Abstract
A metabolite profiling technique for Chlamydomonas reinhardtii cells for multiparallel analysis of low-molecular weight polar compounds was developed. The experimental protocol was optimized to quickly inactivate enzymatic activity, achieve maximum extraction capacity, and process large sample quantities. As a result of the rapid sampling, extraction, and analysis by gas chromatography coupled to time-of-flight mass spectrometry, more than 800 analytes from a single sample could be measured, of which more than 100 could be identified. Analyte responses could be determined mostly with ses less than 10%. Wild-type cells of C. reinhardtii strain CC-125 subjected to nitrogen-, phosphorus-, sulfur-, or iron-depleted growth conditions develop highly distinctive metabolite profiles. Individual metabolites undergo marked changes in their steady-state levels. Compared to control conditions, sulfur-depleted cells accumulated 4-hydroxyproline more than 50-fold, whereas the amount of 2-ketovaline was reduced to 2% of control levels. The contribution of each compound to the differences observed in the metabolic phenotypes is summarized in a quantitatively rigorous way by principal component analysis, which clearly discriminates the cells from different growth regimes and indicates that phosphorus-depleted conditions induce a deficiency syndrome quite different from the response to nitrogen, sulfur, or iron starvation.
With the recent completion of the Chlamydomonas reinhardtii genome project, a systems biology perspective, allowing the understanding of how the numerous components in a living system interact to comprise a functioning whole, comes into focus for this model organism, too. To this end, investigation of the world beyond mRNA abundance will be essential: Genes, proteins, and metabolites are all integrated into a seamless and dynamic network to sustain cellular functions (Kitano, 2002; Stelling et al., 2004).
Metabolomic analysis aims at the unbiased representation and quantitative determination of the suite of metabolites in a biological sample (Fiehn, 2002). It complements transcript profiling and proteomic approaches in functional genomics (Bino et al., 2004; Kromer et al., 2004).
Metabolic profiles reflect the dynamic response of the network of biochemical reactions to environmental, genetic, or developmental signals. As such, they are a valuable integrated measure of how a living system adjusts to a changing environment. This property has led to the successful application of large-scale analysis of metabolites in distinguishing between silent plant phenotypes (Weckwerth et al., 2004), or characterization of freezing tolerance in plants (Cook et al., 2004). It is also of considerable interest in biomarker recovery (Foyer et al., 2003), biotechnological engineering (Buchholz et al., 2002; Sauer and Schlattner, 2004), or disease profiling (Shi et al., 2003). Given the diversity of chemical structures present in the metabolome, each extraction method and each detection technique will yield only a subset of the suite of metabolites present in a biological sample. A variety of technological platforms has been used to measure metabolic profiles. In most cases, hyphenated techniques where chromatographic separation precedes detection of analytes by mass spectrometry have been used (Roessner et al., 2001; Soga et al., 2002; Tolstikov and Fiehn, 2002).
Because of unsurpassed chromatographic separation power, sensitivity, and reproducibility, we have chosen gas chromatography coupled to time-of-flight mass spectrometry (GC-TOF) to establish a method for analysis of metabolite levels of the unicellular green alga C. reinhardtii. Here, we report the detection of several hundred analytes from polar phase extracts of algal cells and the analysis of nutrient deprivation as a test case for metabolite profiling in Chlamydomonas.
RESULTS AND DISCUSSION
A Protocol for Metabolite Analysis in Cell Cultures
To obtain metabolomic data that correctly measure amount of substance and reflect the levels of intracellular metabolites, we followed a multistep procedure that conceptually can be divided into cell harvest and extraction, sample preparation for the acquisition of GC-TOF data, and processing of the analytical signal obtained.
