Abstract
Protoporphyrinogen IX oxidase (PPO) catalyzes the last common step in chlorophyll and heme synthesis, and ferrochelatase (FeC) catalyzes the last step of the heme synthesis pathway. In plants, each of these two enzymes is encoded by two or more genes, and the enzymes have been reported to be located in the chloroplasts or in the mitochondria. We report that in the green alga Chlamydomonas reinhardtii, PPO and FeC are each encoded by a single gene. Phylogenetic analysis indicates that C. reinhardtii PPO and FeC are most closely related to plant counterparts that are located only in chloroplasts. Immunoblotting results suggest that C. reinhardtii PPO and FeC are targeted exclusively to the chloroplast, where they are associated with membranes. These results indicate that cellular needs for heme in this photosynthetic eukaryote can be met by heme that is synthesized in the chloroplast. It is proposed that the multiplicity of genes for PPO and FeC in higher plants could be related to differential expression in differently developing tissues rather than to targeting of different gene products to different organelles. The FeC content is higher in C. reinhardtii cells growing in continuous light than in cells growing in the dark, whereas the content of PPO does not significantly differ in light- and dark-grown cells. In cells synchronized to a light/dark cycle, the level of neither enzyme varied significantly with the phase of the cycle. These results indicate that heme synthesis is not directly regulated by the levels of PPO and FeC in C. reinhardtii.
Hemes and chlorophylls are major tetrapyrrole pigments that are essential for energy metabolism in all photosynthetic organisms. Hemes and chlorophylls are synthesized in a multistep pathway that involves nucleus-encoded enzymes (Beale, 2005). 5-Aminolevulinic acid (ALA) is the earliest universal biosynthetic precursor of tetrapyrroles. The biosynthetic steps from ALA to protoporphyrin IX are common to the synthesis of both chlorophylls and hemes. The enzyme that catalyzes the last shared step of heme and chlorophyll synthesis is protoporphyrinogen IX oxidase (PPO; EC 1.3.3.4). Ferrochelatase (FeC; EC 4.99.1.1) catalyzes the last step in heme biosynthesis: the insertion of a ferrous ion into protoporphyrin IX to form protoheme.
PPO and FeC have been extensively studied, and the crystal structures of both enzymes have been determined, enabling further determination of their reaction mechanisms (Wu et al., 2001; Koch et al., 2004). In mammalian cells, PPO and FeC are both located in the mitochondria. PPO is associated with the mitochondrial inner membrane, and its active site faces the intermembrane space, whereas FeC is present as an integral component of the mitochondrial inner membrane with its active site on the matrix side (Dailey, 2000). The subcellular localizations of PPO and FeC in plant and algal cells are less well characterized (Lister et al., 2001). The presence of chloroplasts in photosynthetic eukaryotes complicates the potential spatial and functional organization of the tetrapyrrole pathway. The enzymes for the early steps up to the formation of protoporphyrinogen IX, as well as the tRNAGlu that is required for ALA synthesis in these organisms, appear to be located exclusively in the chloroplasts (Beale, 1999). The enzymes for all later steps of chlorophyll synthesis beginning with magnesium chelation are also located exclusively within the chloroplasts (Beale, 1999). However, there is conflicting evidence regarding the intracellular location of PPO and FeC. Different isoforms of PPO have been reported to be targeted to either chloroplasts or mitochondria after their synthesis in the cytoplasm (Lermontova et al., 1997; Watanabe et al., 2001). In addition, FeC isoforms that are encoded by two different genes have differential tissue distribution and show differential light regulation (Suzuki et al., 2002). However, so far there is no clarity as to whether the different gene products are differentially targeted to chloroplasts and mitochondria (Lister et al., 2001; Masuda et al., 2003), although this was proposed earlier (Chow et al., 1998).
The unicellular green alga Chlamydomonas reinhardtii is similar to plants in that it contains both a chloroplast and mitochondria. However, unlike plants, C. reinhardtii does not undergo tissue differentiation. We report here that, in contrast to plants, C. reinhardtii contains only one gene each for PPO and FeC, and that the products of these genes are present only in the chloroplast. Our results on the light regulation of PPO and FeC expression and the intracellular location of these proteins lead us to suggest possible roles for the multiple genes encoding these enzymes in plants.
RESULTS
Isolation of C. reinhardtii PPO and FeC cDNAs
C. reinhardtii cDNAs coding for PPO and FeC were isolated by complementation of Escherichia coli hemG and hemH mutant strains, which lack PPO and FeC, respectively. There was a noted difference in the yield of colonies of complemented hemG and hemH cells: the C. reinhardtii cDNA library produced high numbers of complemented hemG colonies, whereas the library produced only a single colony after multiple attempts to complement the hemH strain. Possible reasons for this difference in colony yield include a lower abundance of FeC transcripts compared with those for PPO in the mRNA that was used to construct the cDNA library, and differences in the functional state of the two full-length (unprocessed) translation products in the E. coli cells. Nevertheless, the fact that C. reinhardtii PPO and FeC did complement the E. coli mutant strains indicates that the isolated cDNAs code for functional PPO and FeC proteins. Searches of the C. reinhardtii expressed sequence tag (EST) databases at the Joint Genome Institute (JGI; http://genome.jgi-psf.org/cgi-bin/runAlignment?db=chlre2) and Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/plant/chlamy/EST/blast.html) yielded only one short EST clone for PPO and no matches corresponding to FeC.
PPO and FeC Gene Copy Numbers
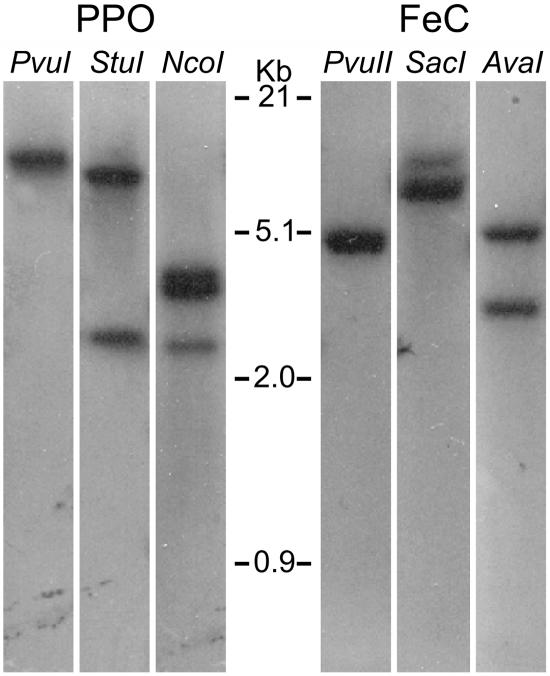
A search of the C. reinhardtii genome sequence database indicated that there is only one gene each encoding PPO (ppx, JGI C_330078; as previously reported by Lohr et al., 2005) and FeC (hem15, JGI C_420005). The sequences of these genes correspond to the cDNAs isolated in this work. However, because the currently available draft database (version 2.0) is not yet complete, it was necessary to confirm the presence of a single gene each for PPO and FeC in C. reinhardtii by Southern-blot analysis. Using as specific probes the regions of each cDNA that encode the almost-complete mature proteins, no evidence for more than one PPO gene and one FeC gene was obtained (Fig. 1). It is therefore concluded that, in C. reinhardtii, PPO and FeC are each encoded by a single gene. This is in clear contrast with plants, where usually two distinct genes exist for each protein.
Figure 1.
Southern-blot hybridization with C. reinhardtii genomic DNA, digested with the indicated restriction endonucleases and hybridized as described in the text. PvuI does not cut, whereas StuI and NcoI cut once and twice, respectively, in the PPO probe sequence; in the FeC probe sequence, PvuII does not cut, whereas SacI and AvaI both cut once.
Analysis of C. reinhardtii PPO and FeC cDNA Products
The sequence of the isolated C. reinhardtii PPO cDNA (2,480 bp) was found to be identical to that previously reported (Randolph-Anderson et al., 1998; GenBank accession no. AF068635). The PPO cDNA encodes a 563-residue 59,802-Mr precursor protein, which is predicted to have a 49-residue presequence that confers protein targeting to the chloroplast, according to analysis using several prediction programs including TargetP (Lohr et al., 2005). The predicted 514-residue 54,897-Mr mature protein has a pI of 9.29. The C. reinhardtii PPO amino acid sequence exhibits an invariant GxGxxG motif near the N terminus that has been proposed to be a dinucleotide-binding motif for binding the FAD cofactor (Dailey and Dailey, 1996a, 1996b). This proposed functional assignment has been partially confirmed by the recently available crystal structure of PPO-II from tobacco (Nicotiana tabacum), which indicates that one of these residues, corresponding to S92 of the C. reinhardtii PPO preprotein, is hydrogen bonded to the cofactor (Protein Data Bank accession no. 1SEZ; Koch et al., 2004).
