Abstract
The extensive data on the transcription of the plant genome are derived primarily from the sporophytic generation. There currently is little information on genes that are expressed during female gametophyte development in angiosperms, and it is not known whether the female gametophyte transcriptome contains a major set of genes that are not expressed in the sporophyte or whether it is primarily a subset of the sporophytic transcriptome. Because the embryo sac is embedded within the maternal ovule tissue, we have utilized the Arabidopsis (Arabidopsis thaliana) mutant sporocyteless that produces ovules without embryo sacs, together with the ATH1 Arabidopsis whole-genome oligonucleotide array, to identify genes that are preferentially or specifically expressed in female gametophyte development. From analysis of the datasets, 225 genes are identified as female gametophyte genes, likely a lower limit as stringent criteria were used for the analysis, eliminating many low expressed genes. Nearly 45% of the identified genes were not previously detected by sporophytic expression profiling, suggesting that the embryo sac transcriptome may contain a significant fraction of transcripts restricted to the gametophyte. Validation of six candidate genes was performed using promoter∷β-glucuronidase fusions, and all of these showed embryo sac-specific expression in the ovule. The unfiltered expression data from this study can be used to evaluate the possibility of female gametophytic expression for any gene in the ATH1 array, and contribute to identification of the functions of the component of the Arabidopsis genome not represented in studies of sporophytic expression and function.
The plant life cycle alternates between a haploid gametophytic phase and a diploid sporophytic phase. In angiosperms, gametophyte development takes place within the sporophyte. While the mature male gametophyte, pollen grains, are released from the anthers, the mature female gametophyte, also called embryo sac or megagametophyte, remains embedded within the maternal ovule tissues, making it difficult to access the female gametophyte for study. In Arabidopsis (Arabidopsis thaliana), the female gametophyte is a seven-cell structure consisting of four cell types: three antipodal cells, two synergid cells, one egg cell, and one central cell. Relatively little information is available on the genes expressed in the female gametophyte, since embryo sacs are small and it is difficult to isolate them free of maternal tissue contamination.
Recently, we identified 130 female gametophytic mutants from a population of insertional mutants that were analyzed in their genetic and molecular characteristics (Pagnussat et al., 2005). However, it is estimated that several thousand genes could be required for female gametophyte development (for review, see Drews and Yadegari, 2002), and alternate strategies are needed to identify additional genes involved in this developmental process. In animals, the oocyte transcriptome has been studied in humans, Caenorhabditis elegans, mice, bovine, and rainbow trout using cDNA or oligonucleotide arrays (Miller et al., 2003; Dobson et al., 2004; Hamatani et al., 2004; Yao et al., 2004; von Schalburg et al., 2005). In Arabidopsis, the Affymetrix ATH1 oligonucleotide chip, which contains probe sets for 22,591 annotated genes, has been used to study expression profiles at the whole-genome level. While genomic expression profiles have been used to identify genes related to reproduction, including seed development, floral organs (petal, sepal, stamen, and carpel), and male gametophytes by cDNA, oligonucleotide arrays (Honys and Twell, 2003, 2004; Köhler et al., 2003; Hennig et al., 2004; Wellmer et al., 2004), or peptide sequencing (Mayfield et al., 2001), a study of the female gametophyte transcriptome has not been performed at the whole-genome level.
We have previously described a gene called SPOROCYTELESS (SPL), which we showed to be required for initiation of both microsporogenesis and megasporogenesis in Arabidopsis (Yang et al., 1999). The SPL gene, which is identical to the NOZZLE gene (Schiefthaler et al., 1999), encodes a putative transcription factor expressed in the sporogenous cells of the anther and the ovule. Overexpression of SPL in wild-type flowers has no phenotype (Yang et al., 1999; Ito et al., 2004), but in agamous mutant flowers ectopic SPL can induce sporogenesis in petals (Ito et al., 2004). In this study, we identify genes involved in female gametophyte development through gene expression by comparison of wild-type ovules (embryo sac+) versus spl mutant ovules (embryo sac−) at the whole-genome level using the Affymetrix ATH1 oligonucleotide array. In addition, we determined the expression patterns of selected genes using promoter∷β-glucuronidase (GUS) fusions and found that the predictions based on the microarray analysis are in good agreement with the actual expression patterns. Our study provides a dataset of genes that are likely to be specific or preferentially expressed components of the female gametophyte transcriptome.
RESULTS
Sample Preparation to Analyze the Transcripts Related to Female Gametophyte Development
To characterize genes involved in embryo sac development, we set out to identify the genes having specific or increased expression in the ovules of heterozygous siblings (spl/SPL) compared to those of homozygous spl mutants (spl/spl) using the Affymetrix ATH1 oligonucleotide array. The spl mutation was identified by its complete male and female sterility as a single recessive mutation (Yang et al., 1999). The megasporocyte development in ovules of homozygous spl mutants is arrested at the archesporial cell and fails to undergo meiosis. Although megasporocytes are not formed and nucellus are arrested until the completion of integuments development, both inner and outer integuments and endothelium differentiated normally as in wild-type ovules (Fig. 1). Two ovule samples were collected according to different floral stages (Table I). The “early staged ovule” sample was collected from flowers of late 11 stage that have green anthers, and petals and long stamens of the same length, and mid-12 stage that have yellow anthers and in which the length of petal is longer than that of long stamen. The “late staged ovule” sample was obtained from flowers of late 12 stage that show protruded stigma from green bud, and 13 stage that have white bending petals and in which the length of stigma and long stamen is the same (Bowman, 1994). Additional precautions were taken when late staged ovules of heterozygous plants were collected to avoid pistil pollination. We detached anthers from flowers of early 12 stage and collected ovules after the flowers reached the appropriate floral stages (late 12 to 13). The collected ovules of each sample were randomly cleared, and the embryo sac stages were confirmed under the differential interference contrast microscope. The early staged ovules from heterozygotes contained functional megaspores or two- or four-nucleated embryo sacs (FG1–FG4), and the late staged ovules contained eight-nucleated embryo sacs or mature embryo sacs (FG5–FG7). The experiments were performed in triplicate. The three samples of each stage consisting of about 2,500 to approximately 3,000 ovules were independently collected from at least six different plants to minimize any effects of plant-to-plant transcriptional variations, and labeled RNAs made from each sample were hybridized separately with an Affymetrix ATH1 array. These ovule populations were enough to make the RNA probe for direct microarray hybridization without any amplification step. In the case of late staged ovules, we prepared two additional spl ovule samples from spl emasculated flowers to verify that detaching the anthers would not significantly affect the expression profile of the ovules.
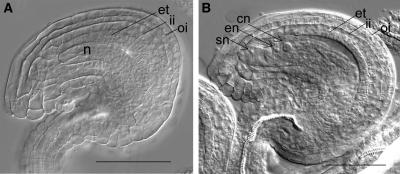
Figure 1.
Comparison of a homozygous spl mutant ovule with sibling heterozygous ovule, both at floral stage 13. A, spl mutant ovule showing the complete absence of the embryo sac but presence of all maternal cell types. B, Heterozygous spl plant ovule showing mature embryo sac. Abbreviations: cn, central cell nuclei; en, egg cell nucleus; et, endothelium; ii, inner integument; n, nucellus; oi, outer integument; sn, synergid nucleus. Scale bars represent 50 μm.
Table I.
The ovule sample collected on the basis of stages of flowers and embryo sacs
| Sample Division | Floral Stagea | Embryo Sac Stageb |
|---|---|---|
| Early staged ovule | Late 11 | FG1 (one nucleate) |
| Early 12 | FG2 (early two nucleate) | |
| FG3 (late two nucleate) | ||
| Mid 12 | FG4 (four nucleate) | |
| Late staged ovule | Late 12 | FG5 (eight nucleate) |
| 13 | FG6 (seven celled) | |
| FG7 (four celled) |
Floral stages are described by Bowman (1994).
Embryo sac stages are summarized from Christensen et al. (1997).
Statistical Data Analysis of the Genes Involved in Female Gametophyte Development
Arabidopsis ATH1 whole-genome arrays, which contain oligos for 24,000 Arabidopsis gene sequences, were used to study comparative gene expressions during embryo sac development of heterozygous siblings (spl/SPL) and spl mutants (spl/spl). Signal intensities were calculated by a perfect match (PM)-only model (Li and Wong, 2001a) that uses only PM probes to calculate all-positive expression values as a statistical algorithm using the dChip program (version 1.3) freely available at http://www.dchip.org/ (Li and Wong, 2001b). The median intensities and Present call% obtained by the dChip program were used to assess the overall quality of the arrays or differences between arrays, and hybridization controls such as bioB, bioC, bioD, and cre and internal control genes such as actin and GAPDH were used to evaluate sample hybridization efficiency to gene expression arrays and RNA sample quality. As a result, all analyzed arrays were judged to have high quality (data not shown).
We first calculated the correlation coefficient to ensure the reliability and determine the reproducibility of the microarray analysis using average difference based on the signal intensities between arrays using statistic package R program (version 1.6.1; http://www.r-project.org/; Ihaka and Gentleman, 1996). All 14 arrays had high correlation coefficients of >0.94, which suggested an excellent reproducibility among individual samples. Especially strong positive correlation between samples of the same kind was demonstrated with the correlation coefficients of ≥0.98 (Supplemental Table I).
The signal values of data were not normalized, and these raw signal data were used for data analysis without any filtering methods, such as corrections for between-chip heterogeneity and eliminations of backgrounds, because we did not want to remove potential genes of interest (Thomas et al., 2001). The following three screening criteria were applied for data analysis.
First, we applied the conventional t test to the raw signal data. This t test provides the probability (P) that a difference in gene expression occurred by chance. The P values were calculated by Student's t test for two-sample equal variance (homoscedastic; Devore and Peck, 1997; Pan, 2002). The genes were judged significantly changed when they were assigned a P value of <0.005. The signal values from ovules of spl mutants were used as a baseline. In late staged ovule data, signal values of emasculated spl and that of nonemasculated spl were combined and used as signal values of spl mutants at late stage. A total of 508 genes at early staged ovule and 1,015 genes at late staged ovule satisfied this criterion.
Second, we applied a 2-fold cutoff for the genes with <0.005 P value and retained only the genes that showed 2-fold or greater expression in heterozygous ovules (embryo sac+) as compared to spl mutant ovules (embryo sac−). According to these criteria, 107 genes at early staged ovule and 248 genes at late staged ovule were identified.
