Menand et al., 2002). Bearing this in mind, we carried out an analysis of rapamycin-mediated effects in Chlamydomonas cells.
This work reports TOR signaling inactivation by rapamycin in a photosynthetic organism. We have identified and characterized two components of TOR signaling in Chlamydomonas, the TOR kinase and the FKBP12 immunophilin, which we will refer to as FKB12, as previously proposed for this alga (Vallon, 2005). Our findings indicate that rapamycin inhibits growth of Chlamydomonas cells. Unlike the plant homolog, Chlamydomonas FKB12 mediates rapamycin action and interacts with the FRB domain of the Chlamydomonas TOR (CrTOR) kinase in the presence of rapamycin.
RESULTS
Rapamycin Inhibits Chlamydomonas Cell Growth
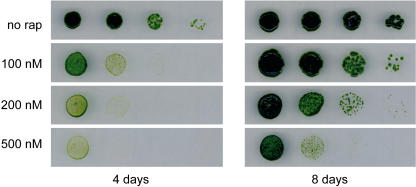
To investigate the sensitivity of Chlamydomonas to rapamycin, cells were spotted onto acetate-containing medium supplemented with different concentrations of the drug. After 4 d of incubation we found that 100 nm rapamycin inhibited cell growth (Fig. 1). The negative effect of rapamycin on cell growth was more pronounced at higher concentrations such as 500 nm rapamycin (Fig. 1), which is about 5 times the concentration inhibiting yeast growth. We found a similar growth-inhibiting effect of rapamycin in minimal medium (data not shown). Interestingly, we found that growth of Chlamydomonas cells is not fully arrested by rapamycin. After longer incubation we still detected slow growth of Chlamydomonas cells on rapamycin-containing plates (Fig. 1). This is in contrast to yeast cell cycle, which is fully arrested in the G1 phase upon rapamycin treatment (Barbet et al., 1996). Our results show that rapamycin inhibits growth of a photosynthetic organism and strongly suggest the presence of a TOR signaling cascade in Chlamydomonas.
Figure 1.
Rapamycin inhibits growth of Chlamydomonas. Wild-type Chlamydomonas cells were subjected to 10-fold serial dilutions and spotted onto TAP plates containing the indicated concentrations of rapamycin. Plates were incubated at 25°C under continuous illumination.
Cloning of Chlamydomonas FKB12 cDNA
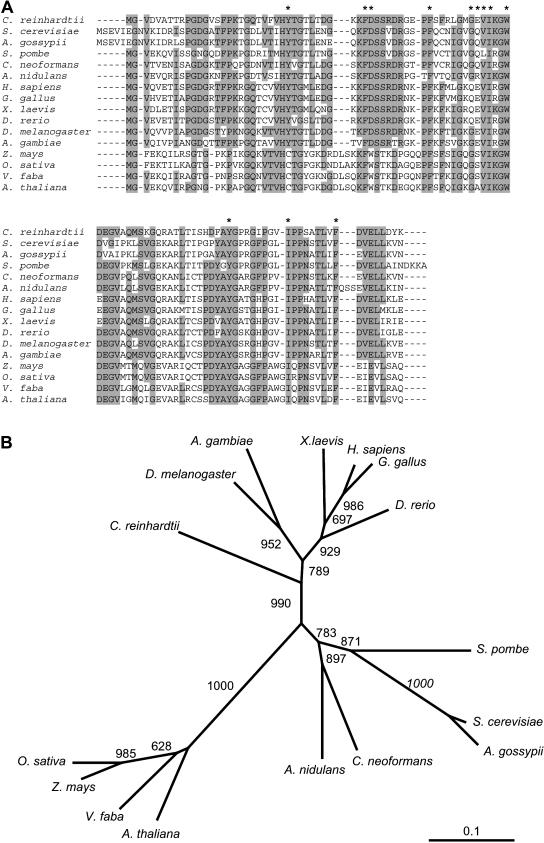
The recent release by the Joint Genome Institute of a draft sequence of the Chlamydomonas nuclear genome allowed us to identify the gene encoding FKB12. TBLASTN analysis using S. cerevisiae FKBP12 as the query identified Chlamydomonas FKB12 on the version 2.0 draft genome sequence (http://genome.jgi-psf.org/chlre2/chlre2.home.html). The complete FKB12 cDNA was cloned by PCR from a Chlamydomonas cDNA library (see “Materials and Methods”). The amino acid sequence deduced from the cDNA is shown in Figure 2A. The drug-binding pocket formed by several hydrophobic residues is conserved in Chlamydomonas FKB12, and, with the exception of Gln-53, the amino acids that establish hydrogen bonds with rapamycin (Asp-37, Glu-54, Ile-56, and Tyr-82) are also conserved (Fig. 2A). Chlamydomonas FKB12 was aligned to several FKBP12s from other eukaryotic organisms, and the highest homology was found to human FKBP12 protein (Fig. 2A), as recently reported in a complete study of Chlamydomonas immunophilins (Vallon, 2005). The identity between the Chlamydomonas protein and homologs in other organisms ranges from 45% to 73% at amino acid level. Interestingly, the evolutionary analysis evidenced that Chlamydomonas FKB12 does not group with plant homologs, although it is closer to higher eukaryotes than other unicellular organisms like fungi (Fig. 2B).
Figure 2.
Phylogenetic analysis of Chlamydomonas FKB12. A, Amino acid sequence of Chlamydomonas FKB12 compared to the FKBP12 protein from other representative organisms. Identical residues in at least nine of the 16 sequences are shaded in gray. Asterisks indicate residues from human FKBP12 that interact with rapamycin (Choi et al., 1996). B, Neighbor-Joining tree of FKBP12s. Bootstrap values from 1,000 replicates are shown. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
TOR Is Conserved in Chlamydomonas
The sensitivity of Chlamydomonas cells to rapamycin prompted us to investigate the presence of a TOR homolog in this organism. A survey of the Version 1.0 draft genome for TOR homologs revealed a DNA region in scaffold 207 whose translation products were highly conserved (more than 50% identity) in AtTOR. The putative CrTOR gene was not fully sequenced neither in version 1.0 nor version 2.0 draft genome sequences. The information available on version 1.0 covered only about 30% of the gene coding sequence, whereas this information was even reduced in version 2.0. Therefore, we proceeded to obtain the nucleotide sequence of the entire TOR gene taking advantage of data available on version 1.0.
