Abstract
The characterization of in vitro xylogenic cultures of zinnia (Zinnia elegans) has led to major discoveries in the understanding of xylem formation in plants. We have constructed and characterized a subtractive library from zinnia cultures enriched in genes that are specifically expressed at the onset of secondary wall deposition and tracheary element (TE) programmed cell death. This Late Xylogenesis Library (LXL) consisted of 236 nonredundant cDNAs, 77% of which encoded novel sequences in comparison with the 17,622 expressed sequence tag sequences publicly available. cDNA arrays were constructed to examine dynamic global gene expression during the course of TE formation. As a first step in dissecting auxin and cytokinin signaling during TE differentiation, macroarrays were probed with cDNAs from cells cultured in different hormonal conditions. Fifty-one percent of the LXL genes were induced by either auxin or cytokinin individually, the large majority by auxin. To determine the potential involvement of these categories of genes in TE differentiation, multiplex in situ-reverse transcription-PCR was performed on cells for two genes encoding putative cell wall proteins: Gibberellin stimulated transcript-1, induced by auxin alone, and expansin 5, induced by cytokinin alone. All transcriptionally active TEs expressed both genes, indicating that, although these genes may not be considered as specific markers for TE differentiation per se, they are nevertheless an integral part of TE differentiation program. Among the non-TE population, four different gene expression-based cell types could be distinguished. Together, these results demonstrate the underlying complexity of hormonal perception and the existence of several different cell types in in vitro TE cell cultures.
The formation of xylem, or xylogenesis, constitutes one of the most spectacular forms of cell differentiation in plants. Xylem, initiating from meristematic procambial or cambial cells, is a heterogeneous tissue composed of nonconducting cells including parenchyma and fibers, and conducting cells or tracheary elements (TEs; for recent review, see Ye, 2002). In angiosperms, TEs are essential for the transport and storage of water and nutrients in land plants. Although it is clear that TE function is of prime importance in plant development, our knowledge of the cascade of cellular and molecular events from procambium and cambium formation to the initiation of xylem differentiation, cell elongation, secondary wall deposition, and programmed cell death (PCD), is at best fragmentary. Our lack of knowledge is due to the high degree of organizational complexity of vascular tissues throughout the plant and the limited number of cells actually undergoing differentiation at a given time. As a result, the accessibility of vascular cells is limited and makes experimentation difficult in planta.
To gain a more in-depth knowledge of xylem formation, two approaches have been largely exploited. The genetic dissection of Arabidopsis (Arabidopsis thaliana) mutants with altered xylem differentiation and/or organization has provided insight as to the regulatory mechanisms underlying various aspects of vascular development from the formation of proper, continuous networks of veins in leaves (polaris [Casson et al., 2002], scarface [Deyholos et al., 2000], cvp1 and 2 [Carland et al., 1999], and van [Koizumi et al., 2000]), overall vascular bundle organization in leaves and stems (avb1 [Zhong et al., 1999]), to secondary cell wall (SCW) formation (irx1-5 [for review, see Turner and Sieburth, 2002]). More recently, a myb transcription factor controlling phloem-xylem patterning has been identified (Bonke et al., 2003). In the apl mutant, asymmetric cell divisions and subsequent phloem differentiation were impaired. Moreover, ectopic expression of APL inhibited xylem development. These studies suggest a dual role of APL in the promotion of phloem and the repression of xylem differentiation. As expected from earlier physiological studies pointing to the role of hormones in vascular differentiation, many vascular mutants are also affected in their sensitivity and/or response to hormones, and more particularly auxin (axr6 [Hobbie et al., 2000], mp [Hardtke and Berleth, 1998], and pin1 [Vernoux et al., 2000]). To date, mutants that are completely blocked in xylem differentiation have not been isolated, presumably because this defect would be lethal. This implies that a strictly genetic approach may not be sufficient to completely unravel xylogenesis.
Another powerful approach relies on large-scale cDNA sequencing of expressed sequence tags (ESTs) of developing xylem from poplar (Populus spp.; Sterky et al., 1998) and pine (Pinus taeda; Allona et al., 1998). Recently, microarray analysis was performed along a precisely defined developmental gradient of secondary xylem in poplar, revealing strict stage-specific transcriptional regulation of many genes, several of which encode gene products of unknown function (Hertzberg et al., 2001). Although these studies provide an impressive amount of information as to genes involved in xylem formation, the existence of numerous cell types that comprise secondary xylem does not allow us to establish a direct link between a given genetic program and a specific cell type such as TEs.
To learn more about TE formation, complementary genomic approaches have been applied to the in vitro xylogenesis system in zinnia (Zinnia elegans). In this model system, single isolated mesophyll cells transdifferentiate into TEs within 3 d when cultured in the presence of auxin and cytokinin (Fukuda and Komamine, 1980). Early studies based on differential screening methods resulted in the isolation and characterization of several stage-specific marker genes of TE formation including the TE differentiation (TED) genes (Demura and Fukuda, 1993, 1994), Cys proteases (Ye and Varner, 1996), and a pectate lyase (Domingo et al., 1998). More recently, high-throughput strategies have been applied to zinnia TEs. These include systematic EST sequencing coupled to microarray analysis (Demura et al., 2002) and a cDNA-amplified fragment length polymorphism approach (Milioni et al., 2002).
As each strategy of gene discovery possesses its unique technical basis, we predicted that a complementary approach would generate a novel set of genomic data to significantly contribute to our growing knowledge of TE formation. In this paper, by using the suppression subtractive hybridization (SSH) technique (Diatchenko et al., 1996) applied to zinnia TEs, we were able to identify hundreds of novel putative xylogenesis markers in comparison with the 17,622 ESTs isolated from two previous functional genomic studies in zinnia (Demura et al., 2002; Milioni et al., 2002). Moreover, as a complementary approach to a study performed on deciphering early signaling events of TE formation (Milioni et al., 2002), we focused our effort on genes induced at the onset of secondary wall formation and PCD. Comprehensive macroarray analysis of these genes was performed to determine global temporal expression during TE differentiation. As both auxin and cytokinin are strict requirements for TE differentiation, genes were also classified as a function of their regulation by auxin and/or cytokinin alone, constituting a first step in dissecting putative auxin and cytokinin cross talk pathways during TE differentiation. The results presented herein confirm the assertion that using complementary time points and methodologies will generate complementary genomic results, demonstrate that the use of SSH libraries coupled to cDNA arrays is a viable approach to dissecting TE development in zinnia, and integrate gene expression data, via multiplex in situ-reverse transcription-PCR (IS-RT-PCR), at the single cell level in xylem development in vitro and in planta.
RESULTS
Characterization of an SSH Library Enriched in Genes Involved in Secondary Wall Deposition and Cell Death during TE Formation in Zinnia
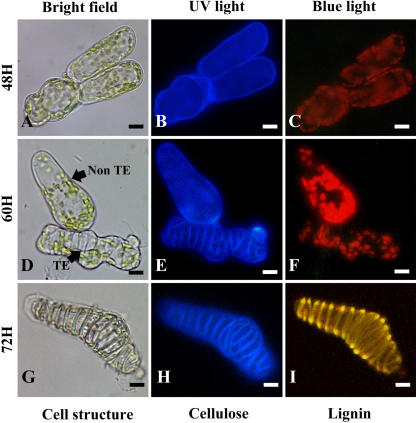
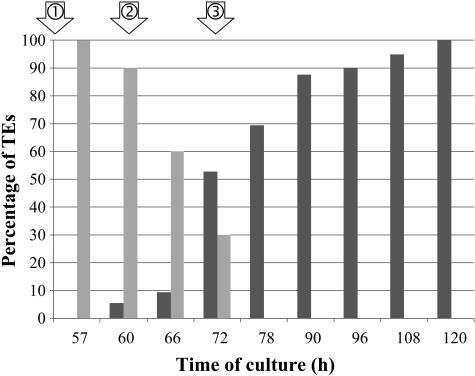
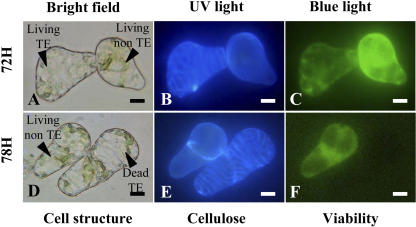
To isolate genes specifically involved in secondary wall deposition and autolysis during TE formation, cells were harvested at three selected time points during the course of TE differentiation: secondary wall-associated cellulose deposition, lignification, and autolysis. Throughout this study, the time points indicate the number of hours after culture initiation (T0). Since judicious sampling is one of the major keys to constructing a high quality library, we developed a double-staining procedure to histochemically characterize the SCW from zinnia TE cultures. By using a combination of calcofluor and auramine-O, we were able to simultaneously visualize cellulose and lignin, respectively, in zinnia TE cultures (Fig. 1). At 48 h, TEs were not yet visible (Fig. 1, A–C). All cells, regardless if they were to become TEs or not, were characterized by calcofluor staining of the primary cell wall (Fig. 1B). As expected, no lignin was apparent as indicated by the absence of yellow-green auramine-O staining under blue light (Fig. 1C). This stage was considered as the presecondary cellulose deposition stage. At 60 h, the TE SCWs were readily visible using both bright-field microscopy (Fig. 1D) and calcofluor (Fig. 1E). At this stage, only 5% of the TEs were lignified as indicated by the number of TEs exhibiting positive auramine-O staining (Fig. 2). This stage corresponded to the onset of lignification. At 72 h, 52% of the TEs were lignified as determined by positive calcofluor and auramine-O staining of SCWs (Figs. 1, G–I, and 3). Interestingly, this double-staining procedure enabled us to detect cellulose associated with secondary wall thickenings, even in highly lignified cells. Concomitantly, another double-staining procedure combining calcofluor and fluorescein diacetate (FDA) was also employed to assess TE viability (Groover and Jones, 1999; Kuriyama, 1999). At 72 h, 30% of the TEs were still viable as indicated by positive FDA staining (Figs. 2, A–C, and 3). At 78 h, all TEs were dead (Figs. 2, D–F, and 3). Therefore, 72 h was considered as the most representative time point for the autolytic stage. RNA from three time points (48, 60, and 72 h) were pooled, and the resulting cDNAs (tester population) were subjected to a subtraction procedure against a pool of cDNAs extracted from cells cultured in the absence of hormone and in the presence of either auxin or cytokinin alone (driver population) at the same time points. All three driver populations exhibited a similar morphology and were not significantly different (intact chloroplasts and turgid vacuoles) as compared to freshly isolated mesophyll cells. The resulting subtractive library was named the Late Xylogenesis Library (LXL).