Harvest and extraction essentially comprise the injection of the cell suspension into a quenching solution to stop enzymic reactions, recovery of the cells by centrifugation, homogenization of the material, and metabolite extraction. GC-TOF samples are then prepared by further separating the crude extract into a lipophilic and a polar phase. Metabolites contained in the latter are concentrated and derivatized to increase compound volatility for separation of the complex mixture by gas chromatography. Once the samples have been measured by GC-TOF, the raw data chromatograms are processed to find analyte peaks, compare individual samples, and identify analytes by mass spectral comparison with custom mass spectral libraries of genuine compounds. The initial peak lists are filtered for false negatives, artifact peaks, and contaminants. Normalization to the amount of sample material and internal standards enables comparison and evaluation of the metabolic profiles.
Metabolite Analysis in Cell Cultures Needs Rapid Quenching of Metabolism before Extraction
Steady-state metabolite pools are the result of an equilibrium of catalyzed reactions. Environmental stimuli of any kind provoke rapid changes in metabolite concentrations, especially for intermediates that participate in reactions with high turnover rates. To generate an authentic picture of the metabolic composition of a biological system, it is therefore crucial to stop biochemical activity instantly as the material is removed from the original growth conditions for harvest. Whereas plant material can be flash-frozen in liquid nitrogen directly, the situation is different for suspension cultures. Although freezing of entire microbial cultures with subsequent analysis of the metabolite composition have been reported (Barsch et al., 2004), this approach suffers from severe drawbacks, as no distinction can be made between intracellular and excreted compounds and the large excess of constituents of the growth medium is not compatible with the use of highly sensitive chromatographic separation methods. Separation of the cells from the medium is usually achieved by centrifugation or filtration. Centrifugation requires handling of cultures outside their growth environment in the range of minutes, subjecting the cells to conditions drastically different from the growth settings under investigation. Although filtration can be done within a few seconds, it is not suited to process a large number of samples or large volumes of culture as only one sample can be processed at a time and only up to 5 mL of log-phase algal culture could be efficiently filtered with a variety of filters of different material and pore sizes (data not shown).
Taking up an approach originally suggested for rapid sampling of yeast cultures (de Koning and van Dam, 1992), we decided prior to collecting the cells to stop metabolic activity by injecting the cell suspension into an excess of cold (−25°C) solution of methanol-water supplied with Tris-acetate phosphate (TAP) macro salts for isosmotic conditions. Methanol concentration and ratio of cell suspension to quenching solution were optimized to allow rapid cooling and handling of cells at temperatures below −20°C throughout the procedure, while simultaneously minimizing methanol concentration to ensure integrity of the cells after injection.
Chlamydomonas Cells Are Resistant to Quenching
To demonstrate that the quenching procedure does not compromise the integrity of the cells to an extent where significant leakage of intracellular matter into the quenching solution occurs, cells of a late log-phase culture were cultivated in TAP medium in the presence of labeled [U-14C]acetic acid as reduced carbon source. After 3 h of cultivation with the labeled substrate, cells were harvested and the culture medium containing the labeled substrate carefully removed. After equilibration of the cells for 15 min in TAP medium without labeled acetate, cells were subjected to the quenching procedure. To account for leakage due to centrifugation itself, aliquots of the equilibrated cell suspensions were centrifuged and the supernatants treated in parallel with the samples containing whole suspension aliquots. Both the walled wild-type strain CC-125 and the wall-less strain CC-400 were tested in this way (Table I).
Table I.
Resistance of C. reinhardtii suspended in quenching solution to leakage
Activities refer to 1 mL of cell suspension and for the quench samples to the volume of quench supernatant corresponding to 1 mL of cell suspension. The quenching was performed in triplicates. Mean and se for the measured activities are indicated; percentage values refer to means.
