Abstract
The genome sequencing of Arabidopsis (Arabidopsis thaliana) has revealed that secondary metabolism plant glycosyltransferases (UGTs) are encoded by an unexpectedly large multigenic family of 120 members. Very little is known about their actual function in planta, in particular during plant pathogen interactions. Among them, members of the group D are of particular interest since they are related to UGTs involved in stress-inducible responses in other plant species. We provide here a detailed analysis of the expression profiles of this group of Arabidopsis UGTs following infection with Pseudomonas syringae pv tomato or after treatment with salicylic acid, methyljasmonate, and hydrogen peroxide. Members of the group D displayed distinct induction profiles, indicating potential roles in stress or defense responses notably for UGT73B3 and UGT73B5. Analysis of UGT expression in Arabidopsis defense-signaling mutants further revealed that their induction is methyljasmonate independent, but partially salicylic acid dependent. T-DNA tagged mutants (ugt73b3 and ugt73b5) exhibited decreased resistance to P. syringae pv tomato-AvrRpm1, indicating that expression of the corresponding UGT genes is necessary during the hypersensitive response. These results emphasize the importance of plant secondary metabolite UGTs in plant-pathogen interactions and provide foundation for future understanding of the exact role of UGTs during the hypersensitive response.
Plant resistance to pathogen infection is often associated with the hypersensitive response (HR) characterized by localized cell death at the site of infection allowing the restriction of pathogen spread. Main events occurring during the HR are the rapid production of reactive oxygen species (ROS), ion fluxes across the plasma membrane, and transcriptional activation of defense genes as those involved in production of secondary metabolites like salicylic acid (SA) and phytoalexins (Hammond-Kosack and Jones, 1996). Secondary metabolites perform multiple functions in plants, including roles in UV protection, lignification, herbivore protection, and disease resistance (Vet and Dicke, 1992; Li et al., 1993; Dixon, 2001; Anterola and Lewis, 2002). These compounds rarely accumulate in their free form but are often conjugated to sugars, particularly Glc, through the action of glycosyltransferases (UGTs; EC 2.4.1.128; Vogt and Jones, 2000). Glc conjugation is thought to regulate bioactivity of aglycones, to enhance their solubility, to protect their reactivity toward cellular oxidases, and to alter their transport properties throughout the whole plant (Jones and Vogt, 2001).
UGTs are encoded by a ubiquitous gene family (family 1 of UGTs; Lim and Bowles, 2004) and are as plentiful as they are diverse. The Arabidopsis (Arabidopsis thaliana) genome contains 120 UGT genes classified into 14 groups (A–N) according to their sequence similarity (Ross et al., 2001). Some of them contain enzymes sharing similar catalytic properties. For instance, enzymes forming SA Glc ester and indole-3-acetic acid Glc ester in tobacco (Nicotiana tabacum) and maize (Zea mays) exhibit a high sequence homology to group L from Arabidopsis, which comprises enzymes producing hydroxycinnamoyl Glc esters (Jackson et al., 2001; Lim et al., 2001). However, group D encompasses enzymes glucosylating substrates as diverse as the steroid solanidin (Moehs et al., 1997), the betalain betanidin (Vogt et al., 1999), the hydroxycoumarin scopoletin (Fraissinet-Tachet et al., 1998), the flavonoid baicalein (Hirotani et al., 2000), and the sesquiterpenoid abscisic acid (Xu et al., 2002; Fig. 1), and no definite conclusion can be drawn on the function of a UGT from its primary sequence. Determination of UGT substrate specificities in vitro has been intensively studied in many plants for decades, but the characterization of their target in planta is still in its infancy. Therefore, their physiological roles in plants have been inferred mainly from their substrate specificity in vitro and partially from their activity in planta.
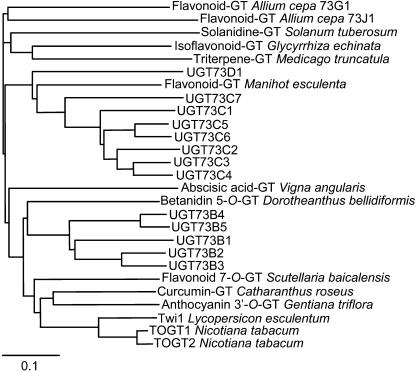
Figure 1.
Neighbor-joining distance tree of Arabidopsis UGTs and other plant glucosyltransferases (GT) belonging to the group D. The amino acid sequences were aligned using ClustalX (Thompson et al., 1997). The Arabidopsis Genome Initiative codes corresponding to Arabidopsis UGTs are given in Table II. The GenBank accession numbers of other plant GTs are: Allium cepa 73G1 and 73J1, AAP88406 and AAP88407, respectively (Kramer et al., 2003); Catharanthus roseus, BAD29722 (Kaminaga et al., 2004); Dorotheanthus bellidiformis, CAB56231 (Vogt et al., 1999); Gentiana triflora, BAC54092 (Fukuchi-Mizutani et al., 2003); Glycyrrhiza echinata, BAC78438 (Nagashima et al., 2004); tomato, CAA59450 (O'Donnell et al., 1998); Manihot esculenta, CAA54610 (Hughes and Hughes, 1994); Medicago truncatula, AAW56091 (Achnine et al., 2005); tobacco, TOGT1 and TOGT2, AAK28303 and AAK28304, respectively (Fraissinet-Tachet et al., 1998); Scutellaria baicalensis, BAA83484 (Hirotani et al., 2000); Solanum tuberosum, AAB48444 (Moehs et al., 1997); and Vigna angularis, BAB83692 (Xu et al., 2002).
The first example of the identification of a UGT function was through the isolation of the maize bronze-1 transposon-tagging mutant, which was altered in a flavonol-3-O-glucosyltransferase gene (Dooner and Nelson, 1977). A clear characterization for the biological role of a UGT acting on the hydroxycoumarin scopoletin was made in tobacco by using transgenic plants harboring the corresponding tobacco glucosyltransferase (TOGT1) gene in antisense and sense orientation. Down-regulation of TOGT1 gene weakened resistance to Tobacco mosaic virus (TMV) during the HR, and the observed phenotype resulted from a decrease in the antiviral secondary metabolite scopoletin, correlated with an increase in the oxidative burst (Chong et al., 2002). Conversely, up-regulation of TOGT1 led to the overaccumulation of scopoletin and its glucoside scopolin, to the early appearance of lesions during the HR to TMV (Gachon et al., 2004a), and to increased resistance against Potato virus Y (Matros and Mock, 2004). In recent years, a number of UGTs were characterized in Arabidopsis (Gachon et al., 2005b); metabolic profiling of T-DNA insertion lines revealed that UGT73C6 and UGT78D1 encode UDP-rhamnose:flavonol-3-O-rhamnosyltransferase and UDP-Glc:flavonol-3-O-glycoside-7-O-glucosyltransferase, respectively (Jones et al., 2003). In the trp1-100 mutant, which is defective in Trp biosynthesis, a loss-of-function mutation in UGT74F2 led to the disappearance of anthranilic acid-Glc, indicating that anthranilic acid is metabolized by this UGT in this particular genetic background (Quiel and Bender, 2003). Likewise, overexpression of UGT84B1 led to the accumulation of 1-O-(indol-3-acetyl)-β-d-Glc, suggesting that indole-3-acetic acid is the main substrate in vivo (Jackson et al., 2002). Recent studies provided evidence for a biological role of UGT74B1 in glucosinolate biosynthesis (Grubb et al., 2004) and showed that UGT78D2 and UGT75C1 encode flavonoid 3-O-glucosyltransferase and anthocyanin 5-O glucosyltransferase, respectively (Tohge et al., 2005).