Third, genes with hybridization signals of <60 in heterozygous ovules were removed on the basis of signal intensities of the poly-A controls (dap, lys, phe, thr, trp) of Affymetrix ATH1.
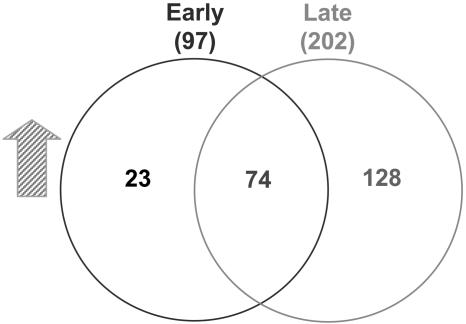
When we applied all three criteria, 23 genes were identified in early staged ovules, 128 genes in late staged ovules, and 74 genes in both early and late staged ovules (Fig. 2). We also compared the signal intensities of late staged nonemasculated (Supplemental Fig. 1) spl ovules with late staged wild-type ovules. We found that the results from this comparison were similar to the comparison of the combined late staged emasculated and nonemasculated spl ovules with the late staged wild-type ovules, indicating that emasculation did not significantly affect the outcome (Fig. 2). The whole data set for 14 arrays is available at the laboratory Web site (http://sundarlab.ucdavis.edu/) and in the supplemental data.
Figure 2.
Identified genes expressed during female gametophyte development. The gene sets included in the Venn diagrams were ≥2-fold higher in wild type than in spl with P values of <0.005 and signal values of >60.
Functional Classification and Identification of the Embryo Sac Genes
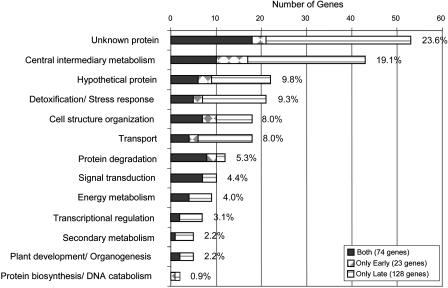
We classified functionally the 225 genes that are predicted to be expressed during megagametogenesis on the basis of the biological or biochemical function of the gene ontology annotation for the Arabidopsis genome provided by The Arabidopsis Information Resource at www.arabidopsis.org. Using the current annotation, 22 (9.8%) of the 225 genes encode predicted hypothetical proteins, and 53 genes (23.6%) encode proteins with an expressed sequence tag match but without any protein match (unknown proteins) or unclassified proteins as shown in Figure 3. These numbers indicate an overrepresentation of the classes of unknown and hypothetical proteins, representing approximately 33% of the female gametophyte genes in this study, as compared to approximately 21% for the whole Arabidopsis genome. The remaining genes were distributed across all the major classification groups from central metabolism, detoxification/stress response, cell structure organization, and transport, to protein degradation, signal transduction, and transcriptional regulation. The detailed gene information for genes within each classification group is provided in Table II.
Figure 3.
Distribution and classification of the female gametophyte genes.
Table II.
Identification of embryo sac genes
Genes were selected on the basis of the P value of <0.0050, fold change of >2.0, and signal intensity of >60.0 at wild-type siblings. WT1–3, Mean of signal intensities for arrays 1 to 3 of wild-type siblings. spl1–3, Mean of signal intensities for arrays 1 to 3 of spl mutants. WTd1–d3, Mean of signal intensities for arrays 1 to 3 of emasculated wild-type siblings. spl1–3, d1–d2, Mean of signal intensities for arrays 1 to 3 of nonemasculated spl, and arrays 1 and 2 of emasculated spl.
| Affymetrix Codea
|
Gene IDb
|
Description
|
Early Staged Ovule
|
Late Staged Ovule
|
Divisione
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT1–3 | spl1–3 | FCc | P Valued | WTd1–d3 | spl1–3, d1–d2 | FCc | P Valued | ||||
| Unknown protein | |||||||||||
| 261271_at | At1g26795 | Self-incompatibility protein related | 769 | 30 | 25.3 | 0.000083 | 761 | 27 | 28.2 | 0.000000 | Both |
| 265133_s_at | At1g51250 | Expressed protein | 151 | 25 | 6.1 | 0.001456 | 539 | 19 | 28.2 | 0.000000 | Both |
| 259726_at | At1g60985 | Expressed protein | 98 | 16 | 5.9 | 0.002140 | 349 | 16 | 21.4 | 0.000003 | Both |
| 265762_at | At2g01240 | Reticulon family protein (RTNLB15) | 227 | 15 | 15.1 | 0.000418 | 148 | 14 | 10.3 | 0.000020 | Both |
| 263713_at | At2g20595 | Expressed protein | 278 | 15 | 18.9 | 0.003556 | 1,047 | 25 | 41.4 | 0.000000 | Both |
| 267193_at | At2g30900 | Expressed protein | 97 | 23 | 4.3 | 0.000554 | 108 | 20 | 5.4 | 0.000016 | Both |
| 256719_at | At2g34130 | CACTA-like transposase family (Ptta/En/Spm) | 117 | 11 | 10.3 | 0.000181 | 114 | 10 | 11.8 | 0.000000 | Both |
| 256600_at | At3g14850 | Expressed protein | 261 | 34 | 7.6 | 0.000088 | 294 | 35 | 8.5 | 0.000000 | Both |
| 257889_at | At3g17080 | Self-incompatibility protein related | 187 | 30 | 6.2 | 0.002931 | 342 | 19 | 17.8 | 0.000000 | Both |
| 251698_at | At3g56610 | Expressed protein | 518 | 16 | 32.0 | 0.004682 | 1,444 | 31 | 46.8 | 0.000000 | Both |
| 254257_s_at | At4g23350 | Expressed protein | 104 | 30 | 3.5 | 0.003879 | 335 | 22 | 15.3 | 0.000000 | Both |
| 253164_at | At4g35725 | Expressed protein | 158 | 15 | 10.4 | 0.003044 | 298 | 14 | 21.2 | 0.000024 | Both |
| 250871_at | At5g03930 | Expressed protein | 171 | 24 | 7.0 | 0.001971 | 106 | 14 | 7.5 | 0.000002 | Both |
| 250325_s_at | At5g12060 | Self-incompatibility protein related | 138 | 41 | 3.4 | 0.001255 | 206 | 30 | 6.8 | 0.000001 | Both |
| 249855_at | At5g22970 | Expressed protein | 261 | 25 | 10.6 | 0.003517 | 1,028 | 32 | 31.7 | 0.000001 | Both |
| 249757_at | At5g24316 | Pro-rich family protein | 471 | 17 | 27.7 | 0.001766 | 1,328 | 29 | 45.6 | 0.000000 | Both |
| 249401_at | At5g40260 | Nodulin MtN3 family protein | 566 | 37 | 15.2 | 0.000603 | 427 | 30 | 14.3 | 0.000004 | Both |
| 248284_at | At5g52975 | Expressed protein | 755 | 81 | 9.3 | 0.000353 | 403 | 49 | 8.2 | 0.000001 | Both |
| 262503_at | At1g21670 | Expressed protein | 161 | 41 | 4.0 | 0.000009 | 64 | 45 | 1.4 | 0.031616 | Early |
| 261731_s_at | At1g47780 | Acyl-protein thioesterase related | 84 | 15 | 5.6 | 0.004140 | 60 | 13 | 4.7 | 0.000003 | Early |
| 249375_at | At5g40730 | Arabinogalactan protein (AGP24) | 806 | 129 | 6.2 | 0.000342 | 661 | 575 | 1.1 | 0.245297 | Early |
| 264610_at | At1g04645 | Self-incompatibility protein related | 106 | 18 | 5.7 | 0.157950 | 793 | 21 | 38.1 | 0.000557 | Late |
| 261846_at | At1g11540 | Expressed protein | 145 | 107 | 1.4 | 0.117890 | 170 | 85 | 2.0 | 0.000012 | Late |
| 255908_s_at | At1g18010 | Expressed protein | 128 | 64 | 2.0 | 0.018058 | 93 | 45 | 2.1 | 0.000005 | Late |
| 256079_at | At1g20680 | Expressed protein | 56 | 14 | 3.9 | 0.011798 | 67 | 12 | 5.7 | 0.000019 | Late |
| 263027_at | At1g24010 | Expressed protein | 23 | 12 | 1.9 | 0.256580 | 96 | 22 | 4.3 | 0.000001 | Late |
| 260942_s_at | At1g45190 | Expressed protein | 318 | 6 | 49.3 | 0.007497 | 1,404 | 30 | 47.1 | 0.000000 | Late |
| 265138_at | At1g51300 | Acyl-protein thioesterase related | 192 | 43 | 4.4 | 0.015275 | 450 | 49 | 9.1 | 0.000000 | Late |
| 262314_at | At1g70810 | C2 domain-containing protein | 23 | 17 | 1.4 | 0.182775 | 65 | 17 | 3.8 | 0.000004 | Late |
| 262972_at | At1g75620 | Glyoxal oxidase related | 67 | 49 | 1.4 | 0.205253 | 133 | 38 | 3.5 | 0.000079 | Late |
| 264297_at | At1g78710 | Expressed protein | 120 | 62 | 1.9 | 0.005001 | 144 | 57 | 2.5 | 0.000001 | Late |
| 267241_at | At2g02490 | Hydroxyproline-rich glycoprotein family protein | 45 | 4 | 10.6 | 0.052466 | 395 | 11 | 37.0 | 0.000186 | Late |
| 267218_at | At2g02515 | Expressed protein | 163 | 39 | 4.2 | 0.007206 | 838 | 34 | 24.8 | 0.000001 | Late |
| 265517_at | At2g06090 | Self-incompatibility protein related | 254 | 14 | 17.8 | 0.013973 | 1,109 | 23 | 47.4 | 0.000000 | Late |
| 264590_at | At2g17710 | Expressed protein | 144 | 194 | 0.7 | 0.361347 | 841 | 394 | 2.1 | 0.000010 | Late |
| 263518_at | At2g21655 | Expressed protein | 271 | 17 | 15.8 | 0.015412 | 946 | 25 | 37.6 | 0.000001 | Late |