The C. reinhardtii FeC cDNA sequence was newly obtained and was deposited in the GenBank database under accession number AF332962. The 2,660-bp cDNA contains a 1,479-bp open reading frame (ORF) that encodes a 493-residue 54,433-Mr precursor protein. The TargetP program predicts a 77-residue chloroplast-targeting sequence. The predicted 416-residue 46,100-Mr mature protein has a pI of 5.12. The FeC protein exhibits the invariant His residue that was reported to be required for activity (Wu et al., 2001) at position 282 of the precursor and the typical active site loop QSRVGPAEWL, beginning at residue Q325 of the precursor, which contains all the residues (underlined) that were found to be important for enzymatic activity (Shi and Ferreira, 2004).
The C-terminal portions of FeCs show characteristics specific to certain groups of organisms (Dailey et al., 2000; Dailey and Dailey, 2002). The FeCs of animals, yeasts, and plants contain a C-terminal extension that is absent from most bacterial FeCs (Fig. 2A; Dailey et al., 2000). Animal FeCs, and those of some yeasts and bacteria, are characterized by the presence of a [2Fe-2S] cluster (Dailey et al., 2000; Dailey and Dailey, 2002). Three of the four Cys ligands required to bind the [2Fe-2S] cluster are found at the C terminus of these FeCs (Dailey et al., 1994). No Cys residues for a [2Fe-2S] cluster are found in FeCs of Saccharomyces cerevisiae or photosynthetic eukaryotes. The C terminus is also involved in the dimerization of FeCs, although some FeCs that contain a C-terminal extension are nonetheless monomeric (Wu et al., 2001; Dailey and Dailey, 2002). In common with FeCs of other photosynthetic organisms, C. reinhardtii FeC has a C-terminal extension that lacks the Cys residues that are involved in the binding of a [2Fe-2S] cluster (Fig. 2A).
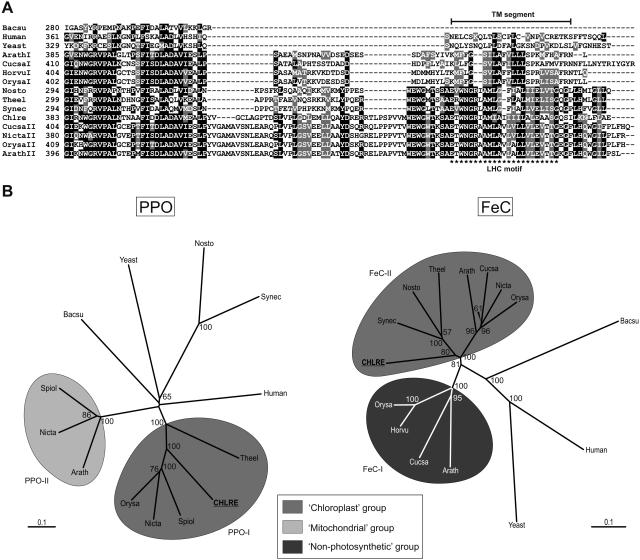
Figure 2.
Analysis of the amino acid sequences of PPO and FeC. A, The C-terminal portion of C. reinhardtii FeC contains an LHC motif that is found in Type II plant FeCs and also in cyanobacterial FeCs. The approximate region of a transmembrane segment (TM segment) as predicted by TMPred is indicated for the sequences of Type II plant FeCs and the FeCs of cyanobacteria and C. reinhardtii. Chlre, C. reinhardtii; Arath, Arabidopsis; Cucsa, cucumber (Cucumis sativus); Glyma, pea; Horvu, barley (Hordeum vulgare); Nicta, tobacco; Orysa, rice (Oryza sativa); Nosto, Nostoc; Synec, Synechococcus; Theel, Thermosynechococcus elongatus; Bacsu, Bacillus subtilus. B, Bootstrapped neighbor-joined rootless trees showing the relatedness of PPO and FeC enzymes from various sources. For the eukaryotic enzymes, full-length preproteins were used in the analyses. Bootstrap values above 50 are shown at the nodes. The bar labeled “0.1” is the branch length representing a mean difference of 0.1 per residue along each branch. C. reinhardtii PPO groups most closely with the Type I chloroplast counterparts in plants, which are found only in photosynthetic tissues. Mitochondrial and nonphotosynthetic refer, respectively, to PPO enzymes reported to be present in mitochondria and to FeCs that are up-regulated in nonphotosynthetic conditions (see text). PPO GenBank accession numbers (clockwise, starting with Chlre): AF068635, BAA96808, Y13465, BAB39760, AAM26644, Y13466, BAB60710, P32397, P40012, NP_484159, BAA18074, NP_000300, BAC07926; FeC GenBank accession numbers (clockwise, starting with Chlre): AF332962, S75788, NP_487791, BAC09768, O04921, BAB20760, CAC50871, XP_475111, P32396, P22830, P16622, P42043, BAA05102, P42045, BAD33213.
In photosynthetic organisms, FeCs that are associated with photosynthetic tissues contain a light-harvesting complex (LHC) motif, a conserved hydrophobic stretch that corresponds to the third transmembrane segment found in the LHC family of proteins and that likely plays a role in anchoring the protein in membranes (Jansson, 1999). The LHC motif is present in plant FeCs that are expressed in photosynthetic tissues but not in the FeCs that are expressed in nonphotosynthetic tissues (Suzuki et al., 2002; Masuda et al., 2003). This difference could be important for targeting the FeC to specific membranes within the chloroplast. In common with plant FeCs that are associated with chloroplasts, the C. reinhardtii FeC does contain an LHC motif (Fig. 2A).
PPO and FeC Phylogenies
In plants, PPO and FeC are each encoded by two different genes that have been referred to as Types I and II. PPO-I is located in chloroplasts, whereas PPO-II was variously reported to be located in mitochondria (Lermontova et al., 1997) or in both organelles (Watanabe et al., 2001). FeC-I is found in the chloroplasts but is mainly expressed in nonphotosynthetic tissues. FeC-II is also found in chloroplasts and is expressed in photosynthetic tissues in a light-responsive manner (Suzuki et al., 2002; Masuda et al., 2003).
BLAST searches revealed that C. reinhardtii PPO exhibits an identity of 55% to 62% with Type I plant PPOs, 40% to 43% with cyanobacterial PPOs, 28% to 30% with Type II plant PPOs, and 32% or less with nonplant mitochondrial and bacterial PPOs. C. reinhardtii FeC shows the highest identity to plant Type II FeCs and cyanobacterial FeCs, being 57% to 69% and 52% to 59% identical, respectively, in overlapping regions. The identity with plant Type I FeCs is 47% to 49%, whereas that with nonplant mitochondrial FeCs is 41% or lower. In phylogenetic analyses, both PPO and FeC of C. reinhardtii group most closely with their counterparts in plants that are expressed in photosynthetic tissues, and away from those enzymes that are active in nonphotosynthetic tissues and which, in the case of PPOs, have been reported to reside in mitochondria (Fig. 2B).
Analysis of PPO and FeC Targeting and Localization Sequences
To obtain information about the chloroplast-targeting peptides of C. reinhardtii PPO and FeC, in silico analysis was conducted. Although the TargetP program predicted chloroplast targeting of both PPO and FeC, the scores and reliability of the predictions are moderate (Fig. 3B), in contrast to those obtained for plant PPO and FeC precursors. In addition, for the FeC precursor, other targeting prediction programs (PSORT, Predotar, listed under expasy.org) did not predict a clear chloroplast targeting of the FeC precursor. Generally, targeting predictions for C. reinhardtii precursors to the chloroplast are weak; this can be explained by the fact that C. reinhardtii chloroplast presequences generally resemble more closely mitochondrial-targeting signals of yeast and animals than plant chloroplast signals (Franzén et al., 1990; Kindle, 1998). Furthermore, the TargetP program is based on known sequences of Arabidopsis (Arabidopsis thaliana) chloroplast proteins (Emanuelsson et al., 2000).