UGT group D includes 13 members in Arabidopsis and contains a number of genes identified in other plant species as being involved in stress responses (Fig. 1). IS5a and IS10a genes from Bright-Yellow 2 tobacco cells were shown to be induced by SA and hydrogen peroxide (H2O2; Horvath and Chua, 1996), and TOGT1 and TOGT2 are highly expressed in tobacco reacting hypersensitively to TMV and in tobacco cell suspension cultures treated with the fungal elicitor β-megaspermin (Fraissinet-Tachet et al., 1998). Tomato wound induced 1 is a wound- and elicitor-inducible gene in tomato (Lycopersicon esculentum; O'Donnell et al., 1998), whereas UGT73B2 is responsive to the mycotoxin fumonisin B1 in Arabidopsis (Asai et al., 2000). Expression of UGT73B5 was induced by 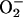 and during the HR of Arabidopsis to Pseudomonas syringae pv tomato (Pst) carrying the AvrRpm1 gene (Pst-AvrRpm1; Mazel and Levine, 2002). In a powdery mildew-resistant mutant, UGT73B2 expression is induced 2-fold higher than in wild-type plants after Erysiphe cichoracearum inoculation (Nishimura et al., 2003). Finally, applications of SA, methyljasmonate (MeJA), or deoxynivalenol, a mycotoxin produced by the plant pathogenic fungus Fusarium, induced the expression of UGT73C5 (Poppenberger et al., 2003). Taken together, these observations point to a particular role for group D UGTs in stress responses. Still, except for UGT73C6 (Jones et al., 2003), none of the endogenous substrates nor the function of these enzymes are known.
and during the HR of Arabidopsis to Pseudomonas syringae pv tomato (Pst) carrying the AvrRpm1 gene (Pst-AvrRpm1; Mazel and Levine, 2002). In a powdery mildew-resistant mutant, UGT73B2 expression is induced 2-fold higher than in wild-type plants after Erysiphe cichoracearum inoculation (Nishimura et al., 2003). Finally, applications of SA, methyljasmonate (MeJA), or deoxynivalenol, a mycotoxin produced by the plant pathogenic fungus Fusarium, induced the expression of UGT73C5 (Poppenberger et al., 2003). Taken together, these observations point to a particular role for group D UGTs in stress responses. Still, except for UGT73C6 (Jones et al., 2003), none of the endogenous substrates nor the function of these enzymes are known.
The aim of this study was to determine which UGT in the group D is the most relevant to plant defense, using the Arabidopsis-Pst interaction as a model system. We first developed an extensive gene expression analysis of 11 UGT genes after Pst infection and treatments with SA and H2O2, two key modulators of HR (Delaney et al., 1994; Levine et al., 1994), or MeJA, a wound-related signal (Liechti and Farmer, 2002). To discriminate between highly similar genes, we used real-time quantitative PCR (RT-qPCR) analysis. UGT genes were shown to be differentially induced during the HR and after elicitation with the signaling molecules. Furthermore, we found that this induction is MeJA independent, but partially SA dependent. Members of this group are also expressed in an organ-specific manner. All these results are further confirmed by data-mining analyses of publicly available microarray datasets. Finally, two T-DNA insertion mutants belonging to group D (ugt73b3 and ugt73b5) show a decreased resistance to Pst-AvrRpm1, suggesting an important role for these two UGTs during the HR.
RESULTS
Group D UGTs Are Differentially Expressed during the HR to Pst-AvrRpm1
To point out candidate genes involved in resistance responses, we decided to gain insight into the transcriptional regulation of group D UGTs during the HR to Pst-AvrRpm1. We chose RT-qPCR to follow the transcriptional changes of UGT genes to ensure the highest possible specificity. The 13 UGT sequences exhibit between 50.8% and 91.3% identity within their coding sequences at the nucleotide level (Table I). Specific primer sets for each gene were carefully designed (Table II). UGT73C4 and UGT73C5 could not be unambiguously targeted with PCR primers and were not studied further. UGT expression levels were normalized over the constitutive tubuline α2 gene (TUBα2; At1g50010; Carpenter et al., 1993), the expression of which was similar throughout the time-course experiments between treated and untreated conditions (Supplemental Fig. 1).
Table I.
Identity/similarity matrix for group D UGTs
Identity (lower triangle) and similarity (upper triangle) percentages were determined using the needle program (http://bioweb.pasteur.fr/seqanal/interfaces/needle.html).
| UGT
|
Nucleotide Identity/Amino Acid Similarity (%)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 73B1 | 73B2 | 73B3 | 73B4 | 73B5 | 73C1 | 73C2 | 73C3 | 73C4 | 73C5 | 73C6 | 73C7 | 73D1 | |
| 73B1 | – | 80.4 | 79.6 | 76.5 | 77.5 | 59.6 | 60.6 | 60.7 | 60.7 | 60.4 | 61.2 | 61.3 | 58.2 |
| 73B2 | 71.5 | – | 90.1 | 78.2 | 78.9 | 57.3 | 58.9 | 59.8 | 60.9 | 58.4 | 59.4 | 60.0 | 57.7 |
| 73B3 | 71.0 | 88.8 | – | 77.5 | 77.9 | 58.5 | 60.2 | 60.2 | 60.3 | 59.3 | 59.3 | 59.6 | 58.8 |
| 73B4 | 69.1 | 71.2 | 69.0 | – | 94.0 | 59.2 | 60.4 | 59.7 | 61.5 | 60.7 | 61.4 | 60.0 | 58.3 |
| 73B5 | 70.4 | 71.2 | 70.2 | 90.9 | – | 60.0 | 59.8 | 59.8 | 61.5 | 59.8 | 61.2 | 61.7 | 59.7 |
| 73C1 | 52.9 | 51.4 | 52.1 | 52.1 | 54.4 | – | 85.7 | 87.5 | 87.5 | 88.3 | 86.3 | 79.0 | 66.3 |
| 73C2 | 53.7 | 52.5 | 53.7 | 55.1 | 53.9 | 79.1 | – | 89.5 | 89.5 | 86.1 | 86.3 | 77.6 | 65.0 |
| 73C3 | 54.7 | 51.7 | 53.8 | 53.8 | 54.3 | 79.5 | 84.8 | – | 93.8 | 88.5 | 88.3 | 77.7 | 68.5 |
| 73C4 | 56.1 | 51.9 | 53.4 | 54.9 | 54.2 | 79.3 | 83.8 | 91.3 | – | 87.7 | 88.3 | 77.9 | 67.1 |
| 73C5 | 53.9 | 54.7 | 55.7 | 54.9 | 54.5 | 79.3 | 81.1 | 84.0 | 83.8 | – | 93.1 | 77.0 | 68.8 |
| 73C6 | 54.0 | 53.9 | 54.0 | 52.9 | 53.8 | 77.6 | 80.4 | 82.6 | 82.0 | 91.0 | – | 77.0 | 68.4 |
| 73C7 | 51.9 | 53.9 | 53.2 | 51.5 | 53.0 | 66.9 | 66.4 | 65.7 | 67.1 | 67.2 | 67.0 | – | 64.7 |
| 73D1 | 50.8 | 51.8 | 53.1 | 52.5 | 55.0 | 53.6 | 55.0 | 54.9 | 54.9 | 57.1 | 55.4 | 54.1 | – |
Table II.