Figure 1.
SCWs of differentiating zinnia TE cell cultures stained with calcofluor and auramine-O to simultaneously detect cellulose and lignin, respectively. Cells were visualized at 48 (A–C), 60 (D–F), and 72 h (G–I) under bright-field conditions (A, D, and G), UV light (B, E, and H), and blue light (C, F, and I). Under UV light, cellulosic primary walls are dark blue (B, E, and H), whereas secondary wall thickenings of TEs appear as light blue (E and H). Under blue light, lignified wall thickenings are yellow (I). These three time points subsequently defined the three stages used to construct the LXL library. Magnification bars = 8 μm.
Figure 2.
Quantification of lignified (dark-gray bars) and living TEs (light-gray bars) during TE differentiation as described by the double-staining procedures in Figures 1 and 3. Results are expressed as a percentage of calcofluor-stained TEs. The time points for the LXL are indicated with arrows. 1, Onset of secondary wall cellulose deposition; 2, onset of lignification; and 3, mid-PCD.
Figure 3.
TE viability visualized by simultaneous calcofluor and FDA staining. Cells were cultured for 72 (A–C) and 78 h (D–F) and visualized under bright-field conditions (A and D), UV light (B and E), and blue light (C and F). TEs are characterized by calcofluor-positive blue SCW under UV light (B and E) and viable cells are stained green (FDA) under blue light (C and F). Magnification bars = 8 μm.
The LXL was comprised of approximately 800 clones ranging in size from 250 to 1,300 bp with an average size of around 500 bp. The clones were systematically characterized by reverse northern analysis by hybridizing duplicate filters containing LXL PCR-amplified inserts, one with the tester cDNAs originally used to construct the library and the other with the driver cDNAs. All of the clones were classified according to their relative expression in TE (tester) versus control (driver) cell cultures. A representative hybridization experiment is shown in Figure 4. Thirty-seven percent of the clones hybridized uniquely with tester cDNA and hence were classified as TE specific (black arrows, Fig. 4), 42% were more highly expressed in TE cultures than in control cultures (gray arrows, Fig. 4), and 12% showed undetectable hybridization signals in both populations, with presumably low expression levels even in TE cultures (striped arrow, Fig. 4). Only 9% had similar-to-identical hybridization signals in TEs as compared to control cultures. These clones were considered as false positives (white arrows, Fig. 4).
Figure 4.
A representative example of reverse northern-blot analysis of LXL cDNA clones. cDNA clones of the LXL were PCR amplified, electrophoresed on agarose gels, and transferred to nylon membranes. Duplicate membranes were hybridized with equivalent amounts of cDNA probe from tester (TE) and driver (control) cultures. Arrows indicate four different expression categories: black, specific expression in tester only; gray, more highly expressed in tester than driver; striped, undetectable expression in both tester and driver; and white, similar-to-identical expression in both tester and driver.
Prior to large-scale sequencing, a pilot experiment consisting of the random sequencing of a handful of genes from the TE-specific and TE up-regulated categories allowed us to identify several known markers of TE differentiation: a Cys protease (ZcP4), a ribonuclease (Rbn I), an endonuclease (Zen1), an unspecific lipid transfer protein (TED4), and a caffeoyl-CoA O-methyl transferase (CCoA-OMT). The identification of previously characterized markers of different stages of xylogenesis confirmed the quality of the LXL and our overall approach for identifying novel genes associated with TE formation.
In-Depth Bioinformatic Analysis of LXL
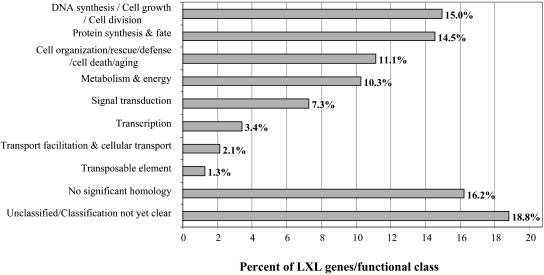
Five hundred forty-four single-pass sequences were generated from all of the above-mentioned expression categories with the exception of those considered as false positives. The average sequence size was 410 ± 180 bp, ranging from 120 to 600 bp. Sequence clusterization was performed using a blastclust program (ftp://ftp.ncbi.nih.gov/) and revealed 238 unique clusters, indicating that the overall redundancy factor of the LXL was 2.4. A comprehensive gene list describing all aspects of the bioinformatic analysis presented in this section is provided in Supplemental Data 1. One hundred eighty-four of the clusters were composed of singletons, whereas the most redundant cluster was comprised of 16 sequences. Functional annotation was performed using a BLASTX program against the integral Arabidopsis protein database (25,545 sequences). All homologies with an expectation value (E value) of <1e−5 were considered significant. A total of 83.8% of the fragments contained a portion of predicted coding region, which enabled homology-based gene identification. The remaining 16.2% of the sequences without a significant hit were presumably composed exclusively of 5′ or 3′ untranslated regions, or were too short to give a significant match. All of the sequences of the LXL were then assigned to functional categories according to the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de; Fig. 5). The major functional groups are involved in DNA synthesis cell growth and cell division (15.0%), protein synthesis (14.5%), and cell organization/rescue/defense/cell death/aging (11.1%), suggesting the importance of these cellular processes during TE differentiation. Interestingly, 18.8% of the sequences share significant similarity to unknown or hypothetical proteins with no function yet assigned. The positioning of zinnia EST sequences with respect to their corresponding Arabidopsis orthologs also allowed us to determine that 56% of the sequences also contained a 5′ and/or 3′ untranslated region. These gene-specific tags provide invaluable tools for the further characterization of individual members of gene families.
Figure 5.
Functional classification of nonredundant LXL cDNA sequences. Functions and categories were established according to MIPs (http://mips.gsf.de).
Comparison of LXL with Other Xylem-Related Functional Genomic Studies
A wealth of genomic information is currently available for secondary wall formation (zinnia TEs, poplar and pine xylem, and Arabidopsis secondary growth) and developmental PCD. We took advantage of this opportunity to determine the extent of similitude/divergence of gene expression in related physiological processes in different species and physiological contexts (see Supplemental Data 1). As a first step, we compared sequences from the LXL with those generated by other functional genomic approaches of zinnia TEs. Towards this end, a BLASTN analysis was performed with each LXL sequence against all publicly available zinnia sequences registered in GenBank databases (to date, a total of 17,622 sequences). Since this comparison was made between ESTs and not between full-length sequences, an overestimation of “no hits” would be likely. A more in-depth comparison was therefore carried out to determine if LXL sequences and publicly available zinnia sequences shared a common Arabidopsis homolog. Based on these comparisons, we conclude that approximately 77.6% of our sequences are, indeed, novel. One of the major deliverables of this type of genomic comparison is the cross identification of genes of unknown function via different approaches. Interestingly, several of the identical sequences encoded genes of unknown function (i.e. unknown gene [UG]-08/DV017539 with AU287424/AU287419, UG-37/DV017393 with AU289520, UG-39/DV017208 with AU293453, and UG-68/DV017575 with AU285513 and AU305636). This finding reinforces the hypothesis that these genes may play a critical role in xylem formation. In addition, one relatively long sequence (UG 36/DV017317, 460 bp) with no significant homology to any other sequence from any other species in the public databases was also identified by Demura et al. (2002), suggesting that it may be a zinnia-specific gene important for xylem formation. Finally, we have identified new members of several multigene families for which only one family member has been described in zinnia up until now. For example, whereas only one CCoA-OMT has been previously characterized in zinnia (Ye et al., 1994), the LXL contains three distinct genes (CCoA-OMT1, DV017314; CCoA-OMT2, DV017250; and CCoA-OMT3, DV017196). Likewise, until now only one TED4 gene has been previously described. The LXL contains at least two different TED4 genes (TED4-1, DV017248 and TED4-2, DV017432). Cysteine proteases were by far the most redundant function with 12 clusters representing at least three different genes. Two different isoforms of Cys protease exhibiting 87% identity had been previously isolated (Ye and Varner, 1996; Demura et al., 2002) from two different zinnia cultivars (cv Peter Pan and cv Canary Bird). Both Cys proteases were present in LXL library and an additional third isoform, CP7 (DV017543; exhibiting 50% identity with previously identified Cys proteases), was also isolated. Two new isoforms of expansins (exp), designated exp4 (DV017234) and exp5 (DV017460), were also identified in the LXL library in addition to the three previously described isoforms (Im et al., 2000). The biological significance of multigene families in the context of xylem formation remains to be elucidated. Together, these results nicely illustrate the importance of carrying out complementary genomic approaches on the same model system in order to provide the most comprehensive view possible of a given physiological process. Zinnia ESTs from the LXL were then compared with ESTs from xylem of pine and poplar using a tBLASTX program. In the case of pine, 209,733 ESTs were publicly available at the time of writing (Allona et al., 1998, and all new GenBank releases since then). A total of 79.4% of zinnia sequences matched with a Pinus sequence with an E value of <1e−5. Again, among the genes of unknown function in zinnia, some exhibited very high homologies with pine sequences. For example, UG-24 (DV017163) matched with CD017099 from a xylem compression wood library, UG-25 (DV017176) with BQ106797 from a xylem late wood library, and UG-42 (DV017236) with AW758726 from a normal xylem wood library. A similar comparison was made with the 272,213 publicly available ESTs from poplar (Sterky et al., 1998, and all sequences released in GenBank since then). A total of 80.9% of zinnia LXL sequences exhibited a match with an E value of <1e−5 (Supplemental Data 1).
Comparison of LXL with Arabidopsis SCW and PCD Microarray Expression Data
A functional comparison of LXL genes was also undertaken by extrapolating microarray data for the closest Arabidopsis sequence of each gene of the zinnia LXL during physiologically related developmental processes in Arabidopsis. Microarray data have been reported for stems undergoing secondary growth (Oh et al., 2003), and various forms of developmental PCD in Arabidopsis cell cultures (Swidzinski et al., 2002). As for secondary growth, which includes substantial secondary xylem formation, only eight cDNAs were up-regulated: two Cys proteases, two lignification genes (trans-cinnamate-4-monooxygenase hydroxylase [C4H] and CCoAOMT1), a polygalacturonase (pg), preprophytosulfokine-1, and two UGs (orthologs of zinnia UG-09 and UG-57 genes; see Supplemental Data 1).