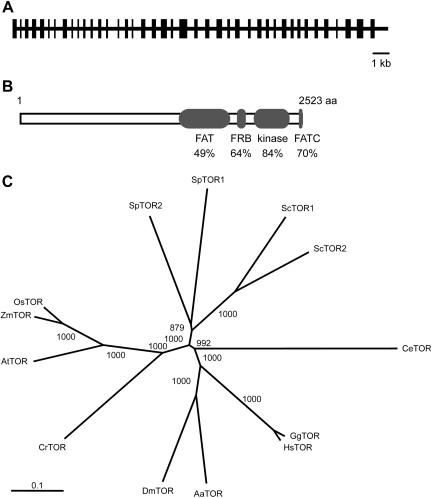
The DNA regions that were not sequenced in version 1.0 were generated by PCR using specific primers and genomic DNA from Chlamydomonas as template (data not shown). Independent PCRs were sequenced for each fragment to correct errors introduced during the PCR reaction. No expressed sequence tags have been reported for CrTOR in the Chlamydomonas expressed sequence tag database. This is consistent with the very low expression detected for CrTOR and our inability to obtain a full-length cDNA of this gene (J.L. Crespo, S. Diaz-Troya, and F.J. Florencio, unpublished data). The coding sequence of the CrTOR gene was deduced from partial cDNA fragments obtained by reverse transcription (RT)-PCR and amino acid sequence comparison to TOR proteins from other organisms. The analysis of the CrTOR gene revealed that this gene spans at least 20 kb of genomic DNA and consists of 41 exons and 40 introns (Fig. 3A). The CrTOR gene encodes 2,523 amino acids with a predicted molecular mass of 277 kD. Highly conserved domains in TOR, such as the FAT and FATC domains, which are characteristic of phosphatidylinositol 3-kinase related kinases (Crespo and Hall, 2002), or the FRB and kinase domains, are conserved in CrTOR (Fig. 3B). A phylogenetic analysis of CrTOR and a number of other TOR proteins revealed that CrTOR is clustered with plant TORs (Fig. 3C), and that AtTOR is the closest homolog with 49% identity.
Figure 3.
TOR is conserved in Chlamydomonas. A, Structure of CrTOR gene predicted from the Chlamydomonas nuclear genome. The intron and exon positions were deduced by comparison of genomic and partial cDNA sequences, and homology analysis with other TOR proteins. The black rectangles indicate the position of the protein-coding regions. B, Comparison of the CrTOR protein sequence to AtTOR. Values indicate the percentage of identity with the corresponding domain sequence of CrTOR. FAT, FRB, kinase, and FATC domains correspond to residues 1,374 to 1,924; 1,961 to 2,055; 2,124 to 2,372; and 2,493 to 2,523, respectively. C, Phylogenetic relationship of TOR from Chlamydomonas and representative organisms. The phylogenetic tree was constructed with full-length TOR amino acid sequences and aligned using the ClustalW program. The bootstrap values represent 1,000 replications. Sc, S. cerevisiae; Sp, Schizosaccharomyces pombe; Os, O. sativa; Zm, Zea mays; At, Arabidopsis; Cr, C. reinhardtii; Dm, Drosophila melanogaster; Aa, Anopheles aedes; Hs, Homo sapiens; Gg, Gallus gallus; Ce, Caenorhabditis elegans.
Isolation and Characterization of a Rapamycin-Resistant Mutant of Chlamydomonas
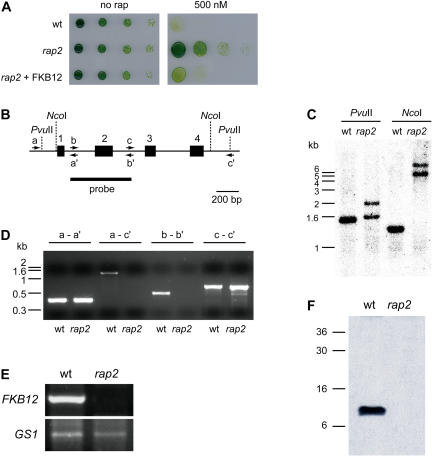
While analyzing the growth-inhibitory properties of rapamycin in Chlamydomonas, we isolated a spontaneous mutant that exhibited complete resistance to the drug. The spontaneous mutant, which we termed rap2, was obtained in Tris-acetate phosphate (TAP) medium containing 500 nm rapamycin at an estimated frequency of 10−8. To investigate the phenotype of the rap2 mutant, we compared growth rates of the wild-type strain with the rap2 mutant in solid and liquid media with and without rapamycin. Whereas no difference was observed between wild-type and rap2 strains in medium containing the drug vehicle, the rap2 mutant displayed full resistance to rapamycin in both solid (Fig. 4A) and liquid media (data not shown). Similar results were obtained with cells grown in minimal medium instead of acetate-containing medium (data not shown).
Figure 4.
Characterization of the Chlamydomonas rap2 mutant strain. A, rap2 mutant cells are rapamycin insensitive. Serial dilutions of wild-type, rap2, and rap2 cells transformed with the pJC20 plasmid on TAP plates containing 500 nm rapamycin (500 nm) or drug vehicle (no rap). Growth was recorded after 4 d incubation at 25°C under continuous illumination. B, Structure of the FKB12 gene from Chlamydomonas. The closed rectangles and numbers (1–4) indicate the four exons of the FKB12 gene. Primers used for the PCR analysis of the FKB12 gene (see D) are represented by arrows and letters. Restriction sites for NcoI and PvuII restriction enzymes are shown by dotted lines. C, Southern-blot analysis of genomic DNA from the rap2 mutant compared to the wild-type strain. Ten micrograms of DNA were digested with NcoI or PvuII. The blot was hybridized with an FKB12 probe obtained by PCR (see B). D, PCR analysis of the FKB12 gene from wild-type and rap2 cells. Letters indicate the different set of primers used for PCRs reactions (see B for the position of different primers). E, RT-PCR analysis of FKB12 in wild-type and rap2 mutant cells. Semiquantitative RT-PCR was terminated after 20 cycles for FKB12 and 24 cycles for GS1. The cytosolic gene, GS1, whose expression is not expected to change in the rap2 mutant, was used as an internal control for RNA level. F, Western blot of FKB12 from wild-type and rap2 strains. Fifteen micrograms of total protein obtained from wild-type and rap2 cells were resolved on 15% polyacrylamide gels, blotted, and incubated with anti-FKB12 antibodies.
Yeast mutants lacking the FPR1 gene (encoding for the FKBP12 protein) display complete resistance to rapamycin (Heitman et al., 1991). Therefore, it is reasonable that the Chlamydomonas rap2 mutant might be impaired in the FKB12 gene. To confirm this hypothesis, we decided to analyze the FKB12 gene in the rap2 mutant. According to the draft nuclear genome sequence, the Chlamydomonas FKB12 gene consists of four exons and three introns (Fig. 4B). To investigate if the rapamycin-resistant phenotype of the rap2 mutant is due to mutations in the FKB12 gene, we examined the integrity of this locus by different approaches. First, DNA gel-blot analysis using either a DNA fragment harboring the second exon of the FKB12 gene (Fig. 4C) or the full-length cDNA (data not shown) as a probe demonstrated that the rap2 mutant presents a reorganization in the FKB12 locus compared to the wild-type strain. Second, PCR experiments performed with several oligonucleotides designed from the sequence of the FKB12 gene strongly suggested that the DNA rearrangement in the FKB12 gene of the rap2 mutant affects a DNA region around exon 2, whereas exons 1, 3, and 4 do not seem to be altered (Fig. 4D). Third, we studied the expression of FKB12 by RT-PCR in wild-type and rap2 mutant cells using the Gln synthetase (GS1) gene as a control of genes whose expression is not expected to be altered in the mutant strain. In contrast to the GS1 gene, no expression of FKB12 was detected in rap2 mutant cells, indicating that this gene is not properly expressed in the rap2 mutant (Fig. 4E). Fourth, we examined the amount of FKB12 protein in wild-type and rap2 mutant cells using antibodies generated against recombinant FKB12 protein from Chlamydomonas (see “Materials and Methods”). As expected, no FKB12 protein was observed in the rap2 mutant strain (Fig. 4F). Finally, we transformed rap2 mutant cells with a plasmid carrying the Chlamydomonas FKB12 gene under the control of the PsaD promoter. As shown in Figure 4A, expression of FKB12 fully restored the rapamycin sensitivity of rap2 mutant cells. Thus, taken together our results indicate that (1) the DNA reorganization detected in the FKB12 gene impairs a correct expression of this gene; (2) the rapamycin-resistant phenotype of the rap2 mutant is exclusively due to the lack of the drug-binding protein FKB12; and (3) similar to yeast cells, Chlamydomonas mutants lacking FKB12 are viable under normal growth conditions. Furthermore, the finding that cell growth is not inhibited by rapamycin in an FKB12 mutant provides strong genetic evidence for the existence of a TOR signaling cascade controlling cell growth in Chlamydomonas.
