Abstract
Sugar-induced anthocyanin accumulation has been observed in many plant species. We observed that sucrose (Suc) is the most effective inducer of anthocyanin biosynthesis in Arabidopsis (Arabidopsis thaliana) seedlings. Other sugars and osmotic controls are either less effective or ineffective. Analysis of Suc-induced anthocyanin accumulation in 43 Arabidopsis accessions shows that considerable natural variation exists for this trait. The Cape Verde Islands (Cvi) accession essentially does not respond to Suc, whereas Landsberg erecta is an intermediate responder. The existing Landsberg erecta/Cvi recombinant inbred line population was used in a quantitative trait loci analysis for Suc-induced anthocyanin accumulation (SIAA). A total of four quantitative trait loci for SIAA were identified in this way. The locus with the largest contribution to the trait, SIAA1, was fine mapped and using a candidate gene approach, it was shown that the MYB75/PAP1 gene encodes SIAA1. Genetic complementation studies and analysis of a laboratory-generated knockout mutation in this gene confirmed this conclusion. Suc, in a concentration-dependent way, induces MYB75/PAP1 mRNA accumulation. Moreover, MYB75/PAP1 is essential for the Suc-mediated expression of the dihydroflavonol reductase gene. The SIAA1 locus in Cvi probably is a weak or loss-of-function MYB75/PAP1 allele. The C24 accession similarly shows a very weak response to Suc-induced anthocyanin accumulation encoded by the same locus. Sequence analysis showed that the Cvi and C24 accessions harbor mutations both inside and downstream of the DNA-binding domain of the MYB75/PAP1 protein, which most likely result in loss of activity.
Sugars have an essential role in general metabolism and energy generation. Moreover, in plants, sugars are essential units for the generation of structural elements. Importantly, sugars have a hormone-like signaling function as well and act as primary messengers in signal transduction processes that regulate many important processes in all phases of the plant life cycle (Smeekens, 2000; Rolland et al., 2002; Rook and Bevan, 2003). Sugar signaling regulates processes such as photosynthesis, nutrient mobilization, and allocation, and stimulates growth and storage of sink tissues (Koch, 1996; Rolland et al., 2002). Elevated sugar levels induce developmental arrest of postgerminated seedlings in Arabidopsis (Arabidopsis thaliana), and this has been one of the principles used to identify mutants in sugar signaling (Smeekens, 2000; Rolland et al., 2002; Rook and Bevan, 2003; Gibson, 2005). Sugars also have effects on floral transition and are involved in the control of leaf senescence (Bleecker and Patterson, 1997; Corbesier et al., 1998; Ohto et al., 2001). Many jasmonate, abscisic acid, stress-inducible, and pathogenesis-related genes are coregulated by sugars as well (Reinbothe et al., 1994; Sadka et al., 1994). Sugar signaling regulates gene expression at different levels, which include transcriptional and posttranscriptional processes (Koch, 1996). Previous research has highlighted the sugar-mediated regulation of mRNA transcription (Koch, 1996) and stability (Chan and Yu, 1998; Cheng et al., 1999), protein translation (Rook et al., 1998; Wiese et al., 2004), and protein stability (Yanagisawa et al., 2003).
Different sugar sensory mechanisms exist in plants that respond to different sugars. Moreover, different systems have been proposed for Glc sensing based on results obtained using various experimental approaches and systems (Smeekens, 2000; Rolland et al., 2002). In Arabidopsis, a hexokinase-dependent pathway was proposed to sense Glc via the AtHXK1 protein. Such an AtHXK1-dependent pathway regulates photosynthesis-related and other genes, including chlorophyll a/b-binding protein (CAB1), enhanced response to ABA (ERA1), plastocyanin (PC), phospholipase (PLD), and small subunit of Rubisco (Xiao et al., 2000). The AtHXK1 has separable catalytic and signaling functions and it was shown that downstream Glc metabolism is not required for hexokinase signaling (Moore et al., 2003). Next to this system, a separate, HXK-independent pathway regulates genes such as PR1, PR5, AGPase, CHS, PAL1, and AS1 (Smeekens, 2000; Rolland et al., 2002).
The presence in plants of a Suc-specific sensing pathway has been proposed as well, even though no information on a putative Suc sensor protein is currently available. In Suc-specific pathways, the effect of Suc cannot or can only partially be mimicked by the Suc breakdown products Glc and Fru, or by other sugars. Suc specifically regulates transcription of patatin, rolC, UDP-Glc pyrophosphorylase, and the BvSUT1 phloem-specific proton-Suc symporter (Wenzler et al., 1989; Jefferson et al., 1990; Yokoyama et al., 1994; Chiou and Bush, 1998; Ciereszko et al., 2001). Interestingly, Suc is an effective regulator of translation of the ATB2/AtbZIP11 transcription factor gene (Rook et al., 1998; Wiese et al., 2004).
Anthocyanins are widely found in plant species and are responsible for the purple coloration of plant parts. Anthocyanins provide color to flowers and fruits needed to attract pollinators and seed-dispersing animals (Winkel-Shirley, 2001). Anthocyanins are also important antioxidant molecules (Gould et al., 2002) and help to protect plants from damage by active oxygen species (Nagata et al., 2003). These properties make them interesting as food ingredients for human and animal nutrition.
The stimulatory effects of sugars on anthocyanin biosynthesis in different organs of several plant species have been reported previously. For example, sugars induce anthocyanin biosynthesis gene transcription and pigment accumulation in developing corollas of Petunia hybrida (Weiss, 2000). Similarly, anthocyanin accumulation is promoted by Suc in Vitis vinifera cells (Larronde et al., 1998) and known cellular signaling intermediates, such as Ca2+ and protein kinases and phosphatases, are involved in this process (Vitrac et al., 2000). Sugars enhance anthocyanin biosynthesis in radish (Raphanus sativus) hypocotyls (Hara et al., 2003). In Arabidopsis, anthocyanins are produced in cotyledons or leaves when growing on a sugar-containing medium, as revealed by purple coloration of the tissue (Tsukaya et al., 1991; Mita et al., 1997b; Ohto et al., 2001). This phenotype is induced by sugars and is not due to osmotic effects (Tsukaya et al., 1991; Martin et al., 2002). Mutants of Arabidopsis that exhibit elevated or reduced β-amylase expression in response to sugars also show elevated or reduced sugar-induced anthocyanin levels (Mita et al., 1997a, 1997b). High sugar-response mutants that exhibit elevated ApL3 expression in response to sugar also show elevated anthocyanin amounts (Baier et al., 2004). Many anthocyanin biosynthetic genes are induced by sugars (Gollop et al., 2001, 2002; Martin et al., 2002; Hiratsu et al., 2003). The expression of the petunia (Petunia hybrida) chalcone synthase (CHS) gene in transgenic Arabidopsis leaves is induced by sugars (Tsukaya et al., 1991). Interestingly, petunia and Arabidopsis CHS genes contain Suc boxes in the 5′-flanking regions of the gene. These Suc boxes were found in the upstream region of sporamin and β-amylase genes, which are induced by Suc (Tsukaya et al., 1991). Glc can induce transcription of CHS via a hexokinase-independent pathway (Xiao et al., 2000).