Primer sequences used in the RT-qPCR experiments
AGI, Arabidopsis Genome Initiative.
| Gene | AGI Code | Primer Pairs |
|---|---|---|
| UGT73B1 | At4g34138 | 5′-TCTGAAACGAGTTCCCAAATTAGC-3′ |
| 5′-AAATGGAGCTTAGAGACTTCGACAG-3′ | ||
| UGT73B2 | At4g34135 | 5′-AGTTAAATTCAAATGGCAGCAACC-3′ |
| 5′-TCTTGAACCATTGATTTTCTCCTAAC3′ | ||
| UGT73B3 | At4g34131 | 5′-ATAGCTTCATTGAAAAGACCTCAGTAAG-3′ |
| 5′-CCAAGACAAAGACTAAGCAGAATCG-3′ | ||
| UGT73B4 | At2g15490 | 5′-GTCCACTTTCACTATCCAACAGAGG-3′ |
| 5′-CTGTTCGTTGGGTAAGCCG-3′ | ||
| UGT73B5 | At2g15480 | 5′-GAGCTGAATGGTAGAAAGTAGAGGAAG-3′ |
| 5′-ATGACATAAAGAACAACTCCAAAGAGG-3′ | ||
| UGT73C1 | At2g36750 | 5′-TGCAGATACTAAAAGCCGGTGTG-3′ |
| 5′-CTTCTTTACTCCTTCTTTATCCACCAG-3′ | ||
| UGT73C2 | At2g36760 | 5′-CAGAAACTGATCGTGCAGGTG-3′ |
| 5′-CTCTCTTTTGCTTCATCACTCTCG-3′ | ||
| UGT73C3 | At2g36780 | 5′-TCTATATTGGAGAAATTTAAATCAGAGCC-3′ |
| 5′-ACTGATTTATCTTCTTCTGATTCATTATCC-3′ | ||
| UGT73C6 | At2g36790 | 5′-TTTTGTCTTCTGTGTGTTAACGTTCTG-3′ |
| 5′-TCTATCAGGAAAATAAGGAACAATGAAG-3′ | ||
| UGT73C7 | At3g53160 | 5′-AGGAGATAGGAGCGATGGTGAG-3′ |
| 5′-TGCTTCTTCACTATCACCCATTAGC-3′ | ||
| UGT73D1 | At3g53150 | 5′-AAGAAACCGAGTGTTGTGAAAGC-3′ |
| 5′-TCATTATCATCATCATTTTCGTCTACAC-3′ | ||
| TUBα2 | At1g50010 | 5′-ATCTCTTGCTTGTGGCGGTAG-3′ |
| 5′-ACCCAGCTTAAATTCAGTTCTTGG-3′ | ||
| PR1 | At2g14610 | 5′-AGGCTAACTACAACTACGCTGCG-3′ |
| 5′-GCTTCTCGTTCACATAATTCCCAC-3′ | ||
| PDF1.2 | At5g44420 | 5′-CTGTTACGTCCCATGTTAAATCTACC-3′ |
| 5′-CAACGGGAAAATAAACATTAAAACAG-3′ | ||
| Iask | At5g26751 | 5′-CTTATCGGATTTCTCTATGTTTGGC-3′ |
| 5′-GAGCTCCTGTTTATTTAACTTGTACATACC-3′ |
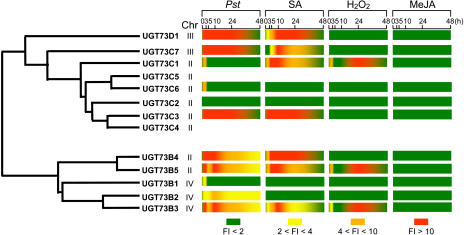
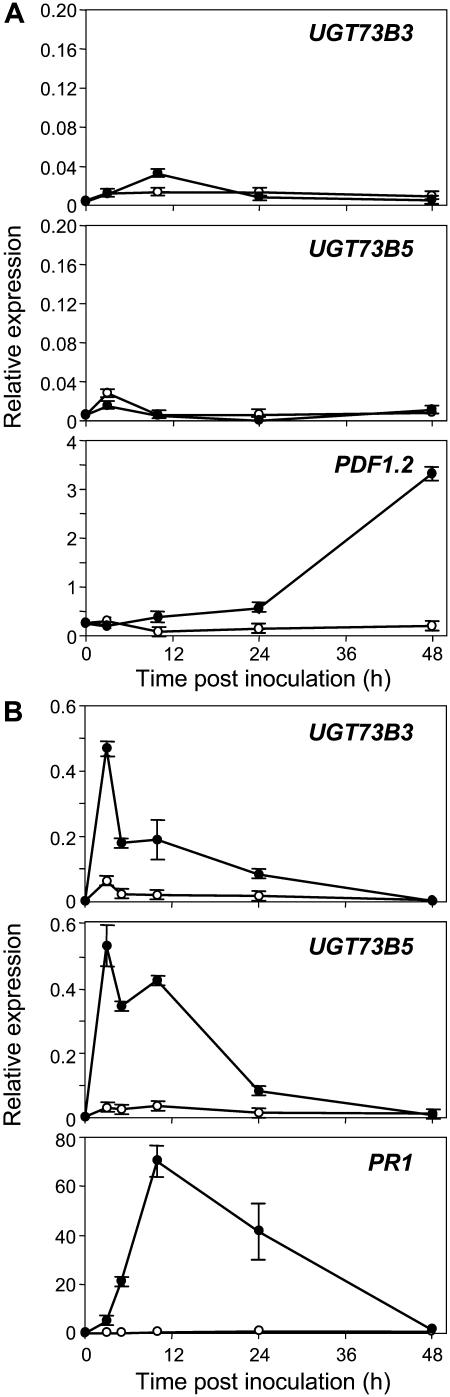
Figure 2 shows the time course of UGT73B3, UGT73B4, UGT73B5, UGT73C1, and PR1 expression during the compatible and incompatible interactions with Pst. PR1 is a well established marker gene for the defense responses of Arabidopsis against Pst (Uknes et al., 1992). Whereas it was barely expressed in leaves inoculated with the virulent bacteria Pst, it was activated to a high level 24 h after inoculation with the avirulent strain Pst-AvrRpm1. Among UGT genes, diverse patterns of expression could be observed. UGT73B3 and UGT73B5, which share 77.9% similarity at the amino acid level (Table I) but do not map on the same chromosome (Fig. 3), presented highly similar expression profiles. Transcripts of UGT73B3 accumulated early and strongly during the incompatible interaction and remained high throughout the time-course experiment. Similarly, UGT73B5 transcripts accumulated early and to higher levels than UGT73B3, peaking at 5 h after Pst-AvrRpm1 inoculation and then decreased to a steady-state level. Leaf inoculation with the isogenic virulent bacteria Pst resulted in a very low induction of UGT73B3 and UGT73B5, indicating that the observed response is specific of HR onset. UGT73B4 was expressed at a lower level compared to UGT73B5 but exhibited a similar pattern of expression in both interactions until 10 h after inoculation. Thereafter, UGT73B4 transcripts decreased to basal level whatever the interaction, whereas UGT73B5 expression remained strongly up-regulated. These two genes share 94% similarity at the amino acid level (Table I) and are mapped in tandem on the same chromosome (Fig. 3). A different picture emerged from UGT73C1 gene expression (Fig. 2). Here the levels of expression were low, and the patterns of induction were the same in both compatible and incompatible interactions. Altogether, these results clearly indicate that members of the group D, except UGT73B1, UGT73C2, and UGT73C6, are responsive to pathogen infection. They display divergent expression patterns after Pst challenge, which are summarized in Figure 3. Thus, genes such as UGT73D1 exhibited a high and persistent expression level, whereas UGT73C2 and UGT73C6 expression was barely detectable (Fig. 3).