Chlamydomonas FKB12 Binds Rapamycin in Vivo
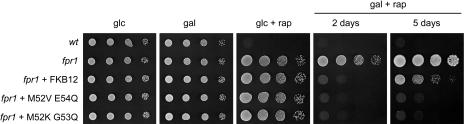
Drug sensitivity assays in yeast cells lacking FKBP12 have been successfully used to assay the function of FKBP12 homologs from other organisms, such as mammals and plants (Koltin et al., 1991; Xu et al., 1998). To investigate whether Chlamydomonas FKB12 can functionally replace yeast FKBP12 in mediating drug action, we performed yeast complementation assays. The wild-type strain JK9-3da and the fpr1 mutant strain were transformed with a plasmid expressing the Chlamydomonas FKB12 gene under the control of the promoter of the GAL1 gene. This promoter is fully repressed in Glc-containing medium and induced when yeast cells are grown in the presence of Gal (Johnston and Davis, 1984). Whereas the fpr1 mutant strain transformed with empty vector displayed normal growth in rapamycin-containing medium, wild-type cells harboring the same plasmid did not grow in the presence of the drug (Fig. 5). However, the fpr1 mutant transformed with the plasmid containing the Chlamydomonas FKB12 gene became sensitive to rapamycin in Gal- but not in Glc-containing medium, indicating that Chlamydomonas FKB12 is able to functionally substitute for yeast FKBP12. Thus, our results indicate that unlike plant FKBP12, Chlamydomonas FKB12 binds rapamycin in vivo.
Figure 5.
Chlamydomonas FKB12 functionally complements a yeast FKBP12 mutant. Wild-type JK9-3da cells were transformed with empty vector (wt). The fpr1 mutant strain lacking the FKBP12 protein was transformed with empty vector (fpr1) or with plasmids expressing wild-type Chlamydomonas FKB12 or M52V E54Q and M52K G53Q mutants. Cultures were normalized, subjected to 10-fold serial dilutions, and spotted onto SD or SGal-Leu plates without rapamycin or containing 200 nm rapamycin. Plates were incubated at 30°C for 2 or 5 d.
Interestingly, growth of fpr1 mutant cells expressing Chlamydomonas FKB12 cannot be fully inhibited by rapamycin. We observed that in contrast to the wild-type strain, fpr1 cells expressing Chlamydomonas FKB12 exhibited very slow growth after longer incubation of these strains on rapamycin-supplemented plates (Fig. 5; data not shown). Therefore, Chlamydomonas FKB12 seems to present an affinity to rapamycin lower than yeast FKBP12. These results are in agreement with our initial observation that, in contrast to what happens in yeasts, rapamycin cannot fully arrest growth of Chlamydomonas cells.
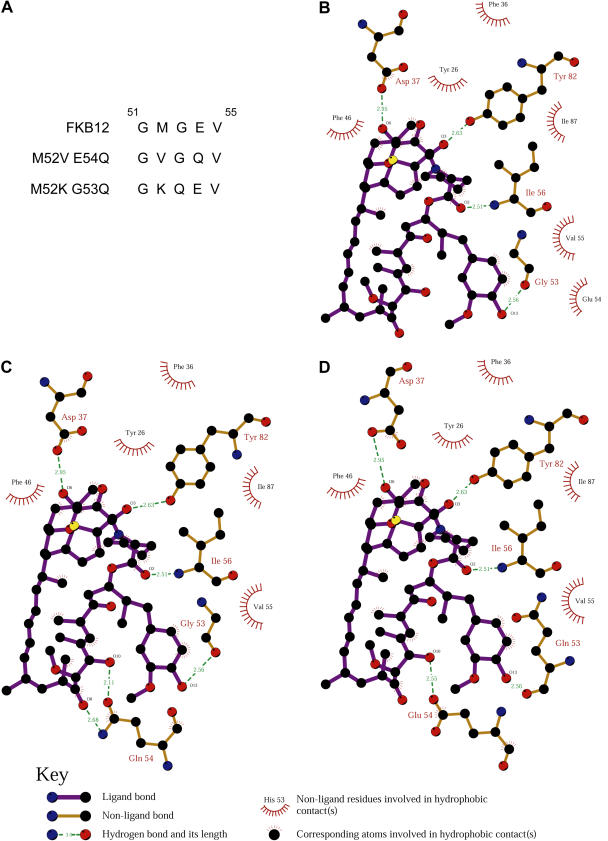
The Presence of a Gln Residue at Position 53 or 54 Increases the Affinity of Chlamydomonas FKB12 to Rapamycin
Crystal structure of the human FKBP12-rapamycin complex revealed that rapamycin binds FKBP12 through hydrophobic contacts and hydrogen bonds with key residues in a hydrophobic pocket of the protein (Choi et al., 1996). Our results suggest that rapamycin binds to Chlamydomonas FKB12 with less efficiency than yeast FKBP12. We found that except for Gln at position 53 (with respect to human FKBP12), all residues that are predicted to establish hydrogen bonds with rapamycin are conserved in FKB12 (Fig. 2A). We reasoned that the absence of this Gln residue in the drug-binding pocket might cause reduced affinity of FKB12 to rapamycin. To confirm this hypothesis, we constructed two different FKB12 mutants with more similarity to yeast and human FKBP12s in the region adjacent to Gln-53. To obtain the same amino acid sequence as yeast FKBP12, mutant FKB12M52V E54Q presented Val and Gln residues instead of Met-52 and Glu-54, respectively. On the other hand, Lys and Gln residues replaced Met-52 and Gly-53, respectively, in FKB12M52K G53Q to get the sequence of human FKBP12 (Fig. 6A). To investigate whether these two mutant forms of FKB12 exhibit more affinity to rapamycin than wild-type FKB12, we performed yeast complementation studies in the fpr1 mutant strain. fpr1 cells transformed with plasmids expressing the FKB12 mutants under the Gal-inducible promoter were spotted onto plates supplemented with rapamycin and incubated at 30°C for 2 and 5 d. After 2-d incubation, both FKB12 mutant forms complemented the rapamycin-resistant phenotype of fpr1 cells similarly to wild-type FKB12 (Fig. 5). However, unlike wild-type FKB12, expression of FKB12 mutants significantly reduced growth of fpr1 cells after longer incubation on rapamycin plates (Fig. 5). These results demonstrated that mutations introduced in FKB12 significantly increased the affinity to rapamycin. Furthermore, our data strongly suggest that the Gln residue that is not conserved in Chlamydomonas FKB12 may play an important role in rapamycin binding.