| Sample
|
CC-125 wt mt+
|
CC-400 cw15 mt+
|
||
|---|---|---|---|---|
| Bq | % | Bq | % | |
| Cell suspension after 3-h labeling | 5,051.6 ± 20.8 | – | 5,558.2 ± 42.5 | – |
| Cells after resuspension in unlabeled medium | 984.1 ± 2.3 | 100.0 | 483.6 ± 1.5 | 100.0 |
| Quenched cells, centrifugation after 3 min | 18.6 ± 0.7 | 1.9 | 23.0 ± 1.8 | 4.8 |
| Quenched cells, centrifugation after 30 min | 24.0 ± 1.4 | 2.4 | 30.2 ± 0.8 | 6.2 |
| Controls for leakage due to harvest | 12.8 ± 0.1 | 1.3 | 9.7 ± 0.2 | 2.0 |
After 3 h of growth, 19.5% and 8.7% of the entire amount of radioactivity remains with the cells of strain CC-125 and CC-400 after removal of the growth medium, washing, and equilibration in normal TAP medium, respectively.
Of the incorporated radioactivity, 1.9% is found in the supernatant of CC-125 cells after quenching and immediate sedimentation of cells. Incubation of the cells in quenching solution for 30 min at −25°C prior to centrifugation increases this value to 2.4%. After centrifugation of the cell suspension and addition of the supernatant to the quenching solution, 1.3% of the activity can be found in the quenched sample.
For the wall-less strain CC-400, 4.8% of the incorporated activity are found in the supernatant of the quenched sample. This value increases to 6.2% when the sample is stored for 30 min at −25°C prior to centrifugation. Addition of the supernatant of the centrifuged cell suspension to quench solution accounts for 2.0% of the activity.
To ensure that the detection limit of 14C-scintillation would allow for spotting metabolite leakage exceeding 1% of the incorporated label, the amount of radioactivity added (20 kBq × mL−1 cell suspension) and the actual volume of cell suspension used for scintillation (125 μL) were chosen accordingly.
These results indicate that even after prolonged handling of cells during quenching, leakage of intracellular substances is below an acceptable level. For the walled strain CC-125, this is well below 2%, whereas with the cell wall-deficient strain CC-400 leakage appears to be slightly higher. Still, also with CC-400 after 30-min suspension in methanol-water at −25°C, less than 5% of the activity taken up by the cells is lost as a result of quenching, if the activity in the supernatant of the quenched sample is corrected by the activity found in the control accounting for leakage due to mechanical stress during harvest.
An Extraction Mixture for Chlamydomonas Samples
Cells are collected by centrifugation at −20°C. After careful removal of the supernatant, the cell pellet is flash-frozen in liquid nitrogen, rendering it biologically inert. Prior to extraction, cells are homogenized by grinding the material under liquid nitrogen. The homogenized material is suspended at −20°C in methanol-chloroform-water (MCW) for extraction.
Composition of the extraction mixture was optimized to maximize extraction capacity, measured as the capacity to quantitatively extract chlorophyll and minimize the volume needed for extraction to concentrate extracted compounds (Fig. 1). As a result, the original extraction formulation of 10:4:4 (v/v/v) MCW, which had been used for extraction of plant material (Roessner et al., 2001), was changed to a composition of 10:3:1 (v/v/v) MCW. The reduced water content of the new formulation compensates for the interstitial liquid contained in the cell pellet that led to phase separation between a chloroform-containing lipophilic phase and polar methanol-water phase already during resuspension of the homogenized material, prior to removal of particulate material from the preparation, when the original formulation was used for algal cells. Moderate reduction of the chloroform content to three volume parts enables complete extraction of cell chlorophyll with only a quarter of extractant compared to the original extraction mix composition and performs equally well for chlorophyll extraction as the standard protocol using 80% (v/v) acetone (Fig. 1). MCW volumes of 2-fold or more extract approximately 2.7 μg chlorophyll per 106 cells and produce completely discolored cell pellets. Re-extraction of the pellets with MCW and acetone produced supernatants that contained 0.037 μg and 0.048 μg chlorophyll per 106 cells, respectively, indicating that residual chlorophyll content was less than 2% of total chlorophyll for the MCW-extracted samples. Under these conditions extraction of smaller polar compounds is surmised to be quantitative, as relative methanol-water content of the extraction mix is even higher than with the original formulation.