| 265674_at | At2g32190 | Expressed protein | 100 | 61 | 1.6 | 0.072973 | 198 | 70 | 2.8 | 0.000267 | Late |
| 265670_s_at | At2g32210 | Expressed protein | 155 | 158 | 1.0 | 0.888298 | 213 | 76 | 2.8 | 0.000186 | Late |
| 265245_at | At2g43060 | Expressed protein | 39 | 45 | 0.9 | 0.564913 | 131 | 62 | 2.1 | 0.000340 | Late |
| 259107_at | At3g05460 | Sporozoite surface protein related | 403 | 43 | 9.4 | 0.008348 | 1,415 | 46 | 31.1 | 0.000069 | Late |
| 258130_at | At3g24510 | Expressed protein | 486 | 10 | 48.6 | 0.012987 | 2,033 | 38 | 53.8 | 0.000000 | Late |
| 252253_at | At3g49300 | Pro-rich family protein | 251 | 42 | 6.0 | 0.007754 | 626 | 32 | 19.3 | 0.000044 | Late |
| 251606_at | At3g57840 | Self-incompatibility protein related | 109 | 39 | 2.8 | 0.058391 | 468 | 38 | 12.4 | 0.000005 | Late |
| 255207_at | At4g07515 | Expressed protein | 453 | 23 | 19.8 | 0.008275 | 859 | 32 | 27.0 | 0.000000 | Late |
| 245424_at | At4g17505 | Expressed protein | 157 | 55 | 2.8 | 0.005099 | 312 | 33 | 9.4 | 0.000003 | Late |
| 254494_at | At4g20050 | Expressed protein | 125 | 85 | 1.5 | 0.069310 | 230 | 59 | 3.9 | 0.000001 | Late |
| 254001_at | At4g26260 | Expressed protein | 174 | 62 | 2.8 | 0.067591 | 149 | 25 | 5.9 | 0.000073 | Late |
| 253724_at | At4g29285 | Expressed protein | 91 | 12 | 7.3 | 0.029036 | 647 | 16 | 39.5 | 0.000000 | Late |
| 253656_at | At4g30090 | Expressed protein | 101 | 85 | 1.2 | 0.276358 | 76 | 37 | 2.1 | 0.000137 | Late |
| 253401_at | At4g32870 | Expressed protein | 88 | 41 | 2.1 | 0.040870 | 165 | 39 | 4.3 | 0.000012 | Late |
| 246641_s_at | At5g34885 | Expressed protein | 216 | 21 | 10.3 | 0.025229 | 631 | 27 | 23.5 | 0.000000 | Late |
| 249179_at | At5g42955 | Expressed protein | 266 | 14 | 19.3 | 0.014861 | 882 | 23 | 38.0 | 0.000000 | Late |
| 248892_at | At5g46300 | Expressed protein | 62 | 5 | 11.8 | 0.014887 | 124 | 5 | 23.1 | 0.000001 | Late |
| Central intermediary metabolism | |||||||||||
| 259786_at | At1g29660 | GDSL-motif lipase/hydrolase family protein | 289 | 84 | 3.4 | 0.003166 | 879 | 425 | 2.1 | 0.000131 | Both |
| 260124_at | At1g36340 | Ubiquitin-conjugating enzyme, E2 | 584 | 38 | 15.3 | 0.001661 | 467 | 34 | 13.8 | 0.000002 | Both |
| 245672_at | At1g56710 | Glycoside hydrolase family 28 protein | 218 | 85 | 2.6 | 0.004220 | 235 | 94 | 2.5 | 0.000298 | Both |
| 257442_at | At2g28680 | Cupin family protein | 316 | 47 | 6.8 | 0.001424 | 303 | 33 | 9.2 | 0.000008 | Both |
| 267408_at | At2g34890 | CTP synthase, putative | 152 | 25 | 6.1 | 0.000651 | 67 | 16 | 4.2 | 0.000011 | Both |
| 257243_at | At3g24230 | Pectate lyase family protein | 332 | 59 | 5.7 | 0.003792 | 198 | 63 | 3.2 | 0.000001 | Both |
| 258763_s_at | At3g30540 | (1-4)-β-Mannan endohydrolase family | 125 | 8 | 15.9 | 0.002254 | 148 | 9 | 17.4 | 0.000000 | Both |
| 252342_at | At3g48950 | Glycoside hydrolase family 28 protein | 348 | 30 | 11.6 | 0.001576 | 603 | 26 | 23.3 | 0.000000 | Both |
| 248925_at | At5g45910 | GDSL-motif lipase/hydrolase-like protein | 279 | 14 | 20.0 | 0.001613 | 1,367 | 26 | 53.5 | 0.000000 | Both |
| 247228_at | At5g65140 | Trehalose-6-phosphate phosphatase | 305 | 38 | 8.0 | 0.000647 | 246 | 92 | 2.7 | 0.000044 | Both |
| 264147_at | At1g02200 | CER1 protein | 206 | 85 | 2.4 | 0.000205 | 40 | 39 | 1.0 | 0.799875 | Early |
| 264146_at | At1g02205 | CER1 protein, At1g02200 | 650 | 279 | 2.3 | 0.000021 | 219 | 225 | 1.0 | 0.820363 | Early |
| 259703_at | At1g77790 | Glycosyl hydrolase family 17 protein | 113 | 16 | 7.0 | 0.000428 | 54 | 11 | 4.8 | 0.000000 | Early |
| 267202_s_at | At2g31030 | Oxysterol-binding family protein | 74 | 10 | 7.3 | 0.000864 | 27 | 7 | 3.8 | 0.000005 | Early |
| 260611_at | At2g43670 | Glycosyl hydrolase family 17 protein | 132 | 53 | 2.5 | 0.002326 | 129 | 75 | 1.7 | 0.000212 | Early |
| 252320_at | At3g48580 | Xyloglucan:xyloglucosyl transferase, putative | 743 | 315 | 2.4 | 0.003795 | 1,176 | 766 | 1.5 | 0.004917 | Early |
| 250082_at | At5g17200 | Glycoside hydrolase family 28 protein | 71 | 17 | 4.2 | 0.002910 | 20 | 11 | 1.8 | 0.000578 | Early |
| 260947_at | At1g06020 | PfkB-type carbohydrate kinase family protein | 58 | 40 | 1.5 | 0.010463 | 75 | 29 | 2.6 | 0.000134 | Late |
| 259391_s_at | At1g06350 | Fatty acid desaturase family protein | 163 | 98 | 1.7 | 0.342723 | 196 | 88 | 2.2 | 0.000376 | Late |
| 255956_at | At1g22015 | Galactosyltransferase family protein | 158 | 43 | 3.7 | 0.007113 | 274 | 34 | 8.0 | 0.000000 | Late |
| 245792_at | At1g32100 | Pinoresinol-lariciresinol reductase, putative | 66 | 66 | 1.0 | 0.993453 | 184 | 68 | 2.7 | 0.000127 | Late |
| 245794_at | At1g32170 | Xyloglucan:xyloglucosyl transferase, putative | 59 | 49 | 1.2 | 0.363717 | 101 | 47 | 2.1 | 0.000150 | Late |
| 256211_at | At1g50960 | Gibberellin 20-oxidase related | 42 | 19 | 2.2 | 0.015340 | 75 | 14 | 5.4 | 0.000000 | Late |
| 260333_at | At1g70500 | Polygalacturonase, putative/pectinase, putative | 581 | 27 | 21.7 | 0.007959 | 674 | 42 | 16.0 | 0.000001 | Late |
| 260066_at | At1g73610 | GDSL-motif lipase/hydrolase family protein | 69 | 24 | 2.9 | 0.035163 | 163 | 20 | 8.0 | 0.000056 | Late |
| 260259_at | At1g74300 | Esterase/lipase/thioesterase family protein | 60 | 57 | 1.1 | 0.690243 | 105 | 47 | 2.2 | 0.000095 | Late |
| 265331_at | At2g18420 | Gibberellin-responsive protein, putative | 27 | 25 | 1.1 | 0.777953 | 73 | 18 | 4.1 | 0.000139 | Late |
| 267607_s_at | At2g26740 | Epoxide hydrolase, soluble (sEH) | 138 | 137 | 1.0 | 0.977676 | 205 | 87 | 2.4 | 0.000242 | Late |
| 267337_at | At2g39980 | Transferase family protein | 93 | 46 | 2.0 | 0.015251 | 69 | 30 | 2.3 | 0.000006 | Late |
| 260559_at | At2g43860 | Polygalacturonase, putative/pectinase, putative | 113 | 38 | 3.0 | 0.005920 | 69 | 30 | 2.3 | 0.000032 | Late |
| 258767_at | At3g10890 | (1-4)-β-Mannan endohydrolase, putative | 100 | 16 | 6.3 | 0.013753 | 698 | 20 | 35.1 | 0.000002 | Late |
| 258151_at | At3g18080 | Glycosyl hydrolase family 1 protein | 199 | 194 | 1.0 | 0.899935 | 384 | 124 | 3.1 | 0.000003 | Late |
| 257065_at | At3g18220 | Phosphatidic acid phosphatase family protein | 45 | 29 | 1.5 | 0.034020 | 63 | 22 | 2.8 | 0.000003 | Late |
| 251491_at | At3g59480 | PfkB-type carbohydrate kinase family protein | 549 | 56 | 9.9 | 0.032782 | 598 | 210 | 2.8 | 0.001882 | Late |
| 255550_at | At4g01970 | Galactinol-raffinose galactosyltransferase, putative | 127 | 41 | 3.1 | 0.006794 | 226 | 83 | 2.7 | 0.000170 | Late |
| 245349_at | At4g16690 | Esterase/lipase/thioesterase family protein | 75 | 40 | 1.9 | 0.193063 | 112 | 53 | 2.1 | 0.000368 | Late |
| 254609_at | At4g18970 | GDSL-motif lipase/hydrolase family protein | 461 | 508 | 0.9 | 0.189942 | 703 | 309 | 2.3 | 0.000085 | Late |
| 246498_at | At5g16230 | Acyl-[acyl-carrier-protein] desaturase, putative | 34 | 26 | 1.3 | 0.219929 | 84 | 23 | 3.6 | 0.001229 | Late |
| 249983_at | At5g18470 | Curculin-like (mannose-binding) lectin family protein | 44 | 35 | 1.2 | 0.486888 | 106 | 48 | 2.2 | 0.001507 | Late |
| 246774_at | At5g27530 | Glycoside hydrolase family 28 protein | 108 | 46 | 2.4 | 0.012277 | 146 | 31 | 4.8 | 0.000001 | Late |
| 249474_s_at | At5g39190 | Germin-like protein (GER2) | 14 | 12 | 1.2 | 0.500732 | 99 | 13 | 7.6 | 0.000004 | Late |
| 248812_at | At5g47330 | Palmitoyl protein thioesterase family protein | 63 | 78 | 0.8 | 0.143704 | 203 | 43 | 4.7 | 0.000004 | Late |
| 248791_at | At5g47350 | Palmitoyl protein thioesterase family protein | 212 | 65 | 3.3 | 0.040644 | 1,167 | 291 | 4.0 | 0.000003 | Late |
| Hypothetical protein | |||||||||||