Figure 6.
Modeling analysis of Chlamydomonas wild-type and mutant FKB12s. A, Amino acid substitutions performed in the M52V E54Q and M52K G53Q mutants compared to wild-type FKB12. Schematic diagrams of wild-type FKB12 (B), and the M52V E54Q (C) and M52K G53Q (D) mutants showing protein-rapamycin interactions. Data were generated using the LIGPLOT program (Wallace et al., 1995).
To further explore this hypothesis, we performed a comparative study of predicted structures of wild-type and mutant forms of Chlamydomonas FKB12. As revealed by crystal structure of the human FKBP12-rapamycin complex, rapamycin establishes five hydrogen bonds with residues Asp-37, Gln-53, Glu-54, Ile-56, and Tyr-82 (Choi et al., 1996). Our modeling analysis of Chlamydomonas FKB12 predicts only four hydrogen bonds between rapamycin and residues Asp-37, Gly-53, Ile-56, and Tyr-82 (Fig. 6B). Interestingly, the presence of a Gln residue either at position 53 (mutant FKB12M52K G53Q) or 54 (mutant FKB12M52V E54Q) allowed rapamycin to establish hydrogen bonds with five residues of the protein (Fig. 6, C and D). Together with the yeast complementation data of wild-type and mutant FKB12s from Chlamydomonas, the modeling analysis of these proteins suggests that mutant FKB12s display higher affinity to rapamycin in vivo due to formation of additional hydrogen bonds between rapamycin and the protein, which might bind rapamycin more tightly in the drug-binding pocket.
Expression of Mutant FKB12s in Chlamydomonas Partially Increases the Sensitivity to Rapamycin
Rapamycin sensitivity of Chlamydomonas cells is particularly interesting since our results indicate that, in contrast to yeast, Chlamydomonas cell growth is not fully arrested by this drug. Partial resistance of Chlamydomonas to rapamycin could be due to reduced affinity of FKB12 to the drug. Indeed, we have shown by yeast complementation assays that, although Chlamydomonas FKB12 is able to mediate rapamycin action, the affinity of this protein to rapamycin is low compared to the yeast homolog. It is therefore feasible that expression in Chlamydomonas of an FKBP12 protein with higher affinity to rapamycin than wild-type FKB12 may increase the sensitivity to the drug.
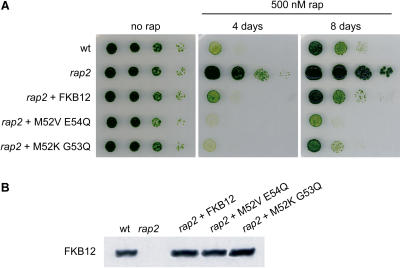
To confirm this hypothesis, we transformed Chlamydomonas cells with plasmids expressing the two mutant forms of FKB12, which bind rapamycin more tightly than the wild-type protein. Endogenous FKB12 is an abundant protein in Chlamydomonas (J.L. Crespo, S. Diaz-Troya, and F.J. Florencio, unpublished data). Therefore, to obtain high expression levels of wild-type and mutant FKB12s, we cloned the genes coding for these proteins in a plasmid where expression is driven by the promoter of the PsaD gene. Chlamydomonas rap2 mutant cells were transformed with these constructs, and transformants were screened for sensitivity to rapamycin. Wild-type FKB12 fully complemented the rapamycin-resistant phenotype of the rap2 mutant (Fig. 7A), indicating that the expressed protein is functional. Expression of FKB12 mutants in the rap2 strain also restored sensitivity to rapamycin (Fig. 7A), although these transformants displayed more sensitivity to rapamycin than wild-type Chlamydomonas cells or rap2 mutant cells transformed with wild-type FKB12 (Fig. 7A). Interestingly, longer incubation of transformants on rapamycin-supplemented plates revealed that cells that express the FKB12M52V E54Q mutant are slightly more sensitive to rapamycin than those that express the FKB12M52K G53Q mutant (Fig. 7A). To confirm that wild-type and mutant FKB12s were expressed at levels similar to endogenous FKB12, we performed immunodetection analysis using anti-FKB12 antibodies. Equal amounts of these proteins were detected in all transformants (Fig. 7B), indicating that different sensitivities to rapamycin displayed by transformants containing wild-type or mutant FKB12s are not due to different amounts of these proteins in the cell. Thus, our results show that, like in yeast cells, expression of mutant FKB12s in Chlamydomonas cells increases the sensitivity to rapamycin. However, we found that these transformants were still able to grow very slowly on rapamycin-supplemented plates (Fig. 7A; data not shown), indicating that rapamycin does not fully arrest growth of Chlamydomonas cells.
Figure 7.
Expression of mutant FKB12s in Chlamydomonas partially increases the sensitivity to rapamycin. A, Rapamycin sensitivity of wild-type and rap2 cells compared to rap2 cells expressing wild-type or mutant FKB12s. Serial dilutions were spotted onto TAP plates supplemented or not with 500 nm rapamycin, and incubated at 25°C under continuous illumination. Growth was recorded after 4 or 8 d. B, Levels of FKB12 protein detected by western blot. About 15 μg of total protein obtained from wild-type (wt) or rap2 mutant cells transformed with the indicated constructs were resolved in 15% polyacrylamide gels, blotted, and incubated with anti-FKB12 antibodies.
CrTOR Interacts with FKB12 in the Presence of Rapamycin
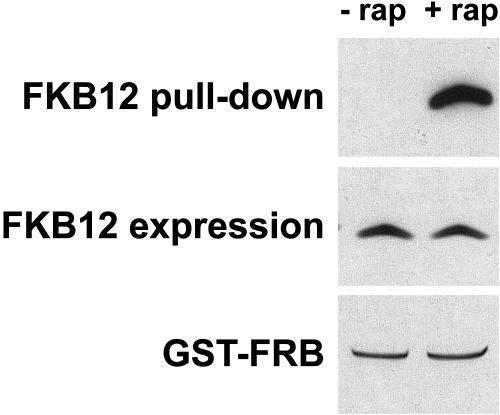
Rapamycin action requires formation of the FKB12-rapamycin-TOR ternary complex. Our data indicate that FKB12 binds rapamycin in vivo. Analysis of the amino acid sequence of the FRB domain of CrTOR shows that critical residues that establish hydrophobic interactions with FKB12-rapamycin are conserved, suggesting that CrTOR might bind the FKB12-rapamycin complex. To investigate if CrTOR is able to interact with this complex, we performed pull-down assays with the FRB domain of CrTOR and the FKB12 protein from total extracts of Chlamydomonas. We expressed the FRB domain as a glutathione S-transferase (GST) fusion protein in Escherichia coli. The soluble fraction of Chlamydomonas protein extracts was incubated with the purified GST fusion protein in the presence of rapamycin or an equal amount of drug vehicle. Glutathione Sepharose beads were then added to immobilize the GST fusion protein. After several washes, the bound fractions were analyzed by immunoblotting using anti-FKB12 antibody (Fig. 8). We observed that the FRB domain of CrTOR interacts with FKB12 in the presence of rapamycin in this in vitro assay, whereas no binding was detected in the absence of the drug. Thus, our data demonstrate that CrTOR is able to bind FKB12 in the presence of rapamycin.