Figure 1.
Extraction capacity of MCW. Changing the ratio of MCW from 10:4:1 (v/v/v; white squares) to a ratio of 10:3:1 (black circles) increases extraction capacity 4-fold, as assessed by extraction of cell chlorophyll. In direct comparison with standard acetone extraction of chlorophyll (white circles), chlorophyll extraction with MCW 10:3:1 performs equally well. Extraction mix volume relates to the use of 3 mL of extraction mix for 1.2×109 cells. Standard errors of triplicate experiments are indicated.
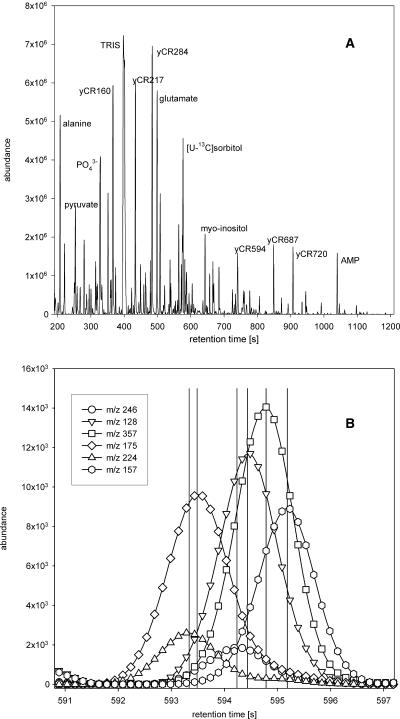
Extracts were used to prepare polar phase samples and acquire GC-TOF data as described previously (Roessner et al., 2001). In chromatograms of C. reinhardtii polar phase metabolite extracts, more than 800 analytes were detected routinely (Fig. 2A). Among the most prominent peaks we found Ala, pyruvate, Glu, glycerolphosphate, and adenosine 5′-monophosphate. Also, isotopically labeled sorbitol, which was introduced as an internal standard, and a number of unidentified analytes produced prominent peaks. Originating from residual culture medium, 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS) and orthophosphate (Pi) were observed with very high detector signal intensities. The former regularly exceeded detector capacity, impeding analysis of analytes coeluting with the TRIS signal. Although reduction of TRIS peak size can be achieved by additional washes of the pelleted cells with quench solution (data not shown), we dispensed with additional washes, as this would require handling of cells for a prolonged period of time before rendering the material inert in liquid nitrogen and still yield chromatograms with a prominent TRIS peak.
Figure 2.
Analytical signal for C. reinhardtii polar phase extract after GC-TOF measurement. A, Total ion chromatogram of wild-type strain CC-125. In a retention time window from 180 s to 1,200 s after sample injection, fragment ion intensities for all m/z in the mass range from 85 to 500 are recorded. The total ion chromatogram is a composite signal of the sum of all m/z values in each recorded spectrum. Identity of prominent peaks is indicated. B, Retention profiles of unique ions of deconvoluted peaks in the retention time interval between 591 and 597 s of the same chromatogram. Peak apices are indicated by black vertical lines.
GC-TOF chromatograms contain detector signal intensities for each mass-to-charge ratio (m/z) value in the chosen mass range for every spectrum acquired during the run. By mass spectral similarity and retention index comparison with custom libraries of authentic compounds, more than 100 of the detected peaks could be assigned a chemical structure, including amino acids, carbohydrates, phosphorylated intermediates, nucleotides, and organic acids. The diverse chemical nature of the compounds identified underlines the usefulness of this technique to detect metabolites in a multiparallel and unbiased manner. However, in such a complex mixture, chromatographic separation does not suffice to resolve all analytes. Instead, additional resolution and mass spectrum purification are achieved by mass spectral deconvolution. The deconvolution algorithm compares retention profiles and position of local maxima of fragment ion intensities to resolve overlapping peaks and computationally derive true mass spectra by identification of unique ions for each analyte peak (Fig. 2B).