Figure 2.
RT-qPCR expression profiles of UGT73B3, UGT73B4, UGT73B5, UGT73C1, and PR1 genes after Pst challenge. Transcript levels were quantified after Pst DC3000 (white circles) or Pst-AvrRpm1 (black circles) inoculation. The transcript level (relative expression) is represented as a ratio of the transcript abundance of the studied gene to the transcript abundance of the TUBα2 gene. Values correspond to the mean and sd of duplicates. The experiments were repeated with similar results.
Figure 3.
Induction of UGT D expression during the HR and after treatments with signaling molecules. UGT D relationship and position on chromosomes are presented on the left. Treatments and kinetics are indicated on the top. Induction of UGT D expression is represented as a ratio (fold induction) of the studied gene relative expression (UGT transcript abundance/TUBα2 transcript abundance) in each inductive condition to its relative expression in the corresponding control (Pst: Pst-AvrRpm1/Pst DC3000; SA: SA/K2HPO4; H2O2: H2O2/water; MeJA: MeJA/air). Fold induction (FI) of gene expression is represented by a color scale.
Group D UGTs Respond Differentially to SA, MeJA, and H2O2
A kinetic analysis of UGT expression was performed after treatments of Arabidopsis leaves with 1 mm SA, MeJA, or 5 mm H2O2. Whereas PDF1.2, a marker gene of the jasmonate pathway, was induced, none of the UGT genes under investigation responded to MeJA treatment (Figs. 3 and 4A). On the contrary, accumulation of UGT73B3 and UGT73B5 transcripts was apparent at 3 h after application of SA (Fig. 4B) and rose continuously until approximately 10 h. Afterward, transcript levels decreased rapidly. This pattern of expression differed from the one observed after bacterial challenge (Fig. 2) where expression was maintained to a high level throughout the time-course experiment. Levels of PR1 transcripts were also determined during the time course. Prior to 5 h, no accumulation of PR1 transcripts was evident, the increase occurred between 10 and 15 h after SA treatment. Surprisingly, UGT73B2 was not induced by SA (Fig. 3), although this gene is the closest relative of the highly SA-responsive UGT73B3 gene. Only three genes, UGT73B3, UGT73B5, and UGT73C1, responded to H2O2 application (Fig. 3). They exhibited a biphasic accumulation of transcripts with an early induction at 3 h, followed by a decrease to the background level. Then transcript levels rose dramatically, peaking at 24 h, and declined at 48 h.
Figure 4.
RT-qPCR expression profiles of UGT belonging to group D following MeJA and SA treatments. A, Transcript levels of UGT73B3, UGT73B5, and PDF1.2 after MeJA treatment (black circles) compared with mock control (white circles). B, Transcript levels of UGT73B3, UGT73B5, and PR1 after SA (black circles) and mock (white circles) infiltration. The transcript level (relative expression) is represented as a ratio of the transcript abundance of the studied gene to the transcript abundance of the TUBα2 gene. Values correspond to the mean and sd of duplicates. SA experiment was repeated with similar results.
Microarray Data Mining Confirms a Stress-Dependent Induction of Group D UGTs
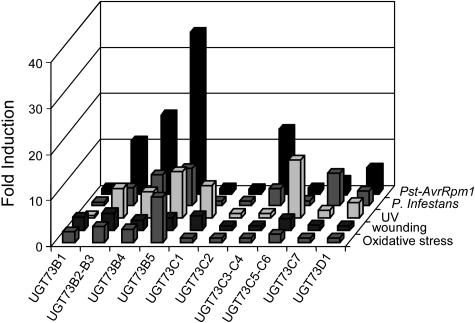
Microarray data obtained under numerous stress conditions were retrieved from the publicly available datasets recently released by the AtGenExpress consortium (http://web.uni-frankfurt.de/fb15/botanik). Analysis of these datasets confirmed that UGT D members were induced differentially under stress conditions (Fig. 5). Data concerning Pst-AvrRpm1 versus Pst DC3000 induction were consistent with our RT-qPCR results confirming that some UGTs belonging to the group D are linked with plant defense responses and particularly UGT73B2-B3 and UGT73B5. Expression of these genes was also highly induced after infection with a biotrophic pathogen, Phytophthora infestans, and after oxidative stress caused either by paraquat application or UV treatment (Fig. 5). Interestingly, UGT73B genes were induced after wounding (Fig. 5), although none of these genes was expressed after MeJA treatment (Fig. 3).
Figure 5.
Stress induction of UGT D revealed by microarray data mining. Induction of UGT, expressed in fold induction, was evaluated 6 h after infection with Pst (Pst-AvrRpm1/Pst DC3000), inoculation with P. infestans (P. infestans/water), and UV-B stress or 12 h after wounding and oxidative stress. Data were obtained from the AtGenExpress microarray datasets. Values correspond to the mean of triplicates.
The analysis of other microarray datasets available in Genevestigator (https://www.genevestigator.ethz.ch; Zimmermann et al., 2004) revealed that some members of the group D, including UGT73B3 and UGT73B5, were induced by the necrotrophic fungus Botrytis cinerea and after ozone treatment (data not shown). Furthermore, most members of group D responded to SA, whereas none of them was induced by MeJA, supporting our RT-qPCR results (data not shown).
Group D UGTs Exhibit a Differential SA Biosynthesis and SA-Signaling Dependence
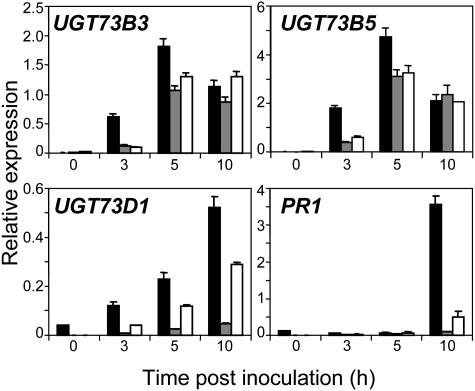
To test whether SA is required for the induction of UGT D genes during the HR, we investigated UGT expression in plants with an altered SA-signaling pathway. NahG plants express constitutively a salicylate hydroxylase enzyme that degrades SA into catechol (Delaney et al., 1994) and npr1-1 mutants are nonresponsive to SA, exhibiting a loss of resistance to Pst and a nonexpression of PR1 after infection (Cao et al., 1994). Figure 6 shows transcript accumulation of UGT73B3, UGT73B5, UGT73D1, and PR1 in wild-type plants, NahG transformants, and npr1-1 mutants. As expected, PR1 expression was abolished in NahG plants and npr1-1 mutants at 10 h after inoculation with Pst-AvrRpm1. UGT73D1 expression was barely detectable after pathogen challenge in NahG plants, whereas it still remained detectable in npr1-1 mutants indicating a SA-dependent and NPR1-independent induction of this gene. UGT73B3 and UGT73B5 were hardly expressed in NahG plants and npr1-1 mutants at 3 h after pathogen challenge, suggesting an SA dependence at this time point. Strikingly, at 5 and 10 h after inoculation, UGT73B3 and UGT73B5 transcript levels were almost similar in wild-type plants, NahG plants, and npr1-1 mutants, suggesting an induction of gene expression independent of SA at these later time points.
Figure 6.
RT-qPCR expression profiles of UGT73B3, UGT73B5, UGT73D1, and PR1 genes in wild-type plants and signaling-pathway mutants after Pst challenge. Transcript levels were quantified in wild-type plants (black bars), NahG plants (gray bars), and npr1-1 mutants (white bars) after Pst-AvrRpm1 inoculation. The transcript level (relative expression) is represented as a ratio of the transcript abundance of the studied gene to the transcript abundance of the TUBα2 gene. Values correspond to the mean and sd of duplicates. Experiment was repeated with similar results.