Figure 8.
The FRB domain of CrTOR interacts with FKB12 in the presence of rapamycin. GST pull-down assays to test the interaction of FKB12 with the FRB domain of CrTOR. Five micrograms of purified GST fusion protein were incubated with 0.5 mg of Chlamydomonas total extract in the presence of 4 μm rapamycin (+rap) or a similar concentration of drug vehicle (−rap). FKB12 and GST fusion protein were detected by immunoblotting using anti-FKB12 antibody and Coomassie staining, respectively.
Rapamycin Treatment Increases Vacuole Size of Chlamydomonas Cells
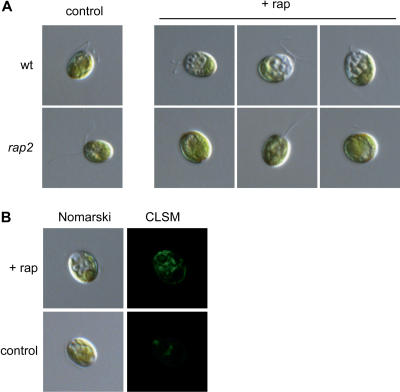
In yeasts, inhibition of the TOR pathway by rapamycin treatment causes several starvation-like phenotypes, including an altered cell morphology due to vacuole expansion (Heitman et al., 1991). To investigate whether rapamycin may induce morphological changes in Chlamydomonas, we performed microscopic studies of rapamycin-treated cells. Since asynchronous cultures of Chlamydomonas cells show disparate size distributions, cells were allowed to synchronize by alternating light and dark cycles before rapamycin addition. The most obvious feature of Chlamydomonas cells treated with rapamycin was a higher degree of vacuolization compared to nontreated cells (Fig. 9A). After 20 to 24 h of rapamycin addition, several vacuole-like organelles, usually three to six, were readily visible at the anterior end of the cell. Rapamycin-induced vacuolization correlated with bleaching around vacuole-like compartments. Similar effects were observed in asynchronous cultures of cells grown under continuous light and treated with rapamycin (data not shown). As expected, Chlamydomonas cells lacking FKB12 did not display vacuole-like organelles upon rapamycin addition (Fig. 9A), indicating that FKB12 mediates rapamycin-induced vacuolization.
Figure 9.
Rapamycin-induced vacuolization in Chlamydomonas. A, Nomarski micrographs of wild-type and rap2 mutant living cells treated with 500 nm rapamycin (+rap) or drug vehicle (control) for 24 h. Cultures were synchronized by alternating light and dark (12 h/12 h) cycles before rapamycin addition. B, Nomarski and CLSM images of a wild-type living cell treated with rapamycin (+rap) or drug vehicle (control) for 24 h. Cells were incubated with the pH-sensitive dye LysoSensor Green DND-189, which produces fluorescent foci mainly in the region of the cell containing vacuoles.
Vacuoles in plant cells, algae, and fungi are storage compartments with an acidic content. To further investigate if the cellular structures observed in rapamycin-treated cells correspond to vacuoles, cells were treated with LysoSensor Green DND-189. This fluorescent dye is an acidotropic probe that accumulates in the membranes of acidic organelles as a result of protonation, and it has been used in plant tissue, Chlamydomonas, and yeast for vacuole analyses (Guttenberger, 2000; Komine et al., 2000; Perzov et al., 2002). In less than 5 min, the dye-labeled structures correlated with cytosolic and contractile vacuoles of untreated cells, as well as the membranes of vacuole-like organelles in rapamycin-treated cells (Fig. 9B), confirming that these structures are indeed acidic vacuoles. Labeling was more intense in rapamycin-treated compared to control cells mainly due to a higher size of vacuoles and possibly to an increase in number of vacuoles upon drug treatment. As expected, incubation of nontreated rap2 mutant cells with LysoSensor Green resulted in a similar staining pattern to wild-type cells (data not shown). Fluorescence was not detected with cells incubated without the dye (data not shown).
DISCUSSION
Plants have been reported to express TOR and FKBP12 proteins (Xu et al., 1998; Menand et al., 2002), although in contrast to other eukaryotes, land plants are naturally insensitive to rapamycin likely due to the inability of plant FKBP12 to bind rapamycin (Xu et al., 1998). This feature of land plants might hamper research on TOR signaling in photosynthetic organisms. Here, we report the identification and characterization of FKB12 and TOR genes in the rapamycin-sensitive green alga Chlamydomonas. Our results show that the CrTOR kinase is structurally conserved and displays higher identity to AtTOR than TOR proteins from nonphotosynthetic organisms. Regions known to be important for TOR function, such as the FAT, FATC, FRB, and kinase domains, are highly conserved in CrTOR, suggesting that certain TOR readouts in Chlamydomonas might be similar to other eukaryotes. The FRB domain mediates the interaction of FKBP12-rapamycin and the subsequent inhibition of certain TOR functions. GST pull-down assays performed with the FRB domain of CrTOR and FKB12 indicated that, contrary to their plant homologs, FKB12 and CrTOR proteins form a complex in the presence of rapamycin. This finding might explain in part the natural sensitivity of Chlamydomonas to rapamycin in contrast to the insensitivity of land plants to this drug.
FKB12 is the minimal peptide sequence harboring peptidyl prolyl cis/trans isomerase activity and drug binding in Chlamydomonas (Vallon, 2005). Southern-blot experiments indicated that FKB12 appears to be a single gene in Chlamydomonas, as proposed in a recent genomic study of Chlamydomonas immunophilins (Vallon, 2005). Results from the in vivo yeast functional assays showed that, in contrast to its plant homolog (Xu et al., 1998), Chlamydomonas FKB12 mediates rapamycin action (Fig. 5). This is consistent with our phylogenetic analysis of FKBP12s, which shows that FKB12 is more distant to plant than human or yeast homologs. Critical residues located in the hydrophobic core of the protein that forms the rapamycin-binding pocket are conserved in Chlamydomonas FKB12 (Fig. 2). However, although FKB12 binds rapamycin in vivo, yeast complementation assays with mutant FKB12s indicated that the presence of Glu either at position 53 or 54 significantly increased drug binding. Modeling studies of wild-type and mutant FKB12s predicts that this Glu residue might establish an additional hydrogen bond with rapamycin, as reported for the crystal structure of the human FKBP12-rapamycin complex (Choi et al., 1996).