The regulatory mechanism involved in sugar induction of anthocyanin biosynthesis is essentially unknown and the aim of this study is to obtain information on this mechanism, especially with respect to the different sugar-signaling pathways. We observed that Suc specifically induces anthocyanin accumulation in Arabidopsis. Moreover, considerable natural variation exists for this trait and we used this observation for quantitative trait loci (QTL) analysis in the Landsberg erecta (Ler)/Cape Verde Islands (Cvi) recombinant inbred line (RIL) population. A major locus involved in sugar-induced anthocyanin accumulation was identified in this way. Using a candidate gene approach, this QTL was identified as encoded by the MYB75/PAP1 regulatory gene.
RESULTS
Suc-Specific Induction of Anthocyanin Accumulation
Different sugars were tested for their ability to induce anthocyanin accumulation in Arabidopsis. Mono and disaccharides tested were Glc, Fru, Gal, Suc, maltose, trehalose, and lactose. Seeds of accession Ler were plated on one-half-strength Murashige and Skoog (MS) agar containing 100 mm of the sugar as indicated. Sorbitol was included in the experiment as an osmotic control. Anthocyanin accumulation in seedlings was measured after 5 d of growth under continuous light. The different sugar treatments resulted in large differences in anthocyanin accumulation. Sorbitol did not induce anthocyanin accumulation. Of the sugars tested, Suc and maltose treatment produced the highest level of anthocyanin. The effect of Glc was significant, although 2-fold less than that of Suc and maltose. Addition of Glc and Fru combined showed at most an additive effect (data not shown). Other sugars did not induce anthocyanin accumulation (Fig. 1A).
Figure 1.
Suc induces anthocyanin accumulation. A, Ability of different sugars to induce anthocyanin accumulation. Ler seedlings were grown on one-half-strength MS medium without sugar (No) or with 100 mm of sorbitol, Glc, Fru, Gal, Suc, maltose, trehalose, lactose, palatinose, turanose, and 3-O-methyl Glc (3OMG) for 5 d. B, Time course of Suc-induced anthocyanin accumulation. Ler seedlings were grown in one-half-strength MS medium without sugar for 3 d and transferred to one-half-strength MS medium with 100 mm of Glc, Fru, Suc, or no sugar, as indicated, and incubated for another 12, 36, and 60 h. C, Anthocyanin accumulation in Col, Ler, and Cvi in response to different Suc concentrations. Col, Ler, and Cvi seedlings were grown on one-half-strength MS medium with 0, 10, 25, 50, 75, and 100 mm Suc for 5 d. The data represented the mean values of three independent experiments.
Two Suc isomers, palatinose and turanose, and the Glc analog, 3-O-methyl-Glc, were also tested. Notably, turanose induced anthocyanin synthesis to the same extent as Glc, whereas palatinose and 3-O-methyl-Glc were essentially ineffective (Fig. 1A).
Uptake of usable sugars always led to considerable growth stimulation. In this study, seedling growth was stimulated differently by the sugars. Suc, Glc, and Fru were effective sugars in stimulating growth (data not shown). Other sugars showed a reduced capacity for growth stimulation, indicating that differences in uptake or metabolism of the sugars exist. Fru stimulated growth but did not induce anthocyanin accumulation, indicating that the growth response did not depend on anthocyanin accumulation. Seedling growth was repressed by the addition of sugar analogs, palatinose, turanose, and 3-O-methyl-Glc (data not shown).
Anthocyanin induction by Suc, maltose, and Glc was further investigated in a time-course experiment. Seedlings were grown on one-half-strength MS agar for 3 d and transferred onto plates with one-half-strength MS medium without sugar or with 100 mm of sugar as indicated. Seedlings were harvested 12, 36, and 60 h after treatment, and the anthocyanin accumulation was determined. Suc induced significant anthocyanin levels within 12 h of transfer, and the anthocyanin level peaked after 36 h (Fig. 1B). Glc had a much weaker effect compared to Suc (Fig. 1B). Interestingly, in this experiment, maltose also showed a weak anthocyanin induction effect, which was similar to Glc (Fig. 1B). In the continuous treatment experiment, maltose was equally effective as Suc in inducing anthocyanin accumulation (Fig. 1A). Apparently, only the continuous presence of maltose results in a high level anthocyanin accumulation. These findings indicate that Suc is the most effective sugar in inducing anthocyanin accumulation.
Next, the Suc concentration needed for induction of anthocyanin accumulation was established. Seeds of accessions Columbia (Col), Ler, and Cvi were grown on one-half-strength MS agar plates in the presence of varying concentrations of Suc. In Col and Ler, already 10 mm of Suc significantly induced anthocyanin accumulation. Here, a near-linear relationship between Suc concentration and anthocyanin level was observed (Fig. 1C). Remarkably, Suc hardly affected anthocyanin levels in Cvi even at a concentration of 100 mm (Fig. 1C). This suggests that natural variation of Suc-induced anthocyanin accumulation exists in Arabidopsis.
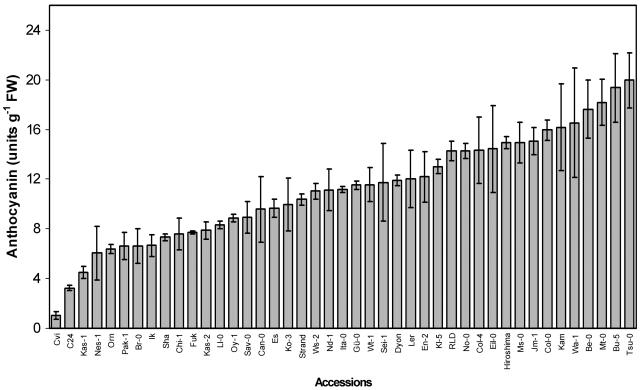
Natural Variation in Suc-Induced Anthocyanin Accumulation among Arabidopsis Accessions
The observation that Suc failed to induce anthocyanin accumulation in Cvi, whereas it was an effective inducer in Ler and Col, led us to investigate natural variation of this trait among Arabidopsis accessions. Anthocyanin content was measured in 43 accessions grown on one-half-strength MS medium with 100 mm of Suc for 5 d. The levels of anthocyanin among these accessions varied greatly (Fig. 2). In our experimental conditions, accession Tsu-0 showed the highest anthocyanin content (19.96 ± 2.19 units/g fresh weight; for unit definition, see “Materials and Methods”). In contrast, Cvi showed the lowest anthocyanin level (1.03 ± 0.31 units/g fresh weight) among the accessions tested. Anthocyanin levels of C24 and Kas-1 were also low (Fig. 2).
Figure 2.
Natural variation of Suc-induced anthocyanin synthesis. Arabidopsis accessions (43) as indicated were grown in one-half-strength MS medium with 100 mm Suc for 5 d and anthocyanin levels were determined. The data represented the mean values of three independent experiments.