| 260318_at | At1g63960 | Hypothetical protein | 62 | 20 | 3.2 | 0.002924 | 72 | 20 | 3.5 | 0.000014 | Both |
| 265579_at | At2g20070 | Hypothetical protein | 661 | 48 | 13.8 | 0.000041 | 487 | 36 | 13.4 | 0.000000 | Both |
| 257434_at | At2g21740 | Hypothetical protein | 128 | 25 | 5.0 | 0.001238 | 301 | 20 | 15.3 | 0.000000 | Both |
| 252753_at | At3g43500 | Hypothetical protein | 80 | 20 | 4.0 | 0.002002 | 131 | 20 | 6.6 | 0.000002 | Both |
| 246859_at | At5g25950 | Hypothetical protein | 70 | 13 | 5.4 | 0.000045 | 131 | 12 | 10.9 | 0.000000 | Both |
| 248225_at | At5g53740 | Hypothetical protein | 168 | 72 | 2.3 | 0.000851 | 111 | 52 | 2.1 | 0.000065 | Both |
| 266706_at | At2g03320 | Hypothetical protein | 97 | 28 | 3.5 | 0.001116 | 47 | 17 | 2.8 | 0.000175 | Early |
| 246866_at | At5g25960 | Hypothetical protein | 131 | 11 | 11.7 | 0.000000 | 41 | 9 | 4.8 | 0.000020 | Early |
| 248396_at | At5g52130 | Hypothetical protein | 111 | 21 | 5.4 | 0.000609 | 47 | 15 | 3.2 | 0.000216 | Early |
| 257468_at | At1g47470 | Hypothetical protein | 147 | 13 | 11.2 | 0.029273 | 1,010 | 25 | 40.1 | 0.000000 | Late |
| 261313_at | At1g52970 | Hypothetical protein | 484 | 49 | 9.9 | 0.007905 | 1,881 | 59 | 32.1 | 0.000000 | Late |
| 259944_at | At1g71470 | Hypothetical protein | 72 | 60 | 1.2 | 0.213121 | 71 | 33 | 2.1 | 0.000104 | Late |
| 263895_at | At2g21920 | Hypothetical protein | 107 | 26 | 4.2 | 0.007542 | 73 | 17 | 4.2 | 0.000906 | Late |
| 258798_at | At3g04540 | Hypothetical protein | 202 | 55 | 3.7 | 0.087047 | 1,542 | 50 | 30.9 | 0.000000 | Late |
| 256773_at | At3g13630 | Hypothetical protein | 31 | 28 | 1.1 | 0.715677 | 60 | 27 | 2.2 | 0.000839 | Late |
| 251607_at | At3g57850 | Hypothetical protein | 73 | 11 | 6.6 | 0.036056 | 370 | 13 | 29.4 | 0.000050 | Late |
| 255029_x_at | At4g09470 | Hypothetical protein | 139 | 8 | 17.2 | 0.012714 | 687 | 16 | 43.5 | 0.000003 | Late |
| 255804_at | At4g10220 | Hypothetical protein | 120 | 31 | 3.9 | 0.009652 | 410 | 34 | 12.0 | 0.000003 | Late |
| 254692_at | At4g17860 | Hypothetical protein | 60 | 35 | 1.7 | 0.028983 | 107 | 28 | 3.9 | 0.000008 | Late |
| 246472_at | At5g17130 | Hypothetical protein | 80 | 83 | 1.0 | 0.853233 | 75 | 34 | 2.2 | 0.000487 | Late |
| 249157_at | At5g43510 | Hypothetical protein | 356 | 13 | 27.3 | 0.012178 | 588 | 29 | 20.1 | 0.000001 | Late |
| 247245_at | At5g64720 | Hypothetical protein | 105 | 15 | 6.9 | 0.010392 | 322 | 13 | 24.5 | 0.000000 | Late |
| Detoxification/stress response | |||||||||||
| 264001_at | At2g22420 | Peroxidase 17 (PER17) | 791 | 121 | 6.5 | 0.003718 | 725 | 120 | 6.1 | 0.000007 | Both |
| 265264_at | At2g42930 | Glycosyl hydrolase family protein 17 | 346 | 55 | 6.3 | 0.003492 | 285 | 34 | 8.5 | 0.000007 | Both |
| 254000_at | At4g26250 | Galactinol synthase induced by water stress | 89 | 18 | 4.8 | 0.003892 | 65 | 15 | 4.2 | 0.000004 | Both |
| 252951_at | At4g38700 | Disease resistance-responsive family protein | 248 | 109 | 2.3 | 0.004228 | 706 | 115 | 6.2 | 0.000000 | Both |
| 247857_at | At5g58400 | Peroxidase, putative | 1,053 | 240 | 4.4 | 0.000004 | 473 | 161 | 2.9 | 0.000023 | Both |
| 261410_at | At1g07610 | Metallothionein-like protein 1C (MT-1C) | 333 | 159 | 2.1 | 0.000832 | 238 | 170 | 1.4 | 0.168590 | Early |
| 266743_at | At2g02990 | Ribonuclease 1 (RNS1) | 67 | 23 | 2.9 | 0.001354 | 140 | 100 | 1.4 | 0.201557 | Early |
| 263026_at | At1g24000 | Bet v I allergen family protein | 43 | 15 | 2.9 | 0.266278 | 250 | 19 | 12.9 | 0.000001 | Late |
| 265920_s_at | At2g15120 | Pseudogene, disease-resistance family protein | 38 | 33 | 1.1 | 0.299880 | 103 | 23 | 4.6 | 0.000933 | Late |
| 266562_at | At2g23970 | Defense-related protein, putative | 68 | 60 | 1.1 | 0.479766 | 194 | 38 | 5.1 | 0.000000 | Late |
| 267138_s_at | At2g38210 | Ethylene-responsive protein, putative | 181 | 86 | 2.1 | 0.091609 | 246 | 76 | 3.2 | 0.001233 | Late |
| 266169_at | At2g38900 | Ser protease inhibitor | 40 | 37 | 1.1 | 0.867313 | 343 | 105 | 3.3 | 0.000002 | Late |
| 260557_at | At2g43610 | Glycoside hydrolase family 19 protein | 94 | 49 | 1.9 | 0.003261 | 76 | 33 | 2.3 | 0.000002 | Late |
| 258791_at | At3g04720 | Hevein-like protein (HEL) | 35 | 35 | 1.0 | 0.937369 | 91 | 34 | 2.6 | 0.003414 | Late |
| 258172_at | At3g21620 | Early responsive to dehydration protein related | 60 | 24 | 2.5 | 0.000597 | 68 | 21 | 3.2 | 0.000002 | Late |
| 254098_at | At4g25100 | Superoxide dismutase (Fe), chloroplast (SODB) | 688 | 313 | 2.2 | 0.060518 | 76 | 14 | 5.6 | 0.000220 | Late |
| 253655_at | At4g30070 | Plant defensin-fusion protein, putative | 99 | 39 | 2.5 | 0.007716 | 167 | 23 | 7.4 | 0.000001 | Late |
| 250200_at | At5g14130 | Peroxidase, putative | 94 | 67 | 1.4 | 0.229669 | 106 | 41 | 2.6 | 0.000012 | Late |
| 250083_at | At5g17220 | Glutathione S-transferase-like protein | 99 | 93 | 1.1 | 0.870900 | 537 | 235 | 2.3 | 0.000020 | Late |
| 249560_at | At5g38330 | Plant defensin-fusion protein, putative | 280 | 26 | 11.0 | 0.008435 | 1,140 | 29 | 39.5 | 0.000000 | Late |
| 249527_at | At5g38710 | Pro oxidase, putative | 70 | 18 | 3.9 | 0.006176 | 74 | 15 | 4.8 | 0.000019 | Late |
| Cell structure organization | |||||||||||
| 260573_at | At2g47280 | Pectinesterase family protein | 227 | 36 | 6.4 | 0.000454 | 139 | 25 | 5.5 | 0.000000 | Both |
| 257878_at | At3g17150 | Invertase/pectin methylesterase inhibitor family | 399 | 26 | 15.3 | 0.004375 | 1,121 | 38 | 29.2 | 0.000000 | Both |
| 258438_at | At3g17230 | Invertase/pectin methylesterase inhibitor family | 434 | 30 | 14.4 | 0.000924 | 758 | 30 | 24.8 | 0.000000 | Both |
| 251748_at | At3g55680 | Invertase/pectin methylesterase inhibitor family | 162 | 51 | 3.2 | 0.000901 | 112 | 39 | 2.8 | 0.000002 | Both |
| 255699_at | At4g00190 | Putative pectinesterase | 86 | 31 | 2.8 | 0.001537 | 90 | 24 | 3.7 | 0.000001 | Both |
| 248823_s_at | At5g46960 | Invertase/pectin methylesterase inhibitor family | 270 | 8 | 35.1 | 0.002890 | 519 | 13 | 39.4 | 0.000003 | Both |
| 247246_at | At5g64620 | Invertase/pectin methylesterase inhibitor family | 747 | 338 | 2.2 | 0.000298 | 870 | 425 | 2.0 | 0.000000 | Both |
| 260802_at | At1g78400 | Glycoside hydrolase family 28 protein | 201 | 23 | 8.8 | 0.000001 | 30 | 18 | 1.7 | 0.001391 | Early |
| 258764_at | At3g10720 | Pectinesterase, putative | 415 | 205 | 2.0 | 0.001974 | 112 | 77 | 1.5 | 0.008263 | Early |
| 253725_at | At4g29340 | Profilin 3 (PRO3) (PFN3) | 63 | 27 | 2.3 | 0.000446 | 26 | 23 | 1.1 | 0.244366 | Early |
| 264500_at | At1g09370 | Pectinesterase inhibitor domain-containing protein | 61 | 10 | 6.0 | 0.123949 | 594 | 14 | 43.8 | 0.000012 | Late |
| 259613_at | At1g48010 | Invertase/pectin methylesterase inhibitor family | 38 | 36 | 1.1 | 0.597625 | 105 | 29 | 3.6 | 0.000000 | Late |
| 262083_at | At1g56100 | Pectinesterase inhibitor domain-containing protein | 128 | 53 | 2.4 | 0.444629 | 2,622 | 746 | 3.5 | 0.000012 | Late |
| 245656_at | At1g56620 | Pectinesterase inhibitor domain-containing protein | 50 | 26 | 1.9 | 0.044275 | 252 | 19 | 13.3 | 0.000000 | Late |
| 257679_at | At3g20470 | Pseudogene, Gly-rich protein | 117 | 99 | 1.2 | 0.700308 | 578 | 267 | 2.2 | 0.001050 | Late |
| 245371_at | At4g15750 | Invertase/pectin methylesterase inhibitor family | 200 | 63 | 3.2 | 0.067454 | 1,378 | 440 | 3.1 | 0.000004 | Late |
| 249962_at | At5g18990 | Pectinesterase family protein | 39 | 19 | 2.0 | 0.028245 | 82 | 15 | 5.5 | 0.000041 | Late |
| 247377_at | At5g63180 | Pectate lyase family protein | 175 | 125 | 1.4 | 0.147262 | 410 | 192 | 2.1 | 0.000518 | Late |
