Abstract
The alternative oxidase (AOX) pathway of plant mitochondria uncouples respiration from mitochondrial ATP production and may ameliorate plant performance under stressful environmental conditions, such as cold temperatures, by preventing excess accumulation of reactive oxygen species. We tested this model in whole tissues by growing AtAOX1a-transformed Arabidopsis (Arabidopsis thaliana) plants at 12°C. For the first time, to our knowledge, in plants genetically engineered for AOX, we identified a vegetative shoot growth phenotype. Compared with wild type at day 21 after sowing, anti-sense and overexpressing lines showed, on average, 27% reduced leaf area and 25% smaller rosettes versus 30% increased leaf area and 33% larger rosette size, respectively. Lines overexpressing a mutated, constitutively active AOX1a showed smaller phenotypic effects. These phenotypic differences were not the result of a major alteration of the tissue redox state because the changes in levels of lipid peroxidation products, reflecting oxidative damage, and the expression of genes encoding antioxidant and electron transfer chain redox enzymes did not correspond with the shoot phenotypes. However, the observed phenotypes were correlated with the amount of total shoot anthocyanin at low temperature and with the transcription of the flavonoid pathway genes PAL1 and CHS. These results demonstrate that (1) AOX activity plays a role in shoot acclimation to low temperature in Arabidopsis, and that (2) AOX not only functions to prevent excess reactive oxygen species formation in whole tissues under stressful environmental conditions but also affects metabolism through more pervasive effects, including some that are extramitochondrial.
The electron transport chain (ETC) of plant mitochondria has unique features compared with that of other eukaryotes, including the ubiquitous presence of a terminal alternative oxidase (AOX) that competes for electrons with the standard cytochrome (Cyt) pathway (for review, see Finnegan et al., 2004). This results in a branching of electron flow at the level of the ubiquinone pool. The partitioning of electrons between the Cyt and AOX pathways has a direct impact on cellular energy balance because the AOX pathway does not contribute to ATP production (Millenaar and Lambers, 2003).
Recently, evidence has accumulated that AOX is crucial in controlling the reduction state of the ubiquinone pool (Millenaar et al., 1998; Wagner et al., 1998), thus preventing overreduction of the ETC. The maintenance of ETC redox balance is critical because electron input in excess of ETC capacity can be responsible for the production of reactive oxygen species (ROS), in particular superoxide and H2O2. Within cells, ROS formation in excess of the detoxification capacity of protective antioxidant enzyme systems (Møller, 2001) has been linked to irreversible DNA damage and to the disruption of membrane lipids through the formation of lipid peroxides (Fridovich, 1978; Scandalios, 1993). In addition, excess ROS can directly impair the function of several mitochondrial enzymes (Gardner et al., 1995; Millar and Leaver, 2000; Sweetlove et al., 2002; Taylor et al., 2002). The finding that tobacco (Nicotiana tabacum) culture cell mitochondria with an anti-sense construct for AOX show elevated basal ROS formation, which is further enhanced in the presence of the ETC inhibitor antimycin A, directly supports the role of AOX in alleviating ROS formation (Maxwell et al., 1999). We have recently demonstrated increased ROS production in whole leaf and root tissue of AOX anti-sense transgenic Arabidopsis (Arabidopsis thaliana) plants in the presence of KCN as an ETC inhibitor (Umbach et al., 2005).
Inhibitors of ETC complexes cause a dramatic and unnatural overreduction of the ETC, but environmental factors encountered by plants in their natural habitats are also likely to affect ETC redox status and result in oxidative stress (Finnegan et al., 2004). It has been proposed that AOX may be an important factor in allowing plants to tolerate chilling-induced damage (Purvis and Shewfelt, 1993). The effects of exposure to chilling temperatures include impaired electron flow through the Cyt pathway, a decrease in Cyt c oxidase subunit II protein accompanied by an increase in H2O2 production (Prasad et al., 1994a, 1994b), and a reduced capacity of the Cyt relative to the AOX pathway (McNulty and Cummins, 1987). In addition, AOX transcripts (Ito et al., 1997; Takumi et al., 2002), protein levels (Stewart et al., 1990; Vanlerberghe and McIntosh, 1992; Gonzàlez-Meler et al., 1999), and AOX capacity (Elthon and McIntosh, 1986; Vanlerberghe and McIntosh, 1992) are responsive to low temperatures in several plant species. However, fewer clear-cut results have been obtained in intact tissues using the oxygen fractionation technique that enables measurement of the in vivo electron partitioning between the Cyt and AOX pathways (Ribas-Carbo et al., 1995). For example, after exposure to low temperature, a chilling-sensitive maize (Zea mays) cultivar showed a higher electron flow through the AOX pathway compared with a chilling-tolerant cultivar (Ribas-Carbo et al., 2000a). Although these studies confirm that AOX activity is responsive to cold stress, they do not indicate a conclusive association between cold tolerance and ROS stress amelioration (Gonzàlez-Meler et al., 1999; Ribas-Carbo et al., 2000a).
To elucidate the function of AOX in Arabidopsis, we generated and characterized AtAOX1a transgenic lines that display a range of AOX protein levels. Initial screenings under nonlimiting growth conditions did not reveal any shoot growth, root growth, or other morphological phenotypes associated with transformation (Umbach et al., 2005). However, given that AOX may alleviate oxidative stress under conditions that lead to overreduction of the ETC, we reasoned that it may be possible to uncover conditional phenotypes under suboptimal growth conditions. To that end, we set out to study shoot responses to low temperature of AtAOX1a transgenics and were able to characterize morphological and growth phenotypes associated with the altered AOX levels.
RESULTS
Effects of Cold Temperature on Growth
To examine in detail the effects of low temperature on growth, we grew transgenic AOX1a plants in Duke University Phytotron chambers. In a preliminary experiment conducted at 12°C, we found a significant difference (one-way ANOVA; P < 0.05) in total leaf blade area at day 21 after germination of five lines with contrasting levels of AOX protein (Supplemental Fig. 1). Subsequently, we measured in two replicate experiments total leaf blade area, number of leaves, and rosette diameter throughout the vegetative phase (21, 28, and 42 d after germination) for multiple independent transgenic lines and compared them with wild-type and empty vector controls both at 23°C and 12°C. Germination and emergence across all the genotypes at either temperature were not significantly different. In addition, at 23°C we did not find significant differences among the lines throughout the vegetative phase in any of the above parameters (data not shown). All genotypes had reduced growth at low temperature. However, a two-way ANOVA of these two experiments revealed a significant genotype × growth temperature interaction (P < 0.01), indicating that growth responses at low temperature depended on the genotype.