Organ-Specific Expression of UGTs Belonging to Group D
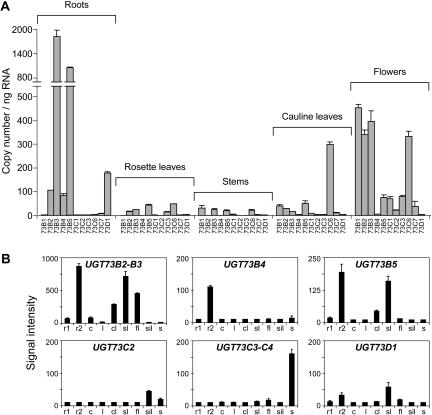
To determine the organ-specific expression pattern of each UGT D member, RT-qPCR was performed on RNA isolated from roots, rosette leaves, stems, cauline leaves, and flowers. It revealed that UGT D members are weakly expressed in rosette leaves, stems, and cauline leaves, with the exception of UGT73C6, which exhibited a high expression in cauline leaves (Fig. 7A). UGT73B2, UGT73B3, and UGT73B5 were abundantly expressed in roots and flowers, whereas UGT73B4 and UGT73D1 transcripts were only detectable in roots. UGT73C6 was expressed in cauline leaves and flowers, and UGT73C1, UGT73C2, and UGT73C7 were undetectable in all organs except in flowers, where low levels of transcripts were found. Microarray data mining showed that UGT D members were mainly expressed in roots and reproductive organs confirming our RT-qPCR analyses (Fig. 7B). UGT73B2, UGT73B3, UGT73B4, and UGT73B5 were highly expressed in roots grown on agar plates containing Murashige and Skoog basal medium. Surprisingly, these genes were barely expressed in soil-grown roots, suggesting a specific induction of gene expression either under light exposure or through medium composition. Furthermore, the HR-responsive UGT73B3, UGT73B5, and UGT73D1 were also highly expressed in developmental cell death as shown in senescent leaves (Fig. 7B).
Figure 7.
Expression of UGT D genes in Arabidopsis tissues. A, RT-qPCR analysis of UGT expression in roots, rosette leaves, stems, cauline leaves, and flowers. The transcript level of each gene is expressed in absolute copy number per ng total RNA. Values correspond to the mean and sd of duplicates. B, Expression of UGT D genes in soil-grown roots (r1), Murashige and Skoog-grown roots (r2), cotyledons (c), leaves (l), cauline leaves (cl), senescent leaves (sl), flowers (fl), siliques (sil), and seeds (s). Gene expression level, obtained from the AtGenExpress microarray datasets, are expressed in an arbitrary unit. Values correspond to mean and sd of triplicates.
ugt73b3 and ugt73b5 Mutants Display Decreased Resistance to an Avirulent Strain of P. syringae
In the Salk library (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al., 2003), we identified putative ugt73b3 and ugt73b5 knockout lines exhibiting a T-DNA insertion. Plants were selfed to obtain homozygous knockout lines. The T-DNA flanking sequences were amplified by PCR and sequenced to confirm the position of the insertion. The ugt73b3 mutant harbors the T-DNA insert 29 bp downstream of the start codon, whereas T-DNA insertion is 164 bp upstream of the start codon in ugt73b5 knockout line. As expected, no induction of the mutated gene could be detected in both mutant lines following Pst-AvrRpm1 challenge (Fig. 8A).
Figure 8.
A, RT-qPCR expression profiles of UGT genes in wild-type plants and ugt mutants after Pst-AvrRpm1 challenge. Transcript levels were quantified for UGT73B3 and UGT73B5 in wild-type plants (black circles) and in ugt73b3 mutant and ugt73b5 mutant, respectively (white circles). Values correspond to the mean and sd of duplicates. B, Bacterial growth of Pst in Arabidopsis leaves of wild-type plants and ugt D mutants. Leaves were infiltrated with 105 cfu mL−1 Pst DC3000 or Pst-AvrRpm1 and harvested 24 and 72 h after inoculation. The bacterial population (log cfu cm−2) was quantified in wild-type plants (black bars), ugt73b3 (gray bars), and ugt73b5 (white bars) mutants. The graph represents the mean and sd of three replicates. In the incompatible interaction, the bacterial amount in mutant plants was significantly higher than that of the wild type at 72 h post inoculation, according to one-tailed Student's t test (P value = 0.03 and 0.0004 for ugt73b3 and ugt73b5, respectively). Experiment was repeated with similar results.
Resistance to pathogens was investigated using inoculation with Pst. After inoculation with a medium titer (105 colony forming units [cfu] mL−1) of the virulent Pst DC3000 strain, ugt73b3 and ugt73b5 mutants developed disease symptoms similar to those observed in wild-type plants (data not shown) and supported the same bacterial growth as in wild-type plants, 24 and 72 h after Pst DC3000 challenge (Fig. 8B). After inoculation with the avirulent Pst-AvrRpm1 strain, bacterial growth did not increase in wild-type plants. Conversely, bacterial population dramatically increased in ugt73b3 and ugt73b5 mutants, indicating an almost complete loss of resistance in these two genotypes.
DISCUSSION
By transferring sugars to a wide range of secondary metabolites, UGTs increase the stability and solubility of aglycones and therefore modify their bioactivity (Lim and Bowles, 2004). In Arabidopsis, 120 UGT genes have been recognized (Paquette et al., 2003) and classified into 14 groups on the basis of their sequence similarity (Ross et al., 2001). Even if UGT substrate specificities have been extensively studied in vitro (Lim et al., 2001, 2002), little functional information is available for the majority of the genes in this family. To extend our knowledge of the physiological role of UGTs in plant-pathogen interactions, we first analyzed the stress-responsive expression of 11 members belonging to the group D, combining RT-qPCR analyses and data mining of public microarray datasets.
Previous studies based on northern- or classical-PCR analyses have revealed that group D UGTs are induced under stress conditions in Arabidopsis and other plant species (Horvath and Chua, 1996; Fraissinet-Tachet et al., 1998; O'Donnell et al., 1998; Asai et al., 2000; Mazel and Levine, 2002; Nishimura et al., 2003; Poppenberger et al., 2003). Although these studies highlight the stress inducibility of UGT D genes, they were limited by a lack of specificity and did not allow the detection of lowly expressed genes. Therefore, we used RT-qPCR, a highly sensitive and specific method to accurately measure transcript levels of UGTs (Gachon et al., 2004b). We first examined the pathogen-responsive expression of some UGTs of group D in time-course experiments after inoculation with the Pst-AvrRpm1 avirulent bacteria (Figs. 1 and 2). Importantly, these genes were not induced during the compatible interaction of Arabidopsis with the virulent Pst, suggesting that these UGTs play a role in resistance and that induction is dependent on the onset of the HR. Our results clearly indicate that some UGTs of group D exhibit all the transcriptional features of HR-inducible genes, like glutathione-S-transferase (GST) genes (Lieberherr et al., 2003). In addition to pathogen signals, eight UGTs were responsive to the defense-associated signal SA and three of them were also responsive to H2O2 (Fig. 4). Furthermore, besides RT-qPCR analysis, mining of microarray data revealed that UGT expression was induced after wounding, B. cinerea and P. infestans inoculation, and during UV, ozone, and oxidative stresses (Fig. 5). Altogether, these results point to a pathogen- and stress-responsive expression of most UGTs belonging to group D.