QTL Analysis of Suc-Induced Anthocyanin Synthesis
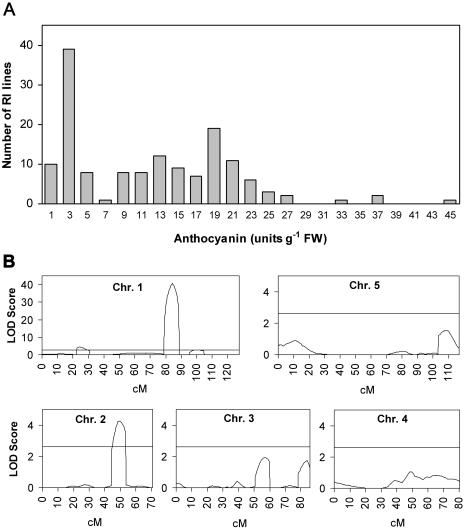
The widely varying Suc-induced anthocyanin content in Ler and Cvi suggested the use of the Ler/Cvi RIL population to identify QTL affecting this trait. The anthocyanin content was measured in seedlings of RILs grown in one-half-strength MS medium containing 100 mm Suc. The averages of two duplicate experiments were used for QTL analysis. Interestingly, the distribution of anthocyanin accumulation in the RIL population was bimodal, suggesting the existence of a major QTL (Fig. 3A).
Figure 3.
QTL analysis of Suc-induced anthocyanin accumulation in the Ler/Cvi RIL population. A, Distribution of anthocyanin content in the RIL population. RILs were grown on one-half-strength MS medium with 100 mm of Suc for 5 d. B, LOD score profile of the QTLs affecting Suc-induced anthocyanin accumulation. The horizontal line represents the LOD threshold.
QTL mapping was performed for anthocyanin accumulation and four QTLs were identified. These were named Suc-induced anthocyanin accumulation (SIAA) loci 1 to 4. SIAA1, SIAA2, and SIAA4 loci were located on chromosome 1, whereas SIAA3 was on located chromosome 2 (Fig. 3B). The additive effects of these four QTLs accounted for 67.0% of the total phenotypic variance. SIAA1 showed the strongest effect, and this locus explained 58.1% of the variation for anthocyanin content. The presence of the Ler genotype at this QTL led to increased anthocyanin content. Similarly, the Ler genotype on SIAA2 also increased the anthocyanin content. However, the Ler genotype at SIAA3 and SIAA4 decreased anthocyanin content (Table I). No significant epistasis was detected between the QTLs identified (P < 0.005).
Table I.
QTLs for Suc-induced anthocyanin accumulation in the Ler/Cvi RIL population
| Chromosome | QTL | Closest Marker | Peak Position | LOD Score | Explained Variance | Additive Effect |
|---|---|---|---|---|---|---|
| cM | ||||||
| 1 | SIAA2 | GD.86L | 24.3 | 4.36 | 3.3 | 1.68 |
| 1 | SIAA1 | EC.88C | 84.4 | 40.59 | 58.1 | 8.23 |
| 1 | SIAA4 | BF.116C | 99.8 | 2.65 | 2.3 | −1.74 |
| 2 | SIAA3 | Erecta/GPA1 | 49.2 | 4.25 | 3.3 | −1.65 |
Genetics Analysis, Fine Mapping, and a Candidate Gene of the SIAA1 Locus
Relationships between alleles of Ler and Cvi on SIAA1 were investigated by testing seedlings derived from seed of crosses between Ler and Cvi. Moreover, RIL Cvl8 was also investigated. Cvl8 contains the Cvi segment of this major QTL region and showed no anthocyanin synthesis when plated on Suc-containing agar. Cvl8 was crossed to Ler and the F2 population was analyzed. Seedlings were examined after growth for 3 to 5 d on one-half-strength MS agar containing 100 mm Suc. Under these conditions, Ler showed a purple coloration of the cotyledons, especially on the abaxial side (Fig. 4A), whereas Cvi did not show coloration (Fig. 4B). Purple cotyledons as in Ler were also observed in F1 seedlings of the Ler×Cvi and Cvi×Ler crosses (Fig. 4, C and D). Seedlings of F2 from Cvl8×Ler could be easily classified into Ler type (with purple cotyledons) and Cvi type (without purple cotyledons) after growth on 100 mm Suc for 3 to 5 d. The numbers of Ler-type and Cvi-type seedlings were 144 and 56, respectively, not deviating significantly from a 3:1 segregation ratio. These results show monogenic inheritance with the Ler allele of SIAA1 dominant over the Cvi allele.
Figure 4.
Genetic analysis and fine mapping of a major QTL in Suc-induced anthocyanin accumulation in the Ler/Cvi RIL population. Coloration of seedlings of Ler (A), Cvi (B), Ler × Cvi (C), Cvi × Ler (D), Cvl11 (E), and Cvl12 (F). Seedlings were grown on one-half-strength MS medium with 100 mm of Suc for 3 to 5 d. Top, Adaxial view of cotyledon; middle, abaxial view of cotyledon; bottom, whole seedling.
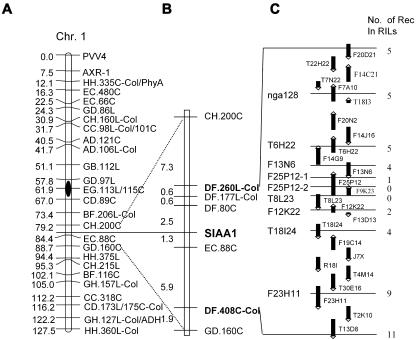
The SIAA1 locus was fine mapped by scoring the anthocyanin phenotype of the cotyledons in the RIL population after growth on 100 mm Suc for 3 to 5 d. RILs with purple cotyledons were classified as Ler type. RILs without the clear purple coloration were classified as Cvi type (Fig. 4, E and F). Thus, the anthocyanin accumulation phenotype could be analyzed as a qualitative trait in the Ler/Cvi RIL population. Initial experiments mapped SIAA1 between CH.200C and EC.88C using a core map with 99 markers and Mapmaker/EXP 3.0 software (Lander et al., 1987; Fig. 5A). Next, all markers in this region (Alonso-Blanco et al., 1998) were used as input data and the location of SIAA1 narrowed down between markers DF.80C and EC.88C with genetic distance of 2.5 and 1.3 cM, respectively (Fig. 5B). Among the amplified fragment-length polymorphism (AFLP) markers in this region, DF.260L-Col and DF.408C-Col were integrated into the physical map by in silico AFLP analysis (Peters et al., 2001). These markers are present on bacterial artificial chromosomes F20D21 and T13D8, respectively (Fig. 5, B and C). The physical distance between these two markers was about 2 Mb, according to the Arabidopsis genomic sequence. Eight new simple sequence-length polymorphism (SSLP) markers were developed for this region and were used for further fine mapping, together with SSLP marker nga128 (see “Materials and Methods”). Finally, SIAA1 was mapped between F25P12-1 and F12K22 in a genomic region of about 200 kb; markers F25P12-2 and T8L23 cosegregated with SIAA1 (Fig. 5C).
Figure 5.
Fine mapping of SIAA1. A, SIAA1 was mapped on the long arm of chromosome 1 (Chr. 1) between DF.80C and EC.88C using the set of 99 core markers. B, Mapping of the major QTL of Suc-induced anthocyanin synthesis with available AFLP markers in this region. C, Fine mapping of the QTL with nga128 and eight further developed SSLP markers. The major QTL was narrowed down to a 210-kb genomic DNA region between markers F25P12-1 and F12K22.