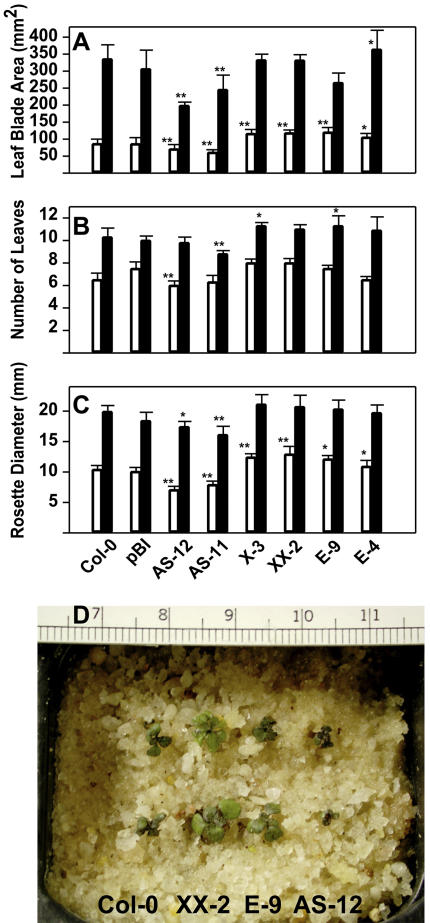
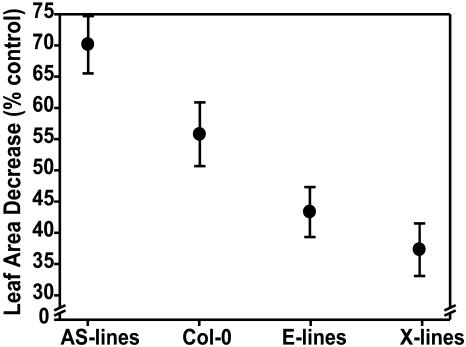
In detail, at 12°C the two anti-sense lines (AS-11 and AS-12) showed significantly reduced total leaf blade area at days 21 and 28, compared with Columbia (Col)-0 and an empty vector control (Fig. 1A; each bar is the average of a total of 12 individuals from two replicate experiments). In contrast, two lines overexpressing wild type (X-3 and XX-2) and two lines overexpressing the mutated, constitutively active AOX1a (E-4 and E-9; see Umbach et al., 2005) had higher total leaf areas at day 21. Importantly, at day 21 the average percentage reduction in total leaf blade area at 12°C relative to 23°C was negatively correlated with amount of AOX protein in the transformation types (Fig. 2), indicating that genetic manipulation of AOX protein level results in an altered (decreased/increased) potential for leaf area expansion at suboptimum temperature. This effect was statistically significant (P < 0.01). At day 28, among all the overexpressors, only the leaf blade area of E-4 was significantly greater compared with wild type, and by day 42 all genotypes had a similar total leaf blade area of about 500 mm2 (Table I), except AS-12. This line had a significantly lower leaf area compared with wild type, suggesting that it had reduced ability to cope with low temperature throughout the whole vegetative growth period due to the reduced amount of AOX protein (see below).
Figure 1.
Low temperature (12°C) growth phenotype of multiple independent Arabidopsis AOX transgenic lines during the vegetative phase. A to C show total leaf blade area (mm2), number of rosette leaves, and rosette diameter (mm), respectively. Bars represent the mean ± se of two replicate experiments. In all sections, the asterisks at the top of the bars indicate P < 0.05 (*) and P < 0.01 (**) relative to wild type and vector control. For each genotype, six rosettes were randomly harvested in each experiment at days 21 (white bars) and 28 (black bars). Col-0, Wild type; pBI, vector control; AS-11 and AS-12, lines transformed with an AOX anti-sense construct; X-3 and XX-2, AOX overexpressors; E-4 and E-9, mutated AOX overexpressors. For rosette diameter, 20 plants in each experiment were measured. D, At day 21, the phenotypes could be visually identified: overexpressors (XX-2 and E-9) had larger rosettes compared with wild-type Col-0. Conversely, anti-sense lines (AS-12) had smaller rosettes and reduced leaf number compared with both Col-0 and overexpressor lines. For each genotype, two representative plants are shown.
Figure 2.
Decrease in leaf area at day 21 caused by growth at low temperature (12°C) relative to control temperature (23°C) for genotypes characterized by high and low AOX protein levels. The data relative to total leaf blade area for day 21 and 12°C were pooled for genotypes with a similar AOX protein level (AS, anti-sense lines, AS-11 and AS-12; E, lines overexpressing mutated AOX, E-9 and E-4; X, lines overexpressing wild-type AOX, X-3 and XX-2). The average percentage decrease ±se relative to the leaf area value calculated at control temperature for each group (=100%) is shown.
Table I.
Total leaf blade area, number of leaves, and rosette diameter at day 42 after germination for wild type and for transgenic genotypes displaying a range of AOX protein levels
Data are averages ± se of two replicated experiments (n = 12). *, P < 0.05; **, P < 0.01.
| Leaf Area | Rosette Diameter | No. of Leaves | |
|---|---|---|---|
| mm2 | mm | ||
| Col-0 | 500 ± 50 | 37 ± 1 | 15 ± 1 |
| pBI | 480 ± 30 | 34 ± 2 | 15 ± 1 |
| AS-11 | 470 ± 40 | 32 ± 1* | 15 ± 1 |
| AS-12 | 430 ± 30* | 29 ± 1** | 15 ± 2 |
| XX-3 | 510 ± 45 | 34 ± 1 | 16 ± 1 |
| XX-2 | 530 ± 20 | 35 ± 2 | 16 ± 1 |
| E-4 | 521 ± 30 | 31 ± 2** | 16 ± 1 |
| E-9 | 520 ± 40 | 32 ± 1* | 16 ± 1 |
Differences in total leaf blade area are the result of changes in the number of leaves and/or changes in lamina expansion of individual leaves. At days 21 and 28, the differences in total leaf blade area were, in part, caused by a decreased leaf number in the anti-sense lines and by an increased leaf number in the overexpressing lines compared with Col-0 and pBI. These values were statistically significant (P < 0.01) for AS-12 (day 21), AS-11 (day 28), and X-3 and E-9 (day 28). By day 42, all genotypes had a similar number of leaves (Table I). The observed differences in total leaf blade area between lines at days 21 and 28 were closely reflected in the rosette diameter (Fig. 1, C and D), and at day 21 the percentage differences in both of these parameters were very similar when results for each genotype class in the two experiments were averaged: Anti-sense line leaf blade area was reduced by 27% and rosette size by 25% compared with Col-0 and the empty vector control (average of these two genotypes = 100%), whereas for lines overexpressing wild-type AOX1a, leaf area was increased by 30% and rosette size by 33%, and in lines overexpressing mutated AOX1a these values were 24% and 27%, respectively.