| Transport | |||||||||||
| 260319_at | At1g63950 | Heavy-metal-associated domain-containing protein | 310 | 28 | 10.9 | 0.002905 | 148 | 19 | 7.9 | 0.000001 | Both |
| 259757_at | At1g77510 | Protein disulfide isomerase, putative | 1,163 | 447 | 2.6 | 0.000344 | 943 | 367 | 2.6 | 0.000170 | Both |
| 258760_at | At3g10780 | Emp24/gp25L/p24 family protein | 435 | 94 | 4.6 | 0.001770 | 400 | 75 | 5.4 | 0.000000 | Both |
| 245892_at | At5g09370 | Protease inhibitor/seed storage/lipid transfer protein | 384 | 57 | 6.8 | 0.004630 | 1,326 | 108 | 12.3 | 0.000002 | Both |
| 263765_at | At2g21540 | SEC14 cytosolic factor, putative | 135 | 64 | 2.1 | 0.000985 | 64 | 42 | 1.5 | 0.002042 | Early |
| 249346_at | At5g40780 | Lys- and His-specific transporter, putative | 783 | 170 | 4.6 | 0.002599 | 877 | 624 | 1.4 | 0.054790 | Early |
| 264520_at | At1g10010 | Amino acid permease, putative | 91 | 35 | 2.6 | 0.005939 | 167 | 34 | 4.8 | 0.000000 | Late |
| 265002_at | At1g24400 | Lys- and His-specific transporter | 156 | 41 | 3.8 | 0.024933 | 311 | 142 | 2.2 | 0.000005 | Late |
| 259580_at | At1g28030 | Oxidoreductase, 2OG-Fe(II) oxygenase family protein | 52 | 44 | 1.2 | 0.276557 | 95 | 32 | 3.0 | 0.000024 | Late |
| 265064_at | At1g61630 | Equilibrative nucleoside transporter, putative (ENT7) | 70 | 40 | 1.7 | 0.013220 | 67 | 29 | 2.3 | 0.000179 | Late |
| 259844_at | At1g73560 | Protease inhibitor/seed storage/lipid transfer protein | 49 | 22 | 2.2 | 0.005234 | 91 | 19 | 4.9 | 0.000122 | Late |
| 264482_at | At1g77210 | Sugar transporter, putative | 73 | 65 | 1.1 | 0.601658 | 118 | 55 | 2.1 | 0.000012 | Late |
| 257366_s_at | At2g03040 | Transmembrane protein related | 159 | 39 | 4.1 | 0.005632 | 184 | 22 | 8.3 | 0.000008 | Late |
| 266276_at | At2g29330 | Tropinone reductase, putative | 68 | 30 | 2.3 | 0.007410 | 68 | 28 | 2.4 | 0.000243 | Late |
| 254453_at | At4g21120 | Amino acid permease family protein | 211 | 64 | 3.3 | 0.007991 | 259 | 88 | 2.9 | 0.000000 | Late |
| 246887_at | At5g26250 | Sugar transporter, putative | 159 | 61 | 2.6 | 0.015475 | 146 | 37 | 4.0 | 0.000013 | Late |
| 248275_at | At5g53520 | Oligopeptide transporter OPT family protein | 43 | 32 | 1.3 | 0.204722 | 80 | 20 | 3.9 | 0.000002 | Late |
| 248019_at | At5g56480 | Protease inhibitor/seed storage/lipid transfer protein | 61 | 50 | 1.2 | 0.206709 | 111 | 38 | 2.9 | 0.000015 | Late |
| Protein degradation | |||||||||||
| 256486_at | At1g31450 | Aspartyl protease family protein | 263 | 18 | 14.8 | 0.001311 | 607 | 17 | 35.1 | 0.000000 | Both |
| 245738_at | At1g44130 | Nucellin protein, putative | 117 | 10 | 12.3 | 0.002988 | 94 | 8 | 11.2 | 0.000067 | Both |
| 259368_at | At1g69100 | Aspartyl protease family protein | 546 | 13 | 42.6 | 0.003756 | 952 | 33 | 29.1 | 0.000000 | Both |
| 252499_s_at | At3g46840 | Subtilase family protein | 389 | 34 | 11.6 | 0.000398 | 443 | 26 | 17.2 | 0.000001 | Both |
| 245589_at | At4g15040 | Subtilase family protein | 171 | 23 | 7.3 | 0.000154 | 66 | 17 | 4.0 | 0.000055 | Both |
| 254336_at | At4g22050 | Aspartyl protease family protein | 681 | 37 | 18.3 | 0.000096 | 714 | 67 | 10.6 | 0.000000 | Both |
| 246684_at | At5g33340 | Aspartyl protease family protein | 98 | 25 | 4.0 | 0.000228 | 189 | 21 | 9.1 | 0.000001 | Both |
| 247798_at | At5g58830 | Subtilase family protein | 73 | 15 | 5.0 | 0.002673 | 235 | 13 | 17.7 | 0.000000 | Both |
| 254237_at | At4g23520 | Cys proteinase, putative | 67 | 28 | 2.4 | 0.000243 | 41 | 22 | 1.9 | 0.000254 | Early |
| 247697_at | At5g59810 | Subtilase family protein | 92 | 43 | 2.1 | 0.002288 | 56 | 44 | 1.3 | 0.003872 | Early |
| 264067_x_at | At2g28010 | Aspartyl protease family protein | 33 | 19 | 1.7 | 0.039844 | 78 | 13 | 5.9 | 0.000004 | Late |
| 250345_at | At5g11940 | Subtilase family protein | 58 | 15 | 3.9 | 0.005591 | 154 | 14 | 11.4 | 0.000000 | Late |
| Signal transduction | |||||||||||
| 263740_at | At2g20660 | Rapid alkalinization factor (RALF) family protein | 82 | 33 | 2.4 | 0.000758 | 90 | 24 | 3.8 | 0.000000 | Both |
| 245158_at | At2g33130 | RALF family protein | 435 | 34 | 12.8 | 0.000989 | 437 | 38 | 11.4 | 0.000000 | Both |
| 266418_at | At2g38750 | Annexin 4 (ANN4) | 274 | 62 | 4.4 | 0.003825 | 220 | 67 | 3.3 | 0.000001 | Both |
| 251514_at | At3g59260 | Pirin, putative | 202 | 33 | 6.1 | 0.001924 | 171 | 30 | 5.7 | 0.000002 | Both |
| 255489_at | At4g02650 | Epsin N-terminal homology domain-containing protein | 448 | 22 | 20.7 | 0.004357 | 1,286 | 32 | 39.8 | 0.000000 | Both |
| 245177_at | At5g12380 | Annexin, putative | 190 | 17 | 11.2 | 0.000393 | 215 | 17 | 12.8 | 0.000000 | Both |
| 249013_at | At5g44700 | Leu-rich repeat transmembrane protein kinase | 81 | 31 | 2.6 | 0.001303 | 75 | 30 | 2.5 | 0.000305 | Both |
| 261285_at | At1g35720 | Annexin 1 (ANN1) | 168 | 122 | 1.4 | 0.273697 | 393 | 108 | 3.6 | 0.000012 | Late |
| 257869_at | At3g25160 | ER lumen protein retaining receptor family protein | 86 | 51 | 1.7 | 0.030431 | 147 | 52 | 2.8 | 0.000020 | Late |
| 250090_at | At5g17330 | Glutamate decarboxylase 1 (GAD 1) | 195 | 21 | 9.2 | 0.005864 | 222 | 25 | 9.0 | 0.000000 | Late |
| Energy metabolism | |||||||||||
| 259268_at | At3g01070 | Plastocyanin-like domain-containing protein | 79 | 28 | 2.8 | 0.002166 | 126 | 26 | 4.8 | 0.000001 | Both |
| 253634_at | At4g30590 | Plastocyanin-like domain-containing protein | 705 | 56 | 12.6 | 0.000057 | 500 | 41 | 12.3 | 0.000005 | Both |
| 252897_at | At4g39490 | Cytochrome P450 family protein, At4g38480 | 62 | 19 | 3.4 | 0.004614 | 134 | 18 | 7.5 | 0.000000 | Both |
| 248236_at | At5g53870 | Plastocyanin-like domain-containing protein | 1,236 | 60 | 20.7 | 0.000394 | 653 | 165 | 4.0 | 0.000009 | Both |
| 262528_at | At1g17260 | ATPase 10, plasma membrane type, putative | 45 | 26 | 1.7 | 0.385722 | 344 | 102 | 3.4 | 0.000002 | Late |
| 261396_at | At1g79800 | Plastocyanin-like domain-containing protein | 49 | 40 | 1.2 | 0.236124 | 69 | 31 | 2.2 | 0.000064 | Late |
| 266563_at | At2g23990 | Plastocyanin-like domain-containing protein | 195 | 47 | 4.2 | 0.011017 | 953 | 41 | 23.3 | 0.000015 | Late |
| 255690_at | At4g00360 | Cytochrome P450, putative | 49 | 23 | 2.1 | 0.018309 | 79 | 29 | 2.7 | 0.000026 | Late |
| 254489_at | At4g20800 | FAD-binding domain-containing protein | 47 | 11 | 4.4 | 0.027803 | 121 | 11 | 11.3 | 0.000000 | Late |
| Transcriptional regulation | |||||||||||
| 267528_at | At2g45650 | MADS-box protein (AGL6) | 245 | 59 | 4.2 | 0.002904 | 359 | 170 | 2.1 | 0.000048 | Both |
| 249338_at | At5g41090 | No apical meristem (NAM) family protein | 166 | 18 | 9.1 | 0.000764 | 77 | 13 | 5.8 | 0.000000 | Both |
| 260212_at | At1g74480 | RWP-RK domain-containing protein | 55 | 36 | 1.5 | 0.071151 | 65 | 29 | 2.2 | 0.001510 | Late |
| 266969_at | At2g39540 | Gibberellin-regulated family protein | 41 | 38 | 1.1 | 0.593532 | 179 | 32 | 5.6 | 0.000003 | Late |
| 254619_at | At4g18770 | Myb family transcription factor (MYB98) | 109 | 59 | 1.8 | 0.063530 | 110 | 41 | 2.7 | 0.000002 | Late |
| 251114_at | At5g01380 | Expressed protein | 25 | 26 | 1.0 | 0.855505 | 62 | 23 | 2.7 | 0.000211 | Late |
| 248240_at | At5g53950 | No apical meristem (NAM) family protein | 224 | 201 | 1.1 | 0.399544 | 269 | 110 | 2.4 | 0.000013 | Late |
| Secondary metabolism | |||||||||||
| 260386_at | At1g74010 | Strictosidine synthase family protein | 110 | 21 | 5.1 | 0.001313 | 368 | 22 | 16.4 | 0.000010 | Both |
| 264401_at | At1g61720 | Dihydroflavonol 4-reductase family (BAN) | 54 | 45 | 1.2 | 0.693582 | 535 | 176 | 3.0 | 0.000017 | Late |