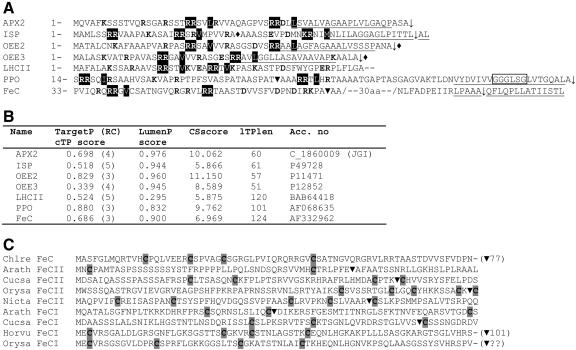
Figure 3.
Sequence analysis of transit peptides. A, Twin Arg-containing presequences of C. reinhardtii chloroplast-targeted precursors and their in silico analysis using TargetP v1.1 and LumenP v1.3. OEE2/3, PSII oxygen-evolving enhancer proteins; APX2, putative ascorbate peroxidase; LHCII, chlorophyll a/b-binding protein. Twin Args and the hydrophobic residue (if present) two or three positions downstream are highlighted in black; note that in ISP, KR was found to be important for luminal targeting instead of RR. Charged residues are in bold, and hydrophobic regions are underlined. The invariant GxGxxG motif, which occurs well into the mature protein sequence of PPO, is boxed. ♦, Experimentally determined cleavage site; ▾, cleavage site predicted by TargetP; ↓, TPP cleavage site predicted by LumenP. B, In silico analysis of C. reinhardtii presequences. TargetP cTP, Chloroplast-targeting peptide; RC, reliability coefficient (1 is high and 5 is low); LumenP prediction cutoff, LumenP score ≥ 0.47 and cleavage site score (CSscore) ≥ 6.8; lTPlen, the predicted luminal-targeting peptide length. Predicted lTPlen for ISP and LHCII are incorrect, as these precursors do not contain TPP cleavage sites. C, Cys residues in the precursor sequences of plant and C. reinhardtii FeCs. The I and II suffixes refer to the isoforms present in photosynthetic and nonphotosynthetic tissues, respectively, as described in the text. ▾, Cleavage sites predicted by TargetP.
Interestingly, the presequences of the C. reinhardtii PPO and FeC precursors show certain characteristics of chloroplast precursors that are targeted to the thylakoid lumen via the twin-Arg translocase (Tat) pathway. The Tat pathway is capable of translocating proteins across the thylakoid membrane in their folded state. Tat domains consist of two adjacent Arg residues followed by a hydrophobic residue two or three residues afterward (Tat motif), a central hydrophobic region (H-region) directly followed by a consensus thylakoid processing peptidase (TPP) cleavage site (AxA↓x; for review, see Robinson and Bolhuis, 2004). The C. reinhardtii PPO and FeC precursors both contain Tat motifs (Fig. 3A) and possible TPP cleavage sites, as found in most lumen-targeted proteins (Mayfield et al., 1989; von Heijne et al., 1989; Merchant et al., 1990). The program LumenP, a neural network predictor of lumen targeting in plants (Westerlund et al., 2003), produced very high scores for known lumenal proteins such as the Rieske iron-sulfur protein (ISP), the PSII oxygen-evolving enhancer proteins OEE2 and OEE3 (Mayfield et al., 1989; Finazzi et al., 2003 and refs. therein), and a putative ascorbate peroxidase (APX2), which is considered very likely to be lumenal because it is found in the lumen in Arabidopsis (NM_116970; Kieselbach et al., 2000; Peltier et al., 2002). Light-harvesting chlorophyll a/b-binding protein (LHCII) receives a low score because it does not contain lumen-targeting sequences (Lamppa, 1988). Although the C. reinhardtii PPO and FeC precursors yielded relatively high LumenP scores (Fig. 3B), the LumenP program returned TPP cleavage site positions well into the mature protein, in the case of the PPO precursor directly after a predicted transmembrane helix, and for the FeC precursor inside a moderately hydrophobic stretch (Fig. 3A). Moreover, the segment that typically comprises the H-region in valid Tat domains, directly following the twin Args, could not be unequivocally identified in the C. reinhardtii precursors. Therefore, although the presence of Tat signals in these proteins is striking, their functional validity is uncertain. However, it should be noted that all of the analyzed C. reinhardtii precursors that are known to be targeted to the lumen exhibit two separate twin Args in their presequences, which is also true for the FeC precursor and potentially also for the PPO precursor, depending on the position of the N terminus of the latter.
Another interesting feature of FeC presequences from photosynthetic eukaryotes, including that of C. reinhardtii, is the presence of several Cys residues (Fig. 3C). A survey of plant chloroplast-targeting peptides indicates that the occurrence of multiple Cys residues is uncommon (data not shown).
Overexpression of C. reinhardtii PPO and FeC in E. coli
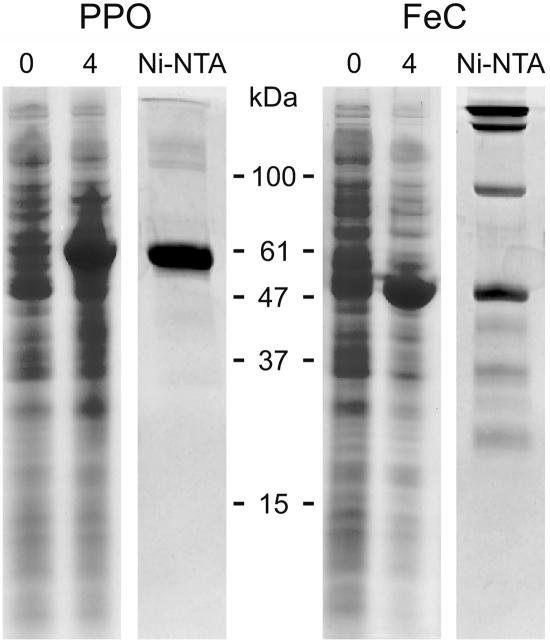
The predicted mature regions of the PPO and FeC were overexpressed in E. coli using the pQE30 expression vector, which introduces an N-terminal His tag. The recombinant proteins were highly expressed (Fig. 4) and invariably found in inclusion bodies (data not shown). These inclusion bodies were dissolved in 6 m guanidinium hydrochloride and the proteins were purified under denaturing conditions. On SDS-PAGE, purified, overexpressed PPO appeared as a single 59-kD band, whereas purified FeC was seen as a 47-kD monomer but also as a set of higher Mr bands even in the presence of 1% (w/v) SDS and 100 mm dithiothreitol, suggestive of a strong tendency of the protein to aggregate, at least under denaturing conditions. Purified PPO and FeC were used to raise antibodies in rabbits. The obtained antisera cross-reacted with their corresponding overexpressed proteins (data not shown) and recognized bands of the appropriate size in C. reinhardtii cell extracts (Fig. 5).
Figure 4.
Expression and purification of cloned C. reinhardtii PPO and FeC. Coomassie Blue-stained SDS-PAGE of His-tagged C. reinhardtii PPO and FeC proteins in extracts of E. coli cells before (0) and after 4 h of induction (4), and after purification of the expressed proteins by N-affinity chromatography (Ni-NTA). Positions of molecular mass marker proteins are indicated.
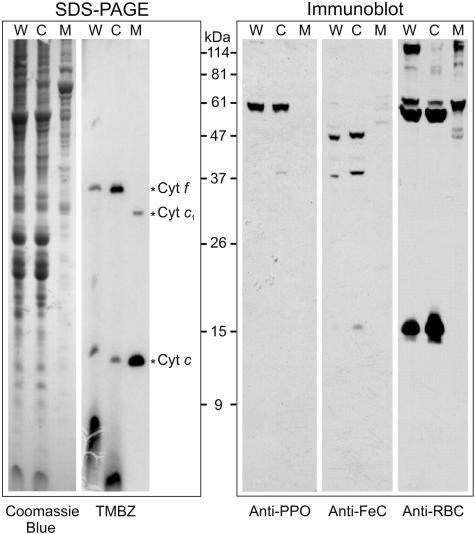
Figure 5.
Cell fractionation and immunoblot hybridization with antibodies raised against the overexpressed PPO and FeC proteins. In FeC immunoblots, a 36-kD contaminant of chloroplast origin was usually observed. TMBZ detected heme-containing proteins indicative of mitochondria (cyt c and cyt c1) and chloroplasts (cyt f). Antibody against Rubisco detected the two subunits of 50 and 15 kD only in cells and the chloroplast fraction. For each sample, 100 μg of total proteins were loaded onto the gel. W, Whole cells; C, chloroplasts; M, mitochondria; RBC, Rubisco.