At day 42, anti-sense lines and overexpressors of mutated AOX1a had significantly smaller rosettes but had a very similar number of leaves compared with Col-0 (Table I). This suggests that the expansion of individual leaves was affected in these lines. Finally, it is likely that differences in petiole length (not measured) unrelated to transformation type were responsible for the lack of correlation between rosette diameter, number of leaves, and leaf blade area in two cases (E9 compared with Col-0 at day 28, Fig. 1, A–C; and AS-11 compared with AS-12 at day 42, Table I).
In summary, these results show that effects of low temperature on growth were present early and for much of the vegetative phase but they diminished as the plants approached flowering. Moreover, these data indicate that genetic modification of AOX protein levels results in an altered sensitivity (leaf area decrease) to low temperature at early developmental stages.
AOX Protein Levels
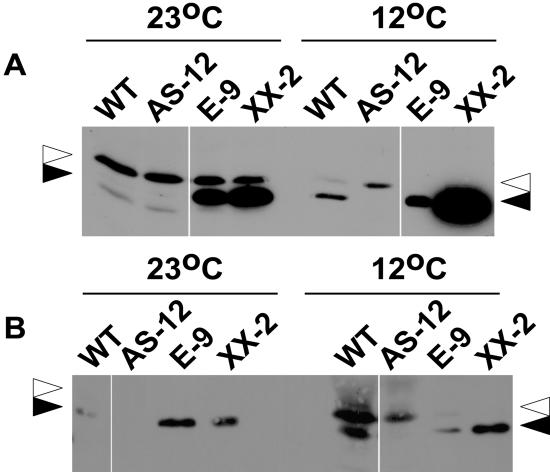
We measured the levels of AOX protein at the end of the vegetative period (day 42) in whole leaves of one anti-sense (AS-12) and two overexpressor lines (XX-2 and E-9) grown at control (23°C) and low temperature (12°C). Immunoblot analysis showed that in wild-type leaves AOX was not detectable at 23°C and was up-regulated at 12°C (Fig. 3A, black arrowhead). In contrast to wild type, AS-12 anti-sense leaves did not up-regulate AOX protein to a detectable level in the cold. The overexpressor lines used in this experiment showed a very large amount of AOX protein at both control and low temperatures. In addition to immunoblots of whole leaf proteins, we repeated these experiments with fractions from 28-d-old leaves enriched in mitochondria (see “Materials and Methods”). The results were nearly identical except that trace amounts of AOX were detectable in wild-type crude mitochondria samples from control-temperature plants (Fig. 3B). Importantly, the elevated AOX at 12°C in wild type appears to be due to posttranscriptional effects because there was no indication that AOX1a or other AOX isoform transcripts were increased at low temperature in leaves of similar age (see below). These findings overall are consistent with our data obtained using isolated mitochondria (Umbach et al., 2005) and indicate that AOX levels in the transgenic lines were essentially unchanged by the low temperature treatment at these developmental stages. In addition, anti-sense silencing of AOX protein synthesis appeared stable throughout the experimental growth period in AS-12 at both temperature regimes.
Figure 3.
Whole leaf and crude mitochondria preparation immunoblots of AOX protein in AOX transgenic plants grown at 23°C and 12°C. A, At day 42 after germination, whole leaves were harvested. Total protein extracts from 50 mg FW of leaf tissue extracted in 150 μL of double-strength sample buffer was separated by SDS-PAGE in the presence of reductant. For wild type (WT) and AS-12, 35 μL of extracts was loaded; for the overexpressors, 15 μL was loaded except for E-9 at 12°C, for which 7 μL was loaded. After transfer to nitrocellulose, the blot was probed with the AOA monoclonal antibody. At the right and left, the black arrowhead shows the position of AOX 34-kD monomer and the white arrowhead the position of a nonspecific band as determined by incubating the blots only with the secondary antibody. The lower band present in trace amounts in wild-type and AS-12 samples at control temperature also is nonspecific. In B, 30 μL of crude mitochondria preparation from 28-d-old leaves was loaded on the gel. Note that E-9 and XX-2 samples were diluted 1:6 with gel sample buffer before loading 30 μL. The black and white arrowheads are the same as for A. Labels for the genotypes (see Fig. 1 legend) and for samples taken at the two growth temperatures are shown at the top of the blot.
Lipid Peroxidation
To investigate whether changes in ROS levels were associated with the low temperature phenotypes of AOX1a anti-sense and overexpressing plants, we measured the extent of lipid peroxidation caused by ROS (day 28). One transgenic line for each class of transformant was used in this experiment. The absolute thiobarbituric acid reactive substances (TBARS) values are consistent with elevated oxidative stress levels in all lines at either temperature (Table II). Experiments conducted with freshly harvested leaves from low light-grown plants of the same genotypes typically yielded TBARS values of about 1 nmol/g fresh weight (FW; data not shown). We did not detect any significant differences in the levels of TBARS of plants grown at 23°C (Table II; one-way ANOVA, P > 0.05). Growth at low temperature resulted in an increase in TBARS values in all lines compared with control temperature at the same date. XX-2 had lower average levels and AS-12 had the highest average values, as it did at 23°C, compared with all the other lines (Table II). Due to a relatively large variability, these values were not significantly different from wild-type and vector control plants (P > 0.05). Comparison of the percentage increase in TBARS for the low temperature plants relative to TBARS at 23°C showed that wild type, the vector control, and the anti-sense lines had similar values, while the TBARS increase was less for the two overexpressor lines (Table II). These results suggest that AOX overexpression may decrease ROS in the cold and partially account for the improved growth phenotype of these plants. However, lack of AOX in the anti-sense line does not appear to increase ROS generation in the cold and therefore cannot alone account for the anti-sense phenotype.
Table II.
TBARS content at day 28 of AOX transgenic plants grown at 23°C (control) or 12°C (low temperature)
Average ± se is shown for all samples (n = 3).
| TBARS (nmol g FW−1)
|
||
|---|---|---|
| 23°C
|
12°C
|
|
| Day 28 | Day 28 | |
| Col-0 | 14.5 ± 1.1 | 24.3 ± 4.4 (68)a |
| pBIb | 13.7 ± 1.4 | 23.7 ± 3.0 (73) |
| E-9 | 15.5 ± 1.0 | 23.9 ± 3.0 (54) |
| XX-2 | 15.4 ± 1.4 | 19.8 ± 3.6 (29) |
| AS-12 | 16.8 ± 2.4 | 29.1 ± 3.0 (73) |
Average percentage increase at 12°C relative to 23°C.
Transgenic line carrying the empty transformation vector.