Identification of corresponding peaks from individual samples involves automatic peak finding and comparison against a reference chromatogram derived from a reference extract containing equal amounts of extract from all experimental groups involved. Peaks from individual chromatograms are assigned to the reference analytes by virtue of their mass spectral similarity and retention index match. Due to the complexity of the chromatograms, for most analytes mass spectra derived by automatic deconvolution differ from one chromatogram to the next to a certain extent. This is especially true when comparing different experimental groups that exhibit large differences in total metabolite composition. When setting rigid thresholds for mass spectrometric similarities and retention index match for peak assignment, this procedure may result in a large number of analytes not being reported from individual samples. To exclude such false negatives and ensure that missing values indeed are the consequence of the absence of any detectable analytical signal, all chromatograms in this study were corrected manually for the occurrence of false negative analytes.
Macronutrient and Iron Deficiency Produce Highly Distinct Metabolic Phenotypes
To demonstrate the relevance of our approach, we decided to analyze how nutrient availability is reflected in metabolite profiles under conditions of nitrogen (N), sulfur (S), phosphorus (P), and iron (Fe) depletion, respectively. Table II shows metabolite peak area normalized to cell number and the internal standard [U-13C]sorbitol for a selection of 77 abundant primary metabolites after 24 h of nutrient starvation and for controls. Each metabolite is quantified by area integration of a characteristic fragment ion trace most suitable to distinguish the peak from local coeluting neighbor peaks. In addition, calibration curves are known to have different slopes (analytical sensitivity) for each metabolite, depending on ionization efficiency and detector response in the mass spectrometer. Therefore, to compare normalized peak areas of different peaks, metabolite-specific calibration curves have to be acquired. For the purpose of this study, comparisons are restricted to the differences of peak areas of the same metabolite in different conditions. A large number of metabolites undergo marked changes in at least one of the conditions applied. In Fe-deficient cells, the levels of succinate, threonic acid, and citrate all rise more than 2-fold, whereas the levels of Phe, Leu, Ser, Asn, Ala, and His, among other metabolites, drop to less than one-half of control levels. The increase of organic acids in roots, leaves, and xylem is a well documented response to Fe deficiency among higher plants. Specifically, the accumulation of citrate is implicated in mechanisms for Fe acquisition, formation