Subcellular Localization of PPO and FeC
To determine whether the products of the single genes encoding PPO and FeC in C. reinhardtii are targeted to specific organelles in the cell, chloroplasts and mitochondria were isolated from mixotrophically grown cells. The subcellular fractions were analyzed on SDS-PAGE, and the proteins were transferred to nitrocellulose for immunoblotting with specific antisera that were raised against the purified, overexpressed PPO and FeC. Coomassie Blue-stained SDS-PAGE gels revealed clearly different polypeptide patterns for the chloroplast and mitochondrial fractions (Fig. 5). In the chloroplast fraction, heme-mediated peroxidase activity staining detected cytochrome (cyt) f, as expected, but also low levels of a 12-kD c-type cyt that was attributed to contamination by mitochondrial cyt c (Fig. 5). In contrast, the mitochondrial fraction contains heme-staining bands at positions corresponding to cyt c and cyt c1, but this fraction contained no evidence for the presence of chloroplast cytochromes. In addition, an anti-Rubisco antibody detected high levels of the large and small Rubisco subunits in the chloroplast fraction. The mitochondrial fraction contained some cross-reacting bands, but none of those bands corresponded to the Rubisco large and small subunits. It can therefore be concluded that the chloroplast fraction might be slightly contaminated by mitochondria whereas the mitochondrial fraction is free of chloroplast contamination. Immunoblot analysis with the anti-PPO antibodies revealed in whole cells a single band of an apparent molecular mass of 59 kD. In the organellar fractions, this protein was detected exclusively in the chloroplast fraction (Fig. 5). Anti-FeC antibodies detected a major band of 47 kD in whole cells and in the chloroplast fraction. Several very faint bands were detected in the mitochondrial fraction, but none were detected at the position of FeC. The apparent molecular mass of the mature PPO was approximately 4 kD higher (59 kD) than predicted; since the overexpressed PPO exhibited the same apparent molecular mass, the observed electrophoretic mobility might be influenced by the high pI value of the PPO protein. The FeC band correlated with the mass calculated for the predicted mature protein. A 36-kD nonspecific band, especially visible in the FeC immunoblot, was caused by a common contaminant in antisera of preimmunized rabbits for C. reinhardtii proteins (data not shown). Altogether, these results indicate that in C. reinhardtii, both PPO and FeC are located exclusively in the chloroplasts.
Chloroplast Membrane Association of PPO and FeC
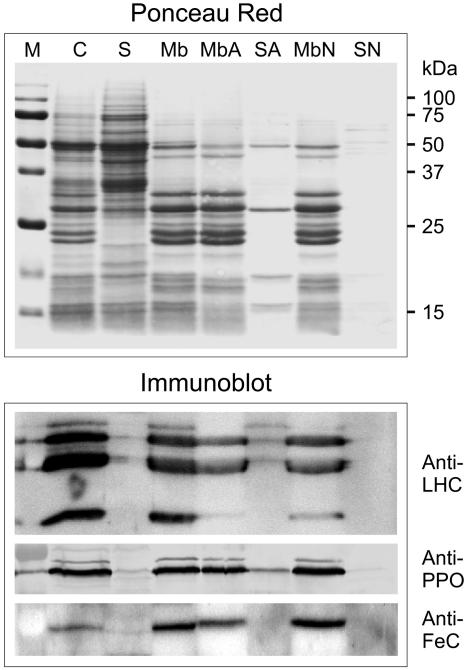
To determine whether C. reinhardtii PPO and FeC proteins are associated with membranes, intact chloroplasts were fractionated into membrane and soluble fractions. Immunoblot analysis revealed that both PPO and FeC were located only in the membrane fraction (Fig. 6). Membranes were further subjected to high salt or alkaline treatment to examine the nature of the membrane association of the enzymes. PPO was not extracted by 1 m NaCl and was mostly resistant to extraction by alkaline treatments to a similar level as the LHC proteins, indicating that the protein is intrinsic to the membrane. No dissociation of FeC by either 1 m NaCl or alkaline treatment could be detected.
Figure 6.
Distribution of PPO and FeC between chloroplast membrane and soluble fractions. Chloroplast membranes were treated either with high pH (20 mm Na2CO3, pH 11.0) or high salt (1 m NaCl) to determine the nature of membrane associations. An anti-LHC antibody was used as control for a set of proteins known to be confined to the membranes. C, Whole chloroplasts; S, soluble fraction; Mb, membrane fraction; MbA and SA, alkaline-treated membranes and supernatant; MbN and SN, high salt-treated membranes and supernatant; M, Mr marker. Blots were stained with Ponceau Red to detect proteins and then treated with the indicated specific antibodies.
Light and Nutritional Regulation of mRNA and Protein Levels
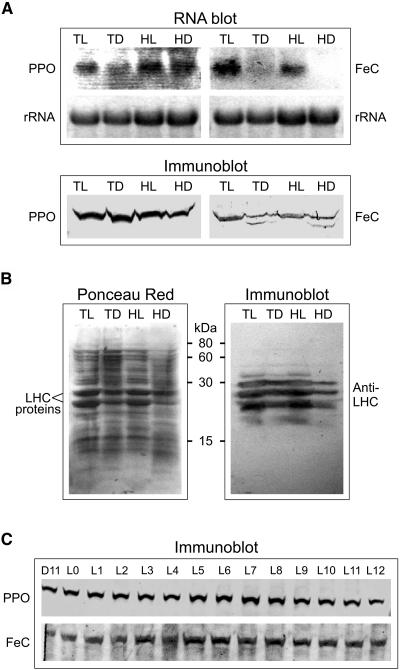
The regulation of PPO and FeC expression in cells growing in continuous light and dark, and with different acetate levels in the growth medium, was investigated by RNA-blot and immunoblot analysis. It was found that the PPO mRNA level was at least 2-fold lower in cells growing in Tris-acetate phosphate (TAP) medium in the dark, compared to cells growing in the other conditions (Fig. 7A). The PPO protein level was not significantly influenced by either light or acetate concentration. In contrast, the levels of FeC mRNA and protein were both lower in cells growing in the dark than in light-grown cells, regardless of the acetate concentration, and the effect was more pronounced on the mRNA level than on the protein level. It was very important to make sure that the cells were in their exponential growth phase in darkness, especially in TAP medium, due to the limited amount of carbon source. On this medium, dark-adapted cells (cell wall-deficient strain) started dying off within 72 h after the initiation of the exponential growth phase (R. van Lis, unpublished data). As an indication for dark adaptation of the C. reinhardtii cells, the levels of LHC proteins were visualized by immunoblotting, using an antibody that recognizes C. reinhardtii LHCII proteins P11, P16, and P17 (Pineau et al., 2004). It is clear that LHC proteins are less abundant in cells growing in the dark than in the light, and least abundant in cells growing in the darkness on high-acetate medium (Fig. 7B). It is known that C. reinhardtii cells retain an operational photosynthetic apparatus even when growing in complete darkness (Harris, 1989), which is consistent with the presence of LHC proteins in dark-grown cells. Our data indicate that there was no correlation between the steady levels of PPO and the varying levels of LHC in cells grown under continuous conditions. A correlation could have been expected because PPO is required for the synthesis of chlorophyll, the major cofactor of LHC.
Figure 7.
Expression of PPO and FeC in cells grown in continuous light and dark and in light/dark-synchronized cells. A, RNA blots and concomitant immunoblots of C. reinhardtii cells grown in continuous light and dark, in either TAP or high-acetate medium. For RNA blots, 10 μg of total RNA was transferred; for immunoblots, 50 μg of protein. TL and TD, Cells grown on TAP medium in continuous light and dark, respectively; HL and HD, cells grown on high-acetate medium in light and dark, respectively. B, Left, Ponceau Red-stained blot after transfer of an SDS-polyacrylamide gel (12% acrylamide) loaded with identical samples as in A; right, immunoblot using an anti-LHC antibody. C, PPO and FeC protein levels in 12-h-light/12-h-dark-synchronized cells grown on TAP medium. The samples were from cells harvested 1 h before the light went on (L11) and then at every hour during the light period (L0–L12); 40 μg protein per lane for the anti-PPO immunoblot and 120 μg per lane for the anti-FeC immunoblot was electrophoresed on 8% polyacrylamide SDS gels. Blots were stained with Ponceau Red to detect proteins and with the indicated specific antibodies.