Leaf Anthocyanin Content
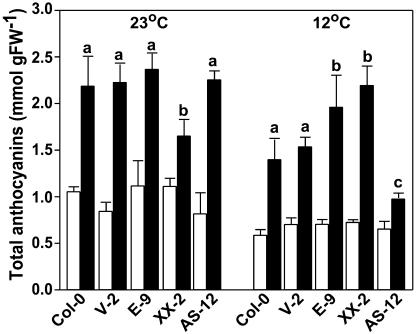
We initially observed that all genotypes evaluated for growth characteristics had purple leaves especially on the abaxial side (Fig. 1D) under the Phytotron environmental conditions (“Materials and Methods”) both at 23°C and 12°C. To test whether the transgenic AOX1a plants accumulated different amounts of pigments compared with wild type, we measured total anthocyanin levels in leaves of five lines representing wild type and empty vector control and the three different classes of AOX1a transgenics (Fig. 4). At either day 21 or 28 at 23°C, there were no significant differences in anthocyanin levels among the genotypes. At day 28, total anthocyanin content increased in all lines to a level more than double that of the previous date (>2 mmol/g FW) with the exception of XX-2, in which the increase was more modest (Fig. 4). At day 21 at 12°C, the anthocyanin content was nearly identical in all genotypes, and on average the values were lower than those at 23°C on the same date. This can be partly explained by the fact that the low temperature-grown plants were developmentally younger than those grown at 23°C at the same time point. Similar to the results at the control temperature, anthocyanin values increased at day 28 in all lines at 12°C. However, anthocyanin content at this date was correlated with AOX expression level; mutated and wild-type AOX overexpressors accumulated pigment to similar high levels relative to day 21 whereas control leaves accumulated less, and this accumulation was almost entirely suppressed in the anti-sense line (Fig. 4). These effects were statistically significant (one-way ANOVA; P < 0.05).
Figure 4.
Total anthocyanin content of leaves of Arabidopsis AOX transgenic plants grown at control (23°C) and low temperature (12°C). The figure shows the total anthocyanin content (anthocyanidin-3-glucoside equivalents) in leaves of AOX transgenic plants grown at control and at low temperature (white bars = day 21; black bars = day 28). Each bar is the mean of three replicate samples ±se. The letters at the top of the bars indicate homogenous subgroups that are significantly different (P < 0.05, one-way ANOVA and Tukey b post-hoc test).
Analysis of Gene Expression
We analyzed transcript levels of all five AOX genes in leaves of one set of selected transformant lines (vector control, E-9, XX-2, AS-12) and wild type harvested at day 22 for 23°C and day 26 for 12°C so that the developmental stages were similar for the two growth conditions. AOX1a transcript levels of wild-type, vector control, and overexpressor plants were similar to those measured for growth room plants (Umbach et al., 2005). All of these lines showed a crossing point increase at 12°C (average increase = 1.02 [se ± 0.11]), equivalent to a 2-fold decrease in transcript amount. Under the conditions used, we were not able to distinguish between sense and anti-sense AOX1a transcripts in AS-12 (Umbach et al., 2005). AOX1b, AOX1c, and AOX2 transcripts were generally of low abundance regardless of growth temperature or line. However, AOX1d transcript levels were greater than previously observed (Thirkettle-Watts et al., 2003; Escobar et al., 2004; Umbach et al., 2005). The crossing point average for all five lines at 23°C was 25.48 (se ± 0.42), compared to an average of 31.23 (se ± 0.27) for the seven lines in table II of Umbach et al. (2005), representing a 54-fold increase. At 12°C, AOX1d expression levels were maintained except for in the anti-sense line, where they were lower (crossing point = 30.01 ± 0.14), perhaps indicating some silencing.
Because low temperature could impact other components of the ETC and the oxidative stress level of tissues, we analyzed expression changes of selected genes representing these two components of metabolism. Within a temperature treatment, most of the 17 genes analyzed (listed in table I of Umbach et al., 2005; see Fig. 5 legend for nomenclature) showed some variation in expression among the lines (≤one crossing point or 2-fold) with no changes relating to transformation. However, ubiquitin and peroxiredoxin IIC (PrxIIC) showed larger variation that corresponded to AOX1a transformation (see below).
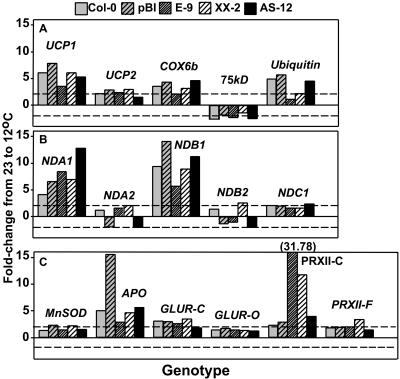
Figure 5.
Difference in leaf transcript levels for selected genes between control and low temperature-grown Arabidopsis transgenic plants. RNA was extracted from leaves of plants grown at either 12°C (day 26) or 23°C (day 22) and used for real-time PCR. Results for one growth experiment are shown using Col-0 (wild type), pBI (vector control), E-9 and XX-2 (overexpressors of mutated and wild-type AOX, respectively), and AS-12 (anti-sense). Differences between raw crossing point values (12°C minus 23°C) for each line were converted to fold-change by exponentiation and are plotted for each gene. Standard errors for crossing point differences are 0.10 for ubiquitin and 0.38 for all other genes. These pooled ses reflect variation due to 15 replicate LightCycler runs for ubiquitin from a single RNA isolation from tissue for each line at each temperature (other genes are measured only once at each temperature). The gene designations (for further details and references, see Umbach et al., 2005) are as follows: A, uncoupling proteins 1 and 2 (UCP1, UCP2), Cyt c oxidase subunit 6b (COX6b), Complex I 75-kD subunit (75 kD), and Ubiquitin 5 (Ubiquitin); B, internal nonphosphorylating mitochondrial NAD(P)H dehydrogenases (NDA1, NDA2), external nonphosphorylating mitochondrial NAD(P)H dehydrogenases (NDB1, NDB2), and mitochondrial NAD(P)H dehydrogenase of unknown location (internal or external; NDC1); C, mitochondrial manganese superoxide dismutase (MnSOD), ascorbate peroxidase, organellar (APO), glutathione reductase, cytoplasmic and organellar (GLUR-C, GLUR-O), PrxIIC (cytoplasmic), and peroxiredoxin IIF (mitochondrial; PrxIIF). Dashed lines, corresponding to a 2-fold (one crossing point) increase or decrease in transcript level, are shown to act as guides with no implication of statistical or physiological significance. Note that data for NDB4 are not included; its transcript levels were too low to be reliably detected (see Umbach et al., 2005).