In mammals, FKBP12 is essential since a knockout mouse dies during embryonic development, probably caused by calcium channel dysfunction (Shou et al., 1998) or cell cycle deregulation (Aghdasi et al., 2001). In Arabidopsis, AtFKBP12 has been shown to interact with AtFIP37, a protein critical for embryo and endosperm development, suggesting that AtFKBP12 might be involved in developmental processes (Faure et al., 1998; Vespa et al., 2004). In contrast to mammals, yeast cells lacking FKBP12 are viable and do not display any growth defect (Heitman et al., 1991; Dolinski et al., 1997). We isolated rap2, a Chlamydomonas FKB12 mutant, as a spontaneous resistant to rapamycin. Characterization of this mutant strain showed that loss of FKB12 has no obvious effect on cell survival (Fig. 4), indicating that, similar to yeasts, FKB12 is not an essential gene in Chlamydomonas.
The finding that Chlamydomonas cells lacking FKB12 are fully resistant to rapamycin demonstrates that, similar to other eukaryotes, FKB12 mediates negative effects of rapamycin on cell growth. However, sensitivity of Chlamydomonas to rapamycin differs from those observed in other organisms. On one hand, the concentration of rapamycin required to potently inhibit growth of Chlamydomonas cells is about 5 times higher than for S. cerevisiae, Candida albicans, or mammalian cells (Vezina et al., 1975; Koltin et al., 1991; Kuo et al., 1992). On the other hand, rapamycin does not fully arrest Chlamydomonas growth, as observed after long incubation on rapamycin-supplemented plates (Figs. 1 and 7). These observations might be explained in part by a reduced affinity of Chlamydomonas FKB12 to rapamycin. Our yeast complementation assays support this hypothesis. We show that although Chlamydomonas FKB12 binds rapamycin in vivo, the drug-protein interaction is lower compared to yeast FKBP12 (Fig. 5). Moreover, the finding that expression of two FKB12 mutants with increased affinity to rapamycin restored complete sensitivity to rapamycin of a yeast strain lacking FKBP12 (Fig. 5) demonstrates that wild-type FKB12 binds rapamycin with less affinity than the yeast homolog. Another hypothesis to explain partial resistance of Chlamydomonas to rapamycin is that TOR function(s) essential for cell growth cannot be inhibited by the FKB12-rapamycin-TOR complex, as proposed for the resistance to rapamycin of plants and fission yeast (Weisman et al., 1997; Menand et al., 2002). Expression of an FKB12 mutant with increased affinity to rapamycin in the rap2 Chlamydomonas strain does not confer complete sensitivity to rapamycin, as it does in yeasts (Figs. 5 and 7). This result supports the hypothesis in which rapamycin-insensitive TOR functions might control Chlamydomonas cell growth. Rapamycin-sensitive and -insensitive TOR functions have already been described in yeasts and mammals (Kunz et al., 1993; Jacinto et al., 2004; Sarbassov et al., 2004). Therefore, it is reasonable that these diverse functions of TOR could also be conserved in photosynthetic organisms such as Chlamydomonas or Arabidopsis.
Inactivation of TOR function by rapamycin results in a nutrient starvation response, suggesting that TOR responds to nutrient availability. Yeast cells treated with rapamycin or limited for nitrogen or carbon undergo a catabolic membrane-trafficking process known as autophagy (Ohsumi, 2001). During this process, a large number of cytoplasmic components are nonselectively enclosed within a double-membrane structure (autophagosome) and delivered to the vacuole for degradation. Autophagy has also been morphologically and genetically described in plants (for a recent review, see Thompson and Vierstra, 2005), although so far no control of this process by TOR signaling has been reported. Chlamydomonas has two contractile vacuoles located at the anterior end and several small vacuoles in the cytoplasm (Sager and Palade, 1957; Harris, 1989). We found that rapamycin-mediated inactivation of TOR signaling in Chlamydomonas cells causes an important enlargement of cytoplasmic vacuoles (Fig. 9). Although our data suggest that rapamycin might induce autophagy in Chlamydomonas, we have no evidence that the rapamycin-induced vacuolization observed in Chlamydomonas is an autophagic process. To our knowledge, autophagy has not been described in Chlamydomonas, although as an essential process in bulk protein turnover in eukaryotes it might be also conserved in algae. Similar to yeasts, it has been proposed that TOR may control autophagy in plants (Thompson and Vierstra, 2005), and therefore it would be reasonable that rapamycin induces autophagy in Chlamydomonas as a result of TOR inhibition. An analysis of the Chlamydomonas draft genome sequence revealed that potential orthologs of yeast autophagy genes, such as ATG1, 3, 4, 5, 7, 8, 12, 13, and VPS30, are conserved in this alga, indicating that Chlamydomonas might recycle intracellular components through autophagic processes similar to those described in yeasts and plants. To verify this model, it would be necessary to characterize these conserved components in the autophagic pathway, as well as to investigate the mechanisms by which TOR controls these proteins.
MATERIALS AND METHODS
Strains and Growth Conditions
Chlamydomonas reinhardtii wild-type strain 6C+ was obtained from the laboratory of J.-D. Rochaix (University of Geneva). Cells were grown as described by Harris (1989) under continuous illumination at 25°C. If required, media (TAP and high-salt minimal medium) were solidified with 1.2% Bacto agar (Difco). Synchronization of Chlamydomonas cells was achieved by alternating light and dark (12 h/12 h) cycles in minimal medium. Saccharomyces cerevisiae wild-type (JK9-3da) and fpr1 mutant (MH324) strains were obtained from the laboratory of M.N. Hall (Biozentrum, University of Basel). Cells were grown in rich (YPD) or synthetic (SD, SGal) media as described previously (Sherman, 1991). Rapamycin was added to Chlamydomonas or yeast media to the indicated concentrations from a 1 mg/mL stock in 90% ethanol-10% Tween20.
Transformation
Nuclear transformation of Chlamydomonas wild-type and rap2 mutant cells was performed as described previously (Kindle, 1998). In brief, about 5 × 107 cells were treated with autolysin and agitated in the presence of 1 μg plasmid DNA linearized with SapI and 0.3 g of 0.5-mm glass beads. Plasmids used for nuclear transformation of Chlamydomonas were derived from the pSL18 plasmid (Falciatore et al., 2005), which contains the AphVIII gene conferring paromomycin resistance. Transformants were selected on solid TAP medium supplemented with paromomycin (10 μg/mL). Individual colonies, visible after 6 to 8 d, were transferred to new medium for further analysis. Yeast cells were transformed by the lithium acetate procedure (Sherman, 1991).
Rapamycin Sensitivity Assays
Growth of Chlamydomonas and yeast cells on media containing the indicated concentration of rapamycin was assayed by spotting 2.5 μL of 10-fold dilutions of normalized cultures of exponentially growing cells. Drug vehicle (90% ethanol, 10% Tween 20) had no significant effect on growth of wild-type or rap2 mutant cells at any of the concentrations tested.