As for PCD, out of the 978 specifically induced cDNA clones identified during heat-induced PCD, five cDNAs were identified: cucumisin (a Ser protease), a bifunctional endonuclease, histone 2A, a peroxidase (prx), preprophytosulfokine-1, and a gene of unknown function (UG-57). Out of the 590 cDNA clones induced during both heat-induced PCD and senescence, only one gene, a stress-induced protein like (sti1), was found in common with zinnia LXL. No common homologs could be identified between zinnia LXL and senescence-specific up-related genes, suggesting very divergent mechanisms involved in different forms of developmental PCD. That said, it is interesting to note that among the zinnia LXL genes, orthologs of both preprophytosulfokine-1 and UG-57 were up-regulated in both secondary xylem and heat-induced PCD in Arabidopsis. Finally, we compared our data with those generated from primary cell walls of Arabidopsis cell cultures using a proteomic approach (Chivasa et al., 2002). No genes were found in common, suggesting that different mechanisms are most likely involved in primary and SCW synthesis.
Dynamic Global Gene Expression during TE Formation Using Macroarray Technology
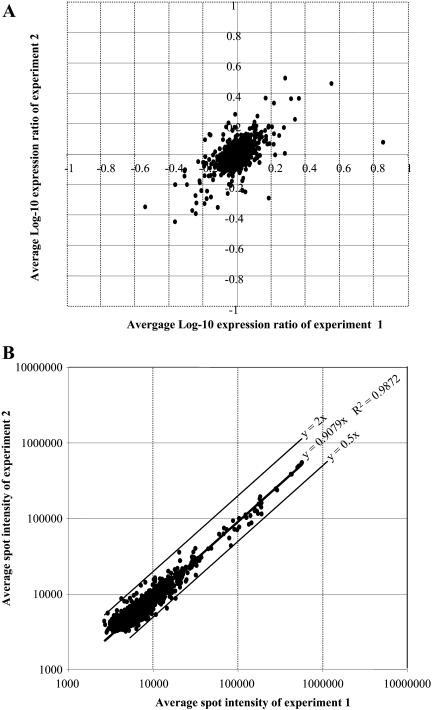
All of the clones were originally classified according to their relative expression in inductive versus noninductive conditions by reverse northern-blot analysis (Fig. 4). However, these experiments did not provide any information as to their temporal gene expression during TE formation since the probe used in hybridizing experiments consisted of pooled cDNAs from the three stages. To determine dynamic gene expression during TE formation, cDNA inserts from the LXL were PCR amplified and spotted onto nylon membranes. Each membrane contained 576 clones, each of which was spotted in duplicate. Various hybridization and background level controls were also spotted (see “Material and Methods”). Preliminary analyses were first performed to assess the innermembrane variation of duplicate spots for each gene on the same membrane and the reproducibility of spot intensity ratios (expressed as a log-10 expression ratio [LR]) resulting from hybridizations of independent membranes using independent probes derived from independent biological samples. The data obtained were highly reproducible within a given membrane and, even more importantly, between two independent membranes (Fig. 6A). In the experiment illustrated in Figure 6A, 89% of the values fell within ±0.176 LR of the mean (equivalent to a 1.5-fold difference) and 97% within ±0.3 LR (equivalent to a 2-fold difference). This variation is similar to those previously observed on microarray experiments (Wang et al., 2000; Kawasaki et al., 2001). In all subsequent experiments, in order for a gene to be considered as differentially expressed between two time points or culture conditions, the difference in spot intensities had to be greater than a 2-fold significance threshold. Macroarray reproducibility was further analyzed by comparing spot intensity values from two independent hybridizations performed on two independent membranes with probes from two independent cultures for each time point (Fig. 6B). Linear coefficients of determination between the two experiments were calculated, thereby defining a high reproducibility between independent hybridizations with 99% of the values confined within a 2-fold limit.
Figure 6.
A, Scatter plot of the LRs of duplicate spots from two independent hybridization experiments. The x axis = LRs of duplicate spots on the membrane used in experiment 1. The y axis = LRs of duplicate spots on the membrane used in experiment 2. Each membrane was hybridized with cDNA probes from an independent, 60-h TE culture. Analyses performed on 16 independent filters hybridized with different probes from different time points showed the same variation (data not shown). B, Scatter plot of spot intensity raw values of two independent experiments probed with cDNAs from 60-h TE cultures. Ninety-nine percent of the cDNA fall within ±2-fold of the mean. Again, data for all other filters fall within the same limits (data not shown).
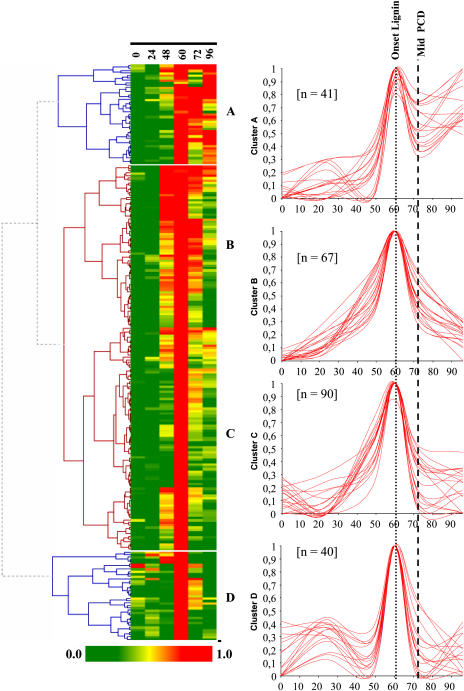
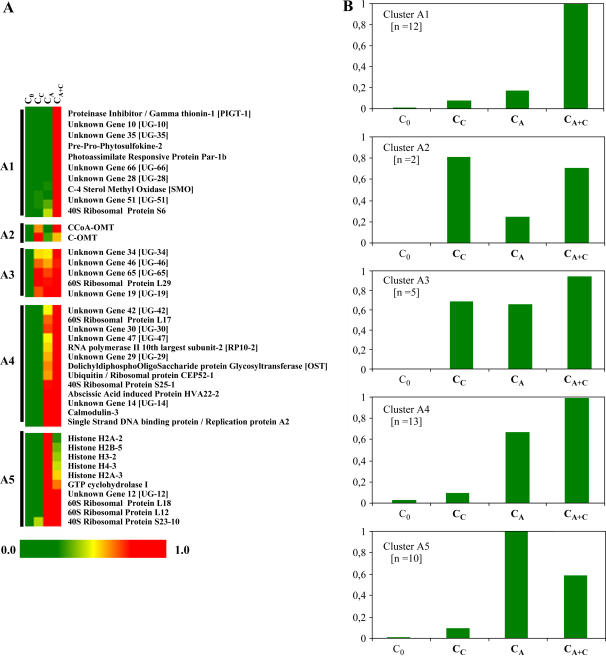
Hybridizations were performed with cDNA probes prepared from cells cultured in TE inductive medium for 0, 24, 48, 60, 72, and 96 h in order to define temporal gene expression dynamics during TE formation. Two hundred thirty-eight average expression profiles, corresponding to the 238 nonredundant cDNA clones of LXL, were calculated. The raw data is presented in Supplemental Data 2. Preliminary pairwise comparisons between time points revealed that most of LXL cDNAs were up-regulated at 60 h. Indeed, 99% of LXL cDNAs were up-regulated at least 2-fold at 60 h as compared to 0 h. Seventy-nine percent and 65% of the cDNAs exhibited 5- and 10-fold difference in expression levels, respectively, for these same time points. In order to visualize gene expression patterns during TE differentiation, complete hierarchical clustering analysis was performed using HCE II software. Four clusters (A–D) of gene expression were identified (Fig. 7). For each cluster, the superposition of 20 representative cDNA expression patterns and the number (n) of genes per cluster are indicated. Cluster A is characterized by a peak of expression at 60 h, with expression levels remaining high even beyond TE PCD when all TEs are dead (at 96 h). Clusters B and C exhibit a peak only at 60 h, the difference being that the overall window of expression of genes belonging to cluster C is slightly narrower over time than cluster B. Genes belonging to clusters B and C are therefore characterized by little-to-no expression beyond TE PCD at 96 h. Many known TE markers fall into clusters B (Zen1, TED4, and ZcP4) and C (Rbn I, CCoAOMT1 and 3, C4H, exp1, and exp3), with very similar expression profiles to those previously reported (Demura et al., 2002). Cluster D was characterized by two peaks of expression, a small peak at 24 h and a major peak at 60 h. One of the most striking observations is that, regardless of the cluster, a large majority of LXL genes are maximally expressed at 60 h.
Figure 7.
Hierarchical clustering of the LXL during TE differentiation. On the basis of the constructed tree, the 238 LXL nonredundant cDNAs were clustered into four expression groups for each expression cluster; a representative sample of 20 independent cDNA expression profiles is illustrated. The number (n) of nonredundant cDNAs for each cluster is indicated. The x axis = time of culture (h), and the y axis = relative expression intensity (%).
Hormonal Regulation of Gene Expression of the LXL
TE differentiation of zinnia mesophyll cells can be triggered only if both auxin and cytokinin are included in the culture medium. How auxin and cytokinin each participate in TE differentiation, to our knowledge, has never been addressed in detail. As a first step in dissecting hormonal signaling pathways involved in differentiation, we determined the action of auxin and cytokinin individually on global gene expression of the LXL. As the majority of LXL genes are the most highly expressed at 60 h, hybridization experiments were carried out with cDNA probes derived from cells cultured for 60 h in the presence of either both hormones (Culture A + C or CA + C), auxin (CA), or cytokinin (CC) alone, or in basal medium without hormones (C0). To classify all of the LXL genes into categories in terms of hormonal regulation, pairwise comparisons were made between the hormonal condition in which the gene was maximally expressed and each of the other individual hormonal conditions. Again, a greater than 2-fold difference in expression was used as the significance threshold. We have annotated as CA + C, genes highly expressed only in CA + C medium; CA + C > CA, genes highly expressed in both CA + C and CA medium with a stronger expression in CA + C medium; CA > CA + C, genes highly expressed in both CA + C and CA medium with a stronger expression in CA medium, and so on (Table I). Four major conclusions may be drawn from this analysis. (1) A large proportion of genes (49%) exhibit a strict requirement for both auxin and cytokinin. (2) Very few genes are expressed in cells cultured in C0 medium. Indeed, 95% and 92% of the cDNAs exhibit a greater than 5- and 10-fold difference, respectively, in expression levels in CA + C versus C0 medium (data not shown). (3) Many genes are induced by either auxin alone or cytokinin alone, with the large majority being up-regulated by auxin (38% are CA as compared to 4% for CC). That said, it is interesting to note that, as a general rule, these genes are even more highly expressed when both hormones are present in the culture medium. (4) In some cases (4%), gene expression levels are higher in the presence of one hormone than in the presence of both hormones (CA > CA + C and CC > CA + C), suggesting that the repression of certain genes may be necessary to promote TE differentiation. This appears to be the case with many of the histone genes.