| 260335_at | At1g74000 | Strictosidine synthase family protein | 144 | 72 | 2.0 | 0.005947 | 104 | 39 | 2.7 | 0.000038 | Late |
| 254283_s_at | At4g22870 | Leucoanthocyanidin dioxygenase, putative | 81 | 85 | 1.0 | 0.880918 | 416 | 183 | 2.3 | 0.000118 | Late |
| 249215_at | At5g42800 | Dihydroflavonol 4-reductase | 31 | 26 | 1.2 | 0.546725 | 301 | 84 | 3.6 | 0.000030 | Late |
| Plant development/organogenesis | |||||||||||
| 262113_at | At1g02820 | Late embryogenesis abundant 3 family protein | 82 | 33 | 2.5 | 0.000057 | 76 | 27 | 2.8 | 0.000001 | Both |
| 262659_at | At1g14240 | Nucleoside phosphatase family protein | 80 | 25 | 3.3 | 0.000911 | 92 | 16 | 5.8 | 0.000000 | Both |
| 262549_at | At1g31290 | PAZ domain-containing protein | 140 | 99 | 1.4 | 0.096043 | 169 | 63 | 2.7 | 0.000011 | Late |
| 252234_at | At3g49780 | Phytosulfokines 3 (PSK3) | 17 | 9 | 2.0 | 0.386279 | 163 | 43 | 3.8 | 0.000014 | Late |
| 251301_at | At3g61880 | Cytochrome P450, putative | 94 | 51 | 1.9 | 0.003809 | 91 | 41 | 2.2 | 0.000032 | Late |
| Protein biosynthesis/DNA catabolism | |||||||||||
| 245883_at | At5g09500 | 40S ribosomal protein S15 (RPS15C) | 85 | 18 | 4.7 | 0.002080 | 54 | 9 | 5.9 | 0.000028 | Early |
| 260438_at | At1g68290 | Bifunctional nuclease, putative | 44 | 28 | 1.6 | 0.093917 | 149 | 25 | 5.9 | 0.000104 | Late |
Affymetrix probe set number.
Arabidopsis Genome Initiative number.
Fold change for signal intensity of wild type/signal intensity of spl.
P values calculated by Student's t test for two-sample equal variance.
Expressed ovule stage.
Promoter∷GUS Fusions Validate Embryo Sac-Specific Expression in Both Early and Late Staged Ovules
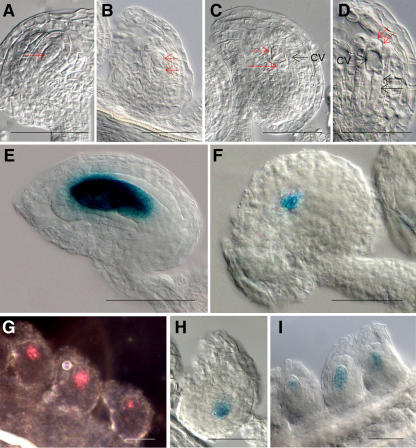
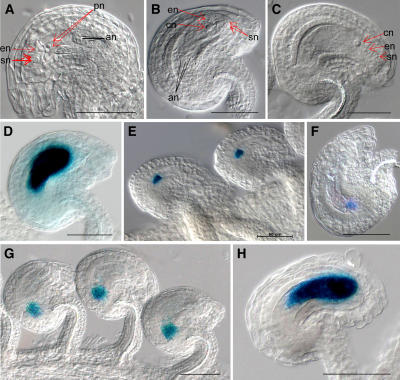
For validation of the microarray analysis results, we examined previously uncharacterized genes from 225 identified embryo sac genes. Six genes were selected to examine their expression pattern during female gametophyte development. To increase the probability of obtaining detectable levels of reporter gene expression in promoter fusion studies, two criteria were used. The first is that they rank in the top 20 when they are arranged by highest signal intensity of early staged wild-type ovule. The second is that they had a greater than 10-fold change for both early and late staged wild-type ovules compared to spl ovules (Table III). When we examined their RNA expression patterns using reverse transcription-PCR, all six genes were detected only in wild-type ovules of both of early and late stage and not in spl mutant ovules (data not shown). Next, we constructed promoter∷GUS cassettes to analyze their expression. All six promoter∷GUS fusions were examined during embryo sac development to determine whether they are expressed only in the embryo sac as predicted. We examined six transgenic lines for each promoter fusion, by analyzing five or more T2 generation plants for each transgenic line to ensure that the expression patterns were reproducible. For these six fusions, we observed reporter GUS expression only in early and late staged embryo sacs and not in leaves, stems, petals, and sepals. The GUS expression of the promoter∷GUS fusion with At1g26795, which encodes a self-incompatibility protein with a transmembrane domain, was detected strongly in dividing nuclei of the embryo sac from FG3 (Fig. 4E). We detected strong GUS expression in the embryo sac, including the egg cell and antipodal cells, from FG5 to FG7 (Fig. 5D). GUS expression of the promoter∷GUS fusion with At1g36340, which encodes a ubiquitin-conjugating enzyme, was detected in the closest nucleus to the chalazal end of the embryo sac at FG4 (Fig. 4F). In the late staged ovule, GUS activity was detected in only the antipodal cells (Fig. 5E) or was observed in the area of the degenerated antipodal cells at FG7. Both of the activities of the promoter∷GUS fusions with At2g20070, encoding a hypothetical protein, and At4g22050, encoding an aspartyl protease family, could be detected in FG1 of embryo sacs (Fig. 4, G and H). In particular, expression of the promoter∷GUS fusion with At2g20070 was detected in dividing nuclei of the embryo sac from FG1 to FG3 and in the chalazal end of the embryo sac at FG4. Expression of promoter∷GUS fusion with At4g22050 was detected in the chalazal end of the embryo sac from FG1. From FG5 to FG7, the activity of the promoter∷GUS fusion with At2g20070 or At4g22050 was detected in only the chalazal ends of the embryo sacs (Fig. 5, F and G). The expression of the promoter∷GUS fusion with At5g40260 was observed in the embryo sac from FG1 to FG7 (Figs. 4I and 5H). We could detect GUS expression of the promoter∷GUS fusion with At5g40260 in the functional megaspore at FG1 and in nuclei of the micropylar end of the embryo sac and the central vacuole from FG2 to FG4. In the late staged ovule, expression of the promoter∷GUS fusion with At5g40260 was detected strongly in the embryo sac. At5g40260 is annotated as encoding a protein of the nodulin MtN3 family expressed during root development of Medicago truncatula, but it also shows similarity with LIM7, a protein that is induced during the meiotic prophase in lily (Lilium longiflorum) microspores (Kobayashi et al., 1994). This gene was previously identified as stamen specific in Arabidopsis (Wellmer et al., 2004), and we have confirmed its expression in pollen (see below). The GUS activity of the promoter∷GUS fusion with At4g30590, encoding a plastocyanin-like domain containing protein, was weak but detectable in developing embryo sacs from FG4 to FG7 in a pattern similar to those of the promoter∷GUS fusions with At1g26795 or At5g40260 (data not shown). In the anthers, the activity of the promoter∷GUS fusion with At5g40260 showed strong staining beginning with floral stage 8, in which locules are visible in the stamen and microsporocytes are conspicuous, to floral stage 13, in which pollen grains are mature, indicating that this gene is expressed strongly in male gametophytes as well (for details, see Supplemental Fig. 2). In addition, GUS expression from the promoter∷GUS fusions with At1g26795, At1g36340, and At4g30590 could also be detected weakly in developing pollen from floral stages 9 to 13 (data not shown). Therefore, these four genes appear to be expressed in both male and female gametophytes.
Table III.
The genes used the promoter∷GUS fusion analysis
Genes were selected on the basis of the P value of <0.0050, fold change of >2.0, and signal intensity of >60.0 at wild-type siblings. Column headings and footnotes are the same as in Table II.
| Affymetrix Codea
|
Gene IDb
|
Description
|
Early Staged Ovule
|
Late Staged Ovule
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT1–3 | spl1–3 | FCc | P Valued | WTd1–d3 | spl1–3, d1–d2 | FCc | P Valued | |||
| 261271_at | At1g26795 | Self-incompatibility protein related | 769 | 30 | 25.3 | 0.0000827 | 761 | 27 | 28.2 | 0.0000000 |
| 260124_at | At1g36340 | Ubiquitin-conjugating enzyme, E2 | 584 | 38 | 15.3 | 0.0016607 | 467 | 34 | 13.8 | 0.0000023 |
| 265579_at | At2g20070 | Hypothetical protein | 661 | 48 | 13.8 | 0.0000412 | 487 | 36 | 13.4 | 0.0000000 |
| 254336_at | At4g22050 | Aspartyl protease family protein | 681 | 37 | 18.3 | 0.0000964 | 714 | 67 | 10.6 | 0.0000000 |
| 253634_at | At4g30590 | Plastocyanin-like domain-containing protein | 705 | 56 | 12.6 | 0.0000568 | 500 | 41 | 12.3 | 0.0000050 |
| 249401_at | At5g40260 | Nodulin MtN3 family protein | 566 | 37 | 15.2 | 0.0006028 | 427 | 30 | 14.3 | 0.0000038 |
Figure 4.