It is also clear that UGTs of group D are differentially regulated after inoculation with Pst-AvrRpm1 or after SA and H2O2 treatments. UGT73B3 and UGT73B5 share 77.9% similarity at the amino acid level and exhibit the same pattern of expression after pathogen challenge and signaling-molecule application (Fig. 3). However, UGT73B1, which shows a comparable level of sequence divergence with UGT73B3 and UGT73B5, does not respond to any treatment (Fig. 3). Moreover, UGT73C3 and UGT73C2, which exhibit 89.5% similarity, present totally different expression profiles. UGT73C3 is induced after Pst-AvrRpm1 infection and SA treatment in Arabidopsis leaves, whereas UGT73C2 expression is barely detectable. On the contrary, UGT73C7 and UGT73D1, which have 64.7% similarity within their coding sequences, present similar transcript profiles after Pst-AvrRpm1 inoculation and SA application (Fig. 3). Despite numerous studies aimed at deciphering the modalities of expression divergence between duplicated genes (Haberer et al., 2004), our data are in line with the idea that at present, no predictive conclusion can be drawn on the regulation of paralogue genes from the comparison of their primary sequence (Duan and Schuler, 2005). As a result, a systematic study of differential regulation within a gene family is required to identify candidates involved in an inducible response and to point out possible redundant genes that would be able to complement a mutation. Among the 11 genes investigated through the RT-qPCR and data mining analyses, UGT73B3 and UGT73B5 appear to be highly responsive to pathogens and therefore are good candidates for playing an effective role in defense responses.
Importantly, their kinetics of expression resemble those of early induced genes such as GSTs (Lieberherr et al., 2003). Some UGTs of group D are up-regulated as soon as 3 h after challenge with Pst-AvrRpm1, whereas the late-induced PR1 gene is peaking at 24 h after inoculation (Fig. 2). Early induced genes serve a function of stress adaptation, intercellular communication, and transcriptional regulation of late genes to coordinate long-term biological responses (Abel and Theologis, 1996). Consistent with this role, the activation of the early induced genes does not require de novo protein synthesis (Horvath and Chua, 1996; Horvath et al., 1998; Uquillas et al., 2004). This early induction of UGTs may indicate a role in stress or defense responses to regulate the activities and subcellular localization of substrates, which may preexist prior to pathogen infection, and the biosynthesis of which does not necessitate induction of biosynthetic pathways.
Most group D UGTs were responsive to SA whereas none of them was induced after exposure to MeJA, suggesting a jasmonate-independent and SA-dependent induction of these UGTs. Likewise, the wound-inducible tomato UGT Tomato wound induced 1, which belongs to group D, was previously shown to be induced by SA but not by MeJA (O'Donnell et al., 1998). Transduction of the SA signal requires the function of the regulatory protein NPR1 to activate PR gene expression and systemic acquired resistance (Dong, 2004). Interestingly, activation of SA early induced genes like GST6 and IS10a, a tobacco GT, was shown to be NPR1 independent (Horvath and Chua, 1996; Uquillas et al., 2004). UGT73D1 was not induced in NahG plants after Pst-AvrRpm1 inoculation. However, transcripts of this gene were still found in npr1-1 mutants at 5 and 10 h after pathogen challenge, suggesting a SA-dependent, NPR1-independent induction, which is the feature of some SA early induced genes (Uquillas et al., 2004). On the contrary, although UGT73B3 and UGT73B5 were hardly expressed in NahG plants at 3 h after Pst-AvrRpm1 inoculation, their transcript levels were similar in wild-type plants, NahG transformants, and npr1-1 mutants at 5 and 10 h after inoculation (Fig. 6). This suggests an SA dependence in the early times of infection and the contribution of other unknown signal(s) later. Such a modular regulation of stress-inducible genes by different transduction pathways has already been shown for GST induction (Lieberherr et al., 2003). UGT73B3 and UGT73B5 were responsive to H2O2, whereas UGT73D1 was not (Fig. 3). ROS and notably H2O2 function as signaling molecules in plants and control various processes, including pathogen defense and programmed cell death (Apel and Hirt, 2004). Given their responsiveness to H2O2 and oxidative stresses, UGT73B3 and UGT73B5 induction might be SA dependent at 3 h after Pst-AvrRpm1 challenge and ROS dependent at later times.
Our data further provide direct evidence of the involvement of UGTs during the HR of Arabidopsis to pathogens. Thus, ugt73b3 and ugt73b5 mutants both exhibited a loss of resistance to Pst-AvrRpm1 (Fig. 8). However, both mutants were not fully susceptible to Pst-AvrRpm1, due to either the presence of other effectors of disease resistance or additional UGT genes partially complementing UGT73B3 and UGT73B5. One question that still remains to be answered is which role UGT73B3 and UGT73B5 could play in this process. Since they are also induced during senescence and after challenge with different pathogens, we can assume that these genes are associated to the cell death process occurring during senescence and HR. Moreover, their responsiveness to H2O2 and paraquat oxidative stress suggests that they may also participate in the maintenance of cellular redox homeostasis. As GSTs, which might use stress metabolites as substrates (Edwards et al., 2000) and were thought to be antioxidant proteins against H2O2 produced during the HR (Levine et al., 1994), UGT73B3 and UGT73B5 might contribute to redox homeostasis. In line with this hypothesis, the expression profiles of UGT73B3 and UGT73B5 display striking similarities with the closely related TOGT gene from tobacco. TOGT-mediated glucosylation was shown to be required for scopoletin accumulation in cells surrounding TMV lesions, where this compound plays a role in ROS buffering (Chong et al., 2002). Finally, UGT73B3 and UGT73B5 transcripts were mainly detected in reproductive organs in the absence of stress (Fig. 7). This suggests that these UGTs could be involved in different tissues both in defense responses and in development. The next step in the determination of UGT73B3 and UGT73B5 function is the identification of their substrates in planta. Scopoletin and its glucoside scopolin have recently been identified in Arabidopsis (Rohde et al., 2004), but they are not detected in leaves inoculated with Pst-AvrRpm1 (F. Bellvert and P. Saindrenan, unpublished data). Therefore, it is less than probable that the function of UGT73B3 and UGT73B5 during the HR is linked to scopoletin glucosylation. Transcripts of UGT73B3 and UGT73B5 were found in Murashige and Skoog-grown roots, whereas they were not expressed in soil-grown roots, suggesting a light induction of these genes (Fig. 7). Interestingly, in Arabidopsis roots, light was recently shown to induce phenylpropanoid metabolism, accumulation of flavonoid glucosides, and high levels of coniferin and syringin (coniferyl and sinapyl-4-O-glycosides; Hemm et al., 2004). The identification of UGT73B3 and UGT73B5 endogenous substrates using metabolic profiling is currently under progress with a special interest in phenylpropanoid and flavonoid compounds. Our working hypothesis is that UGT73B3 and UGT73B5 might contribute to redox homeostasis through the glucosylation of a still-unknown metabolite that would fulfill in Arabidopsis the role of scopoletin in tobacco. To the best of our knowledge, this would represent the first example of closely related enzymes displaying a conserved function and a conserved expression profile but having evolved a new substrate specificity between species.