In C. reinhardtii cells synchronized to a 12-h-light/12-h-dark cycle, no significant changes in protein levels of either PPO or FeC were observed upon onset of the light phase. This suggests a different type of regulation of these two enzymes in the synchronized cells compared to continuous light and dark conditions. The synchronized cells were previously verified to exhibit the typical synchronization behavior of dividing once during the dark phase and not synthesizing chlorophyll in the dark (Nogaj et al., 2005).
Experiments to determine the in vivo turnover of PPO and FeC proteins were done by administering 200 μg/mL of the protein synthesis inhibitor cycloheximide to cultures growing in continuous light, continuous dark, and at the beginning of the light phase in light/dark-synchronized cells. Samples were taken at various times during the next 24 h and the levels of PPO and FeC were quantitated by immunoblotting. Under all growth conditions, both PPO and FeC were relatively stable, with at least 75% of the initial levels remaining at 24 h (data not shown).
Potential Genomic Upstream cis-Regulatory Elements
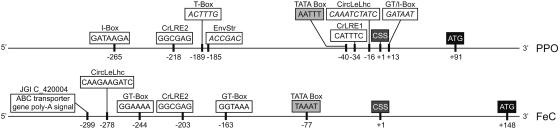
Using the C. reinhardtii genomic sequence (JGI draft sequence, version 2.0), the regions upstream of the nuclear PPO and FeC genes were searched for transcriptional elements that involve light regulation. In addition to putative TATA boxes, the upstream regions of the two genes exhibit several potential light-responsive elements, which for the FeC gene are all located relatively far from the cDNA start site (Fig. 8). C. reinhardtii-specific light-responsive elements (Hahn and Kück, 1999) as well as GT or similar elements (Villain et al., 1996), among others, were found upstream of both genes (Fig. 8).
Figure 8.
Schematic representation of potential transcription factor-binding and/or cis-acting elements present upstream of the C. reinhardtii PPO and FeC ORFs that are potentially implicated in the light responsiveness of the genes. The elements were identified using the PLACE database, and the C. reinhardtii-specific light-responsive elements from Hahn and Kück (1999). S-numbers refer to transcription factor elements in the PLACE database. CSS, cDNA start site; TATA box, putative RNA polymerase-binding site; CrLRE, C. reinhardtii-specific light-responsive element; CircLeLhc, element necessary for circadian expression of tomato (Lycopersicon esculentum) Lhc (S000252); EnvStr, environmental stress including high light (S000402), involves phytochrome in plants (S000153); GT-Box, GT-1-binding sites involved in light-activated transcriptional activation (S000198); I-Box (S000199, S000424), conserved sequence upstream of light-regulated genes; T-box (S000383), found involved in Arabidopsis light-induced gene activation. Also shown is the end of a putative ABC transporter gene (JGI C_420004) upstream of the C. reinhardtii FeC gene.
DISCUSSION
The presence of only one gene each for PPO and FeC made the determination of the predominant organellar targeting in C. reinhardtii a less confounding task than in plants. The antibodies raised against the single forms of both overexpressed enzymes are expected to recognize the proteins regardless their intracellular location. Immunoblots using antibodies against the PPO and FeC detected the proteins only in the chloroplast fraction, which makes it likely that in C. reinhardtii, the last two steps of heme synthesis are confined to the chloroplast.
C. reinhardtii PPO and FeC proteins were both found to be associated with the membrane fraction of the chloroplast. It was not possible to localize these proteins to a specific chloroplast membrane because a protocol to obtain pure envelope and thylakoids from the green alga is not currently available. Although in silico analysis using the program LumenP suggested that both precursor proteins contained Tat domain characteristics that would direct their localization to the lumen, the program seemed to have based its predictions at least in part on H-regions near the N terminus but within the mature proteins, instead of in the presequence, where true H-regions could not be found. Therefore, conclusions based on this analysis should be interpreted with caution, and in any case it is unlikely in either plants or C. reinhardtii that PPO and FeC are located on the lumenal side of the thylakoid. However, the Tat signals might be involved in directing these proteins and inserting them into the thylakoid membranes. Direct determination of the actual N termini of the mature C. reinhardtii PPO and FeC would help to resolve this issue.
An unusually high number of Cys residues is found only in FeC presequences, in C. reinhardtii as well as higher plants. Although the significance of these Cys residues is unknown, it is conceivable that they could be involved in regulation of posttranslational processing or import by the chloroplast redox state, which is strongly influenced by light. It is known that the content of FeCs in plants and in C. reinhardtii is influenced by light (see below).
Several potential transcriptional regulatory elements were found in the genomic regions upstream of C. reinhardtii PPO and FeC. GT-1 transcriptional regulatory elements are ubiquitous (Villain et al., 1996), and GT-like elements have been identified in C. reinhardtii previously (Kucho et al., 2003). cis-Elements involved in circadian gene transcription might be important with respect to variations in mRNA levels in synchronized C. reinhardtii cells. C. reinhardtii-specific light-responsive sequences could also be significant, and they are present upstream of the ORFs of both PPO and FeC. Even though the light regulation of their transcription is in fact different, both genes contain potential upstream light-responsive transcription elements. It is anticipated that our results will aid in eventually achieving a better understanding of the functional status of potential light-responsive elements in C. reinhardtii.
In C. reinhardtii, only one form each of PPO and FeC is present. The expression profile of the genes for PPO and FeC differed in conditions of continuous darkness or continuous light. The cells grown in continuous darkness were truly dark adapted, as judged by the diminished levels of LHCII proteins. The relative lower amount of LHC protein in high-acetate medium as compared to TAP medium in either light or dark can be viewed as being in accordance with Heifetz et al. (2000), who reported that acetate had an adverse effect on carbon fixation/photosynthesis.
In cells growing on TAP medium, PPO mRNA levels were lower in continuous darkness than in light. In cells growing on high-acetate medium, this difference is not observed, which suggests that acetate somehow causes the cells to increase transcription of the PPO gene. However, the levels of PPO protein were comparable in dark- and light-grown cells. Since, unlike in angiosperm plants, chlorophyll synthesis in C. reinhardtii does occur in continuous darkness, it might be expected that light modulation of PPO mRNA and protein levels would be less pronounced than in plants; for example, in tobacco leaves, PPO transcript levels were strongly up-regulated in the light (Lermontova et al., 1997). On the other hand, C. reinhardtii cells grown in continuous darkness, as compared to cells grown in continuous light, do contain lower amounts LHC proteins and the concomitant chlorophyll. Therefore, some level of correlation could have been expected to exist between chlorophyll and PPO levels. C. reinhardtii FeC mRNA levels were markedly lower in dark-grown cells compared to light-grown cells, and FeC protein levels were also lower in the dark compared to the level in light-exposed cells. These results are in accordance with data on light regulation of FeC in plants, at least at the mRNA level (Chow et al., 1998; Suzuki et al., 2002). Light/dark-synchronized cells subjected to an imposed circadian cycle showed no significant variation in the PPO and FeC levels during the light phase. Synchronized cells steadily increase their chlorophyll content throughout the light phase and do not synthesize chlorophyll in the dark phase (Nogaj et al., 2005). It would therefore be expected that the PPO level would increase during the light phase, but this was not observed.
Taken together, the lack of correlation between the PPO level and the rate of chlorophyll synthesis under a variety of growth conditions indicates that the rate of chlorophyll synthesis is not limited by the rate of synthesis of PPO or its cellular abundance. For FeC, it is difficult to interpret the significance of the observed changes in mRNA and protein levels in cells growing under different conditions. If the rate of heme synthesis is reflected by the cellular content of FeC, then it would appear that heme synthesis occurs at a relatively constant rate throughout the light/dark cycle in synchronized cells, but at a lower rate in cells growing in continuous dark than in cells in continuous light. This can be contrasted to the situation in higher plants, where different tissue types can have greatly different needs for heme, which are met by expression of tissue-specific FeC genes. We propose that the existence of multiple genes for PPO and FeC in higher plants fulfils the need for different tissue-specific levels of expression rather than for different organelle-specific targeting of the gene products.