GUS expression patterns of the promoter∷GUS fusion with female gametophyte genes in early staged ovule. A to D, Ovule of wild-type plant at FG1 (A), FG2 (B), FG3 (C), and FG4 (D). A, The arrow points the nucleus of functional megaspore. B, Each arrow points a micropylar nucleus or chalazal nucleus within embryo sac. C, The two red arrows point the micropylar nuclei or chalazal nucleus, and a black arrow points a large central vacuole (cv). D, Embryo sac has two micropylar nuclei (red arrows), two chalazal nuclei (black arrows), and a large central vacuole (cv). E, The GUS activity of the promoter∷GUS fusion with At1g26795 was detected in embryo sac at FG4. F, GUS expression of the promoter∷GUS fusion with At1g36340 was detected in the closest nucleus to the chalazal end of the embryo sac at FG4. G, GUS activity of the promoter∷GUS fusion with At2g20070 was detected in dividing nuclei of the FG1 and FG2 embryo sac stages under the dark-field microscope. H, GUS expression of the promoter∷GUS fusion with At4g22050 was observed in chalazal end of embryo sac at FG4. I, GUS activity of the promoter∷GUS fusion with At5g40260 was detected in embryo sac at FG2. Scale bars represent 50 μm.
Figure 5.
GUS expression patterns of the promoter∷GUS fusion with female gametophyte genes in late staged ovule. A to C, Ovule of wild-type plant at FG5 (A), FG6 (B), and FG7 (C). D, GUS expression of the promoter∷GUS fusion with At1g26795 was detected in the embryo sac at FG5. E, GUS activity of the promoter∷GUS fusion with At1g36340 was detected in the antipodal cells at FG7. F and G, GUS expression of the promoter∷GUS fusion with At2g20070 (F) or At4g22050 (G) was observed in the chalazal end of the embryo sac at FG6. H, GUS activity of the promoter∷GUS fusion with At5g40260 was detected in the embryo sac at FG6. Abbreviations: an, antipodal cells; cn, central cell nucleus; en, egg cell nucleus; pn, polar nuclei; sn, synergid nuclei. Scale bars represent 50 μm.
To examine the activity of the promoter∷GUS fusion with these six genes in sporophytic tissues, we stained 15-d-old seedlings for GUS expression (data summarized in Table IV). The GUS expression of At1g26795, At1g36340, At2g20070, and At4g30590 fusions was not detected in any part of the seedlings. Expression of the At5g40260 fusion could be detected in an occasional trichome, but as this expression was not consistent its significance is not clear. However, the expression of the promoter∷GUS fusion with At4g22050 was reproducibly detected in initiation sites of lateral roots and petioles, indicating that this gene is expressed sporophytically.
Table IV.
The summary for GUS expressions of six promoter∷GUS fusions in seedlings
Fifteen-day-old seedlings were used. To detect weak GUS activity, we used GUS staining solution without potassium ferricyanide/ferrocyanide.
| Gene ID | GUS Expression |
|---|---|
| At1g26795 | None |
| At1g36340 | None |
| At2g20070 | None |
| At4g22050 | Lateral root, petiole |
| At4g30590 | None |
| At5g40260 | None except occasional trichome |
To examine the functions in the embryo sac for these six genes, we analyzed the insertional mutants available from public collections listed in Supplemental Table II. However, we did not observe any embryo sac defects or other mutant phenotypes in these insertional mutants. To summarize, all six genes tested by promoter∷GUS fusions showed female gametophyte-specific expression within the developing ovules; four of the genes showed additional expression in the pollen, and one in sporophytic tissues.
DISCUSSION
Identification of Genes Expressed During Early and Late Developmental Stages of Embryo Sac Development
The female gametophyte in angiosperms is embedded in sporophyte tissue throughout development, making it technically challenging to isolate RNA from the developing gametophytes without extensive contamination from the surrounding sporophytic tissues. Here, we have isolated ovules from spl mutant homozygotes and phenotypically wild-type heterozygous siblings, and compared their expression profiles through the oligonucleotide array analysis. As spl ovules do not contain embryo sacs, genes with significantly higher transcripts in heterozygous sibling ovules than in spl ovules are presumptive embryo sac expressed genes. The RNA extracted from 2,500 to 3,000 ovules yielded enough RNA to perform each microarray experiment without any amplification. We avoided using amplified RNA to ensure comprehensive coverage of the transcriptome (Honys and Twell, 2004). The high correlation coefficient of >0.94 and Present call% of >70% validated good quality RNA used in our experiments.
In this study, two different ovule populations, corresponding to early and late developmental stages of the ovules, were used to perform the microarray assays. We used a set of three stringent criteria to narrow down the dataset to the most probable candidates for embryo sac-specific genes (i.e. <0.005 P value, a 2-fold cutoff, and >60 wild-type signal intensity). Using these criteria, we find 23 and 128 genes were identified in early and late staged ovules, respectively, and an additional 74 genes were expressed in both populations (Fig. 2; Table II). Thus, we identified more embryo sac genes expressed during later stages of megagametogenesis. This difference might arise from two sources. First, it could be due to the different complexities of the two ovule samples used in our study. While the embryo sacs of the early staged ovules undergo only three rounds of mitosis, the embryo sacs of the late staged ovules encompass more complex developmental processes, such as cellularization, nuclear fusion, and cell death, and the preparation during fertilization depends on cell-cell communication, such as pollen tube attraction and guidance and sperm nucleus recognition (Yang and Sundaresan, 2000; Hennig et al., 2004). Consistent with this idea, in a large-scale study of female gametophyte mutants, the majority of the identified genes are required after FG5 or during early embryogenesis, whereas only 16 of the identified genes are necessary before FG4 (Pagnussat et al., 2005). Second, the larger embryo sac of late staged ovules might have a more abundant pool of transcripts and this could facilitate the detection of more expressed genes in comparison with early staged ovules. This difference was even larger when we used a more stringent P value while maintaining the fold change and background signal criteria. For example, with a P value of <0.00001, 117 genes were identified for late staged ovules compared to only four genes for early staged ovules.
Comparison of Genes Identified in Female Gametophyte Development with Previous Sporophytic and Gametophytic Expression Studies
When we compared genes identified from our microarray analysis with genes previously shown to be involved in female gametophyte development through mutant analysis, we found very limited overlap. There may be two possible explanations for this. First, several genes identified through female gametophyte mutants, such as PRL (Springer et al., 1995), GFA2 (Christensen et al., 2002), and NOMEGA (Kwee and Sundaresan, 2003), also exhibit sporophytic expression. Genes with sporophytic expression in maternal ovule tissues were excluded from the gene set in the comparison of expression level of wild-type ovules and spl ovules. Consistent with their predicted sporophytic functions, genes required during megagametogenesis, such as PRL (At4g02060), GFA2 (At5g48030), and NOMEGA (At1g78770), were found to have high signal intensities for both wild-type and spl mutant ovules (Supplemental Table III).
Second, because of the high stringent selection criteria to minimize false positives in our microarray analysis, even genes specifically involved in female gametophyte development and function could be eliminated in our analysis if they do not have high expression in the embryo sacs. For example, the genes of the FIE-FIS-MEA Polycomb Group complex are not present in the set of embryo sac genes identified in this study. It is known that MEA and FIS2 are expressed in polar nuclei and central cell by promoter∷GUS fusion analysis (Ohad et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000). As mentioned above, we applied stringent criteria for the microarray analysis to minimize the number of false positives. MEA (FIS1, At1g02580) and FIS2 (At2g35670) showed higher expression in wild type versus spl mutants in the late staged ovules, but they did not satisfy some criteria, such as fold change of >2.0 in the case of MEA or higher signal intensity than the background signal of the Affymetrix control genes in the case of FIS2 (Supplemental Table III). In the case of FIE (FIS3, At3g20740), because FIE is expressed also in sporophyte tissues (Luo et al., 2000), it is eliminated in our analysis from the final dataset of female gametophyte genes (Supplemental Table III).
When the embryo sac genes were compared with female gametophyte genes obtained through the study using Ds tagging mutants in Arabidopsis (Pagnussat et al., 2005), expressed sequence tag analysis of maize (Zea mays) embryo sac (http://www.pgec.usda.gov/McCormick/McCormick/ResearchTopics/Gametes/Gametesindex.htm), and with carpel-specific genes by transcriptome comparison of floral homeotic mutants (Wellmer et al., 2004), 28 genes matched with our identified genes (Supplemental Table IV). The comparison with our female gametophyte mutant study (Pagnussat et al., 2005) showed that gene At2g34130 corresponded to MEE19, which resulted in maternal effect-arrested zygote and endosperm development, and gene At5g44700 corresponded to EDA23, which resulted in varying arrested stages of embryo sac development. The failure to identify more genes corresponding to the female gametophyte mutants arises from both of the factors discussed above, i.e. sporophytic expression of the gene in maternal ovule tissues and/or very low expression of the gene in the gametophyte.
The genes identified in this study are predicted to be either up-regulated in female gametophytes, as compared to the maternal ovule tissue, or specific for the female gametophyte. The higher fraction of unknown and hypothetical proteins for the genes identified here (33%), as compared to the fraction in the whole genome (21%), suggests that many genes of currently unknown function might have specific functions in the gametophytes. We compared the 225 genes identified here with transcriptome data of sporophytic tissues, using ATH1 Genome Array datasets for seedlings at open cotyledon stage (stage 0.7), leaves (stage 6.0), petiole (stage 3.9), stems (stage 6.1), roots, and root hair zone (stage 1.02) provided by the Nottingham Arabidopsis Stock Centre's microarray database (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl) and reanalyzed by Honys and Twell (2004). As a result, 101 genes (44.9%) of the 225 identified genes were found to have no transcriptome data in sporophytic tissues and appear to be gametophyte-specific genes. Further comparison of the 101 putative gametophyte-specific genes with data from expression studies that contain male gametophytes showed that 17 genes were identified as pollen-specific genes (Honys and Twell, 2004) and nine genes were analyzed as stamen-specific genes representing potential male gametophyte genes (Wellmer et al., 2004). Therefore, of the 101 genes that are not expressed in sporophytic tissues, 19 genes might be transcribed in both gametophytes and 82 genes might be specific to the female gametophyte (Supplemental Table IV). These conclusions are reflected in the results from the promoter∷GUS fusions examined. Out of six promoters that exhibited embryo sac-specific expression within the ovule, five did not show detectable expression in the sporophytic tissues examined, and four (At1g26795, At1g36340, At4g30590, and At5g40260) showed expression in the pollen. Examination of the insertional mutants revealed that none of the six mutant genes resulted in an observable phenotype in the embryo sac. This absence of mutant phenotypes could be due to redundancy. For example, At2g23990, which is the closest relative of At4g30590, was also detected as expressed in embryo sacs in our array analysis (Table II) and therefore might be playing the same role as At4g30590 in the same pathways during embryo sac development. Another of the genes examined, At4g22050, is closely related to two other genes (At1g62230 and At4g04460) of the aspartyl protease family. The gene At5g40260, which we found to be strongly expressed in embryo sacs and pollen, is related to four other Arabidopsis genes (At3g16690, At4g15920, At1g66770, and At5g62850) annotated as members of the nodulin MtN3 family. Similar outcomes were recently obtained by Nawy et al. (2005) in their analysis of genes expressed in the Arabidopsis root quiescent center; they also found that disruptions of identified genes with specific expression in the quiescent center did not yield observable mutant phenotypes, again possibly reflecting the extent of functional redundancy in the Arabidopsis genome.