MATERIALS AND METHODS
Biological Materials and Plant Treatments
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used throughout this study. The transgenic line harboring the NahG gene was obtained from R. Dietrich (Syngenta). The Col-0 mutant npr1-1 was provided by X. Dong (Duke University). T-DNA insertion lines ugt73b3 and ugt73b5 were obtained from the Salk institute (SALK_097487 and SALK_078055, respectively). Plants were grown in soil in individual pots under an 8 h/16 h light/dark photoperiod at 150 μE cm−2 s−1 of light intensity, at 20°C and 75% relative humidity. The virulent strain Pseudomonas syringae pv tomato DC3000 (Pst DC3000) and the avirulent strain Pst DC3000 carrying the avirulence gene AvrRpm1 (Pst-AvrRpm1) were obtained from J. Glazebrook (University of Minnesota). Six- to 7-week-old plants were used for Pst inoculations or signaling-molecule treatments. Half leaves were infiltrated using a 1-mL syringe without a needle. Bacteria were grown over night at 28°C on King B liquid medium containing the appropriate antibiotics (kanamycin, 25 μg mL−1; and rifampycin, 50 μg mL−1). They were collected by centrifugation, resuspended in water, and quantified using a spectrophotometer. Plants were infiltrated with Pst DC3000 or Pst-AvrRpm1 at the concentration of 107cfu mL−1 (A600 = 0.01). For signaling-molecule treatments, mature leaves were infiltrated with SA (1 mm in K2HPO4 5 mm pH 6.5) or H2O2 (5 mm). Control plants were infiltrated with K2HPO4 5 mm and water, respectively. For MeJA treatment, plants were kept in closed 12-L chambers with 10 μL MeJA. For each time point, one leaf of six different treated plants was harvested. The resultant six leaves were pooled and frozen in liquid nitrogen. For tissue-specific expression, roots were obtained from 4-week-old plants grown on Murashige and Skoog medium. Rosette leaves were harvested from 4-week-old plants grown in soil under an 8 h/16 h light/dark photoperiod. Plants were then transferred under a 16 h/8 h light/dark photoperiod and cauline leaves, stems, and flowers were harvested after 2 to 3 weeks.
RNA Extraction and cDNA Preparation
Total RNA were extracted from ground, frozen material using Extract All mix (Eurobio) according to the manufacturer's instructions. Samples were treated with the RNase-free DNase I Amplification Grade (Invitrogen Life Technologies) for 15 min. The reaction was stopped by a phenol-chloroform extraction (Sambrook et al., 1989). Total RNA were loaded on a 0.8% agarose gel to check their integrity, and the amount of total RNA was determined spectrophotometrically. Five micrograms of total RNA were reverse transcribed using the SuperScript II First Strand synthesis system for reverse transcription-PCR kit (Invitrogen) according to the manufacturer's instructions.
RT-qPCR
Nucleotidic sequences were aligned to identify divergent regions between the most closely related genes. In this region, position of the primers were determined so that the size of the PCR product ranges between 50 and 200 bp. Oligo 4.0 software (Rychlik and Rhoads, 1989) and Primer Express 1.0 software (Applied Biosystems) were used to evaluate primer dimer formation and to estimate their melting temperature (59°C ± 1°C). A PCR reaction was then simulated in silico using Amplify 1.2 software (Engels, 1993) to test primers amplification on gene of interest and most closely related gene sequences. The sequences of all primers used are summarized in Table II.
Twenty-five-microliter reactions were prepared by mixing 10-μL samples with 12.5 μL Sybr Green Mastermix (Eurogentec), 1 μL of each primer (final concentration 300 nm), and 0.5 μL water. RT-PCR reactions were run on a GeneAmp 5700 device (Applied Biosystems). After 2 min at 50°C followed by a 10-min denaturation step at 95°C, samples were run for 40 cycles of 15 s at 95°C and 1 min at 60°C. After each run, a dissociation curve was acquired to check for amplification specificity by heating the samples from 60°C to 95°C. Data were analyzed with the GeneAmp 5700 SDS software (Applied Biosystems). Despite RNA treatment with DNase I before cDNA synthesis, contamination by genomic DNA was checked in each sample using primers annealing on Ask alpha gene intron (Table II; Charrier et al., 2002). Serial dilutions of genomic DNA extracted with the DNeasy plant maxikit (Qiagen) were used to trace a calibration curve, which was used to deduct the amount of cDNA for each target gene. Samples were run in duplicates. Experiments were repeated twice independently except for H2O2 infiltration, MeJA treatment, and tissue-specific expression. To standardize the data, the amount of target gene was normalized over the abundance of the constitutive TUBα2 gene except for the organ-specific expression analysis.
Microarray Data Mining
Microarray data were obtained from the publicly available dataset released by the AtGenExpress consortium (http://web.uni-frankfurt.de/fb15/botanik). These data were generated using the Affymetrix ATH1 GeneChip probe array (Redman et al., 2004). The probes corresponding to UGT D are UGT73B1, 253281_at; UGT73B2-B3, 253268_s_at; UGT73B4, 265501_at; UGT73B5, 265499_at; UGT73C1, 265197_at; UGT73C2, 265198_at; UGT73C3-C4, 265199_s_at; UGT73C5-C6, 265200_s_at; UGT73C7, 251971_at; and UGT73D1, 251970_at. Induction of UGT D expression was calculated at 6 h after infection with Pst (Pst-AvrRpm1/Pst DC3000) and Phytophthora infestans (P. infestans/water), and exposure to UV-B, or at 12 h after wounding and oxidative stress. To avoid aberrant fold induction based on unreliable very low expression data, the minimum expression level (Affymetrix average difference) of each probe was set to 10 before calculating the expression fold change (Gachon et al., 2005a). For organ-specific expression, data were obtained from the AtGenExpress development baseline experiment. Details on organs, growth conditions, and development stages are available at http://www.weigelworld.org/resources/microarray/AtGenExpress/AtGE_dev_samples.pdf. Datasets included in our study are soil-grown roots, ATGE_3; Murashige and Skoog-grown roots, ATGE_98; cotyledons, ATGE_1; rosette leaves, ATGE_90; cauline leaves, ATGE_26; senescent leaves, ATGE_25; flowers, ATGE_39; siliques, ATGE_77; and seeds, ATGE_82.
ugt D Mutant Characterization
Salk T-DNA insertion lines in the Col-0 background were screened in silico for ugt D mutants. Two lines (SALK_097487 and SALK_078055) were identified to carry a T-DNA insertion at the UGT73B3 and UGT73B5 loci, respectively. The position of the T-DNA insert was confirmed by PCR and DNA sequencing using primers LB (5′-GGACCGCTTGCTGCAACT-3′) and B3F (5′-CCACATCATTCAACACGACAAG-3′) for ugt73b3, and LB and B5R (5′-GACGGTTTCTTGTCTGGATTGG-3′) for ugt73b5. Homozygous plants were identified by PCR, by amplification of the mutant allele using primers LB and B3F for ugt73B3, and LB and B5R for ugt73b5, and by the absence of amplification of the wild-type allele using primers B3F and B3R (5′-GATTTCGAAACTCGGATTCAGG-3′) for ugt73b3 and B5F (5′-GTCTTCTTCAACGTGCACACG-3′) and B5R for ugt73b5.
Determination of Bacterial Growth in Plants
Wild-type plants and ugt73b3 and ugt73b5 mutants were inoculated with a medium titer (105 cfu mL−1) of Pst DC3000 or Pst-AvrRpm1. Whole leaves of 6- to 7-week-old plants were infiltrated using a 1-mL syringe without a needle. Leaf discs (0.5 cm2 each) were harvested from inoculated leaves at 24 and 72 h after infiltration. For each time point, three samples were made by pooling two leaf discs from different treated plants. Bacterial growth was assessed by homogeneizing leaf discs in 400 μL of water, plating appropriate dilutions on solid King B medium containing Kanamycin and Rifampicin and quantifying colony numbers after 2 to 3 d. Statistical analyses of the differences between two means of log-transformed data were performed according to one-tailed Student's t test.