When expression levels were compared between the two growth temperatures, differences in gene expression common to all lines were evident (Fig. 5, A–C). An up-regulation of expression well above a change of one crossing point (2-fold) for all or most of the lines occurred for seven genes: UCP1, COX6b, Ubiquitin, NDA1, NDB1, PrxIIC, and APO (see Fig. 4 legend for nomenclature). For APO, note that the large increase in transcript in the vector control (Fig. 5C, pBI) was due to an unusually low level of transcript in this line at 23°C, whereas the transcript level was comparable among all the lines at 12°C. Among the apparently up-regulated genes, UCP1 transcripts have previously been shown to increase under low temperature conditions (Maia et al., 1998). However, in tobacco, NDA1 and NDB1 transcripts were down-regulated and the 75-kD subunit of Complex I was up-regulated in the cold (Svensson et al., 2002). The Arabidopsis plants exhibited the opposite response, with the 75-kD Complex I subunit being the only gene to show a small down-regulation in all lines (Fig. 5A), perhaps due to different cold sensitivities of the two species or to different experimental conditions.
Ubiquitin and PrxIIC gene expression in AOX1a overexpressors was distinct from other lines. Ubiquitin was up-regulated similarly at 12°C in wild type, vector control, and AS-12, but this increase was less in both E-9 and XX-2 (Fig. 5A), possibly due to a less-stressed physiological state in the overexpressors or to a slight shift in developmental stage. In apparent contrast with the view that AOX functions to decrease cellular oxidative stress, PrxIIC transcript at 12°C showed the greatest transcript level increase in both overexpressor lines (Fig. 5C). This was in part due to a lower level of expression in these two lines at 23°C (data not shown).
When the seven cold-induced genes were analyzed in a replicate set of older tissue (day 28 harvest at both growth temperatures), only UCP1 consistently showed a greater than 2-fold up-regulation at 12°C in all the lines (data not shown).
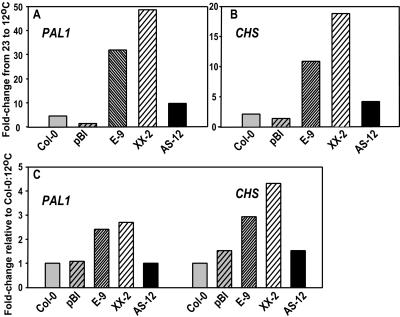
To determine whether they correlated with the line-specific differences in total anthocyanin accumulation, we measured transcript levels of Phe ammonia lyase (PAL1) and chalcone synthase (CHS), key enzymes in the flavonoid biosynthetic pathway. Up-regulation of both PAL1 and CHS expression occurred in the plants at 12°C (day 26) relative to 23°C (day 22) (Fig. 6, A and B), consistent with previous work showing an increase in their transcripts in the cold (Leyva et al., 1995). This effect was particularly large for the two overexpressing lines, in part because PAL1 and CHS transcript levels at 23°C were markedly lower in these lines relative to the others (data not shown). Comparison within the low temperature treatment showed smaller differences in expression among the lines relative to wild type, but the overexpressors still had the most elevated transcript levels (Fig. 6C). The expression of both genes in the anti-sense line was comparable to that of wild type and vector control.
Figure 6.
Relative transcript levels of two genes in the anthocyanin biosynthesis pathway in low temperature-grown Arabidopsis transgenic plants. A and B, Change in expression of PAL1 and CHS transcripts with growth at low temperature. Plants and methods are as described for Figure 5. Values are derived from two replicate LightCycler runs. Pooled ses for the crossing point differences for each line and each gene, as described in Figure 5, are 0.27. Note different y-axis scaling for PAL1 versus CHS. C, Expression levels of PAL1 and CHS in transformed lines at 12°C relative to wild type at 12°C. A fold-difference of 1.0 is equivalent to the expression level of wild type.
DISCUSSION
Previously, we generated and characterized multiple independent AOX transgenic lines in Arabidopsis. We concluded that the hypothesis that AOX may prevent overreduction of the ETC and limit ROS production in isolated cells and mitochondria (Maxwell et al., 1999; Taylor et al., 2002) applies equally to whole plant tissues. However, varying AOX levels, as well as the AOX activation state, did not result in any chronic changes in tissue oxidation state among the transformant lines, based on transcript and lipid peroxidation analysis (Umbach et al., 2005). Further, transgenic plants had no morphological phenotype when they were grown under nonlimiting conditions either in agar plates or on soil. This lack of phenotypic effects is not unexpected because silent phenotypes are frequent for metabolic traits (Rohde et al., 2004; Weckwerth et al., 2004).
In this work, we set out to identify conditional phenotypes under environmental conditions that are known to impair the ETC of plant mitochondria and in which AOX may play a fundamental role (e.g. low temperature; Gonzàlez-Meler et al., 1999). We found that growth at low temperature (12°C) produces a shoot growth phenotype closely associated with genetic manipulation of AOX levels. To our knowledge, this is the first time that phenotypic effects at the whole plant level have been described for AOX transgenic plants. Previous studies using whole plants showed that the response of AOX activity to low temperature is markedly species specific (Gonzàlez-Meler et al., 1999) and does not necessarily correlate with low temperature tolerance (Ribas-Carbo et al., 2000a), leaving the exact role of AOX uncertain. Therefore, demonstration of a whole plant phenotype is of great relevance to unravel the function of AOX, particularly because it avoids extrapolation from data based on in vitro assays and on the use of respiratory inhibitors. Our data suggest that decreased AOX levels in the anti-sense lines affect Arabidopsis growth at low temperature throughout the vegetative growth period, resulting in impaired leaf area development and smaller rosette size. One silenced line (AS-12) that was unable to accumulate AOX protein to detectable levels in the cold (Fig. 3) had significantly smaller rosettes and leaves at all time points examined (i.e. from 3 weeks after germination until bolting onset; Fig. 1, A–C). In contrast, plants overexpressing AOX1a had increased leaf area in early growth stages (through day 21) compared with wild-type plants when grown at low temperature. Because phenotypic effects across all lines generally became less when plants approached flowering, we conclude that AOX plays a role in acclimation to low temperature in Arabidopsis at relatively early growth stages. This is particularly interesting when viewed in a developmental framework, in which acclimation is maximized by the development of new leaves having an altered enzymatic capacity in their respiratory system in response to low temperature (Atkin and Tjoelker, 2003). Consistent with this, the phenotypic difference between control lines and those overexpressing wild-type AOX was smaller at 28 d and minimal at 42 d, probably because the control lines were able to accumulate AOX protein (Fig. 3) over this growth period.
Interestingly, plants overexpressing a mutated AOX lacking the regulatory Cys (Umbach et al., 2002) displayed a variable growth phenotype, resembling overexpressors, controls, or anti-sense lines at different time points. This variability provides evidence that a constitutively active form of the AOX is not equivalent to the wild-type enzyme in vivo and that biochemical regulation of AOX is needed for optimal function. An unregulated AOX may decrease the control of metabolite flux through respiration, offsetting any protective effects derived from its overexpression. There are other possible explanations for the effects of the mutant AOX. One is that the mutated AOX did not integrate normally in mitochondria. However, this is unlikely because the protein is properly targeted and processed (figure 3 in Umbach et al., 2005) and its expression in Escherichia coli resulted in activity levels similar to wild-type AOX activated by α-keto acids (Umbach et al., 2002). Alternatively, because overexpression of AOX protein was less abundant in the roots of mutant AOX transformants (Umbach et al., 2005), it cannot be excluded that the variable shoot growth phenotype of these plants was related to root processes where the absence of additional AOX could be critical and was not due to the presence of the mutation alone.