Transcription of many of the genes for pigment biosynthesis in C. reinhardtii is regulated by light (Matters and Beale, 1995; Lohr et al., 2005; Nogaj et al., 2005), even though the enzyme products of the light-regulated genes are comparably abundant in the cells irrespective of the light conditions (Nogaj et al., 2005; this work) and/or are not rate limiting for end product formation. The rate of pigment synthesis catalyzed by the expressed proteins appears to be regulated by posttranslational processes such as allosteric enzyme activity modulation, rather than by changes in enzyme abundance. These observations suggest that the physiological rationale for light-regulated transcription of the genes is not directly to control the rate of pigment biosynthesis. Instead, light-regulated transcription of the genes might be important for coordinating gene transcription with other cellular processes. For example, in light/dark-synchronized cells, the DNA is unavailable for transcription during synchronized mitosis. Also, transcription of different genes at different times in the light/dark cycle might be important for efficient use of limited cellular resources available for transcription and translation.
Heme is required in chloroplasts mainly for cofactors of PSII and the cyt b6/f complex. Heme is also needed for components of the mitochondrial electron transport chain and as a cofactor of various nonchloroplast hemoproteins (e.g. catalase, peroxidases). Although all of the steps of heme synthesis leading to protoporphyrinogen IX are generally believed to occur exclusively in the chloroplasts in plants, the site(s) of the last two steps of heme synthesis in plants have not been well understood. Many plants have been found to contain two PPO-encoding genes. In tobacco, the products of two genes, PPO-I and PPO-II, are specifically targeted to chloroplasts and mitochondria, respectively (Lermontova et al., 1997). Spinach (Spinacia oleracea) also contains two PPO-encoding genes, and PPO-I is targeted to the chloroplast thylakoids, but PPO-II is dually targeted to the chloroplast envelope and mitochondria by alternative translation initiation at two in-frame start codons (Watanabe et al., 2001). For FeC in plants, reports of mitochondrial localization have been disputed, and the situation remains unresolved (Chow et al., 1998; Lister et al., 2001; Cornah et al., 2002; Masuda et al., 2003). It is difficult to understand a biosynthetic role for mitochondrial PPO if FeC is absent from mitochondria. Because our results clearly show that the single PPO and FeC genes of C. reinhardtii encode proteins that are targeted only to the chloroplast, the chloroplast must be capable of supplying all cellular hemes in this eukaryotic photosynthetic organism. Therefore, alternative explanations for the inconsistent results reported for plants must be considered. It is possible that in some experiments, the mitochondrial fraction might be contaminated with proplastids, which could contain the PPO and FeC that are attributed to mitochondria. The proplastids might even have different isoforms of these enzymes than mature chloroplasts, which could explain the existence of different forms of the enzymes in chloroplast and mitochondrial fractions. Because C. reinhardtii contains only one large chloroplast per cell and proplastids are absent, it is possible to obtain clearer results with C. reinhardtii cell fractions than can be obtained with plant cells.
A hypothetical alternative possibility is that plant mitochondria are devoid of FeC but do contain a PPO that has a nonbiosynthetic role. Tetrapyrroles have been proposed as signaling molecules that convey information about the status of chloroplasts to the rest of the cell, although the mechanisms involved in tetrapyrrole signaling remain unknown (Strand et al., 2003; Strand, 2004). In this regard, it is noteworthy that the PPO reaction produces 3 mol of hydrogen peroxide (H2O2) per mol of protoporphyrinogen IX oxidized (Ferreira and Dailey, 1988). Whereas in the chloroplasts the H2O2 would be quickly dissipated by the abundant stromal AXP, in the mitochondria the H2O2 might be able to participate in peroxide signaling, which has been reported to be involved in regulating a diverse range of responses including development, stress acclimation, and programmed cell death (Gechev and Hille, 2005). In contrast, in C. reinhardtii the mitochondria would not be likely to play a role in peroxide signaling because C. reinhardtii mitochondria, unlike those of plants, contain an active catalase (Kato et al., 1997).
If all cellular hemes are synthesized exclusively in chloroplasts, export of heme from plastids to mitochondria and the cytoplasm must occur. Heme export from chloroplasts is poorly understood, but in vitro export of heme from isolated pea (Pisum sativum) chloroplasts has been described (Thomas and Weinstein, 1990). Two pathways of heme transport from plastids to mitochondria can be envisioned. One is active transport via the cytosol. Another is through a channel formed at the junctions between mitochondria and chloroplasts. In C. reinhardtii, mitochondria are mostly closely associated to the chloroplast, which occupies 60% of the cell volume. This close association could facilitate transport between the organelles. A putative heme chaperone has been described in Arabidopsis and proposed to function in mitochondrial heme import (Spielewoy et al., 2001). It is also worth exploring a possible role for the product of a C. reinhardtii gene that encodes a putative mitochondrial ABC transporter (JGI C_420004) and immediately precedes the FeC gene. This transporter might be a potential partner in the transport of heme into the mitochondrion. Although it is not common for functionally related nuclear genes to occur in clusters in C. reinhardtii, examples do exist, including clustered genes involved in uniparental inheritance (Armbrust et al., 1993) and genes for histones (Walther and Hall, 1995), hydrogenase cofactor assembly (Posewitz et al., 2004), and components of the nitrate assimilation system (Quesada et al., 1993).
MATERIALS AND METHODS
Cell Cultures
Wild-type Chlamydomonas reinhardtii strain CC124 was grown with orbital agitation at 25°C in TAP medium (Harris, 1989) or high-acetate medium (H3 medium), which contains 50 mm acetic acid with the initial pH adjusted to 7.0 (with KOH). For experiments involving organelle isolation, cell wall-deficient strain CC400 was grown on TAP medium supplemented with 1% (w/v) sorbitol. Light intensity was of 5,000 lux for both continuous light and synchronized cells. Cells in continuous dark were first dark adapted in TAP medium for 2 to 4 d until the cells resumed growing, and then transferred to either TAP or H3 medium. The cells were subsequently harvested in their exponential growth phase. Synchronized cells were grown under a 12-h-light/12-h-dark cycle and were checked for synchronization as described (Nogaj et al., 2005).
cDNA Isolation
A cDNA library, constructed in lZAPII phagemid (Stratagene) with mRNA from C. reinhardtii cells grown in light with 5% (v/v) CO2, was obtained from J. Davies (Iowa State University, Ames, IA). A sample of this library was excised with helper phage VCMS13 in XL1-Blue MRF′ Escherichia coli cells, and recovered as pBluescript plasmids. Batches of electro-competent E. coli PPO-deficient hemH (Sasarman et al., 1993) and FeC-deficient hemG (Miyamoto et al., 1991) strains were transformed with 1 μg of plasmid and selected for hemin-independent growth on Luria-Bertani-ampicillin plates.
Expression Constructs
The region of PPO cDNA (corresponding to translated residues I85 to K558) was amplified by PCR using the primers 5′-GACGAGCTCATCGTGGTCGGTGGAGGTC-3′ and 5′-CTGAAGCTTCTTGGACACGCTCTTGGC-3′. The FeC cDNA region (corresponding to translated residues K87–L491) was amplified using the PCR primers 5′-GACGAGCTCAAGGTCGGCGTTCTGCTG-3′ and 5′-CTGAAGCTTGCCAGGAACAGGTTTTTGAGG-3′. The restriction sites are underlined. The template cDNA was subjected to 30 cycles of PCR amplification (1 min at 94°C, 1 min at 60°C, and 2 min at 72°C) using Taq DNA polymerase (Qiagen). The amplified products were first ligated into pGEM-T-easy (Promega) and then digested with SacI and HindIII and ligated into expression vector pQE30 (Qiagen) that was predigested with the same enzymes.
Protein Purification and Antibody Preparation
E. coli strain XL1-Blue MRF′ (Stratagene) was transformed with the PPO- and FeC-pQE30 constructs and induced for 4 h at 37°C after adding 1 mm isopropylthio-β-galactoside to the medium (Luria-Bertani). The cells were harvested and disrupted by sonication, and the pellet containing PPO or FeC recombinant protein was dissolved in 6 m guanidinium chloride and used for nickel-nitrilotriacetic acid agarose affinity chromatography under denaturing conditions, according to Qiagen product manual. Antibodies were raised against the purified PPO and FeC proteins in rabbits (Animal Pharm Services).