The 200-kb genomic region between markers F25P12-1 and F12K22 contains 124 annotated genes (from At1g56590 to At1g57790). Of these, 81 are tRNA genes (from At1g56730 to At1g57530) and therefore unlikely to explain the observed variation, leaving 43 candidate genes. Annotations of these genes and results from the literature pointed our attention to a MYB transcription factor gene, the MYB75/PAP1 (At1g56650). MYB75/PAP1 is a positive regulator of anthocyanin synthesis (Borevitz et al., 2000), which induced the expression of genes involved in anthocyanin synthesis (Tohge et al., 2005). Moreover, the transcription of MYB75/PAP1 is induced by Suc (Kranz et al., 1998). Transgenic Arabidopsis plants that expressed MYB75/PAP1 fused to the SRDX repression domain showed a repression of anthocyanin synthesis in the presence of Suc, whereas wild-type seedlings accumulated anthocyanin (Hiratsu et al., 2003). Therefore, MYB75/PAP1 was considered to be a candidate gene for SIAA1.
The myb75 Null Allele Lacks Suc-Induced Anthocyanin Accumulation
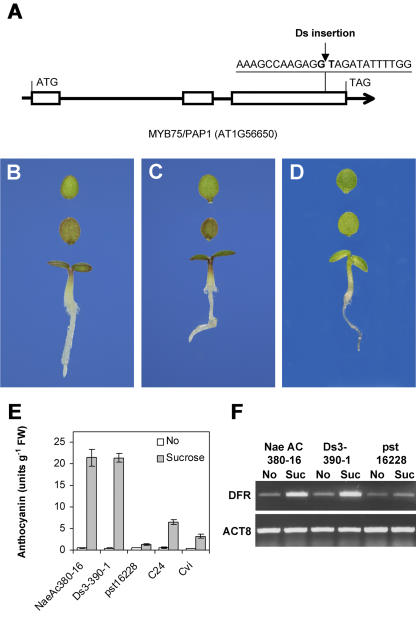
The hypothesis that MYB75/PAP1 is the gene underpinning SIAA1 was tested using a laboratory-generated mutant line in which a dissociation (Ds) transposon was inserted in MYB75/PAP1 (line pst16228 or 13-3235-1 from RIKEN BioResource Center [http://rarge.gsc.riken.jp/dsmutant/index.pl]; Kuromori et al., 2004). The Ds transposon was inserted in the third exon of the MYB75/PAP1 gene in the accession Nossen, and was confirmed by sequencing PCR fragments of both Ds-flanking segments (Fig. 6A). The insertion resulted in an interruption of the coding sequence from amino acid 213 onward. Seedlings of both parents of pst16228, NaeAc380-16 (N8538), and Ds3-390-1 (N8521) accumulated anthocyanin when grown on Suc-containing media (Fig. 6, B and C). Homozygous insertion lines grown on Suc lacked the purple-coloration phenotype, similar to what is observed in the Cvi accession (Fig. 6D). In Suc-grown pst16228, the anthocyanin content was less than in the Cvi accession (Fig. 6E).
Figure 6.
The myb75 Ds insertion allele lacks Suc-induced anthocyanin accumulation. A, Insertion position of Ds transposon in pst16228. Coloration of seedling of NaeAc380-16 (B), Ds3-390-1 (C), and pst16228 (D). Top, Adaxial view of cotyledon; middle, abaxial view of cotyledon; bottom, whole seedling. E, Absence of Suc-induced anthocyanin accumulation in pst16228. Seedlings of NaeAc380-16, Ds3-390-1, pst16228, Cvi, and C24 were grown on one-half-strength MS medium without sugar (No) or with 100 mm Suc for 5 d. The data represent mean values of three independent experiments. F, Suc does not induce the DFR gene in mutant pst16228. Reverse transcription-PCR was performed using RNA isolated from seedlings of NaeAc380-16, Ds3-390-1, and pst16228 grown on one-half-strength MS medium without sugar (No) or with 100 mm Suc.
Plating of 200 seeds from plants heterozygous for the insertion (as identified by PCR analysis of leaf material; see “Materials and Methods”) on Suc media resulted in seedlings of which 154 showed purple cotyledons, whereas 46 showed no coloration, in agreement with a 3:1 segregation ratio. In this seedling population, hygromycin resistance encoded on the Ds element was also investigated. Of the 154 seedlings with purple cotyledons, 56 showed hygromycin sensitivity, not differing significantly from a 2:1 segregation ratio. No hygromycin-sensitive seedlings were present among the 46 seedlings without anthocyanin. Thus, a total of 144 seedlings showed hygromycin resistance, indicative for the presence of a single Ds insertion (3:1 ratio) in this line. The genotypes of insertions in these 144 hygromycin-resistant plants were also established by PCR. All plants with a Cvi-like phenotype carried the homozygous insertion in MYB75/PAP1, whereas all plants that showed purple coloration were heterozygous. These results demonstrate that the Ds insertion is tightly linked to the phenotype.
An active DFR enzyme is essential for anthocyanin biosynthesis. The Suc-induced expression of DFR was tested and it was observed that DFR is expressed at elevated levels in both NaeAc380-16 and Ds3-390-1 grown on Suc (Fig. 6F). No Suc-enhanced expression of DFR was detected in the homozygous insertion plants grown under the same condition (Fig. 6F).
The SIAA1 Locus Represents MYB75/PAP1
Seedlings derived from reciprocal crosses between Cvi and pst16228 carrying the homozygous Ds insertion in MYB75/PAP1 were tested for complementation. In both crosses, no complementation was observed, revealing that the two recessive alleles are allelic (Fig. 7, A and B). As expected, F1 seedlings of a cross between Cvi and either one or both insertion-line parents, NaeAc380-16 and Ds3-390-1 (Fig. 7, C and D), show Suc-induced anthocyanin accumulation, as do F1 seedlings of a cross between pst16228 and Ler (Fig. 7E). Therefore, we conclude that the SIAA1 locus represents MYB75/PAP1.
Figure 7.
Genetic analysis shows that the SIAA1 locus represents MYB75/PAP1. Coloration of seedlings derived from crosses between pst16228 × Cvi (A), Cvi × pst16228 (B), Cvi × NaeAc380-16 (C), Cvi × Ds3-390-1 (D), and pst16228 × Ler (E). Seedlings were grown on one-half-strength MS medium containing 100 mm Suc for 3 to 5 d. Top, Adaxial view of cotyledon; middle, abaxial view of cotyledon; bottom, whole seedling.
The MYB75/PAP1 Allele in the Arabidopsis C24 Accession Is Responsible for Low Suc-Induced Anthocyanin Levels
Low Suc-induced anthocyanin levels (Figs. 2 and 6E) and weak purple coloration of cotyledons were also observed in the C24 accession (Fig. 8A). Genetic analysis showed that this phenotype is also due to a recessive allele of MYB75/PAP1. In the selfed progeny of the Ler×C24 cross, we detected 141 seedlings with anthocyanin and 46 without on one-half-strength MS medium plus 100 mm Suc, consistent with a 3:1 segregation ratio. These results indicated that a single major recessive gene is responsible for the lack of induction of anthocyanin accumulation in C24.