Table I.
The distribution (%) of the LXL library as a function of expression levels under different hormonal conditions
Data were obtained by performing macroarray experiments with cells cultured for 65 h in the presence of both auxin and cytokinin (CA + C), auxin alone (CA), cytokinin alone (CC), or in the absence of hormones (C0). For each gene, pairwise comparisons of expression signal intensity were carried out between the hormonal condition of maximal expression and each of the other conditions individually. For each pairwise comparison, if the maximal intensity was equal to or greater than 2-fold, the other condition examined was considered as not expressed. If the expression intensity value was less than 2-fold, the gene was considered as expressed for the given condition. CA + C, Genes up-regulated only in CA + C medium; CA + C > CA, genes up-regulated in both CA + C and CA medium with a stronger expression in CA + C medium; CA > CA + C, genes up-regulated in both CA + C and CA medium with a stronger expression in CA medium, and so on.
| Hormonal Expression | % of LXL Genes |
|---|---|
| CA + C | 49 |
| CA + C > CA | 38 |
| CA + C > CC | 4 |
| CA + C > CA > CC | 3 |
| CA + C > CA > CC > C0 | 2 |
| CA > CA + C | 3 |
| CC > CA + C | 1 |
To determine whether a correlation exists between hormone regulation and temporal gene expression, a complete hierarchical clustering of the hormone expression data was incorporated into the previously determined temporal expression clusters (Supplemental Data 3 and Fig. 8 as an example of cluster A). Each temporal expression cluster, from A to D, was divided into four to five subclusters based on hormone-induced gene expression. For example, subclusters annotated 1 are composed of CA + C-specific genes; 2, of CA + C > CC genes; 3, of CA + C > CA > CC genes; 4, of CA+ C > CA genes; and 5, of CA > CA + C genes (the difference with 4 is that expression is higher in CA than CA + C). It is interesting to note that the four temporal clusters, A to D, all contain genes with different hormonal profiles, thereby reflecting the complexity of hormone perception and signaling during TE differentiation.
Fine Tuning of Expression Profiles via RT-PCR
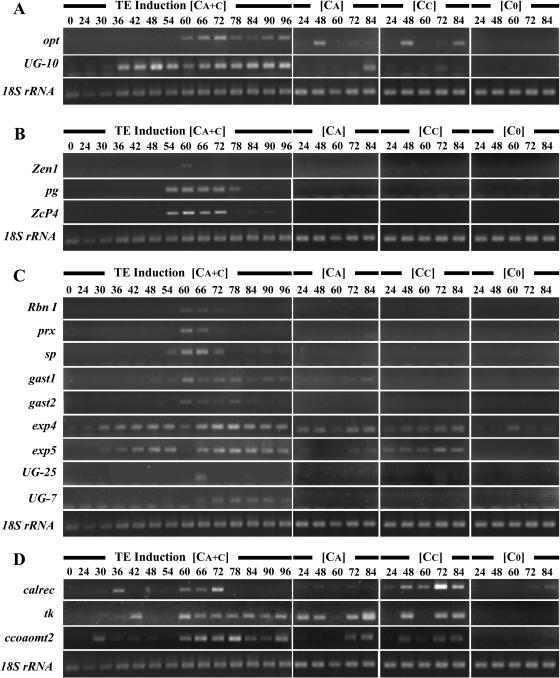
To complete our knowledge of the temporal and hormonal regulation of LXL gene expression during TE differentiation, RT-PCR was carried out for several members of each temporal expression cluster selected both at random and, in some cases based on gene function (Fig. 9). As expected, members of expression cluster A, oligopeptide transporter (opt) and UG-10, are expressed beyond TE death (Fig. 9A). Although these genes were considered to have a strict requirement for both auxin and cytokinin from array data, RT-PCR indicated that both were also transiently expressed in auxin-only and cytokinin-only cultures, however, not at the 60-h time point used for arrays. Interestingly, temporal expression is altered when either one or both hormones are present in the medium. For example, opt is induced by auxin or cytokinin alone within 48 h, whereas in inductive TE cultures, opt is turned on later, starting at 60 h, and remains on after that. On the contrary, UG-10 is expressed at 30 h when both hormones are present in the culture medium, whereas it is induced much later in the presence of each hormone individually (84 and 72 h for auxin and cytokinin, respectively). The expression of members of cluster B (pg, endonuclease [Zen1], and Cys protease [ZcP4]) was restricted to the lifespan of TEs (72 h; Fig. 9B). RT-PCR indicated that these genes were indeed specific to inductive medium, as predicted by array data, and were not expressed in cells cultured in the presence of either auxin or cytokinin throughout the entire time course. Members of expression cluster C (Rbn I, prx, Ser protease [sp], GA acid-stimulated transcript [gast1, gast2], exp4, exp5, UG-25, and UG-7) were also more or less restricted to the TE lifespan (Fig. 9C). It is interesting to note that the timing of gene expression may, in some cases, be extremely precise (i.e. UG-25 is detectable only at 66 h, and prx from 54–66 h). The majority of the genes are expressed only if both auxin and cytokinin are present in the medium. However, there were some exceptions: gast1 is expressed in auxin-only cultures, whereas exp5 is present in cytokinin cultures. In agreement with macroarray data, members of expression cluster D (calreticulin [calrec], thymidylate kinase [tk], and CCoAOMT2) clearly exhibited two peaks of expression (Fig. 9D). RT-PCR was also performed for all of the above-mentioned genes in roots, hypocotyls, epicotyls, cotyledons, and leaves of 3-week-old plantlets (data not shown). Interestingly, many genes were undetectable at the organ level, suggesting a high degree of gene specificity presumably in xylem tissue/cell types.
Figure 9.
RT-PCR expression analysis of genes from the four different temporal expression clusters. Genes from cluster A are presented in A, genes from cluster B in B, and so on. Cells were cultured for different amounts of time in TE induction medium (CA + C), basal medium without hormones (C0), or in the presence of either naphthylacetic acid (CA), or benzylaminopurine alone (CC).
Multiplex IS-RT-PCR of gast1 and exp5 Reveals Multiple Cell Types
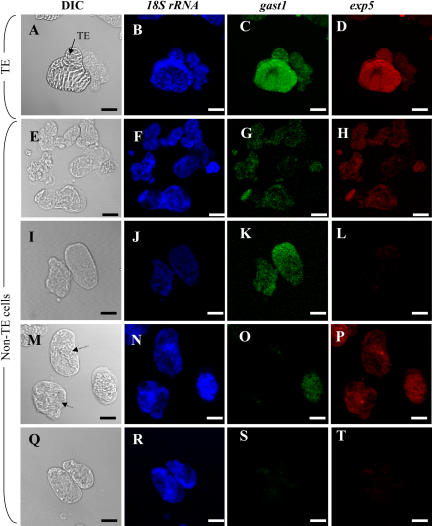
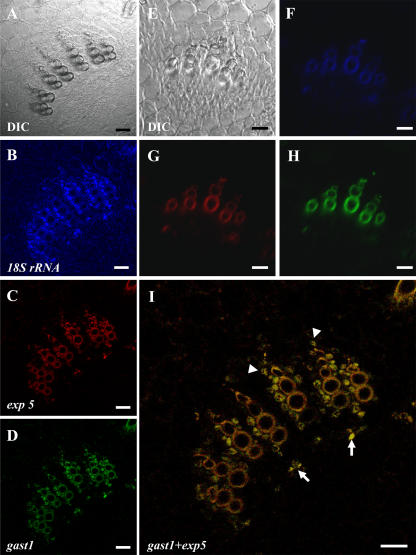
As demonstrated above, in addition to identifying genes with a strict auxin/cytokinin requirement for expression, the LXL also contained many genes that were induced by either auxin or cytokinin alone. To address the biological significance of this finding with regards to TE differentiation, i.e. to determine if genes falling into these categories could also be an integral part of the TE genetic program, multiplex in situ RT-PCR was performed on cells cultivated for 65 h in TE-inductive medium to simultaneously localize two genes with different hormone expression profiles. Two genes encoding putative cell wall proteins from cluster C were selected for this purpose: gast1, induced by auxin alone, and exp5, induced by cytokinin alone (Fig. 10). Interestingly, 100% of the transcriptionally active TEs (assessed by positive 18S expression), expressed both gast1 and exp5, suggesting that both these genes are integral parts of the genetic TE program. These results suggest that genes that do not exhibit a strict auxin/cytokinin requirement may be as essential for TE formation as genes that do. As for the non-TE population, the situation was far more complex. Four transcriptionally active (positive 18S expression) cell types, each characterized by a different expression profile, could be distinguished: cells that expressed both gast1 and exp5 (50.2%), cells that expressed only gast1 (9.3%), cells that expressed uniquely exp5 (1.3%), and cells that expressed 18S but did not express either gast1 or exp5 (39.3%; Fig. 10).
Figure 10.
Multiplex IS-RT-PCR of 18S rRNA, gast1, and exp5 in 65-h TE cultures. A, E, I, M, and Q, Differential interference contrast (DIC) image; B, F, J, N, and R, 18S rRNA expression; C, G, K, O, and S, gast1 expression; D, H, L, P, and T, exp5 expression. A to D, TE expressing 18S/gast1/exp5. E to H, Non-TEs expressing 18S/gast1/exp5. I to L, non-TEs expressing 18S/gast1 and not exp5. M to P, Non-TEs expressing 18S/exp5 but not gast1 (indicated by arrows in M). Q to T, Non-TEs expressing uniquely 18S. Magnification bars = 10 μm.
To investigate the spatial expression of gast1 and exp5 in planta, multiplex IS-RT-PCR was performed in 3-week-old epicotyl sections of zinnia (Fig. 11). Both gast1 and exp5 were exclusively expressed in vascular bundles, whereas 18S was localized in all cell types throughout the section. gast1 and exp5 colocalized in cambium cells directly above each vessel strand and in xylem parenchyma cells immediately surrounding the vessels (Fig. 11I). Contrary to observations made in in vitro TE cultures, cells that expressed uniquely gast1 or exp5 were not detected.
Figure 11.
Multiplex IS-RT-PCR of 18S rRNA, gast1, and exp5 in zinnia epicotyl cross sections. A close up of a vascular bundle is shown. A, DIC image; B, 18S expression; C, exp5 expression; D, gast1 expression. I, merged image of exp5 and gast1 from C and D. In I, the arrows indicate the colocalization of gast1/exp5 in cambium cells situated above vessel strands and arrowheads indicate the colocalization of gast1/exp5 in xylem parenchyma cells surrounding the vessels. A control IS-RT-PCR experiment in which PCR was performed but the RT step was omitted is indicated in E to H. E, DIC image; F, PCR with 18S primers; G, PCR with exp5 primers; H, PCR with gast1 primers. Note the background autofluorescence of dead xylem vessel cell walls in F to H. Magnification bars = 20 μm.