CONCLUSION
Our study uses a comparative expression profiling strategy to provide a window into the female gametophyte transcriptome in Arabidopsis and to identify a subset of genes comprising this transcriptome. Nearly one-half of the identified genes have not previously been identified in expression profiling of sporophytic tissues, and there is only a partial overlap with genes identified from the expression profile of pollen. Mutational analysis of these genes will be important to undertake in future studies to understand their precise roles in embryo sac development and function. The low-level signals for many genes, due to the limiting amounts of plant material as well as low expression levels, suggest that a much larger number of genes may be found to be specific for the female gametophyte that are excluded from the list of genes presented here by the stringent criteria used to compile the final gene set. However, the complete unfiltered dataset that has been generated, which is provided as supplemental material and is also available from the laboratory Web site online (http://sundarlab.ucdavis.edu/), can be used to examine the possibility of female gametophyte expression for any specific gene in the Arabidopsis genome. The biological functions of most Arabidopsis genes remain to be determined, and the expression data in this study can assist in identifying putative functions in the gametophyte that would be missed in studies of sporophytic expression.
MATERIALS AND METHODS
Plant Material and Plant Growth Condition
Seeds of Landsberg erecta were directly or after spreading on Murashige and Skoog plates transferred to soil with 16-h-light/8-h-dark cycle at 22°C with 60% humidity. After the spl mutants were selected on Murashige and Skoog kanamycin plates, they were grown on the soil under the condition as described previously (Yang et al., 1999). The maintenance of the spl line and segregation analysis were carried out according to Sundaresan et al. (1995).
Genotyping and Ovule Collection of spl/spl and spl/SPL Plants
To confirm the spl homozygous or heterozygous plants, genotyping was performed using SPL primers SPL-F (T5′-GGCGAGATCCGGACAGAGAC-3′) and SPL-R (5′-AGAAGCGTTAAACATTTGAGGATT-3′) and Ds primers DS 3-3A (T5′-TCGTTTCCGTCCCGCAAGT-3′) or DS 5 to 3A (5′-CGGTCGGTACGGGATTTTCC-3′). PCR was performed in a total volume of 10 μL containing 1× PCR buffer (Fisher Scientific) with 1.5 mm MgCl2, 0.2 mm of each dNTP, 5 pmol of each primer, and 2 units of Taq DNA polymerase for 34 cycles at an annealing temperature of 55°C.
The ovule samples were dissected from placenta of ovaries with needles and collected under dry ice on the basis of the floral stage. The collected ovules were randomly cleared overnight with Hoyer's solution (Liu and Meinke, 1998) to confirm the embryo sac stages. The cleared whole-mount preparations and observation were performed as described by Pagnussat et al. (2005).
RNA Extraction, Probe Preparation, and Array Hybridization
Total RNA was extracted from ovule samples collected on the basis of the developmental stages using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocols. The yield and RNA purity were determined by the ratio of absorbencies at 260 nm/280 nm wavelengths. RNA integrity was checked by running 1 μL of every RNA preparation on the glyoxylated RNA gel (Sambrook and Russell, 2001). The first- and second-strand cDNAs were synthesized from 5 μg total RNA using the MessageAmp RNA kit (Ambion), which is based on the manual of Dr. James Everwine (Van Gelder et al., 1990). Biotin-labeled target cRNAs were prepared by cDNA in vitro transcription using the BioArray High-Yield RNA transcript labeling kit (Enzo Biochem) and cleaned using GeneChip Sample Cleanup Module (Affymetrix). The 15 μg labeled target cRNAs were fragmented and hybridized with Arabidopsis ATH1 Genome Arrays for 16 h at 45°C as described in the Affymetrix GeneChip expression analysis technical manual. The hybridizations and scanning were carried out at the University of California Davis Microarray Facility.
Data Acquisition and Statistical Analysis
Raw data were processed with Affymetrix Microarray Suite 5.0. The raw data were converted into numbers by calculations of the PM-only model (Li and Wong, 2001a) using dChip program version 1.3 (http://www.dchip.org/). Pearson's correlation coefficient (r) was calculated to examine the relation between microarrays using package R program version 1.6.1 (Ihaka and Gentleman, 1996). Microsoft Excel (Microsoft) was used in the calculation of P value by Student's t test and management of the microarray data.
Plant Transformation
To construct the promoter∷GUS cassettes for six genes (At1g26795, At1g36340, At2g20070, At4g22050, At4g30590, and At5g40260), promoter fragments were amplified by PCR using primers (Supplemental Table V) and inserted into EcoRI and NcoI sites upstream of the GUS gene (Jefferson, 1987) of pRITA. The NotI fragments from these plasmids were subcloned into pMLBART for transferring into plants. For plant transformation, the vectors were transferred into Agrobacterium tumefaciens strain ASE (Fraley et al., 1985). Transformations were performed on wild-type Landsberg erecta or Columbia-0 by floral dip procedures (Clough and Bent, 1998). The seeds obtained from the T0 promoter∷GUS transformants were selected by spraying 0.1% Basta, and Basta-resistant plants of the T1 generation were analyzed for the presence of the transgene by PCR using the primers of Basta-resistance gene, BA-F (5′-CCGTACCGAGCCGCAGGAAC-3′) and BA-R (5′-CAGATCTCGGTGACGGGCAGGAC-3′), and were self-crossed to collect seeds of T2 generation.
GUS Assays and Image Processing
For the expression analysis of GUS fused with promoter, the seeds of the T2 generation from each transformant were germinated on soil without sterilization, and then they were selected with 0.1% Basta. Inflorescences from soil-grown plants were transferred to microtiter well containing 1 mL of GUS staining solution (50 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 2 mm potassium ferricyanide, 2 mm potassium ferrocyanide, 1 mg/mL X-Gluc). The microtiter dish was placed under vacuum for 10 min in desiccators. After release of the vacuum, the dish was covered with aluminum foil and incubated at 37°C overnight (Sundaresan et al., 1995). The solution was removed, and the tissues were cleared in 0.85% sodium chloride/70% ethanol at room temperature. For detection of weak GUS expression, GUS staining was performed under the absence of potassium ferricyanide and ferrocyanide, too. To see clear and specific GUS expression, the potassium ferricyanide and ferrocyanide concentrations were readjusted in the range of 1.3 to approximately 10 mm. The pistils were dissected with needles and cleared in Hoyer's solution (Liu and Meinke, 1998), and the cleared ovules were observed under a Zeiss Axioplan imaging 2 microscope under differential interference contrast optics. Images were captured on an Axiocam HRC CCD camera (Zeiss) using the Axiovision program (version 3.1). All images were processed for publication using Adobe Photoshop CS (Adobe Systems).
Supplementary Material
Acknowledgments
We thank Dr. D. St. Clair and Dr. S. Subrahmanyan for invaluable advice on the statistical analysis; Dr. J. Bowman, Dr. C. Gasser, and D. Skinner for helpful discussions; and the University of California Davis School of Medicine Microarray Core Facility for microarray hybridizations and scanning.
This work was supported by the National Science Foundation (NSF2010 program grant no. 0313501 to V.S.). H.-J.Y. received fellowship support from Korea Science and Engineering Foundation (KOSEF).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Venkatesan Sundaresan (sundar@ucdavis.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067314.
References
- Bowman JL (1994) Arabidopsis: An Atlas of Morphology and Development, Ed 1. Springer-Verlag, New York
- Christensen CA, Gorsich SW, Brown RH, Jones LG, Brown J, Shaw JM, Drews GN (2002) Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14: 2215–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Devore J, Peck R (1997) Statistics: The Exploration and Analysis of Data, Ed 3. Duxbury Press, Pacific Grove, CA
- Dobson AT, Raja R, Abeyta MJ, Taylor T, Shen S, Haqq C, Pera RA (2004) The unique transcriptome through day 3 of human preimplantation development. Hum Mol Genet 13: 1461–1470 [DOI] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick JS, Fink CL, Hoffmann NL, Sanders PR (1985) The SEV system: a new disarmed Ti plasmid vector system for plant transformation. Biotechnology 3: 629–635 [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS (2004) Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet 13: 2263–2278 [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135: 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Statist 5: 299–314 [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Kobayashi T, Kobayashi E, Sato S, Hotta Y, Miyajima N, Tanaka A, Tabata S (1994) Characterization of cDNAs induced in meiotic prophase in lily microsporocytes. DNA Res 1: 15–26 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee HS, Sundaresan V (2003) The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J 36: 853–866 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001. a) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2: RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH (2001. b) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield JA, Fiebig A, Johnstone SE, Preuss D (2001) Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292: 2482–2485 [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D (2003) An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev 17: 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pan W (2002) A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics 18: 546–554 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schiefthaler U, Balasubramanian S, Siebe P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RATH (1995) Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268: 877–880 [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Thomas JG, Olson JM, Tapscott SJ, Zhao LP (2001) An efficient and robust statistical modeling approach to discover differentially expressed genes using genomic expression profiles. Genome Res 11: 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 87: 1663–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U (1999) Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schalburg KR, Rise ML, Brown GD, Davidson WS, Koop BF (2005) A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary. Biol Reprod 72: 687–699 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Sundaresan V (2000) Genetics of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol 3: 53–57 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ren X, Ireland JJ, Coussens PM, Smith TP, Smith GW (2004) Generation of a bovine oocyte cDNA library and microarray: resources for identification of genes important for follicular development and early embryogenesis. Physiol Genomics 19: 84–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.