Figure 8.
C24 harbors a weak allele of MYB75/PAP1. Coloration of seedlings of C24 (A) and of seedlings derived from crosses between C24×Ler (B), Ler×C24 (C), pst16228×C24 (D), C24×pst16228 (E), C24×NaeAc380-16 (F), C24×Ds3-390-1 (G), C24×Cvi (H), and Cvi×C24 (I). Seedlings were grown on one-half-strength MS ;medium containing 100 mm Suc for 3 to 5 d. Top, Adaxial view of cotyledon; middle, abaxial view of cotyledon; bottom, whole seedling.
The possibility that an allele of the MYB75/PAP1 is responsible for the low induced anthocyanin content in C24 was investigated. F1 seedlings derived from the reciprocal crosses of C24×pst16228 and C24×Cvi showed very weak purple coloration, showing that they failed to complement each other (Fig. 8, D, E, H, and I). NaeAc380-16 and Ds3-390-1 lines do complement C24 (Fig. 8, F and G). Therefore, we conclude that the low level of Suc-induced anthocyanin accumulation observed in the C24 accession is due to a recessive MYB75/PAP1 allele, apparently with a reduced function in inducing anthocyanin accumulation on a Suc-containing medium.
Natural Variation in the MYB75/PAP1 Amino Acid Sequence
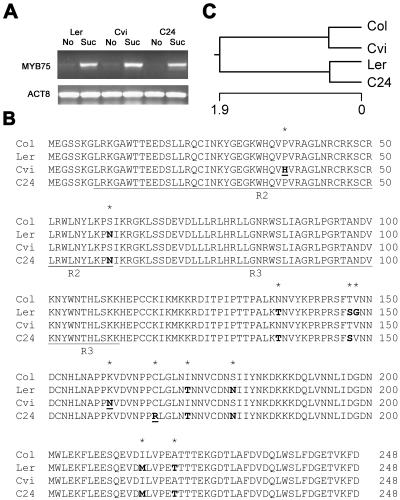
In both Cvi and C24 accessions, MYB75/PAP1 mRNA levels are normally induced by Suc as observed in Ler (Fig. 9A). Therefore, it is likely that the observed variation in Suc-induced anthocyanin accumulation results from different activities of the MYB75/PAP1 proteins present in the different accessions. The sequences of MYB75/PAP1 in Ler, Cvi, and C24 were determined and the sequence of Col was retrieved from The Arabidopsis Information Resource database. The MYB75/PAP1 coding regions of these four accessions showed a total of 15 single-nucleotide polymorphisms (SNPs) in a sequence of 747 nucleotides. Eleven of these SNPs were nonsynonymous substitutions, leading to a changed amino acid residue (Fig. 9B). The deduced amino acid sequences of these four alleles were aligned with ClustalW (Thompson et al., 1994). The phenotypes suggest that Col and Ler harbor functional protein, whereas Cvi and C24 might encode for weak or loss-of-function proteins. Surprisingly, C24 clusters with Ler, whereas Cvi clusters with Col (Fig. 9C). There were eight amino acid substitutions observed between Col and Ler, but only two amino acid substitutions between Col and Cvi and between Ler and C24 (Fig. 9B).
Figure 9.
Natural variation in the MYB75/PAP1 amino acid sequence. A, Suc induces mRNA accumulation of MYB75/PAP1 in Ler, Cvi, and C24 accessions. B, Amino acid sequence of MYB75/PAP1 from Col, Ler, Cvi, and C24 were aligned using ClustalW. The R2 and R3 DNA-binding domains are underlined. Asterisks (*) indicate positions with amino acid substitutions. Boldface letter indicates amino acid substitutions compared to Col. Boldface and underlined letters indicate the amino acid substitutions compared to Col and Ler. C, A phylogenetic tree of MYB75/PAP1 amino acid sequences derived from Col, Ler, Cvi, and C24.
Comparison of the amino acid sequence of the functional proteins from Col and Ler to the sequence of Cvi showed amino acid substitutions for Pro-37 to His-37 (CCT to CAT) and Lys-160 to Asn-160 (AAA to AAT). The Pro-37 to His-37 exchange occurs in the MYB R2 DNA-binding domain (Jia et al., 2003). Since a Pro residue is involved, this mutation likely results in a conformational change in this domain that might affect its DNA-binding ability. The Lys-160 amino acid is positioned outside the R2, R3 DNA-binding domain, but this region probably is important for the function of the protein as well. This region also contains the Cys-167 amino acids, and comparison of the Col and Ler sequence to that of C24 shows a Cys-167 to Arg-167 (TGC to CGC) amino acid substitution. Thus, in C24, the MYB75/PAP1 protein shows reduced activity due to a mutation at amino acid position 167.
DISCUSSION
Anthocyanin accumulation can be induced by sugars in many plant species. Arabidopsis mutants with altered responses to sugar also show altered sugar-induced anthocyanin accumulation (Mita et al., 1997a, 1997b; Baier et al., 2004). Arabidopsis plants defective in the SUC2 Suc transporter gene accumulate large amounts of soluble sugars in leaves. Microarray analysis of suc2/pho3 mutant plants shows that the PAP1/MYB75 and PAP2/MYB90 genes are strongly induced, as well as seven genes of the anthocyanin biosynthesis pathway (Lloyd and Zakhleniuk, 2004). Signaling intermediates, such as Ca2+, protein kinases, and protein phosphatases, are involved in sugar-induced anthocyanin accumulation (Vitrac et al., 2000). The signaling pathway for sugar-induced anthocyanin accumulation is independent of the known Glc sensor protein HXK1 (Xiao et al., 2000). In this study, we discovered that a Suc-dependent signaling pathway induces anthocyanin accumulation in a process that depends on a functional MYB75/PAP1 protein. Moreover, naturally occurring allelic variation was observed for this anthocyanin accumulation trait.
A Suc-Dependent Mechanism Induces Anthocyanin Accumulation
Suc acts as a signaling molecule independent of other neutral sugars, notably Glc and Fru. Suc specifically repressed steady-state mRNA levels as well as Suc transport activity of the proton-Suc symporter in excised sugar beet (Beta vulgaris) leaves (Vaughn et al., 2002). Similarly, Suc controls expression of the AtbZIP11/ATB2 gene via a posttranscriptional mechanism. Translation of the AtbZIP11 mRNA is repressed specifically by Suc; other sugars are less effective (Rook et al., 1998; Wiese et al., 2004). These findings point to the operation of Suc-induced signaling systems in plants, which is separate from the hexose-signaling systems. In this study, it was found that Suc specifically induces the anthocyanin biosynthetic pathway. A number of metabolically active sugars showed different effects on the induction of anthocyanin accumulation, indicating that sugar-induced anthocyanin accumulation is independent of general sugar metabolism. Of the sugars tested, Suc most rapidly and strongly induces anthocyanin accumulation in a concentration-dependent manner (Fig. 1). Maltose effectively induces anthocyanin accumulation when continuously present (Fig. 1A). However, in a time-course experiment, maltose has no effect (Fig. 1B). Possibly, maltose is efficiently metabolized to Suc when present continuously. A similar incubation time-dependent effect of maltose was observed for the bZIP11 translational control system mentioned above.