of stable water-soluble complexes, cation/anion homeostasis, or supply of reducing power for ferric chelate reduction (for review, see Abadia et al., 2002). C. reinhardtii was shown to exhibit a similar response to Fe deficiency as in strategy I plants that involves an inducible Fe3+-reductase activity and regulated Fe uptake (Eckhardt and Buckhout, 1998; Lynnes et al., 1998; Weger, 1999). The finding of elevated levels of organic acids is consistent with these earlier observations. Given the variety of habitats of C. reinhardtii and the restricted bioavailability of Fe in both terrestrial and aquatic environments (Guerinot and Yi, 1994; Behrenfeld and Kolber, 1999), the Fe starvation-induced increase in succinate and citrate could well be related to their Fe-chelating properties. In N-depleted cultures, 2-amino-adipic acid, an important metabolite in Lys biosynthesis and catabolism, accumulates to over 9-fold of control levels. Trp, Ile, and Thr show more than 2-fold accumulation. Most of the metabolites listed in Table II were found largely reduced in their levels in N-depleted cells. N-Acetyl-Orn, present in all other experimental groups, could not be detected at all in N-depleted samples. Both increased levels of select amino acids and decreased levels of important amino group mediators like N-acetyl-Orn and Glu/Gln indicate a massive incapability of maintaining homeostasis of amino acid biosynthesis under N stress. Phosphate removal results in a significant rise in the intracellular Cys pool that is 25-fold higher than with cells cultured in standard TAP medium. S-depleted cells exhibited the largest increase of any single compound: Here, 4-Hyp accumulated more than 50-fold compared to control conditions while undergoing only slight changes with the other treatments. Hyp is believed to be formed only in proteins and is a prominent constituent in the Hyp-rich glycoprotein framework forming the Chlamydomonas cell wall (Imam et al., 1985; Voigt et al., 1991; Voigt and Frank, 2003). Cell wall proteins are known to be extensively sulfated and to be rearranged during S starvation (Takahashi et al., 2001; Hallmann, 2003) while several prolyl 4-hydroxylases are down-regulated (Zhang et al., 2004). Thus, the rise in 4-Hyp could be the result of enhanced degradation of cell wall proteins. A number of phosphorylated intermediates, such as glycerate-3-phosphate, sn-glycerol-3-phosphate, Glc-6-phosphate and Fru-1,6-bisphosphate increased more than 3-fold under S-depleted conditions. Malic, citramalic, and fumaric acid each were depleted to less than 10% of their control levels. Cys levels decreased to 25% while Met, as well as prephenate, decreased below the limit of detection in S-depleted cells. Cystine and sedoheptulose-1,7-bisphosphate, although detectable in the control group, could not be