Whether AOX may be considered an antioxidant enzyme regulating the redox state of plant mitochondria has been a matter of debate (Moore et al., 2002). An important finding of this study is that oxidative stress only partially correlates with phenotypic differences between the AOX transgenic lines. Low temperature did affect levels of lipid peroxidation less in overexpressor lines. However, although the AOX anti-sense lines are more prone to oxidative stress (lipid peroxidation) under severe, nonphysiological conditions (see Umbach et al., 2005), the effect of low temperature on their lipid peroxidation levels was comparable to its effect on wild type (Table II) during a phase (day 28 after germination) in which phenotypic differences among the genotypes are significant. Further, while transcript analysis showed that the plants did respond to low temperature by up-regulation of some genes, there was little evidence of any significant increased or decreased oxidative stress or perturbations in the ETC specific to any transformed lines, relative to wild-type and vector control plants. These results are in marked contrast with data obtained using cell suspensions (Maxwell et al., 1999) and isolated mitochondria (Pastore et al., 2001; Purvis, 2001), and suggest that AOX may not function exclusively to prevent excess formation of ROS in whole tissue under more natural and long-term stress conditions.
Although our analysis showed generally a lack of between-line differences at the transcript level, the analysis did not address the possibility of transient transcript changes during the long-term experiments, so some line-specific differences may have been missed. That transcript levels did change with time is suggested by PCR data from the older tissue samples where differences in expression levels between the two temperatures were few. In addition, long-term acclimation to a specific environment and to AOX transformation may come about through posttranscriptional and posttranslational regulation as well, and these changes would not be detected by a transcript analysis (for further discussion, see Umbach et al., 2005). The increased level of AOX protein in wild-type tissue at 12°C, without a concomitant increase in AOX1a transcript, is an example. Results that particularly address transcriptional control can be complemented in the future by detailed studies at the protein level to further clarify potential effects of low temperature and AOX expression on enzyme activity and mechanisms of biochemical regulation.
Altered AOX levels may have far-reaching effects and important adjustments could have occurred outside the mitochondria, as suggested by the microarray study performed with AOX anti-sense plants under nonlimiting environmental conditions (see Umbach et al., 2005). Accordingly, it is intriguing that the single oxidative stress-related gene whose transcript levels showed any effect of transformation was PrxIIC, which encodes a cytoplasmic enzyme. More striking, we found a correlation between AOX transgenic type and anthocyanin levels at low temperature, including enhanced transcription in the AOX1a overexpressors of PAL1 and CHS, key enzymes of the flavonoid pathway. Although transcripts for these genes were at wild-type levels in the anti-sense plants, these plants did not have wild-type levels of anthocyanin at low temperature at day 28, further indicating AOX function is in some way associated with the synthesis of these compounds. It is interesting that a recent study of Arabidopsis PAL1 and PAL2 mutants revealed far-reaching effects on the transcriptome and metabolome encompassing both amino acid and carbohydrate metabolism (Rohde et al., 2004). Future detailed biochemical and physiological studies may establish the nature of the metabolic link between changes in PAL expression, availability of respiratory substrates, and AOX activity.
Finally, although unconnected to low temperature, an unexpected change in gene expression was the relatively high level of AOX1d transcript in the Phytotron plants. Previously, AOX1d transcript levels had been found to be very low (Thirkettle-Watts et al., 2003; Escobar et al., 2004; Umbach et al., 2005). AOX transcript and protein levels in potato (Solanum tuberosum; Svensson and Rasmusson, 2001) and activity in soybean (Glycine max; Ribas-Carbo et al., 2000b) were stimulated by light and it is possible that AOX1d expression was in response to the high light used in the Phytotron experiments. Alternatively, AOX1d could be responding to the nutritional status of the leaf tissue, for example, to the nitrogen source (Baurain et al., 2003). These and other possibilities can be distinguished by further study. Because AOX1d transcript was depressed in the anti-sense line at low temperature, it cannot be ruled out that at least some of the phenotypic and metabolic effects seen in these plants were due to deficiency in AOX1d as well as AOX1a protein.
In conclusion, we have shown that altered levels of AOX protein result in leaf growth phenotypes in Arabidopsis plants grown at low temperature. AOX did not appear to act primarily through local effects in mitochondria or solely through reduction in ROS formation, but rather through more pervasive metabolic effects, some of which appear to be linked to anthocyanin production. These results contribute new insight into the function of AOX of plant mitochondria and offer a new perspective for future studies of the effects of low temperature on plant respiration and on the acclimation of plants to abiotic stresses.
MATERIALS AND METHODS
Plant Material and Growth Conditions
In this study, we used seeds of Arabidopsis (Arabidopsis thaliana) wild-type Col-0; empty transformation vector controls (pBI and V-2); AOX1a anti-sense lines (AS-11 and AS-12); and overexpressors of wild type (X-3 and XX-2) and mutated AOX1a (E-4 and E-9). All gene constructs were under the control of the 35S cauliflower mosaic virus promoter and were obtained and selected as described by Umbach et al. (2005). T3 seeds were used for all experiments. Seeds were surfaced sterilized overnight in a solution containing 2% (v/v) PPM (Preservative for Plant Tissue Culture Media; Plant Cell Technology), half-strength Murashige and Skoog salts (Sigma-Aldrich), and 1% (w/v) Suc, rinsed with sterile water, and resuspended in 0.1% (w/v) agarose. Before sowing, seeds were placed at 4°C for 48 h. All experiments were conducted at the Duke University National Phytotron in walk-in growth chambers with the following environmental conditions: 23°C ± 1°C (control temperature) and 12°C ± 1°C day, 8°C night (low temperature); 600 ± 20 μmol m−2 s−1 photosynthetically active radiation; photoperiod 14 h; and relative humidity percentage 75 ± 5. Seeds were sown in 60 × 60 × 100 mm pots (1 seed per pot) containing turfice:gravel:vermiculite 1:1:1 (v:v:v) and fertilized weekly with half-strength Hoagland solution. These growth conditions were chosen to produce plants phenotypically similar to field-grown winter annual Arabidopsis (Heidel et al., 2004). A total of 96 plants per genotype in each experiment were used. To minimize positional effects due to microenvironmental conditions in the growth chambers, flats were rotated regularly. We performed a total of three experiments, a preliminary one using four lines representing the four classes of AOX-transformed plants, and two more comprehensive ones each using eight lines including multiple independent transformant lines.