The nonmetabolizable Suc analogs palatinose and turanose were used to study disaccharide sensing independent of metabolism. In potato (Solanum tuberosum) tuber discs, palatinose mimics the stimulatory effect of Suc on starch synthesis (Fernie et al., 2001). In barley (Hordeum vulgare) embryos, both palatinose and turanose mimic the repressive effect of Suc on α-amylase expression (Loreti et al., 2000). In this study, continuous incubation with turanose induced anthocyanin accumulation to about one-half the level observed with Suc, whereas incubation with palatinose did not. Interestingly, the AtSUC2 Suc transporter can transport turanose, whereas palatinose is not transported (Chandran et al., 2003).
MYB75/PAP1 Plays an Essential Role in Suc-Induced Anthocyanin Synthesis
A major QTL affecting Suc-induced anthocyanin accumulation was identified in a RIL population derived from a cross between Ler and Cvi. The gene responsible for this QTL effect was identified and shown to be the MYB75/PAP1 gene. This gene has previously been identified as a positive regulator of anthocyanin biosynthesis (Borevitz et al., 2000; Tohge et al., 2005). The identity of this MYB75/PAP1 gene was confirmed by this analysis of natural allelic variation and analysis of the laboratory-generated gene insertion line.
Many flavonoid biosynthesis structural and regulatory genes have been isolated using different methods in maize (Zea mays), snapdragon (Antirrhinum majus), petunia, and Arabidopsis (Holton and Cornish, 1995; Shirley et al., 1995; Mol et al., 1998). Genes encoding enzymes for the whole biosynthetic pathway have been cloned, as well as those of many regulatory loci (Winkel-Shirley, 2001). Anthocyanin synthesis is regulated by a heterotrimeric MYB-basic helix-loop-helix (bHLH)-WD40 protein complex (Zhang et al., 2003; Baudry et al., 2004; Zimmermann et al., 2004). This complex can harbor different members of the MYB and bHLH protein families. The TTG1 gene encodes the WD40 protein present in the complex. This gene has other functions as well. Next to its involvement in the regulation of anthocyanin synthesis, TTG1 is, for example, involved in the development of trichomes, root hair, and seed coat mucilage (Walker et al., 1999; Ramsay and Glover, 2005). TTG1 is essential for Suc-induced anthocyanin accumulation, and in ttg1 mutants Suc is incapable of inducing anthocyanin (Shirley et al., 1995). Three bHLH genes, GL3, EGL3, and TT8, participate in the regulation of anthocyanin synthesis. Analysis of single, double, and triple mutants indicates that these three bHLH genes are partially redundant in regulating the anthocyanin pathway (Zhang et al., 2003).
Two MYB genes, MYB75/PAP1 and MYB90/PAP2, are involved in the regulation of anthocyanin synthesis (Borevitz et al., 2000). Here we show that Suc-induced anthocyanin synthesis depends on a functional MYB75/PAP1 gene. Moreover, Suc-induced expression of a gene encoding a key enzymatic function, DFR, is lost in the myb75/pap1 mutant. Thus of the two MYB genes, only MYB75/PAP1 is essential for Suc induction of the anthocyanin pathway. A Suc-induced signaling pathway induces the expression of MYB75/PAP1 and, as a consequence, the anthocyanin synthesis pathway is triggered.
Natural Variation of MYB75/PAP1 Genes among Arabidopsis Accessions
Several genes responsible for natural variation have been identified at the molecular level in Arabidopsis. For some of these, the molecular polymorphisms underlying the phenotypic variation have been elucidated. These include SNPs that generate single amino acid substitutions, small deletions that produce truncated protein or altered expression level, large deletions eliminating the complete gene, and large transposon-related insertions in noncoding regulatory regions that alter the expression level (Koornneef et al., 2004).
Extensive natural variation exists in Suc-induced anthocyanin accumulation among the accessions of Arabidopsis, with Cvi and C24 accessions showing very low induced anthocyanin content (Figs. 2 and 6E). Suc induces MYB75/PAP1 transcripts in Cvi and C24 to a similar level as in Ler (Fig. 9A). Therefore, low activity of the Cvi and C24 alleles is due to polymorphisms in the coding region. In the four accessions studied, 15 SNPs were uncovered in the 747-bp MYB75/PAP1 coding region. Eleven of them are nonsynonymous substitutions that change the amino acid residue. Col and Ler possess functional alleles, whereas Cvi and C24 have either very weak or loss-of-function alleles. Surprisingly, the amino acid sequence of C24 is more similar to Ler, whereas Cvi is more similar to Col. Apparently, general similarity in amino acid sequence in this gene does not relate to the functional properties of the protein. Comparison of the Col and Ler alleles with the weak or null alleles of Cvi and C24 suggests that the Pro-37 to His-37 (CCT to CAT) and Lys-160 to Asn-160 (AAA to AAT) substitution in Cvi, and the Cys-167 to Arg-167 (TGC to CGC) substitution in C24 are responsible for the mutant allele phenotype. SNPs are responsible for these amino acid substitutions. Therefore, these nucleotides represent quantitative trait nucleotides.
The physiological significance of Suc-induced anthocyanin accumulation is currently not understood. Similarly, the ecological significance of natural variation of the trait is unclear. Possibly, stressful growth conditions that allow Suc synthesis, but are inhibitory to its utilization, will result in increased Suc levels, which induce anthocyanin accumulation that is helpful in negating the stress imposed. For example, cold-acclimated plants sustain high levels of photosynthesis and have much higher Suc content than plants grown under normal temperatures (Strand et al., 1999). Such high Suc levels might induce anthocyanin important for photoprotection. In such a scheme, Suc acts as a stress signal. Already very low Suc concentrations stimulate anthocyanin accumulation, which increases in a linear way with the Suc level.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) accessions listed below were used for analysis of natural variation of Suc-induced anthocyanin accumulation. They were Be-0 (N964), Br-0 (N994), Bu-5 (N1014), C24 (N906), Can-0 (N1064), Chi-1 (N1074), Col-0 (N1092), Col-4 (N993), Cvi (N8580), Dyon (W10159), Eil-0 (N6693), En-2 (N1138), Es (N1144), Fuk (W10219), Gü-0 (N1212), Hiroshima (N1677), Ik (W10223), Ita-0 (N1244), Jm-1 (N1260), Kam (Steno), Kas-1 (N903), Kas-2 (N1264), Kl-5 (N1284), Ko-3 (N1290), Ler (N8581), Ll-0 (N1338), Ms-0 (N905), Mt-0 (N6799), Nd-1 (N1636), Nes-1 (W10042), No-0 (N1394), Orn (Steno), Oy-1 (N1643), Pak-1 (W10212), RLD (N913), Sav-0 (N1514), Sei-1 (N1504), Sha (N929), Strand (Steno), Tsu-0 (N1564), Wa-1 (N6885), Ws-2 (N1601), and Wt-1 (N1604). The Nottingham Arabidopsis Stock Centre (NASC) numbers (NXXXX) are indicated between parentheses. Others (WXXXXX or Steno) were from the Wageningen University Stock Centre.