Subcellular Fractionation
C. reinhardtii chloroplasts and mitochondria were isolated from CC400 cells grown on TAP medium in continuous light. Cells were harvested by centrifugation at 2,000g for 10 min, resuspended in nebulizing buffer (50 mm MES, 50 mm Tris, 0.25 m sorbitol, 10 mm MgCl2, 1 mm MnCl2, 3 mm KH2PO4, 2 mm EDTA, pH 7.2) and disrupted using the BioNeb cell disruption system (Glas-Col) with N2 (20 psi; Bollivar and Beale, 1996). The cell lysate was briefly centrifuged in a GSA rotor (Du Pont-Sorvall) by accelerating the rotor to 5,000 rpm and then immediately stopping the run. The pellet was used to obtain intact chloroplasts and the supernatant to obtain mitochondria. The pellet was resuspended in nebulizing buffer and intact chloroplasts were recovered at the interface of a 45/75% (v/v) Percoll step gradient in nebulizing buffer after centrifugation in an HB4 swinging bucket rotor (Du Pont-Sorvall) at 6,000g for 20 min at 4°C. Isolated chloroplasts were washed three times by sedimenting in nebulizing buffer and then were resuspended in 10 mm Tricine-NaOH, pH 8.0 in the presence of protease inhibitors (200 μm phenylmethylsulfonyl fluoride, 1 mm benzamidine, 5 mm 6-aminocaproic acid). Mitochondria were isolated from the supernatant of the first, brief centrifugation after cell rupture. This supernatant was first centrifuged at 1,500g for 10 min in an SS34 rotor (Du Pont-Sorvall), and then the supernatant was centrifuged at 10,000g for 10 min. The pellet from this centrifugation was used for further purification on Percoll gradients, as described by Eriksson et al. (1995).
Chloroplast membrane and soluble fractions were obtained after centrifugation of sonicated chloroplasts at 100,000g for 30 min. Membranes were resuspended in 10 mm Tricine-NaOH, pH 8.0, in the presence of protease inhibitors and supplemented with 1 m NaCl or 20 mm Na2CO3 (pH 11.0; Pierre et al., 1995). These samples were subjected twice to a 10-min incubation at room temperature followed by freezing and thawing, with vortex-mixing after every step, and then centrifuged for 15 min at 100,000g. The pellet was washed in the same buffer (containing NaCl or Na2CO3) and again centrifuged for 15 min at 100,000g. The soluble fraction from the first centrifugation and the pellet from the second centrifugation were analyzed by SDS-PAGE and immunoblotting.
Heme Detection in Polyacrylamide Gels
SDS-PAGE gels were stained for heme-associated peroxidase activity by the method of Thomas et al. (1976). A 6.3 mm solution of 3,3′,5,5′-tetramethylbenzidine (TMBZ) was freshly prepared in methanol. Immediately before use, three parts of the TMBZ solution was mixed with seven parts of 0.25 m sodium acetate, pH 5.0. The SDS-PAGE gel was immersed in this mixture and kept at room temperature in the dark with gentle shaking. After 1 h, H2O2 was added to a final concentration of 30 mm. The staining was visible within 3 min.
Nucleic Acid Analyses
For Southern blotting, genomic DNA (10 μg) was digested with various restriction endonucleases, electrophoresed on 1% (w/v) agarose gels and blotted onto Hybond-N nylon membranes (Amersham). Southern hybridization at 65°C was performed as described (Sambrook and Russell, 2001). The DNA probes specific for C. reinhardtii PPO and FeC genes were released from the abovementioned pQE30 expression vectors for PPO and FeC, by digestion with SacI and HindIII. For RNA blotting, total RNA was isolated from C. reinhardtii cells using the RNeasy kit (Qiagen). RNA (10 μg) was electrophoresed on 1% (w/v) formaldehyde agarose gels and transferred to nylon membranes as described in the RNeasy manual. The pQE30 constructs were digested with SacI (PPO) and PstI (FeC) to yield DNA fragments of 920 bp and 515 bp, respectively, that were used as probes. Blots were prehybridized for 2 h in UltraHyb buffer (Ambion) at 48°C, after which the labeled probes were added and hybridization was allowed to proceed. Probes were labeled with 32P using the random priming kit (Invitrogen) and purified on Sephadex G-25 spin columns (Bio-Rad).
Protein Analysis
Proteins from various extracts were separated on SDS-PAGE gels and then electroblotted onto nitrocellulose membranes. The membranes were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline supplemented with 0.05% (v/w) Tween 20. The blocked membranes were incubated first with the different specific antibodies and then with either horseradish peroxidase-conjugated anti-rabbit IgG (Pierce) and detected using ECL western-blotting detection reagents (Amersham), or alkaline phosphatase-conjugated anti-rabbit IgG (Sigma) and detected with the NBT/BCIP Liquid Substrate system (Sigma). In Figure 5, PPO antibodies were used at a dilution of 1:5,000 and FeC antibodies at 1:2,000, with enhanced chemiluminescence detection. In Figures 6 and 7, PPO antibodies were diluted 1:500 and detection was done using the alkaline phosphatase system. In Figure 6, FeC antibodies were used at a dilution of 1:1,000 and detected using the enhanced chemiluminescence system with freshly prepared solutions to obtain maximum sensitivity to compensate for the deterioration of the signal over time (Durrant, 1990). For Figure 7, the FeC antiserum was used at a dilution of 1:50, and detection was done using the alkaline phosphatase system. The antibody against C. reinhardtii Rubisco was obtained from J.V. Morony (Louisiana State University, Baton Rouge, LA) and that against C. reinhardtii LHCII from O. Vallon (Institut de Biologie Physico-Chimique, Paris).
Analytical Software
Multiple sequence alignments were done using the ClustalW 1.82 program (Thompson et al., 1994), and bootstrap neighbor-joining trees were constructed using the ClustalX 1.83 package (Thompson et al., 1997) with default settings (1,000 trials) and visualized with the TreeView program (version 1.6.6; http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Searches for cis-acting regulatory DNA elements in promoter regions were done using the PLACE database (www.dna.affrc.go.jp/PLACE; Prestridge, 1991; Higo et al., 1999). The sequences of the genomic regions upstream of C. reinhardtii PPO and FeC and of other enzymes of the tetrapyrrole synthesis pathway in C. reinhardtii were obtained from the JGI draft sequence, version 2.0 (http://genome.jgi-psf.org/chlre2). Transmembrane protein-embedding sequences were predicted by the TMPred program (expasy.org). Predictions for chloroplast targeting and transit peptide cleavage sites were obtained from the TargetP program (cbs.dtu.dk/services/TargetP/; Nielsen et al., 1997; Emanuelsson et al., 2000). Predictions for chloroplast lumen targeting signal peptides were obtained with the LumenP program (version 1.3; Westerlund et al., 2003), available directly from the authors (olof@csb.yale.edu).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF332962.
Acknowledgments
We thank O. Emanuelsson for the use of the LumenP program, J. Davies for providing the C. reinhardtii cDNA library, H.A. Dailey for providing the E. coli hemG and hemH strains, J.V. Morony for the antibody against C. reinhardtii Rubisco, and O. Vallon for the antibody against C. reinhardtii LHCII. The gene sequence data were produced by the U.S. Department of Energy JGI, http://www.jgi.doe.gov/ and are provided for use in this publication/correspondence only.
This work was supported by the National Science Foundation (grant no. MCB–9808578 to S.I.B.) and the U.S. Department of Energy (grant no. DE–FG02-88ER13918 to S.I.B.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Samuel I. Beale (sib@brown.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.069732.