Sequence Alignment, Phylogenetic, and Gene Structure Analysis
Alignments of the Chlamydomonas FKB12 and TOR proteins with the homologs from other organisms were performed with ClustalW (Thompson et al., 1994). Phylogenetic trees were constructed by the Neighbor-Joining method (Saitou and Nei, 1987) using the Clustal program. Bootstrap analysis was performed with 1,000 replications and the tree was visualized using TreeView 32 software. The intron and exon positions depicted in Figure 3 were predicted by comparing partial cDNA sequences of CrTOR with their corresponding genomic sequences, using GeneMark software, and by homology analysis with TOR proteins from other selected organisms.
Plasmids
The full-length cDNA for Chlamydomonas FKB12 was amplified by PCR from a Chlamydomonas cDNA library using the following primers: FKB12-5′ (5′-CCCTGCAGATGGGTGTCGACGTCGCG-3′) and FKB12-3′ (5′-CCCTGCAGTTACTTGTAGTCCAGCAGCTC-3′). The amplified product was digested with PstI and cloned into the PstI site of pSL18 (rendering pJC20) for expression in Chlamydomonas cells, or cloned into the PstI site of pTB328 (rendering pJC6) for expression in yeast cells. The pSL18 plasmid (Falciatore et al., 2005) contains both PsaD 5′- and 3′-untranslated regions flanking the multicloning site and the AphVIII gene conferring paromomycin resistance. Site-directed mutants were generated by PCR and cloned into the PstI site of pSL18 and pTB328.
Nucleic Acid Analyses
Total DNA from Chlamydomonas cells was prepared using a miniprep procedure adapted from Rochaix et al. (1987). Cells were harvested by low-speed centrifugation and resuspended in 250 μL of DNA extraction buffer (10 mm Tris-HCl, pH 8.0, 10 mm EDTA, 10 mm NaCl). SDS was added to 1%, proteinase K to 200 μg/mL, and the mixture was incubated at 50°C for 2 h. DNA was phenol extracted and precipitated with 1.5 volume 95% ethanol in the presence of 0.2 m NaCl. After resuspension in 100 μL of water, DNA was digested with 20 μg/mL ribonuclease A at 37°C for 15 min. DNA gel-blot analyses were performed using standard methods (Sambrook et al., 1989). Radioactive DNA probes were prepared by the random priming technique, using the Ready-To-Go kit (Amersham) and [α-32P]dCTP (Amersham).
PCR analysis of the FKB12 locus was performed with AccuTaq polymerase (Sigma) using the following primer sets: a, 5′-ACAGGCATGTTGGTAACAATG-3′, and a′, 5′-AGCAATACTCTGGAGCAAAGC-3′; b, 5′-GCTTTGCTCCAGAGTATTGC-3′, and b′, 5′-CAGGTTGTTGTGCAGGTTTC-3′; and c, 5′-AAACCTGCACAACAACCTGC-3′, and c′, 5′-TCAAGAACACGCGAGCTTTTG-3′.
Genomic DNA from Chlamydomonas cells was amplified by PCR using the GC-Rich PCR system (Roche) according to manufacturer indications.
Total RNA from about 108 cells was isolated upon resuspension of frozen pellets in 500 μL of RNA lysis buffer (10 mm EDTA, pH 8, 100 mm Tris-HCl, pH 8.0, 600 mm NaCl, 4% SDS) and 500 μL of phenol/chloroform, and extensive organic extraction. RNA was precipitated first in 2 m LiCl overnight at 4°C, and then in 70% ethanol for 30 min at −20°C. For semiquantitative RT-PCR analysis, first-strand cDNA was produced using 2 μg total RNA, oligo(dT) primer, and 100 units of SuperScript II RNase H− reverse transcriptase (Invitrogen) in a 20-μL reaction. The resulting cDNA was used as template in 50 μL of PCR reaction using gene-specific primers as follows: FKB12, 5′-CCCTGCAGATGGGTGTCGACGTCGCG-3′ and 5′-CCCTGCAGTTACTTGTAGTCCAGCAGCTC-3′; and GS1, 5′-TGGCCGCGGGCGTTAACATCA-3′ and 5′-CAGAAGGATGGTGGTCTCCAC-3′. A total of 20 μL of PCR samples was separated by agarose gel electrophoresis and visualized with ethidium bromide staining.
Preparation and Analysis of Proteins
Cells from liquid cultures in late log phase were collected by centrifugation (3,000g, 5 min), washed once in 10 mm sodium phosphate (pH 7.0), and resuspended in a minimal volume of the same solution. Cells were lysed by two cycles of slow freezing to −80°C followed by thawing to room temperature. The soluble cell extract was separated from the insoluble fraction by centrifugation (15,700g, 10 min) in a microcentrifuge at 4°C, and stored at −80°C until they were analyzed. Proteins were quantitated with the Coomassie dye-binding method as described by the manufacturer (Bio-Rad). For immunoblot analysis, proteins were fractionated on 15% SDS-PAGE and immunoblotted with the anti-FKB12 antibody (1:2,000). The ECL Plus immunoblotting system (Amersham) was used to detect FKB12 with anti-rabbit secondary antibodies.
Anti-FKB12 Antibody Production
For antiserum production, the entire FKB12 cDNA was cloned into the pGEX-4T-1 (Amersham) and introduced into Escherichia coli DH5α. Expression of the GST fusion protein was induced by adding 1 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. The recombinant protein was further purified on Glutathione Sepharose 4B (Amersham), eluted by glutathione incubation, and resuspended in 50 mm Tris-HCl, pH 8.0, buffer. Anti-FKB12 antisera were obtained according to standard immunization protocols by injecting the recombinant protein in a rabbit.
GST Pull-Down Assays
A cDNA fragment containing the FRB domain (residues 1,961–2,055) of CrTOR was obtained by RT-PCR using primers FRB-5′ (5′-GGCCGGATCCCTGTGGCACGAGATGTGG-3′) and FRB-3′ (5′-GGCCCTCGAGTTACTGCTTGTTGATGCGCTT-3′). The PCR product was digested with XhoI and BamHI, and cloned into the pGEX-4T-1 expression vector (Amersham) for expression in bacteria as GST fusion. For the assay, 5 μg of GST fusion protein was first incubated with 0.5 mg of Chlamydomonas protein extracts (soluble fraction) and 4 μm rapamycin (or an equal amount of drug vehicle) for 40 min in 0.4 mL of phosphate-buffered saline, and then immobilized on Glutathione Sepharose 4B beads. After 4 h incubation at 4°C on a rotary incubator, beads were washed four times with the same buffer, resuspended in 30 μL of SDS gel-loading buffer, and resolved by SDS-PAGE (15%).