DISCUSSION
The formation of xylem involves several fundamental cellular processes including cell division, intercellular signaling, cell elongation, cell wall synthesis and deposition (cellulose and lignin), and vacuole-mediated cell death. In order to accomplish these functions in a coordinated manner, the expression of several hundred or perhaps thousands of genes is required. Significantly, headway in elucidating the underlying molecular mechanisms controlling xylem formation has been made by creating EST collections from woody xylem tissue in pine (Allona et al., 1998, and recent GenBank entries) and poplar (Sterky et al., 1998, and recent GenBank entries). More recently, large numbers of ESTs have been generated from the zinnia TE system (Demura et al., 2002; Milioni et al., 2002). Although there are actually 17,622 TE-related zinnia accessions deposited in the public databases, we have shown here that by the judicious use of subtractive suppression hybridization as a complementary technique to obtaining genomic information, we were able to significantly enrich our knowledge of TE formation. Indeed, 77% of our sequences are novel, illustrating that our knowledge of TE formation was far from complete before our contribution and most likely still is. This underlines the necessity of using complementary genomic strategies of a given model system, especially for a system as powerful as zinnia, to obtain a comprehensive picture of a given physiological process.
Cell Wall-Related Genes
When considering the formation of TEs, one would predict that the LXL would contain secondary wall biosynthetic enzymes, structural proteins, enzymes involved in remodeling wall architecture, and hydrolytic enzymes. In terms of wall-synthetic enzymes, no genes for polysaccharide synthesis were identified. This was unexpected since cellulosic secondary walls are deposited in zinnia cultures between 48 and 60 h (Fig. 1). Moreover, Arabidopsis mutants, irx3 (AtCesA7), irx1 (AtCesA8), and irx5 (AtCesA4) affected in SCW-specific cellulose synthases have been identified (Taylor et al., 1999, 2000, 2003). That said, it is interesting to note that out of the 17,622 zinnia EST sequences, only six cellulose synthases have been identified. Two ESTs (AJ409101 and AJ409099) encoding putative cellulose synthases have been reported to be specifically induced 30 min after auxin and cytokinin induction (Milioni et al., 2002), and out of the 9,131 microarray expression profiles provided by Demura et al. (2002), only one cellulose synthase, ZeCesA-1, was induced 20-fold at 60 h compared to 0 h. As for lignin biosynthetic genes, several genes were identified, including C4H, and three CCoAOMTs. These genes are members of cluster B, C, and D and therefore seem to be expressed during TE lifespan. However, a closer examination of ccoaomt2 (Fig. 9) and other lignin-associated genes (data not shown) using RT-PCR indicated that these genes were expressed beyond TE death. These results suggest that non-TE cells could also participate in TE lignification by producing monolignol precursors. This finding supports the theory of cell cooperation in lignin biosynthesis, even in in vitro cultures (Hosokawa et al., 2001). Two gene candidates for lignin polymerization were also identified: a laccase and a prx. The prx gene DV017586 had a very narrow peak of expression occurring at the onset of lignification. In an independent study, several laccase genes were also expressed at the onset of secondary wall deposition (Pesquet et al., 2001). To date, the nature of the enzyme(s) involved in this polymerization step is still unknown (Grima-Pettenati and Goffner, 1999).
Among the most highly represented gene families involved in wall remodeling in the LXL are the expansins. Im et al. (2000) identified three distinct expansins characterized by differential polarized mRNA transcript localization in zinnia xylem cells in planta. We report here an additional two expansins, which we have named exp4 (DV017234) and exp5 (DV017460). Recently, Gray-Mitsumune et al. (2004) identified several expansin genes associated with xylem formation in poplar. Interestingly, the exp4 and exp5 genes identified herein have the highest degree of homology with PttEXP1, the most highly expressed expansin in the cambial region. On the contrary, previously identified ZeExp1 and ZeExp2/3 are most homologous to PttExp2 and PttExp3, respectively, which are most highly expressed in developing poplar leaves. Herein, we localized exp5 in TEs, pointing to a putative role of the expansin gene family and in particular exp5 in cell expansion prior to TE formation.
The degradation of primary wall components by hydrolytic enzymes is an integral part of TE formation. Various enzymatic activities have been detected in differentiating zinnia cultures, pectins being one of the major substrates (Stacey et al., 1995; Ohdaira et al., 2002). A pectate lyase gene has been previously isolated (Domingo et al., 1998) but, by itself, could not explain the total pectolytic activity in TE cultures (Ohdaira et al., 2002). The LXL library provides two new pectin-related genes, an polygalacturonase (pg, DV017558) and a pectin methyl esterase (DV017467) as potential actors involved in TE pectin degradation. Recently, another novel polygalacturonase, ZePG1, was detected in TE SCW in addition to primary walls of non-TEs (Nakashima et al., 2004). All together, these data suggest that a multienzymatic pectolytic mechanism, including both TEs and non-TEs, may function in a cooperative manner to ensure TE pectin degradation.
Gene Regulation by Auxin and Cytokinin: A First Step in Unraveling Cross Talk during Differentiation
It is well established that auxin and cytokinin are required for vascular development. In the zinnia TE system, if one of the two hormones is omitted, mesophylls cells will not differentiate into TEs. In earlier studies, Church and Galston (1988a, 1988b) showed that TE differentiation could be induced by several different molecular forms of auxin and cytokinin and that an initial 24-h exposure to cytokinin and a 56-h exposure to auxin were sufficient for TE differentiation at 72 h, reflecting the complex relationship between hormones and TE differentiation. More recently, Milioni et al. (2001) have shown that a 10-min exposure to both hormones at 48 h is sufficient to induce TE differentiation, indicating that downstream events are independent of exogenous auxin and cytokinin. Using a cDNA-amplified fragment-length polymorphism PCR approach, these authors identified several genes exhibiting significant homologies with known auxin response genes including members of the indole-3-acetic acid/auxin and GH3 families. In addition, Demura et al. (2002) also identified actors involved in auxin signal transduction pathways including nine auxin response factors (one of which is a monopteros homolog) and five auxin response genes during TE differentiation. The importance of hormones on in vitro TE differentiation is not restricted to the zinnia system. They are also essential for the formation of TEs in Pinus radiata callus cultures (Moller et al., 2003) and Tcyt tobacco (Nicotiana tabacum) cell cultures (Blee et al., 2001), in which differentiation occurs due to abnormally high endogenous cytokinin levels resulting from Agrobacterium Tcyt transgene expression.
As a first step in dissecting auxin and cytokinin function during differentiation, we analyzed hormone response of LXL genes, not only in the presence of both hormones together, but also in cells cultured in media containing each hormone independently. Indeed, many genes were induced by either hormone alone. In this study, the large majority were up-regulated by auxin alone (38%) as compared to cytokinin alone (4%). This is not the first report demonstrating that genes induced during TE differentiation may also be induced, albeit to a lesser extent, by either auxin and/or cytokinin individually (Ye and Varner, 1993; Milioni et al., 2002). However, to our knowledge, the significance of this finding has never been addressed in any detail. These results stimulated us to ask the question as to whether these genes could also be part of the TE differentiation program. To address this issue, we performed multiplex IS-RT-PCR on a gene up-regulated in the presence of auxin alone (gast1) together with a gene up-regulated in the presence of cytokinin alone (exp5). The fact that both are systematically expressed in all living TEs strongly suggests that these putative cell wall genes are indeed likely to play a role in some aspect of TE differentiation. This finding must be taken into account when interpreting genomic data from zinnia, since the natural tendency would be to consider as important, genes that exhibit a strict auxin/cytokinin requirement for expression.
One of the more interesting findings presented herein is the fact that non-TE cells with different expression profiles vis-à-vis gast1 and exp5 could be distinguished. These results suggest that cellular perception of auxin and cytokinin is not homogenous throughout the cell population. This is perhaps not surprising considering the heterogeneous nature of the population of cells used to initiate cultures (from first leaves of 2-week-old plants). Finally, from a qualitative point of view, temporal gene expression is altered when either one or both hormones is present in the culture medium. For example, during TE differentiation, CCoAOMT2 possesses two peaks of activity, the first one at 24 h and the second beginning at 57 h (Fig. 9). In the presence of cytokinin only, this gene is expressed within 24 h and strongly expressed throughout the time course. A similar example may be found in the comprehensive study by Demura et al. (2002). Z3895, a gene encoding a receptor-like kinase, exhibits two peaks of expression in inductive medium (the first at 24 h and the second at 48–60 h) and decreases thereafter; whereas in the presence of auxin, Z3865 is strongly induced early on and continues to be so throughout the time course. It is clear that the level of gene expression in the presence of auxin and cytokinin does not simply reflect the sum of each hormone independently. Together, these results reflect the highly complex, intricate relationship that exists in hormone cross talk and signaling pathways during TE formation.
Other Hormones and TE Differentiation
In addition to auxin and cytokinin, other hormones have been shown to be required for xylem formation (Fukuda, 1996). For example, in zinnia cultures, brassinosteroids (BRs) are required for xylogenesis as indicated by inhibitor treatment studies coupled to stage-specific marker gene expression data (Yamamoto et al., 1997). Subsequently, it was shown that BR content in both cells and culture medium increased prior to visible TE morphogenesis (Yamamoto et al., 2001). Interestingly, the production and secretion of BRs continued beyond TE cell death, leading the authors to suppose an auto/paracrine mechanism of BR perception.
Earlier physiological studies in planta have also pointed to the role of GAs in xylem formation (Aloni, 1987; Savidge, 1996). We have identified herein three different gast genes (GAST1, DV017152; GAST2, DV017224; and GAST3, DV017194), one of which, gast1, was localized in both TE and non-TE cells in vitro and in cambial cells and xylem parenchyma cells in planta. Interestingly, promoter-β-glucuronidase fusions performed with RSI-1 and GASA1, GAST homologs in tomato (Lycopersicon esculentum; Taylor and Scheuring, 1994) and Arabidopsis, respectively (Raventos et al., 2000), exhibited vascular-specific expression. All together, these results suggest that GA may be a downstream signaling component of TE formation. Moreover, modulation of GEG expression, a GAST homolog in Gerbera, led to a reduction in carpel cell length implying a direct relationship between GAST and cell expansion (Kotilainen et al., 1999). Recently, the relationship between different forms of GA and lignification has been demonstrated (Biemelt et al., 2004). The overexpression of GA 20-oxidase in transgenic tobacco led to significantly greater amounts of xylem.