The RIL population used for QTL analysis was derived from a cross between Ler and Cvi and consisted of 162 RILs. These RILs were previously characterized genetically using AFLP and cleaved amplified polymorphic sequence markers (Alonso-Blanco et al., 1998).
The RIKEN line pst16228 has a transposon inserted in the third exon of MYB75/PAP1 (RIKEN BioResource Center [http://rarge.gsc.riken.jp/dsmutant/index.pl]; Kuromori et al., 2004). This line and the parental lines NaeAc380-16 (N8538) and Ds3-390-1 (N8521) were used to analyze the role of MYB75/PAP1 in Suc-induced anthocyanin synthesis and for genetic confirmation of the major QTL. All these lines are in Nossen background.
Ler, which is a parent of the Ler/Cvi RIL population, was used for testing sugar specificity.
Growth of Seedling and Anthocyanin Measurement
Seeds were plated on one-half-strength MS medium, pH 5.8, including vitamins. This medium was solidified with 0.8% plant agar. The different sugars (Suc, Glc, Fru, Gal, lactose, trehalose, and lactose) and sugar analogs (palatinose, turanose, and 3-O-methyl-Glc) were added as indicated. Seeds were surface sterilized in chlorine gas for 3.5 h and placed in a laminar flow for at least 1 h for air ventilation. Chlorine gas production was initiated by mixing 100 mL of commercial chlorine bleach (Glorix; Unilever) and 4 mL of hydrochloric acid. About 100 seeds were plated for each line per treatment. Seeds were stratified on plates for 4 d at 4°C in the dark. Next, plates were incubated at 22°C under continuous fluorescent light.
Anthocyanin content of seedlings was determined using the protocol of Mita et al. (1997b). Frozen, homogenized seedlings (20 mg) were extracted for 1 d at 4°C in 1 mL of 1% (v/v) hydrochloric acid in methanol. The mixture was centrifuged at 13,000 rpm for 15 min and the absorbance of the supernatant was measured at 530 and 657 nm. Relative anthocyanin concentrations were calculated with the formula [A530 − (1/4 × A657)]. The relative anthocyanin amount was defined as the product of relative anthocyanin concentration and extraction solution volume. One anthocyanin unit equals one absorbance unit [A530 − (1/4 × A657)] in 1 mL of extraction solution.
QTL Analyses and Fine Mapping
A set of 99 markers evenly distributing and covering most of the Arabidopsis genome was selected from the Ler/Cvi RIL map (Alonso-Blanco et al., 1998). These markers spanned 482 cM, with an average interval of 5 cM and the largest genetic distance being 12 cM.
The computer software package MapQTL, version 4.0 (van Ooijen, 2000), was used to identify and locate QTLs on the linkage map. For this, interval mapping and multiple QTL model mapping was performed, as described in the reference manual (http://www.plant.wageningen-ur.nl/products/mapping/mapqtl). Log of the odds (LOD) threshold values applied to declare the presence of a QTL were estimated by the permutation tests. The quantitative trait data of the RIL population were permutated 10,000 times over the genotypes and empirical LOD thresholds corresponding to a genome-wide significance value of 0.05 were estimated to be 2.6 for this dataset. The estimated additive genetic effect and the percentage of variance explained by each QTL were obtained with MapQTL in the final multiple QTL model in which one cofactor marker was fixed per QTL. A positive additive effect implies that the Ler genotypes increase the anthocyanin level, while negative effects indicate that the Cvi genotypes increase the anthocyanin level.
Two-way interactions among the QTLs identified were tested by ANOVA using the corresponding two markers as random factors.
Eight new SSLP markers, located between DF.260L-Col and DF.408C-Col, were developed according to the sequence information of Insertion/Deletion from The Arabidopsis Information Resource (http://www.arabidopsis.org/Cereon/index.jsp). The sequences of primers used are shown in Table II. The PCR program used was as follows: (1) 1 cycle of 94°C for 2 min; (2) 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and (3) 72°C for 5 min. Following PCR, polymorphisms were detected on a 3% agarose gel buffered with 0.5×Tris-borate/EDTA.
Table II.
List of the SSLP molecular markers used for fine mapping of the SIAA1 major QTL
| Marker Name | Forward Sequence | Reverse Sequence |
|---|---|---|
| nga128 | 5′-ATCTTGAAACCTTTAGGGAGGG-3′ | 5′-GGTCTGTTGATGTCGTAAGTCG-3′ |
| T6H22 | 5′-GCCAAATCAATGCAGTCTCTG-3′ | 5′-TTGCAGCTTTGAAAATCCAG-3′ |
| F13N6 | 5′-CAGGGTGTGTTTACCCCAAG-3′ | 5′-GGGTCACAACAAAACACTAGAGA-3′ |
| F25P12-1 | 5′-TCTTTCACTTGGTCAACACA-3′ | 5′-AGGAACATGCATTCAAAAGT-3′ |
| F25P12-2 | 5′-GACACGTGGCACGATCCTAT-3′ | 5′-ACGCGAGGAATGAAGAGGTA-3′ |
| T8L23 | 5′-ATGTCGTGCCCTTGACTGA-3′ | 5′-TGTATAGGGAGATTGGTCAATTACA-3′ |
| F12K22 | 5′-CGGCTATTTTGAAGCCCTAA-3′ | 5′-CATCGCATGCATACACCTTC-3′ |
| T18I24 | 5′-CCTACGAAGAGCTTTGTGAGTGT-3′ | 5′-CCATGCACCAACAGAAATGA-3′ |
| F23H11 | 5′-GCTCACCTGTCGAAACCACT-3′ | 5′-TGCACTGCACATGCACAAC-3′ |
Analysis of the MYB75/PAP1 Structure Gene and Insertion Flanking Sequences
The MYB75/PAP1 structural gene region was amplified by PCR from accessions Ler, Cvi, and C24. The primers used were: forward, 5′-TGGATATCAAACATGCACGTCACTTCCT-3′ and reverse, 5′-CCAATGAGTAGACTACTCAA-3′ (for Ler and C24) and 5′-CTTCAGTACCAAACCTTCTCTACCGACC-3′ (for Cvi). The SIGnAL T-DNA verification primer design (http://signal.salk.edu/tdnaprimers.2.html) was used to design primer sequences for amplification of the flanking fragments and genotypes of the insertion lines. These were: left primer (LP), 5′-TGGTTTTGTAGGGCTAAACCG -3′ and right primer (RP), 5′-AAACACCGGATACATACCTTTTTC-3′. To amplify the flanking fragment, LP was combined with Ds5-3 (5′-TACCTCGGGTTCGAAATCGAT-3′) and RP was combined with Ds3-2a (5′-CCGGATCGTATCGGTTTTCG-3′). For genotyping the insertion, primers LP, RP, and Ds 3-2a were used. The wild-type line produces a PCR product of about 900 bp (from LP to RP). Lines carrying the homozygous insertion produce a band of about 500 bp (from RP to Ds 3-2a). Heterozygous lines produce both bands. The PCR program was as follows: (1) 1 cycle of 94°C for 2 min; (2) 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 3 min; and (3) 72°C for 10 min. All PCR products ware analyzed on a 1% agarose gel.