Microscopy
For in vivo observations, Chlamydomonas cells were immobilized in TAP or high-salt minimal media containing 1.5% low melting agarose. Untreated or rapamycin-treated cells were observed with an inverted differential contrast-interference (Nomarski) microscope (Leica DMRE) with a HCX PL AP0 × 63, 1.4 N.A. objective. For vacuole staining, cells (2 × 106/mL) were treated with LysoSensor Green DND-189 (Molecular Probes) for 5 min at 30°C. Fluorescence dye was added to a final concentration of 2 μm from a stock solution of 1 mm in dimethyl sulfoxide. Cells were examined and images collected with a Leica DMRE (Leica Imaging System) confocal laser scanning microscope (CLSM) with a HCX PL AP0 × 63, 1.4 N.A. objective. All CLSM observation was performed with a laser excitation wavelength filter set at 488 nm, and fluorescent light was collected between 510 and 550 nm to reduce interference by chlorophyll.
Modeling of Chlamydomonas FKB12
Models of wild-type and site-directed mutants of the Chlamydomonas FKB12 protein were generated using the SWISS-MODEL protein structure homology-modeling server (http://swissmodel.expasy.org/; Guex et al., 1999) and different protein structures of human FKBP12 (3FAP, 1EYM, 1NSG, 1FKL) available in the RCSB Protein Data Bank (http://www.rcsb.org/pdb/). Energy minimization was performed for all models. Schematic diagrams of Chlamydomonas FKB12s showing FKB12-rapamycin interactions were generated using the LIGPLOT program (Wallace et al., 1995).
Sequence data from this article can be found in the GenBank data library under accession number DQ230829.
Acknowledgments
We thank Jean-David Rochaix, Michel Goldschmidt-Clermont, and Michael N. Hall for providing strains and plasmids; Olivier Vallon, Anna Lindhal, and José C. Reyes for helpful discussions; and Setefilla Molina for technical assistance with FKB12 modeling.
This work was supported by the Spanish Ministry of Science and Technology (grant no. BMC 2001–2635 and a Ramon y Cajal contract to J.L.C.) and the Junta de Andalucia (predoctoral fellowship to S.D.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José L. Crespo (crespo@ibvf.csic.es).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070847.
References
- Aghdasi B, Ye K, Resnick A, Huang A, Ha HC, Guo X, Dawson TM, Dawson VL, Snyder SH (2001) FKBP12, the 12-kDa FK506-binding protein, is a physiologic regulator of the cell cycle. Proc Natl Acad Sci USA 98: 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692 [DOI] [PubMed] [Google Scholar]
- Bjornsti MA, Houghton PJ (2004) The TOR pathway: a target for cancer therapy. Nat Rev Cancer 4: 335–348 [DOI] [PubMed] [Google Scholar]
- Brillantes AB, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, Marks AR (1994) Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77: 513–523 [DOI] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J (1996) Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273: 239–242 [DOI] [PubMed] [Google Scholar]
- Crespo JL, Hall MN (2002) Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev 66: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo JL, Powers T, Fowler B, Hall MN (2002) The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA 99: 6784–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K, Muir S, Cardenas M, Heitman J (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94: 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A, Merendino L, Barneche F, Ceol M, Meskauskiene R, Apel K, Rochaix JD (2005) The FLP proteins act as regulators of chlorophyll synthesis in response to light and plastid signals in Chlamydomonas. Genes Dev 19: 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JD, Gingerich D, Howell SH (1998) An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J 15: 783–789 [DOI] [PubMed] [Google Scholar]
- Fruman DA, Burakoff SJ, Bierer BE (1994) Immunophilins in protein folding and immunosuppression. FASEB J 8: 391–400 [DOI] [PubMed] [Google Scholar]
- Guex N, Diemand A, Peitsch MC (1999) Protein modelling for all. Trends Biochem Sci 24: 364–367 [DOI] [PubMed] [Google Scholar]
- Guttenberger M (2000) Arbuscules of vesicular-arbuscular mycorrhizal fungi inhabit an acidic compartment within plant roots. Planta 211: 299–304 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego
- He Z, Li L, Luan S (2004) Immunophilins and parvulins: superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol 134: 1248–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Inoki K, Ouyang H, Li Y, Guan KL (2005) Signaling by target of rapamycin proteins in cell growth control. Microbiol Mol Biol Rev 69: 79–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Johnston M, Davis RW (1984) Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol 4: 1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL (1998) Nuclear transformation: technology and applications. In JD Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplast and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 41–61
- Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP (1991) Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol 11: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Eggink LL, Park H, Hoober JK (2000) Vacuolar granules in Chlamydomonas reinhardtii: polyphosphate and a 70-kDa polypeptide as major components. Planta 210: 897–905 [DOI] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN (1993) Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596 [DOI] [PubMed] [Google Scholar]
- Kuo CJ, Chung J, Fiorentino DF, Flanagan WM, Blenis J, Crabtree GR (1992) Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature 358: 70–73 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Perzov N, Padler-Karavani V, Nelson H, Nelson N (2002) Characterization of yeast V-ATPase mutants lacking Vph1p or Stv1p and the effect on endocytosis. J Exp Biol 205: 1209–1219 [DOI] [PubMed] [Google Scholar]
- Rochaix JD, Mayfield S, Goldschmidt-Clermont M, Erickson J (1987) Molecular biology of Chlamydomonas. In CH Shaw, ed, Plant Molecular Biology: A Practical Approach. IRL Press, Oxford, pp 253–275
- Sager R, Palade GE (1957) Structure and development of the chloroplast in Chlamydomonas: the normal green cell. J Biophys Biochem Cytol 3: 463–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN (2000) TOR, a central controller of cell growth. Cell 103: 253–262 [DOI] [PubMed] [Google Scholar]
- Schreiber SL (1991) Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science 251: 283–287 [DOI] [PubMed] [Google Scholar]
- Schreiber SL (1992) Immunophilin-sensitive protein phosphatase action in cell signaling pathways. Cell 70: 365–368 [DOI] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL (2000) Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr Biol 10: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Shou W, Aghdasi B, Armstrong DL, Guo Q, Bao S, Charng MJ, Mathews LM, Schneider MD, Hamilton SL, Matzuk MM (1998) Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature 391: 489–492 [DOI] [PubMed] [Google Scholar]
- Sigal NH, Dumont FJ (1992) Cyclosporin A, FK-506, and rapamycin: pharmacologic probes of lymphocyte signal transduction. Annu Rev Immunol 10: 519–560 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon O (2005) Chlamydomonas immunophilins and parvulins: survey and critical assessment of gene models. Eukaryot Cell 4: 230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M (2004) The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol 134: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN (1975) Rapamycin (AY-22,989), a new antifungal antibiotic: taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 28: 721–726 [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Wang T, Donahoe PK, Zervos AS (1994) Specific interaction of type I receptors of the TGF-beta family with the immunophilin FKBP-12. Science 265: 674–676 [DOI] [PubMed] [Google Scholar]
- Wang T, Li BY, Danielson PD, Shah PC, Rockwell S, Lechleider RJ, Martin J, Manganaro T, Donahoe PK (1996) The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell 86: 435–444 [DOI] [PubMed] [Google Scholar]
- Weisman R, Choder M, Koltin Y (1997) Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J Bacteriol 179: 6325–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Liang S, Kudla J, Luan S (1998) Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J 15: 511–519 [DOI] [PubMed] [Google Scholar]