The role of ethylene in xylem formation has also been suggested by physiological studies (Aloni, 1987; Savidge, 1996). Herein, we have identified a gene encoding a biosynthetic enzyme of ethylene, 1-amino-cyclopropane-1-carboxylic acid synthase (DV017373). The genomic information presented herein supports the idea that both GA and ethylene hormones may also be signaling intermediates required for TE determination/differentiation. The ease with which the zinnia system may be manipulated enables the testing of such hypotheses and opens new prospects to elucidating hormone signaling during TE formation.
How Many Different Cell Types Are in Zinnia TE Cultures?
Up until now, zinnia cultures are usually described as having TEs and non-TEs. This is based on readily observable, gross morphological criteria. With the recent advent of a multiplex IS-RT-PCR protocol that can be performed in tissues and heterogeneous cell populations (Pesquet et al., 2004), we are now able to distinguish among different cell types based on gene expression at the cellular level. In this study, the strict colocalization of the gene couple, gast1/exp5, in all transcriptionally active TEs did not allow us to discriminate different TE cell types at the gene expression level. However, on the other hand, the analysis of non-TE cells in TE-inductive medium indicated that, by using only one gene pair, we were able to identify at least four different expression-based cell types: cells expressing both gast1 and exp5, cells expressing uniquely gast1, cells expressing uniquely exp5, and cells expressing neither gast1 nor exp5. Although it is tempting to speculate that these cells also participate in TE differentiation, functional proof must be provided to confirm this hypothesis.
From a functional standpoint, a number of studies suggest that proper TE formation is dependant on cell cooperation events between TE and non-TE cells not only in planta but also in in vitro xylogenic cultures of zinnia. These events include TE lignification (Hosokawa et al., 2001) and TED4-mediated survival of non-TE cells after TE PCD (Endo et al., 2001). One assumption that has been made is that in planta xylem parenchyma cells may have functional analogs in in vitro cultures (Motose et al., 2001; Fukuda, 2004). Based on comparative localization of gast1 and exp5 gene expression in planta and in vitro, we provide an additional argument in favor of the presence of xylem parenchyma equivalents in vitro by associating gene expression of gast1/exp5-expressing TEs in vitro with cambium cells in planta, and a subgroup of gast1/exp5-expressing non-TEs in vitro with xylem parenchyma cells in planta. In addition, the localization of gast1 and exp5 in planta was restricted to cambial cells immediately above vessel strands, implying that the cambium is a heterogeneous cell layer composed of cells characterized by different gene expression patterns, despite the lack of any major morphological differences among the cells (Pesquet et al., 2003). In conclusion, we show that the judicious combination of macroarrays and multiplex IS-RT-PCR on plant cells and organs opens new prospects in the quest to associate gene expression and cell fate at the single cell level in the context of xylem differentiation.
MATERIALS AND METHODS
Plant Material and Xylogenic Cell Cultures
The first pair of leaves from 14-d-old seedlings of Zinnia elegans cv Envy (Hem Zaden BV) were used to isolate mesophyll cells for xylogenic cell suspension cultures according to the method of Fukuda and Komamine (1980). Cells were cultured in induction medium (CA + C) containing 0.1 mg/L alpha-naphtylacetic acid and 0.2 mg/L benzyladenine (Sigma-Aldrich). Cells were also cultured in control media without hormones (C0), or in the presence of either auxin (CA) or cytokinin (CC ) as indicated.
SCW and Viability Staining of TE Cultures
To simultaneously detect cellulose and lignin in SCWs of developing TEs, 100 μL of cell culture was first stained with 10 μL of 0.01% calcofluor (fluorescent brightener 28; Sigma-Aldrich), and then, after 30 s, 10 μL of 0.001% solution of auramine-O (Sigma-Aldrich) was added.
For viability staining, 100 μL of cell culture was stained with 10 μL of 0.01% solution of calcofluor and then with 2 μL of 0.5% FDA (Sigma-Aldrich) solubilized in acetone. Cells were mounted on glass slides and observed using an inverted microscope (DMIRBE, Leica) with bright-field optics or epifluorescence illumination.
For FDA and auramine-O staining, cells were observed in the blue excitation range (excitation filter BP 450–490 nm, suppression filter LP 515 nm) and calcofluor in the UV excitation range (excitation filter BP 270–380 nm, emission filter BP 410–580 nm). Image acquisition was performed using a CCD camera (Color Coolview, Photonic Science).
RNA Extraction
Cell suspensions were harvested by centrifugation for 5 min at 150g. Culture medium was removed with a Pasteur pipette and 1 mL of Extract-All solution (Eurobio) was added to the cell pellet. Cells resuspended in Extract-All solution were then frozen in liquid N2 and conserved at −80°C for further use. Total RNA was isolated according to the manufacturer's instructions and subjected to DNA digestion with 5 units of ribonuclease-free deoxyribonuclease I (Promega) for 1 h at 37°C. A second round of RNA extraction was carried out as indicated above. RNA was quantified using an RNA Biophotometer (Eppendorf), and visualized after electrophoresis on 1.5% agarose gels. The lack of DNA contamination was confirmed by performing PCR on RNA with 18S rRNA PCR primers. Under these conditions, a band was not detected.
Construction of an LXL by SSH
The LXL was constructed using a SMART-PCR cDNA synthesis kit (CLONTECH) for cDNA synthesis and a PCR-Select cDNA subtraction kit (CLONTECH) for the subtraction step. First-strand cDNAs were prepared from 1 μg of pooled total RNA from zinnia cell suspensions cultured for 48, 60, and 72 h (approximately 350 ng each for tester population) and pooled total RNA from control cultures at the same time points in the absence of hormone or in the presence of one hormone only in the same way (approximately 150 ng each for driver population). RNA from freshly isolated mesophyll cells was also included in controls (driver population). Double-stranded cDNA were obtained by PCR according to the manufacturer's instructions. cDNAs, before and after digestion with RsaI, were purified on Microcon PCR columns (Amicon, Millipore). In two separate ligation reactions, tester cDNA was ligated to adapters 1 and 2. In the first hybridization, an excess of driver cDNA was hybridized at 68°C for 8 h with tester cDNA ligated to both adapters. In the second hybridization, ligation reactions 1 and 2 were hybridized together in the presence of fresh driver cDNA at 68°C overnight. The subtracted fragments were amplified by PCR using oligonucleotides complementary to adapters 1 and 2. PCR was performed according to the following parameters: 75°C for 5 min and 27 cycles at 94°C for 30 s, 66°C for 30 s, and 72°C for 1.5 min. A nested PCR was then performed as follows: 12 cycles at 94°C for 30 s, 66°C for 30 s, and 72°C for 1.5 min. The resulting PCR products corresponded to the gene population specific/up-regulated in TE cultures. PCR products were cloned into a pGEM-T vector (Promega). Colonies were grown on selective medium containing Luria-Bertani agar, 100 μg/mL ampicillin (Eurobio), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; Promega), and isopropyl-β-d-thiogalactopyranoside (Promega). A total of 800 recombinant bacterial clones were plated out, ordered into 96-well microplates, and stored at −80°C in 35% glycerol.
Screening of the Subtractive Clones by Reverse Northern-Blot Analysis
All 800 clones were transferred from 96-well microplates using a 96-steel tip microplate replicator onto Luria-Bertani agar plates containing 100 μg/mL ampicillin and incubated at 37°C overnight. Colonies were transferred using the 96-steel tip microplate replicator comb into 96-well PCR plates containing 5.9 μL of ultrapure water per well. Each PCR reaction contained 1.25 μL of 10× PCR buffer (Promega), 1.25 μL of 25 mm MgCl2, and 4 μL of dNTP (200 μm each), 1 μL of nested primer 1 and nested primer 2R (5 μm), and 0.1 μL of 5 units/μL Taq DNA Polymerase (Promega). PCR was performed according to the following parameters: 95°C for 30 s and 30 cycles at 95°C for 10 s, 65°C for 10 s, and 72°C for 2 min. Five microliters of PCR products were analyzed by electrophoresis on 2% agarose gels. Agarose gels were blotted onto a Hybond N+ nylon membrane (Amersham Pharmacia) by direct capillary transfer in a 0.8 m NaOH/3 m NaCl solution. Membranes were then washed in 100 mm Tris-HCl/150 mm NaCl solution and UV crosslinked 1,200 × 100 μJ/cm2 in a UV crosslinker (Amersham).
For cDNA probe synthesis, 1 μL of SMART double-stranded cDNA from either the tester and driver sample were labeled with [α-32P]dCTP (Amersham Pharmacia) using the Prime-A-Gene labeling kit (Promega) according to the manufacturer's instructions. Probes were purified on G50 Sephadex columns, and the first eluted peak was quantified by Cerenkov effect using a liquid scintillation analyzer Tri-Carb 1900-TR (Canberra-Packard). Membranes were prehybridized at 65°C for 2 h in 20 mL of 3× SSC containing 0.2% low-fat milk and 0.5% SDS. Duplicate membranes were then hybridized separately (one with the tester probe and the other with the driver) with the same amount of cpm of denatured 32P-labeled probes in 5 mL of fresh hybridization solution overnight at 65°C. Membranes were washed twice for 15 min in 3× SSC/0.5% SDS, once in 1× SSC/0.5% SDS, and once in 0.3× SSC/0.5% SDS at 65°C. The membranes were then exposed to BioMax MR x-ray film (Kodak) at −80°C overnight.
LXL Database Construction
All sequence chromatograms were transformed using chromas software (http://www.technelysium.com.au/) batch export in a fasta format database. Nucleotide and protein searches were performed using BLASTZ pack (ftp://ftp.ncbi.nih.gov/) on downloaded databases: NR (nonredundant total protein sequences: 907,641 sequences), FAA (Arabidopsis [Arabidopsis thaliana] protein sequences: 25,545 sequences), FNA (Arabidopsis genome: five sequences), poplar EST (Populus balsamifera subsp. Trichocarpa; EST sequences: 272,213 sequences), pine EST (Pinus taeda; EST sequences: 209,733 sequences), plant DNA (nonredundant total transcript sequences: 983,935 sequences), and zinnia EST (zinnia EST sequences: 17,622 sequences). Elimination of pGEM-T vector sequences was carried out by BLASTN analysis on raw sequence databases with linker 1 and 2R sequences flanking SSH fragments. SSH fragment sizes were estimated by electrophoresis on agarose gels. Sequence clusterization was performed using blastclust analysis and resulted in 238 clusters. Sequence annotation was made using BLASTX and tBLASTX analysis on NR protein and Plant DNA databases respectively. The closest Arabidopsis orthologs were searched using BLASTX analysis on Arabidopsis protein database FAA and tBLASTX analysis on pine EST and poplar EST databases to search for woody xylem homologs. A functional classification for each gene was determined according to the MIPS protein class criteria (http://mips.gsf.de/). Results were centralized on a spreadsheet using a Microsoft Excel program (Supplemental Data 1). Sequences determined were deposited in dbEST, GenBank, National Center for Biotechnology Information (accession nos.DV017146–DV017591).