The PCR production of the MYB75/PAP1 structural gene region from accessions Ler, Cvi, and C24, and the flanking fragments of insertion were purified from the agarose gel for sequence analysis.
Reverse Transcription-PCR
Total RNA was isolated using the RNeasy plant mini kit (Qiagen). The quality and the quantity of the RNA were analyzed by electrophoresis on 1.2% agarose and spectrometry, respectively. RNA was DNase-treated (Fermentas) and a 1-μg aliquot was reverse transcribed using Maloney murine leukemia virus (Promega). The PCR reaction mixture with a total volume of 50 μL consisted of Taq buffer (Fermentas), 50 pmol of each primer, 20 nmol of dATP, dCTP, dGTP, and dTTP, 2.5 units of Taq, and cDNA equivalent to 50 ng of RNA. The PCR program was as follows: (1) 1 cycle of 94°C for 2 min; (2) N cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 3 min; and (3) 72°C for 10 min. For act8, N is 30; for MYB75/PAP1, N is 40; for DFR, N is 27. The primers for MYB75/PAP1 were as follows: forward, 5′-GCTCTGATGAAGTCGATCTTC-3′ and reverse, 5′-CTACCTCTTGGCTTTCCTCT-3′; for DFR: forward, 5′-ATGGTTAGTCAGAAAGAGACCG-3′ and reverse, 5′-GTCTTATGATCGAGTAATGCGC-3′ (Nesi et al., 2000); for ACT8: forward, 5′-ATGAAGATTAAGGTCGTGGCA-3′ and reverse, 5′-CCGAGTTTGAAGAGGCTAC-3′. The PCR products ware analyzed on a 1.5% agarose gel.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ222404 (Ler), DQ222405 (Cvi), and DQ222406 (C24).
Acknowledgments
We are grateful to the RIKEN BioResource Center for providing myb75 knockout mutant and the Nottingham Arabidopsis Stock Centre for providing wild-type lines. We thank Dr. Ton Peeters and Dr. Basten Snoek, Department of Plant Ecophysiology, Utrecht University, for contributing plant lines and for helpful suggestions.
This work was supported by the The Netherlands National Genomics Programme QTL express (050–10–029), by the Centre for Biosystems Genomics, and by the European Union program NATURAL (contract no. QLG2–CT–2001–01097).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sheng Teng (s.teng@bio.uu.nl).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066688.
References
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW (2004) Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiol 134: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MT, Yu SM (1998) The 3′ untranslated region of a rice alpha-amylase gene mediates sugar-dependent abundance of mRNA. Plant J 15: 685–695 [DOI] [PubMed] [Google Scholar]
- Chandran D, Reinders A, Ward JM (2003) Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem 278: 44320–44325 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1999) Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc Natl Acad Sci USA 96: 10512–10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95: 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Kleczkowski LA (2001) Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem J 354: 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G (1998) The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206: 131–137 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Roessner U, Geigenberger P (2001) The sucrose analog palatinose leads to a stimulation of sucrose degradation and starch synthesis when supplied to discs of growing potato tubers. Plant Physiol 125: 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI (2005) Control of plant development and gene expression by sugar signaling. Curr Opin Plant Biol 8: 93–102 [DOI] [PubMed] [Google Scholar]
- Gollop R, Even S, Colova-Tsolova V, Perl A (2002) Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot 53: 1397–1409 [PubMed] [Google Scholar]
- Gollop R, Farhi S, Perl A (2001) Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci 161: 579–588 [Google Scholar]
- Gould KS, McKelvie J, Markham KR (2002) Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ 25: 1261–1269 [Google Scholar]
- Hara M, Oki K, Hoshino K, Kuboi T (2003) Enhancement of anthocyanin biosynthesis by sugar in radish (Raphanus sativus) hypocotyl. Plant Sci 164: 259–265 [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R, Goldsbrough A, Bevan M (1990) Transcriptional regulation of a patatin-1 gene in potato. Plant Mol Biol 14: 995–1006 [DOI] [PubMed] [Google Scholar]
- Jia L, Clegg MT, Jiang T (2003) Excess non-synonymous substitutions suggest that positive selection episodes occurred during the evolution of DNA-binding domains in the Arabidopsis R2R3-MYB gene family. Plant Mol Biol 52: 627–642 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11,800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Larronde F, Krisa S, Decendit A, Cheze C, Merillon JM (1998) Regulation of polyphenol production in Vitis vinifera cell suspension cultures by sugars. Plant Cell Rep 17: 946–950 [DOI] [PubMed] [Google Scholar]
- Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Loreti E, Alpi A, Perata P (2000) Glucose and disaccharide-sensing mechanisms modulate the expression of α-amylase in barley embryos. Plant Physiol 123: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Hirano H, Nakamura K (1997. a) Negative regulation in the expression of a sugar-inducible gene in Arabidopsis thaliana — a recessive mutation causing enhanced expression of a gene for β-amylase. Plant Physiol 114: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K (1997. b) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Nagata T, Todoriki S, Masumizu T, Suda I, Furuta S, Du ZJ, Kikuchi S (2003) Levels of active oxygen species are controlled by ascorbic acid and anthocyanin in Arabidopsis. J Agric Food Chem 51: 2992–2999 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K (2001) Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol 127: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, Gerats T (2001) A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol 127: 1579–1589 [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Mollenhauer B, Reinbothe C (1994) JIPs and RIPs: the regulation of plant gene expression by jasmonates in response to environmental cues and pathogens. Plant Cell 6: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Bevan MW (2003) Genetic approaches to understanding sugar-response pathways. J Exp Bot 54: 495–501 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Sadka A, DeWald DB, May GD, Park WD, Mullet JE (1994) Phosphate modulates transcription of soybean VspB and other sugar-inducible genes. Plant Cell 6: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Strand A, Hurry V, Henkes S, Huner N, Gustafsson P, Gardestrom P, Stitt M (1999) Acclimation of Arabidopsis leaves developing at low temperatures. Increasing cytoplasmic volume accompanies increased activities of enzymes in the Calvin cycle and in the sucrose-biosynthesis pathway. Plant Physiol 119: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y (1991) Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol 97: 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW (2000) MapQTL Version 4.0: Usefriendly Power in QTL Mapping: Addendum to the Manual of Version 3.0. Plant Research International, Wageningen, The Netherlands
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99: 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitrac X, Larronde F, Krisa S, Decendit A, Deffieux G, Merillon JM (2000) Sugar sensing and Ca2+-calmodulin requirement in Vitis vinifera cells producing anthocyanins. Phytochemistry 53: 659–665 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D (2000) Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiol Plant 110: 152–157 [Google Scholar]
- Wenzler H, Mignery G, Fisher L, Park W (1989) Sucrose-regulated expression of a chimeric potato-tuber gene in leaves of transgenic tobacco plants. Plant Mol Biol 13: 347–354 [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao WY, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Hirose T, Fujii N, Aspuria ET, Kato A, Uchimiya H (1994) The rolc promoter of Agrobacterium-Rhizogenes Ri plasmid is activated by sucrose in transgenic tobacco plants. Mol Gen Genet 244: 15–22 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]