Morphometric Measurements
At days 21, 28, and 42 after germination, the rosette diameter and total number of leaves of each of six randomly chosen plants per genotype were measured and then harvested individually. The leaf blades of each rosette were dissected and total leaf blade area was measured with a calibrated leaf area meter (LI-3000A; LI-COR). To check the accuracy of the area measurement of small leaves, digital pictures of a two-replicate series of Col-0 leaves (n = 10) were taken and total leaf area was measured with public domain image analysis software (ImageJ version 1.32; http://rsb.info.nih.gov/ij/). Comparison of the same data set with the two methodologies by linear regression yielded a regression coefficient of 0.92. Rosette diameter was estimated on 20 plants in each experiment and for each time point.
AOX Western Blots
About 50 mg of fully expanded leaves from control and cold-treated plants were harvested at day 42 after germination (Col-0, XX-2, E-9, AS-12), frozen in liquid N2, and kept at −80°C. Protein extraction, SDS-PAGE, protein transfer, and immunoblotting with the AOA antibody (Elthon et al., 1989) were conducted as described by Umbach et al. (2005) for the analysis of whole tissue samples, except that 100 mm dithiothreitol was present in the sample buffer. For the isolation of a membrane fraction (crude mitochondria preparation), 100 mg of leaves from 28-d-old plants were chilled on ice and homogenized in a mortar with sand in 2 mL of a grinding buffer, pH 7.5 (300 mm Suc, 25 mm tetrasodium-pyrophosphate, 10 mm KH2PO4, 2 mm Na-EDTA, 1% [w/v] PVP-40, 1% [w/v] bovine serum albumin, 20 mm ascorbate). Samples were centrifuged at 1,000g for 5 min at 4°C. The supernatant was centrifuged at 11,000g for 5 min at 4°C. The pellets were resuspended in double-strength gel sample buffer containing 100 mm dithiothreitol. Thirty microliters of sample was loaded on the gel. SDS-PAGE, protein transfer, and immunoblotting were performed as described previously (see Umbach et al., 2005), except that the AOA antibody dilution was 1:500 and secondary antibody (horseradish peroxidase-conjugated anti-mouse antibody; Santa Cruz Biotechnology) was 1:5,000.
Determination of TBARS
One hundred milligrams of leaf tissue was harvested at day 28 after germination for control and low temperature plants. TBARS assays were performed according to Hodges et al. (1999) and Taylor et al. (2002) and as described in detail by Umbach et al. (2005). TBARS levels (nmol g FW−1) were calculated according to Hodges et al. (1999) using the extinction coefficient for malondialdehyde at 532 nm (0.157 mol L−1).
Anthocyanin Determination
Total anthocyanins were estimated following Hodges et al. (1999). Approximately 0.1 g FW of leaves of control and cold-grown plants (Col-0, V2, AS-12, XX-2, E-9) was homogenized in 5 mL of methanol-1% HCl. Samples were centrifuged to remove cell debris and 1 mL of the supernatant appropriately diluted in methanol-1% HCl was used for analysis. Total anthocyanins were determined spectrophotometrically as the difference between absorbance at 536 and 600 nm (to correct for phaeophytin absorbance; Hodges and Nozzolillo, 1996). The assay was conducted for samples at day 21 and day 28. Results were expressed as cyanidin 3-glucoside equivalents (mmol g−1 FW−1) using an extinction coefficient of 0.449 mol L−1 at 536 nm and represent the mean ± se of three biological replicates for each genotype and time point.
Real-Time PCR
Leaf tissue for RNA extraction was harvested from two separate sets of plants grown at both low temperature and the control temperature as described in “Plant Material and Growth Conditions.” One experimental set consisted of the lines Col-0, pBI, E-9, XX-2, and AS-12. The second set was the same, except V2 was used instead of pBI for the vector control transformant line. For set 1, the treatment and control plants were of different chronological ages at harvest (22 d for the control, 26 d for the low temperature plants) but were at developmentally similar stages. For set 2, plants were harvested at the same time point, 28 d. For each sample, rosette leaves of different ages were collected. Transcript levels of selected genes were measured by the method described previously (Umbach et al., 2005) using a Roche LightCycler for real-time PCR. Primers for specific gene transcripts were as listed in table I of Umbach et al. (2005). Two additional primer pairs were used for PAL1 (At2g37040; forward 5′-TGTAGCGCAACGTACC; reverse 5′-GTTCGGGATAGCCGATG) and for CHS (At5g13930; forward 5′-AGTCCCTAAGCTAGGCA; reverse 5′-AGGTAACGGCTGTGAT). The amplification efficiencies for these primer pairs, determined as by Umbach et al. (2005), were 1.8 and 1.9, respectively. For these tissue samples, ubiquitin 5 was judged unsuitable as a “housekeeping gene” for data normalization. Its transcript levels varied far more between lines and between temperatures than those of a number of other genes (see “Results”). A true housekeeping gene, particularly for use in comparisons between tissues that are likely to be at different metabolic poises, is probably a nonexistent entity (Bustin, 2002; Vandesompele et al., 2002; Dheda et al., 2004). Therefore, we used the PCR data in unadjusted form to assess changes in transcript levels, relying on equal RNA being used in the reverse transcription step, and limited our conclusions to only qualitative differences among samples. This approach is justified in part because of the data for manganese superoxide dismutase. This gene's transcription has been shown to be largely invariant under a number of conditions (Kliebenstein et al., 1998), and we found that to be essentially the case for our samples (Fig. 5). Ubiquitin was nevertheless included in every PCR run for each sample, providing a measure for run to run variability.
Statistical Analysis
For real-time PCR data in Figures 5 and 6, we analyzed crossing points using mixed-model analysis of variance techniques (SAS Proc Mixed; SAS Institute). We regarded temperature, line, and their interaction as fixed effects, and LightCycler run and interactions with it as random effects. Because there is a single RNA isolation at each temperature, the analysis can only assess variability induced by repeated LightCycler runs. For ubiquitin, there are 15 LightCycler runs at each temperature for each line, whereas there are one or two runs for the other genes. Therefore, estimates of variation for all genes were based on the observed variation for ubiquitin. Crossing point values met the normality and constant-variance assumptions of the analysis.
All other statistical analysis was performed with SPSS 12.0 for Windows software. Before performing analysis of variance, the data were tested for homogeneity of variance (Levene test), and they did not require transformation.
Supplementary Material
Acknowledgments
We thank Lina Taneva for her contribution to early stages of this work, and the Duke Phytotron staff for help with plant and growth chamber maintenance. We also thank Tom Elthon for providing the AOA antibody and David Umbach for statistical analysis of the real-time PCR data.