Macroarray Construction
SSH cDNA fragments were PCR amplified (95°C for 30 s and 40 cycles at 95°C for 10 s, 65°C for 10 s and 72°C for 2 min) in 96-well PCR plates in a total volume of 100 μL per well consisting of 10 μL of 10× PCR buffer (Promega), 10 μL of 25 mm MgCl2, and 40 μL of dNTP (200 μm each), 4 μL of nested primer 1 and nested primer 2R (10 μm), 1 μL of 5 units/μL Taq DNA Polymerase (Promega), and 35 μL of ultrapure water. PCR products were precipitated in isopropanol overnight at −20°C. PCR plates were then centrifuged for 30 min at 4,000 rpm, excess medium was removed, and pellets were washed twice in 70% ethanol. Pellets were vacuum dried and resuspended in 40 μL of TE (pH 8.0). Two microliters of each PCR product were confirmed for the presence of a single band, quantified by electrophoresis on 2% agarose gels, and diluted to a final concentration of 1 μg/μL. PCR products were then denatured in 50% dimethyl sulfoxide and transferred to 384-well plates. Controls were added in a separate 384-well plate including 30 NPT II fragments (positive hybridization control), eight pUC19:35S-β-glucuronidase-Tnos (unspecific hybridization control), and 32 TE, pH 8.0 (blank background control). Fragments were then spotted onto a 20 × 20 cm Nytran SuPerCharge nylon membrane (Schleicher and Schuell) using a BioGrid spotting robot (BioRobotics) in a 4 × 4 grid organization (duplicates for every spot). Resulting macroarrays were UV crosslinked at 1,200 × 100 μJ/cm2 before use.
Macroarray cDNA Probe Synthesis
First-strand cDNAs were synthesized from 5 μg of total RNA per time point or hormonal condition. Total RNA and 1 μL of 500 ng/μL of an NNVdT(20) primer (anchored oligodT primer) adjusted to 8 μL with ultrapure water were denatured for 10 min at 70°C and cooled down on ice for 5 min. Four microliters of MuMLV-RT reverse transcriptase 5× buffer (Promega), 2 μL of 5 mm dNTP (except for dCTP), 40 μCi of [α-33P]dCTP (Amersham Pharmacia), and 400 units of MuMLV-RT reverse transcriptase (Promega) were added to the denatured RNA and incubated at 37°C for 1 h. Two hundred units of MuMLV-RT reverse transcriptase were then again added to the labeling mixture for 30 min. RNA was hydrolyzed with 1 μL of 1% SDS, 1 μL of 0.5 m EDTA, pH 8.0, and 3 μL of 3 m NaOH for 30 min at 65°C and then left for 15 min at room temperature. Probes were neutralized with 10 μL of 1 m Tris-HCl, pH 8.0, and 3 μL of 2 n HCl and quantified with a liquid scintillation analyzer to determine total amount of initial cpm. cDNAs were precipitated by adding 5 μL of 3 m sodium acetate, pH 5.3, 2 μL of mussel glycogen (Sigma-Aldrich) and 60 μL isopropanol, and incubated for at least 1 h at −20°C. After centrifugation at 13,000 rpm and two washes in 70% ethanol, the amount of incorporated cpm was quantified. Labeling efficiency was determined by the ratio of probe-incorporated cpm/total initial amount of cpm (typically around 10%). The specific activity of all probes was adjusted to equal amounts of cpm/μL with ultrapure water, supplemented with 0.1 per thousand of individually labeled cDNA corresponding to the NPT II gene as a positive hybridization control, and incubated 5 min at 65°C. Probes were denatured at 100°C for 5 min and placed on ice for 5 min prior to hybridization.
Membrane Hybridization
Prior to all hybridization experiments, membranes were incubated twice for 15 min at 99°C in 0.1% SDS to minimize unspecific binding. Membranes were prehybridized for at least 3 h in 3× SSC/0.5% SDS/0.2% low-fat milk/20% polyethylene glycol 6000 at 65°C in a rotating incubator. Membranes were then hybridized with cDNA probes of identical specific activity in 2 mL of fresh hybridization solution and incubated at 65°C overnight in a rotating incubator. Membranes were then washed twice for 15 min in 3× SSC/0.5% SDS, and once for 15 min in 1× SSC/0.5% SDS, wrapped in cellophane, and placed in a PhosphoImager cassette (Molecular Dynamics, Amersham Pharmacia) for 72 h. Image scanning was performed at 50 μm/pixel by a Strom 820 scanner (Amersham Pharmacia).
Macroarray Data Analysis
Macroarray gridding and gene expression levels were measured with ImageQuant 5.0 software (Molecular Dynamics, Amersham Pharmacia) using 4 × 4 grids. Gridding was performed manually and tested in regards to the distance of the maximal value to the centroid of the measured area on the x and y axes. Expression data for all gene sequences were analyzed using Microsoft Excel. Normalization between samples was established using the linear slope defined by the blank background, unspecific hybridization, and positive hybridization controls (Tris-EDTA, pH 8, pUC 19, and NPT II, respectively) in the different samples. A total of 16 LXL membranes were hybridized with probes from the differentiation time course (0, 24, 48, 72, and 96 h) and under different hormonal conditions (C0, CA, and CA + C), and the slope defined by the controls indicated a linear correlation factor of R2 = 1. The significance threshold was determined by the analysis of the log10 duplicate ratio for each gene under all hybridizing conditions as shown in Figure 6A. Reproducibility of hybridization was estimated by comparing raw signal intensity values from duplicate experiments. For these experiments, cDNA was synthesized from two independent cultures and hybridized to two different membranes. A coefficient of determination was calculated between duplicate experiments. When six sets of duplicate membranes were hybridized with independent probes for each time point during differentiation, a variation ranging from R2 = 0.944 to 0.989 was observed. The average signal value from the duplicate set and sds were calculated for each gene. The genes whose sds exceed their average signal values were eliminated from the Microsoft Excel spreadsheet gene list. Expression data was then transformed from raw intensity values to percentage of expression by dividing the values for each gene by its maximal expression either during the TE differentiation time course or during hormonal treatment. Genes that were localized to different positions on the array due to the LXL redundancy were averaged together. All redundant genes showed similar expression profiles during the differentiation time course and hormonal treatments. Genes that showed greater than a 2-fold difference in average signal values for each comparison were defined as differentially expressed genes. Gene clustering was performed according to their expression level using HCE2 software (http://www.cs.umd.edu/hcil/multi-cluster/).
RT-PCR
Total RNA from cells harvested at different time points during the culture period was isolated as described above. cDNA synthesis and RT-PCR was performed according to Pesquet et al. (2004). The PCR primer combinations for each gene were as follows: opt (DV017591), reverse 5′-GTTCACAACACTCGTTTCCG-3′, forward 5′-CTGGGTGTTTGTAAAAACCC-3′; UG-10 (DV017178), reverse 5′-AGAACATTTGCCAAAGGTC-3′, forward 5′-CACCCTTACATTTCCATCAC-3′; 18S rRNA (AB089282), reverse 5′-TGTCACTACCTCCCCGTGTC-3′, forward 5′-TGCTACTCGGATAACCGTAG-3′; endonuclease (DV017394), reverse 5′-CTTGAGCTCCCGAAAACCCG-3′, forward 5′-CATCAGCCGATGCATGTTGG-3′; endopolygalacturonase (DV017558), reverse 5′-ACTCCATCTCCGGCTGCCCC-3′, forward 5′-CCGTTCTTGTAATCTTGCTA-3′; Cys protease (DV017552), reverse 5′-CACATGTGCCTTCACTCATG-3′, forward 5′GGCGAGCTCGCTGAGAGGAA-3′; Rbn1 (DV017482), reverse 5′-CAATCCGTCGCCACTTGGGC-3′, forward 5′-ACATGGAAGCACATTGCTAG-3′; prx (DV017586), reverse 5′-CTCTATTTCACCATTTTCCGCC-3′, forward 5′-GATCTCTAGCGGCGATGGCT-3′; sp (DV017578), reverse 5′-TGAGACCTAATTGACAGTAT-3′, forward 5′-CTGGGTTGGTTGTGAAGGTT-3′; GAST1 (DV017152), reverse 5′-GTAACAGTTAGACCCTTCAAC-3′, forward 5′-GTTGCAGAGCAGGAGAATGGC-3′; GAST2 (DV017224), reverse 5′-TCATAAACACCACGCGCGAG-3′, forward 5′-CAATGAAGCCAATTGTTGCAA-3′; exp4 (DV017234), reverse 5′-CATGTTAGTTGTGACTCGTG-3′, forward 5′-CAGGAAGTGATCGCCGCACT-3′; exp5 (DV017460), reverse 5′-CGGCCCTTTTTAAATCAAGCC-3′, forward 5′-GTGGTCCCAGCCGGTTGGTC-3′; UG-25 (DV017176), reverse 5′-ATCGTTGTTGAAGATGGCGG-3′, forward 5′-TCACCAGCTTCATGTGAACC-3′; UG-7 (DV017396), reverse 5′-CATGGCACAATGTATCTCCC-3′, forward 5′-CGCGCCCAAGTGTCTATGTG-3′; calrec (DV017386), reverse 5′-GCAATTAAGACGGGGGGCAG-3′, forward 5′-CGTCTTGATATGCGATGACC-3′; TK (DV017557), reverse 5′-TGATCATCCAGCTGGGATTG-3′, forward 5′-CTGATCCAAGCTAGGGTTCA-3′; and CCoAOMT2 (DV017250), reverse 5′-GAAATCTCTGTAATAACGGA-3′, forward 5′-ATCAAATGATCGCTGATGGA-3′. All primers were synthesized by Sigma-Genosys.