Note Added in Proof
Poppenberger et al. (2005) recently reported that UGT73C5 glucosylates brassinosteroids (Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, et al [2005] The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102: 15253–15258).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4g34138 (UGT73B1), At4g34135 (UGT73B2), At4g34131 (UGT73B3), At2g15490 (UGT73B4), At2g15480 (UGT73B5), At2g36750 (UGT73C1), At2g36760 (UGT73C2), At2g36780 (UGT73C3), At2g36770 (UGT73C4), At2g36800 (UGT73C5), At2g36790 (UGT73C6), At3g53160 (UGT73C7), and At3g53150 (UGT73D1).
Supplementary Material
Acknowledgments
We are grateful to R. Dietrich and X. Dong for providing seeds of NahG plants and npr1-1 mutants and to the AtGenExpress consortium for making their microarray data freely available. Special thanks to Annaïck Mingam and to Floriant Bellvert for helpful discussions.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Patrick Saindrenan (saindrenan@ibp.u-psud.fr).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067223.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41: 875–887 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel FM (2000) Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter JL, Kopczak SD, Snustad DP, Silflow CD (1993) Semi-constitutive expression of an Arabidopsis thaliana alpha-tubulin gene. Plant Mol Biol 21: 937–942 [DOI] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Downregulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1249 [DOI] [PubMed] [Google Scholar]
- Dixon RA (2001) Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Dooner HK, Nelson OE (1977) Genetic control of UDPglucose:flavonol 3-O-glucosyltransferase in the endosperm of maize. Biochem Genet 15: 509–519 [DOI] [PubMed] [Google Scholar]
- Duan H, Schuler MA (2005) Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol 137: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V (2000) Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Engels WR (1993) Contributing software to the Internet: the Amplify program. Trends Biochem Sci 18: 448–450 [DOI] [PubMed] [Google Scholar]
- Fraissinet-Tachet L, Baltz R, Chong J, Kauffmann S, Fritig B, Saindrenan P (1998) Two tobacco genes induced by infection, elicitor and salicylic acid encode glucosyltransferases acting on phenylpropanoids and benzoic acid derivatives, including salicylic acid. FEBS Lett 437: 319–323 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Okuhara H, Fukui Y, Nakao M, Katsumoto Y, Yonekura-Sakakibara K, Kusumi T, Hase T, Tanaka Y (2003) Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol 132: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C, Baltz R, Saindrenan P (2004. a) Over-expression of a scopoletin glucosyltransferase in Nicotiana tabacum leads to precocious lesion formation during the hypersensitive response to tobacco mosaic virus but does not affect virus resistance. Plant Mol Biol 54: 137–146 [DOI] [PubMed] [Google Scholar]
- Gachon C, Mingam A, Charrier B (2004. b) Real-time PCR: what relevance to plant studies? J Exp Bot 55: 1445–1454 [DOI] [PubMed] [Google Scholar]
- Gachon CMM, Langlois-Meurinne M, Henry Y, Saindrenan P (2005. a) Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol Biol 58: 229–245 [DOI] [PubMed] [Google Scholar]
- Gachon CMM, Langlois-Meurinne M, Saindrenan P (2005. b) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10: 542–549 [DOI] [PubMed]
- Grubb DC, Zipp BJ, Ludwig-Muller J, Masuno MN, Molinski TF, Abel S (2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40: 893–908 [DOI] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136: 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JD (1996) Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C (2004) Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J 38: 765–778 [DOI] [PubMed] [Google Scholar]
- Hirotani M, Kuroda R, Suzuki H, Yoshikawa T (2000) Cloning and expression of UDP-glucose:flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210: 1006–1013 [DOI] [PubMed] [Google Scholar]
- Horvath DM, Chua NH (1996) Identification of an immediate-early salicylic acid-inducible tobacco gene and characterization of induction by other compounds. Plant Mol Biol 31: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Horvath DM, Huang DJ, Chua NH (1998) Four classes of salicylate-induced tobacco genes. Mol Plant Microbe Interact 11: 895–905 [DOI] [PubMed] [Google Scholar]
- Hughes J, Hughes MA (1994) Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq 5: 41–49 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ (2001) Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Jones P, Vogt T (2001) Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213: 164–174 [DOI] [PubMed] [Google Scholar]
- Kaminaga Y, Sahin FP, Mizukami H (2004) Molecular cloning and characterization of a glucosyltransferase catalyzing glucosylation of curcumin in cultured Catharanthus roseus cells. FEBS Lett 567: 197–202 [DOI] [PubMed] [Google Scholar]
- Kramer CM, Prata RT, Willits MG, De Luca V, Steffens JC, Graser G (2003) Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry 64: 1069–1076 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberherr D, Wagner U, Bubuis PH, Métraux JP, Mauch F (2003) The rapid induction of glutathione S-transferases AtGSTF2 and AtGSTF6 by avirulent Pseudomonas syringae is the result of combined salicylic acid and ethylene signaling. Plant Cell Physiol 44: 750–757 [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE (2002) The jasmonate pathway. Science 296: 1649–1650 [DOI] [PubMed] [Google Scholar]
- Lim EK, Bowles DJ (2004) A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J 23: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ (2002) The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277: 586–592 [DOI] [PubMed] [Google Scholar]
- Lim EK, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Matros A, Mock HP (2004) Ectopic expression of a UDP-glucose phenylpropanoid glucosyltransferase leads to increased resistance of transgenic tobacco plants against infection with potato virus Y. Plant Cell Physiol 45: 1185–1193 [DOI] [PubMed] [Google Scholar]
- Mazel A, Levine A (2002) Induction of glucosyltransferase transcription and activity during superoxide-dependent cell death in Arabidopsis plants. Plant Physiol Biochem 40: 133–140 [Google Scholar]
- Moehs CP, Allen PV, Friedman M, Belknap WR (1997) Cloning and expression of solanidine UDP-glucose glucosyltransferase from potato. Plant J 11: 227–236 [DOI] [PubMed] [Google Scholar]
- Nagashima S, Inagaki R, Kubo A, Hirotani M, Yoshikawa T (2004) cDNA cloning and expression of isoflavonoid-specific glucosyltransferase from Glycyrrhiza echinata cell-suspension cultures. Planta 218: 456–459 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Truesdale MR, Calvert CM, Dorans A, Roberts MR, Bowles DJ (1998) A novel tomato gene that rapidly responds to wound- and pathogen-related signals. Plant J 14: 137–142 [DOI] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S (2003) On the origin of family 1 plant glycosyltransferases. Phytochemistry 62: 399–413 [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G (2003) Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem 278: 47905–47914 [DOI] [PubMed] [Google Scholar]
- Quiel JA, Bender J (2003) Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J Biol Chem 278: 6275–6281 [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim EK, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2: 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik W, Rhoads RE (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res 17: 8543–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Thompson JD, Gibson TJ, Plewnak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uquillas C, Letelier I, Blanco F, Jordana X, Holuigue L (2004) NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol Plant Microbe Interact 17: 34–42 [DOI] [PubMed] [Google Scholar]
- Vet LEM, Dicke M (1992) The ecology of infochemical use by natural enemies of herbivores in a tritrophic context. Annu Rev Entomol 37: 141–172 [Google Scholar]
- Vogt T, Grimm R, Strack D (1999) Cloning and expression of a cDNA encoding betanidin 5-O-glucosyltransferase, a betanidin- and flavonoid-specific enzyme with high homology to inducible glucosyltransferases from the Solanaceae. Plant J 19: 509–519 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Xu ZJ, Nakajima M, Suzuki Y, Yamaguchi I (2002) Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol 129: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.