References
- Armbrust EV, Ferris PJ, Goodenough UW (1993) A mating-type linked gene-cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell 74: 801–811 [DOI] [PubMed] [Google Scholar]
- Beale SI (1999) Enzymes of chlorophyll biosynthesis. Photosynth Res 60: 43–73 [Google Scholar]
- Beale SI (2005) Green genes gleaned. Trends Plant Sci 10: 309–312 [DOI] [PubMed] [Google Scholar]
- Bollivar DW, Beale SI (1996) The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase: characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803. Plant Physiol 112: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KS, Singh DP, Walker AR, Smith AG (1998) Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and subcellular targeting of the precursor proteins. Plant J 15: 531–541 [DOI] [PubMed] [Google Scholar]
- Cornah JE, Roper JM, Singh DP, Smith AG (2002) Measurement of ferrochelatase activity using a novel assay suggests that plastids are the major site of haem biosynthesis in both photosynthetic and non-photosynthetic cells of pea (Pisum sativum L.). Biochem J 362: 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA (2000) Terminal steps of haem biosynthesis. Biochem Soc Trans 30: 590–595 [DOI] [PubMed] [Google Scholar]
- Dailey HA, Dailey TA (1996. a) Protoporphyrinogen oxidase of Myxococcus xanthus: expression, purification, and characterization of the cloned enzyme. J Biol Chem 271: 8714–8718 [DOI] [PubMed] [Google Scholar]
- Dailey HA, Dailey TA, Wu C-K, Medlock AE, Wang K-F, Rose JP, Wang B-C (2000) Ferrochelatase at the millennium: structures, mechanisms and [2Fe-2S] clusters. Cell Mol Life Sci 57: 1909–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey TA, Dailey HA (1996. b) Human protoporphyrinogen oxidase: expression, purification, and characterization of the cloned enzyme. Protein Sci 5: 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey TA, Dailey HA (2002) Identification of [2Fe-2S] clusters in microbial ferrochelatases. J Bacteriol 184: 2460–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey TA, Meissner P, Dailey HA (1994) Expression of a cloned protoporphyrinogen oxidase. J Biol Chem 269: 813–815 [PubMed] [Google Scholar]
- Durrant I (1990) Light-based detection of biomolecules. Nature 346: 297–298 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Gardeström P, Samuelsson G (1995) Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol 107: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira GC, Dailey HA (1988) Mouse protoporphyrinogen oxidase: kinetic parameters and demonstration of inhibition by bilirubin. Biochem J 250: 597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G, Chasen C, Wollman F-A, de Vitry C (2003) Thylakoid targeting of Tat passenger proteins shows no ΔpH dependence in vivo. EMBO J 22: 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén L-G, Rochaix JD, von Heijne G (1990) Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences. FEBS Lett 260: 165–168 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Kück U (1999) Identification of DNA sequences controlling light- and chloroplast-dependent expression of the lhcb1 gene from Chlamydomonas reinhardtii. Curr Genet 34: 459–466 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego
- Heifetz PB, Förster B, Osmond CB, Giles LJ, Boynton JE (2000) Effects of acetate on facultative autotrophy in Chlamydomonas reinhardtii assessed by photosynthetic measurements and stable isotope analyses. Plant Physiol 122: 1439–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4: 236–240 [DOI] [PubMed] [Google Scholar]
- Kato J, Yamahara T, Tanaka K, Takio S, Satoh T (1997) Characterization of catalase from green algae Chlamydomonas reinhardtii. J Plant Physiol 151: 262–268 [Google Scholar]
- Kieselbach T, Bystedt M, Hynds P, Robinson C, Schröder WP (2000) A peroxidase homologue and novel plastocyanin located by proteomics to the Arabidopsis chloroplast thylakoid lumen. FEBS Lett 480: 271–276 [DOI] [PubMed] [Google Scholar]
- Kindle KL (1998) Amino-terminal and hydrophobic regions of the Chlamydomonas reinhardtii plastocyanin transit peptide are required for efficient protein accumulation in vivo. Plant Mol Biol 38: 365–377 [DOI] [PubMed] [Google Scholar]
- Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A (2004) Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis. EMBO J 23: 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Yoshioka S, Taniguchi F, Ohyama K, Fukuzawa H (2003) Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 133: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamppa GK (1988) The chlorophyll a/b-binding protein inserts into the thylakoids independent of its cognate transit peptide. J Biol Chem 263: 14996–14999 [PubMed] [Google Scholar]
- Lermontova I, Kruse E, Mock H-P, Grimm B (1997) Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc Natl Acad Sci USA 94: 8895–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Rudhe C, Lee M-N, Whelan J (2001) Arabidopsis thaliana ferrochelatase-I and -II are not imported into Arabidopsis mitochondria. FEBS Lett 506: 291–295 [DOI] [PubMed] [Google Scholar]
- Lohr M, Im C-S, Grossman AR (2005) Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol 138: 491–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Suzuki T, Shimada H, Ohta H, Takamiya K (2003) Subcellular localization of two types of ferrochelatase in cucumber. Planta 217: 602–609 [DOI] [PubMed] [Google Scholar]
- Matters GL, Beale SI (1995) Blue-light-regulated expression of genes for two early steps of chlorophyll biosynthesis in Chlamydomonas reinhardtii. Plant Physiol 109: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield SP, Schirmer-Rahire M, Frank G, Zuber H, Rochaix J-D (1989) Analysis of the genes of the OEE1 and OEE3 proteins of the photosystem II complex from Chlamydomonas reinhardtii. Plant Mol Biol 12: 683–693 [DOI] [PubMed] [Google Scholar]
- Merchant S, Hill K, Kim JH, Thompson J, Zaitlin D, Bogorad L (1990) Isolation and characterization of a complementary DNA clone for an algal pre-apoplastocyanin. J Biol Chem 265: 12372–12379 [PubMed] [Google Scholar]
- Miyamoto K, Nakahigashi K, Nishimura K, Inokuchi H (1991) Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol 219: 393–398 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Nogaj LA, Srivastava A, van Lis R, Beale SI (2005) Cellular levels of glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase do not control chlorophyll synthesis in Chlamydomonas reinhardtii. Plant Physiol 139: 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier J-B, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre Y, Breyton C, Kramer D, Popot JL (1995) Purification and characterization of the cytochrome b6/f complex from Chlamydomonas reinhardtii. J Biol Chem 270: 29342–29349 [DOI] [PubMed] [Google Scholar]
- Pineau B, Girard-Bascou J, Eberhard S, Choquet Y, Trémolières A, Gérard-Hirne C, Bennardo-Connan A, Decottignies P, Gillet S, Wollman F-A (2004) A single mutation that causes phosphatidylglycerol deficiency impairs synthesis of photosystem II cores in Chlamydomonas reinhardtii. Eur J Biochem 271: 329–338 [DOI] [PubMed] [Google Scholar]
- Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML (2004) Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem 279: 25711–25720 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Quesada A, Galvan A, Schnell RA, Lefebvre PA, Fernandez E (1993) Five nitrate assimilation-related loci are clustered in Chlamydomonas reinhardtii. Mol Gen Genet 240: 387–394 [DOI] [PubMed] [Google Scholar]
- Randolph-Anderson BL, Sato R, Johnson AM, Harris EH, Hauser CR, Oeda K, Ishige F, Nishio S, Gillham NW, Boynton JE (1998) Isolation and characterization of a mutant protoporphyrinogen oxidase gene from Chlamydomonas reinhardtii conferring resistance to porphyric herbicides. Plant Mol Biol 38: 839–859 [DOI] [PubMed] [Google Scholar]
- Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694: 135–147 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sasarman A, Letowski J, Czaika G, Ramirez V, Nead MA, Jacobs JM, Morais R (1993) Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can J Microbio1 39: 1155–1161 [DOI] [PubMed] [Google Scholar]
- Shi Z, Ferreira GC (2004) Probing the active site loop motif of murine ferrochelatase by random mutagenesis. J Biol Chem 279: 19977–19986 [DOI] [PubMed] [Google Scholar]
- Spielewoy N, Schulz H, Grienenberger JM, Thöny-Meyer L, Bonnard G (2001) CCME, a nuclear-encoded heme-binding protein involved in cytochrome c maturation in plant mitochondria. J Biol Chem 276: 5491–5497 [DOI] [PubMed] [Google Scholar]
- Strand Å (2004) Plastid-to-nucleus signalling. Curr Opin Plant Biol 7: 621–625 [DOI] [PubMed] [Google Scholar]
- Strand Å, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Masuda T, Singh DP, Tan F-C, Tsuchiya T, Shimada H, Ohta H, Smith AG, Takamiya K (2002) Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: their difference in phylogeny, gene expression, and localization. J Biol Chem 277: 4731–4737 [DOI] [PubMed] [Google Scholar]
- Thomas J, Weinstein JD (1990) Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol 94: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Ryan D, Levin W (1976) An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem 75: 168–176 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain P, Mache R, Zhou D-X (1996) The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem 271: 32593–32598 [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Walther Z, Hall JL (1995) The uni chromosome of Chlamydomonas: histone genes and nucleosome structure. Nucleic Acids Res 23: 3756–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Che F-S, Iwano M, Takayama S, Yoshida S, Isogai A (2001) Dual targeting of spinach protoporphyrinogen oxidase II to mitochondria and chloroplasts by alternative use of two in-frame initiation codons. J Biol Chem 276: 20474–20481 [DOI] [PubMed] [Google Scholar]
- Westerlund I, von Heijne G, Emanuelsson O (2003) LumenP: a neural network predictor for protein localization in the thylakoid lumen. Protein Sci 12: 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-K, Dailey HA, Rose JP, Burden A, Sellers VM, Wang B-C (2001) The 2-Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol 8: 156–160 [DOI] [PubMed] [Google Scholar]