detected in any of the nutrient stress situations. Shikimic acid was below the detection limit in each of the three macronutrient-depleted cultures and was reduced to just 40% of control in Fe-depleted cultures. Taken together, many important primary metabolic modules are affected in each of the macronutrient stress condition, but for S stress specifically carbohydrate metabolism seemed to be more dramatically affected, providing evidence for our hypothesis that a plethora of metabolic responses would result from nutrient depletion.
Table II.
Levels, response ratios, and PCA loadings of metabolites in Fe-, N-, S-, and P-limited and TAP control conditions
Mean and se (n = 5) of metabolite levels in arbitrary units are shown. Response ratios above 2.5 or below 0.4 are in bold. Loadings for principal components 1 and 2 are given in powers of 10−2, i.e. 1.23 designates 0.0123. LOD indicates response below limit of detection. NA indicates not applicable.
| Metabolite | TAP | −Fe | −N | −S | −P | −Fe/TAP | −N/TAP | −S/TAP | −P/TAP | Loading PC1 | Loading PC2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1,3-Dihydroxyacetone | 0.48 ± 0.04 | 0.05 ± 0.003 | 0.097 ± 0.005 | 0.062 ± 0.006 | 0.17 ± 0.01 | 0.10 | 0.20 | 0.13 | 0.36 | −8.93 | 7.67 |
| 2-Aminoadipate | 0.026 ± 0.001 | 0.024 ± 0.002 | 0.248 ± 0.004 | 0.202 ± 0.005 | 0.045 ± 0.003 | 0.90 | 9.42 | 7.66 | 1.69 | 9.00 | −2.00 |
| 2-Ketoglutaric acid | 0.49 ± 0.02 | 1.11 ± 0.05 | 0.223 ± 0.005 | 0.136 ± 0.004 | 1 ± 0.4 | 2.28 | 0.46 | 0.28 | 2.03 | −6.92 | −4.71 |
| 2-Ketovaline | 0.44 ± 0.03 | 0.65 ± 0.04 | 0.034 ± 0.006 | 0.008 ± 0.008 | 0.042 ± 0.006 | 1.47 | 0.08 | 0.02 | 0.10 | −6.39 | 9.32 |
| 2-Phosphoglycolic acid | 0.014 ± 0.002 | 0.009 ± 0.003 | 0.011 ± 0.002 | 0.0111 ± 0.0008 | 0.016 ± 0.002 | 0.61 | 0.79 | 0.79 | 1.14 | −7.13 | −3.18 |
| 3-Hydroxybutyric acid | 2.72 ± 0.04 | 1.64 ± 0.04 | 0.077 ± 0.005 | LOD | 2.6 ± 0.1 | 0.60 | 0.03 | NA | 0.96 | −12.69 | −0.70 |
| 3-Hydroxypyridine | 10.5 ± 0.8 | 11 ± 1 | 7.9 ± 0.4 | 11.9 ± 0.8 | 10.3 ± 0.9 | 1.08 | 0.76 | 1.14 | 0.98 | −8.05 | 0.31 |
| 3-Isopropylmalic acid | 5.3 ± 0.1 | 2.39 ± 0.04 | LOD | LOD | 2.53 ± 0.1 | 0.45 | NA | NA | 0.47 | −11.72 | 6.74 |
| 4-Hydroxybenzoic acid | 0.103 ± 0.01 | 0.086 ± 0.01 | 0.053 ± 0.003 | 0.073 ± 0.008 | 0.104 ± 0.009 | 0.84 | 0.51 | 0.70 | 1.01 | −11.23 | 0.30 |
| 4-Hyp | 0.24 ± 0.01 | 0.202 ± 0.007 | 0.201 ± 0.005 | 12.6 ± 0.4 | 0.27 ± 0.03 | 0.84 | 0.84 | 52.27 | 1.11 | 9.10 | 1.79 |
| 5-Oxoproline | 0.83 ± 0.05 | 0.64 ± 0.02 | 0.19 ± 0.03 | 0.315 ± 0.006 | 0.71 ± 0.02 | 0.77 | 0.23 | 0.38 | 0.85 | −12.29 | 1.13 |
| 6-Phosphogluconic acid | 0.8 ± 0.2 | 0.4 ± 0.1 | 0.047 ± 0.007 | 0.23 ± 0.05 | 0.15 ± 0.02 | 0.50 | 0.06 | 0.29 | 0.19 | −7.41 | 11.02 |
| Adenine | 4.32 ± 0.07 | 1.8 ± 0.2 | 0.39 ± 0.01 | 0.57 ± 0.04 | 2.51 ± 0.07 | 0.43 | 0.09 | 0.13 | 0.58 | −11.88 | 5.22 |