Multiplex IS-RT-PCR
Multiplex IS-RT-PCR was performed according to the protocol described by Pesquet et al. (2004) on cells cultured in TE-inductive medium for 65 h and hand-made epicotyl cross sections of 3-week-old zinnia plants. PCR was carried out with 5′-labeled fluorescent-forward PCR primers, for gast1, 5′-[fluorescein isothiocyanate]-TACAGTTTGGTTACGCCGGT-3′; exp5, 5′-[TMRA]-TACAGTTTGGTTACGCCGGT-3′; and 18S, 5′-[Cy-5]-TGCTACTCGGATAACCGTAG-3′, and unlabeled reverse primers, for gast1, 5′-GTAACAGTTAGACCCTTCAAC-3′; exp5, 5′-CGGCCCTTTTTAAATCAAGCC-3′; and 18S, 5′-TGTCACTACCTCCCCGTGTC-3′. Fluorescein-, TMRA-, and Cy5-labeled samples were observed using the 488-nm, 543-nm, and 633-nm ray line of a HeNe laser, respectively. The emitted light was collected between 500 to 530 nm (fluorescein isothiocyanate), 550 to 580 nm (TMRA), and 650 to 680 nm (Cy5). To determine the spectral range to be used for image acquisition, emission spectra were collected for each fluorochrome using the lambda scan module of the SP2 confocal microscope. Quantification of gast1 and exp5 gene expression (expressed as a percentage for transcriptionally active, 18S-expressing cells) was carried out on a total of 321 cells (62 TEs and 259 non-TEs).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DV017146 to DV017591.
Supplementary Material
Figure 8.
Hierarchical clustering as a function of hormonal expression of LXL cDNAs belonging to temporal expression cluster A. A, The green-to-red color scale indicates the relative expression (with 1 = value of maximal intensity). B, Average expression profile of each hormonal cluster group. The x axis = hormonal treatment, and the y axis = relative average expression intensity.
Acknowledgments
The authors wish to thank N. Ladouce and V. Le Berre for help in spotting macroarrays, the Genopôle de Toulouse for spotting facilities, and F. Legeai and D. Samson of Génoplante Info for help in bioinformatics analysis.
This work was supported by the Centre National de la Recherche Scientifique, the Université Paul Sabatier, and the Génoplante program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Deborah Goffner (goffner@scsv.ups-tlse.fr).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.064337.
References
- Allona I, Quinn M, Shoop E, Swope K, St Cyr S, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, et al (1998) Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA 95: 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R (1987) Differentiation of vascular tissues. Annu Rev Plant Physiol 38: 179–204 [Google Scholar]
- Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blee KA, Wheatley ER, Bonham VA, Mitchell GP, Robertson D, Slabas AR, Burrell MM, Wojtaszek P, Bolwell GP (2001) Proteomic analysis reveals a novel set of cell wall proteins in a transformed tobacco cell culture that synthesizes secondary walls as determined by biochemical and morphological parameters. Planta 212: 404–415 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Carland FM, Berg BL, Fitzgerald JN, Jinamornphongs S, Nelson T, Keith B (1999) Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 11: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Chilley PM, Topping JF, Evans IM, Souter MA, Lindsey K (2002) The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14: 1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S, Ndimba BK, Simon WJ, Robertson D, Yu XL, Knox JP, Bolwell P, Slabas AR (2002) Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 23: 1754–1765 [DOI] [PubMed] [Google Scholar]
- Church DL, Galston AW (1988. a) Kinetics of determination in the differentiation of isolated mesophyll cells of Zinnia elegans to tracheary elements. Plant Physiol 88: 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Galston AW (1988. b) Hormonal induction and antihormonal inhibition of tracheary element differentiation in Zinnia cell cultures. Phytochemistry 27: 2435–2439 [DOI] [PubMed] [Google Scholar]
- Demura T, Fukuda H (1993) Molecular cloning and characterization of cDNAs associated with tracheary element differentiation in cultured Zinnia cells. Plant Physiol 103: 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Fukuda H (1994) Novel vascular cell-specific genes whose expression is regulated temporally and spatially during vascular system development. Plant Cell 6: 967–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y, et al (2002) Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll into xylem cells. Proc Natl Acad Sci USA 99: 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Cordner G, Beebe D, Sieburth LE (2000) The SCARFACE gene is required for cotyledon and leaf vein patterning. Development 127: 3205–3213 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93: 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Roberts K, Stacey NJ, Connerton I, Ruíz-Teran F, McCann MC (1998) A pectate lyase from Zinnia elegans is auxin inducible. Plant J 13: 17–28 [DOI] [PubMed] [Google Scholar]
- Endo S, Demura T, Fukuda H (2001) Inhibition of proteasome activity by the TED4 protein in extracellular space: a novel mechanism for protection of living cells from injury caused by dying cells. Plant Cell Physiol 42: 9–19 [DOI] [PubMed] [Google Scholar]
- Fukuda H (1996) Xylogenesis: initiation, progression and cell death. Annu Rev Plant Physiol Plant Mol Biol 47: 299–325 [DOI] [PubMed] [Google Scholar]
- Fukuda H (2004) Signals that control plant vascular cell differentiation. Nat Rev Mol Cell Biol 5: 379–391 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Komamine A (1980) Establishment of an experimental system for the tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol 52: 57–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzell A, Sterky F, Blomqvist K, McQueen-Mason S, Teeri TT, Sundberg B (2004) Expansins abundant in secondary xylem belong to subgroup A of the alpha-expansin gene family. Plant Physiol 135: 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima-Pettenati J, Goffner D (1999) Lignin genetic engineering revisited. Plant Sci 145: 51–65 [Google Scholar]
- Groover A, Jones AM (1999) Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol 119: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, Bandyopadhyay A, Estelle M (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Suzuki S, Umezawa T, Sato Y (2001) Progress of lignification mediated by intercellular transportation of monolignols during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol 42: 959–68 [DOI] [PubMed] [Google Scholar]
- Im KH, Cosgrove DJ, Jones AM (2000) Subcellular localization of expansin mRNA in xylem cells. Plant Physiol 123: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ (2001) Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Sugiyama M, Fukuda H (2000) A series of novel mutants of Arabidopsis thaliana that are defective in the formation of continuous vascular network: calling the auxin signal flow canalization hypothesis into question. Development 127: 3197–3204 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH (1999) GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell 11: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama H (1999) Loss of tonoplast integrity programmed in tracheary element differentiation. Plant Physiol 121: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioni D, Sado PE, Stacey NJ, Domingo C, Roberts K, McCann MC (2001) Differential expression of cell-wall-related genes during the formation of tracheary elements in the Zinnia mesophyll cell system. Plant Mol Biol 47: 221–238 [PubMed] [Google Scholar]
- Milioni D, Sado PE, Stacey NJ, Roberts K, McCann MC (2002) Early gene expression associated with the commitment and differentiation of a plant tracheary element is revealed by cDNA-amplified fragment length polymorphism analysis. Plant Cell 14: 2813–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller R, McDonald AG, Walter C, Harris PJ (2003) Cell differentiation, secondary cell wall formation and transformation of callus tissue of Pinus radiata D. Don. Planta 217: 736–747 [DOI] [PubMed] [Google Scholar]
- Motose H, Fukuda H, Sugiyama M (2001) Involvement of local intercellular communication in the differentiation of zinnia mesophyll cells into tracheary elements. Planta 213: 121–31 [DOI] [PubMed] [Google Scholar]
- Nakashima J, Endo S, Fukuda H (2004) Immunocytochemical localization of polygalacturonase during tracheary element differentiation in Zinnia elegans. Planta 218: 729–739 [DOI] [PubMed] [Google Scholar]
- Oh S, Park S, Han K (2003) Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot 393: 2709–2722 [DOI] [PubMed] [Google Scholar]
- Ohdaira Y, Kakegawa K, Amino S, Sugiyama M, Fukuda H (2002) Activity of cell-wall degradation associated with differentiation of isolated mesophyll cells of Zinnia elegans into tracheary elements. Planta 215: 177–184 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Barbier O, Ranocha Ph, Jauneau A, Goffner D (2004) Multiple gene detection by in situ RT-PCR in isolated plant cells and tissues. Plant J 39: 947–959 [DOI] [PubMed] [Google Scholar]
- Pesquet E, Jauneau A, Digonnet C, Boudet AM, Pichon M, Goffner D (2003) Zinnia elegans: the missing link from in vitro tracheary elements to xylem. Physiol Plant 119: 463–468 [Google Scholar]
- Pesquet E, Pichon M, Pineau C, Ranocha Ph, Digonnet C, Jauneau A, Boudet AM, Fukuda H, Demura T, Goffner D (2001) Xylem formation and lignification in trees and model species. In N Morohoshi, A Komamine, eds, Molecular Breeding of Woody Plants. Elsevier Science Publishing, New York, pp 11–18
- Raventos D, Meier C, Mattsson O, Jensen AB, Mundy J (2000) Fusion genetic analysis of gibberellin signaling mutants. Plant J 22: 427–38 [DOI] [PubMed] [Google Scholar]
- Savidge RA (1996) Xylogenesis: genetic and environmental regulation. IAWA J 17: 269–310 [Google Scholar]
- Stacey NJ, Roberts K, Carpita NC, Wells B, McCann MC (1995) Dynamic changes in cell surface molecules are very early events in the differentiation of mesophyll cells from Zinnia elegans into tracheary elements. Plant J 8: 891–906 [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, et al (1998) Gene discovery in the wood forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA 95: 13330–13335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 30: 431–446 [DOI] [PubMed] [Google Scholar]
- Taylor BH, Scheuring CF (1994) A molecular marker for lateral root initiation: the Rsi-1 gene of tomato (Lycopersicon esculentum Mill.) is activated in early lateral root primordia. Mol Gen Genet 243: 148–157 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci USA 100: 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR (2000) Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S, Sieburth LE (2002) Vascular patterning. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0009, http://www.aspb.org/publications/arabidopsis
- Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J (2000) PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127: 5157–5165 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Demura T, Fukuda H (1997) Brassinosteroids induce entry into the final stage of tracheary element differentiation in cultured Zinnia cells. Plant Cell Physiol 38: 980–983 [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Fujioka S, Demura T, Takatsuto S, Yoshida S, Fukuda H (2001) Brassinosteroid levels increase drastically prior to morphogenesis of tracheary elements. Plant Physiol 125: 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH (2002) Vascular tissue differentiation and pattern formation in plants. Annu Rev Plant Biol 53: 183–202 [DOI] [PubMed] [Google Scholar]
- Ye ZH, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Varner JE (1993) Gene expression patterns associated with in vitro tracheary element formation in isolated single mesophyll cells of Zinnia elegans. Plant Physiol 103: 805–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Varner JE (1996) Induction of cysteine and serine proteases during xylogenesis in Zinnia elegans. Plant Mol Biol 30: 1233–1246 [DOI] [PubMed] [Google Scholar]
- Zhong R, Taylor JJ, Ye ZH (1999) Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol 120: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.