This work was supported by the National Science Foundation (grant no. MCB–0091080 to J.N.S. and A.L.U.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ann L. Umbach (umbacha@duke.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070789.
References
- Atkin OK, Tjoelker MG (2003) Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci 8: 343–351 [DOI] [PubMed] [Google Scholar]
- Baurain D, Dinant M, Coosemans N, Matagne RF (2003) Regulation of the alternative oxidase Aox1 gene in Chlamydomonas reinhardtii. Role of the nitrogen source on the expression of a reporter gene under the control of the Aox1 promoter. Plant Physiol 131: 1418–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29: 23–39 [DOI] [PubMed] [Google Scholar]
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Lumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37: 112–119 [DOI] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, McIntosh L (1986) Characterization and solubilization of the alternative oxidase of Sauromatum guttatum mitochondria. Plant Physiol 82: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Franklin KA, Svensson ÅS, Salter MG, Whitelam GC, Rasmusson AG (2004) Light regulation of the Arabidopsis respiratory chain. Multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol 136: 2710–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Soole KL, Umbach AL (2004) Alternative mitochondrial electron transport proteins in higher plants. In DA Day, AH Millar, J Whelan, eds, Plant Mitochondria: From Gene to Function, Vol 17, Advances in Photosynthesis and Respiration. Kluwer, Dordrecht, The Netherlands, pp 163–230
- Fridovich I (1978) The biology of oxygen radicals. Science 201: 875–880 [DOI] [PubMed] [Google Scholar]
- Gardner PR, Raineri I, Epstein LB, White CW (1995) Superoxide and iron modulate aconitase activity in mammalian cells. J Biol Chem 270: 13399–13405 [DOI] [PubMed] [Google Scholar]
- Gonzàlez-Meler MA, Ribas-Carbo M, Giles L, Siedow JN (1999) The effect of growth and measurement temperature on the activity of the alternative respiratory pathway. Plant Physiol 120: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Clarke JD, Antonovics J, Dong X (2004) Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Hodges DM, Nozzolillo C (1996) Anthocyanin and anthocyanoplast content of cruciferous seedlings subjected to mineral nutrient deficiencies. J Plant Physiol 147: 749–754 [Google Scholar]
- Ito Y, Saisho D, Nakazono M, Tsutsumi N, Hirai A (1997) Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203: 121–129 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde R-A, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Jarillo A, Salinas J, Martinez-Zapater JM (1995) Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol 108: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia IG, Benedetti CE, Leite A, Turcinelli SR, Vercesi AE, Arruda P (1998) AtPUMP: an Arabidopsis gene encoding a plant uncoupling mitochondrial protein. FEBS Lett 429: 403–406 [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty AK, Cummins WR (1987) The relationship between respiration and temperature in leaves of the arctic plant Saxifraga cernua. Plant Cell Environ 10: 319–325 [Google Scholar]
- Millar AH, Leaver CJ (2000) The cytotoxic lipid peroxidation product, 4-hydroxy-2-nonenal, specifically inhibits decarboxylating dehydrogenases in the matrix of plant mitochondria. FEBS Lett 481: 117–121 [DOI] [PubMed] [Google Scholar]
- Millenaar FF, Benschop JJ, Wagner AM, Lambers H (1998) The role of the alternative oxidase in stabilizing the in vivo reduction state of the ubiquinone pool and the activation state of the alternative oxidase. Plant Physiol 118: 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millenaar FF, Lambers H (2003) The alternative oxidase: in vivo regulation and function. Plant Biol 5: 2–15 [Google Scholar]
- Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Moore AL, Albury MS, Crichton PG, Affourtit C (2002) Function of the alternative oxidase: Is it still a scavenger? Trends Plant Sci 7: 478–481 [DOI] [PubMed] [Google Scholar]
- Pastore D, Trono D, Laus MN, Di Fonzo N, Passarella S (2001) Alternative oxidase in durum wheat mitochondria. Activation by pyruvate, hydroxypyruvate and glyoxylate and physiological role. Plant Cell Physiol 42: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR (1994. a) Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Stewart CR (1994. b) Acclimation, hydrogen peroxide, and abscisic acid protect mitochondria against irreversible chilling injury in maize seedlings. Plant Physiol 105: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis AC (2001) Reduction of superoxide production by mitochondria oxidizing NADH in the presence of organic acids. J Plant Physiol 158: 159–165 [Google Scholar]
- Purvis AC, Shewfelt RL (1993) Does the alternative pathway ameliorate chilling injury in sensitive plant tissues? Physiol Plant 88: 712–718 [DOI] [PubMed] [Google Scholar]
- Ribas-Carbo M, Aroca R, Gonzalez-Meler MA, Irigoyen JJ, Sanchez-Diaz M (2000. a) The electron partitioning between the cytochrome and the alternative respiratory pathways during chilling recovery in two cultivars of maize differing in chilling sensitivity. Plant Physiol 122: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Berry JA, Yakir D, Robinson SA, Lennon AL, Siedow JN (1995) Electron partitioning between the cytochrome and alternative pathways in plant mitochondria. Plant Physiol 190: 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Robinson SA, Gonzalez-Meler MA, Lennon AM, Giles L, Siedow JN, Berry JA (2000. b) Effects of light on respiration and oxygen isotope fractionation in soybean cotyledons. Plant Cell Environ 23: 983–989 [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios JG (1993) Oxygen stress and superoxide dismutases. Plant Physiol 101: 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, Martin BA, Reding L, Cherwick S (1990) Seedling growth, mitochondrial characteristics, and alternative respiratory capacity of corn genotypes differing in cold tolerance. Plant Physiol 92: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson ÅS, Johansson FI, Møller IM, Rasmusson AG (2002) Cold stress decreases the capacity for respiratory NADH oxidation in potato leaves. FEBS Lett 517: 79–82 [DOI] [PubMed] [Google Scholar]
- Svensson ÅS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73–82 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Heazlewood JL, Herald V, Holtzapffel R, Day DA, Leaver CJ, Millar AH (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Takumi S, Tomioka M, Eto K, Naydenov N, Nakamura C (2002) Characterization of two non-homologous nuclear genes encoding mitochondrial alternative oxidase in common wheat. Genes Genet Syst 77: 81–88 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277: 42663–42668 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133: 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed]
- Umbach AL, Gonzalez-Meler MA, Sweet CR, Siedow JN (2002) Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim Biophys Acta 1554: 118–128 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034 [DOI] [PMC free article] [PubMed]
- Vanlerberghe GC, McIntosh L (1992) Lower growth temperature increases alternative pathway capacity and alternative oxidase protein in tobacco. Plant Physiol 100: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AM, Wagner MJ, Moore AL (1998) In vivo ubiquinone reduction levels during thermogenesis in araceae. Plant Physiol 117: 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Ehlers Loureiro M, Wenzel K, Fiehn O (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.