| Ala | 9.9 ± 0.4 | 2.6 ± 0.09 | 0.61 ± 0.03 | 2.57 ± 0.06 | 6 ± 0.2 | 0.26 | 0.06 | 0.26 | 0.61 | −11.13 | 4.04 |
| Asn | 4 ± 0.1 | 1.29 ± 0.06 | 0.73 ± 0.03 | 2.27 ± 0.1 | 3.1 ± 0.1 | 0.32 | 0.18 | 0.57 | 0.78 | −10.59 | 2.27 |
| Asp | 26.9 ± 0.4 | 11.4 ± 0.2 | 16.8 ± 0.3 | 50 ± 2 | 40 ± 2 | 0.42 | 0.63 | 1.85 | 1.49 | −2.99 | −8.65 |
| Benzoic acid | 0.99 ± 0.05 | 0.91 ± 0.03 | 0.72 ± 0.03 | 0.53 ± 0.03 | 1.05 ± 0.06 | 0.92 | 0.73 | 0.54 | 1.07 | −11.12 | −2.58 |
| Citramalic acid | 1.56 ± 0.05 | 0.74 ± 0.02 | 0.115 ± 0.003 | 0.077 ± 0.003 | 1.09 ± 0.07 | 0.47 | 0.07 | 0.05 | 0.70 | −12.42 | 3.29 |
| Citric acid | 21.6 ± 0.6 | 53 ± 1 | 12.19 ± 0.07 | 11.4 ± 0.1 | 88 ± 2 | 2.45 | 0.56 | 0.53 | 4.06 | −8.10 | −12.25 |
| Citrulline | 1.4 ± 0.3 | 1.4 ± 0.3 | 0.33 ± 0.03 | 2.4 ± 0.4 | 1.1 ± 0.2 | 0.97 | 0.23 | 1.74 | 0.76 | −3.47 | 6.51 |
| Cys | 0.243 ± 0.006 | 0.13 ± 0.03 | 0.33 ± 0.04 | 0.06 ± 0.01 | 6.1 ± 0.4 | 0.54 | 1.35 | 0.25 | 25.13 | −5.84 | −14.85 |
| Cystine | LOD | LOD | LOD | LOD | 43 ± 2 | NA | NA | NA | NA | −5.73 | −14.96 |
| d-Fru-1,6-bisphosphate | 0.064 ± 0.007 | 0.08 ± 0.03 | 0.043 ± 0.003 | 0.18 ± 0.05 | 0.23 ± 0.02 | 1.21 | 0.67 | 2.90 | 3.61 | −3.18 | −12.61 |
| d-Fru-6-phosphate | 0.86 ± 0.07 | 0.92 ± 0.04 | 0.55 ± 0.02 | 1.2 ± 0.07 | 0.85 ± 0.05 | 1.07 | 0.64 | 1.40 | 0.99 | −8.13 | 2.13 |
| d-Glc-6-phosphate | 3.2 ± 0.3 | 3.9 ± 0.2 | 2.87 ± 0.03 | 10.2 ± 0.2 | 3 ± 0.2 | 1.21 | 0.89 | 3.17 | 0.95 | 8.30 | 3.72 |
| Dihydroxyacetone phosphate | 0.22 ± 0.01 | 0.13 ± 0.006 | 0.056 ± 0.003 | 0.13 ± 0.01 | 0.108 ± 0.003 | 0.58 | 0.25 | 0.59 | 0.48 | −9.71 | 9.95 |
| d-Rib-5-phosphate | 0.0145 ± 0.001 | 0.016 ± 0.002 | 0.014 ± 0.002 | 0.025 ± 0.003 | 0.025 ± 0.002 | 1.13 | 0.99 | 1.72 | 1.70 | −4.23 | −10.32 |
| Fru | 0.58 ± 0.07 | 0.13 ± 0.02 | LOD | 0.15 ± 0.02 | 0.46 ± 0.09 | 0.22 | NA | 0.27 | 0.79 | −10.31 | 1.16 |
| Fumaric acid | 13.2 ± 0.2 | 6.1 ± 0.1 | 2.07 ± 0.05 | 0.58 ± 0.07 | 7.6 ± 0.2 | 0.46 | 0.16 | 0.04 | 0.57 | −12.11 | 5.26 |
| Galactinol | 0.94 ± 0.05 | 0.45 ± 0.03 | 0.87 ± 0.03 | 2.1 ± 0.2 | 0.85 ± 0.05 | 0.47 | 0.92 | 2.22 | 0.90 | 5.89 | 0.86 |
| Galactonic acid | 0.144 ± 0.006 | 0.186 ± 0.007 | 0.0155 ± 0.0008 | 0.024 ± 0.003 | 0.165 ± 0.008 | 1.29 | 0.11 | 0.17 | 1.15 | −11.27 | −1.01 |
| Glc-1-phosphate | 7.7 ± 0.4 | 8.3 ± 0.3 | 4.7 ± 0.2 | 6.9 ± 0.5 | 5.8 ± 0.3 | 1.07 | 0.61 | 0.89 | 0.75 | −9.25 | 7.18 |
| Glu | 10.2 ± 0.2 | 8.5 ± 0.2 | 1.8 ± 0.06 | 3.3 ± 0.1 | 8.4 ± 0.2 | 0.83 | 0.18 | 0.32 | 0.83 | −12.30 | 2.05 |
| Gln | 40 ± 2 | 28 ± 1 | 4.1 ± 0.1 | 21.8 ± 0.9 | 47 ± 2 | 0.70 | 0.10 | 0.55 | 1.18 | −11.75 | −2.61 |
| Glyceraldehyde-3-phosphate | 0.101 ± 0.002 | 0.123 ± 0.004 | 0.1 ± 0.01 | 0.05 ± 0.003 | 0.21 ± 0.01 | 1.22 | 0.98 | 0.50 | 2.05 | −8.29 | −11.32 |
| Glycerate-3-phosphate | 2.07 ± 0.09 | 5.7 ± 0.3 | 3.3 ± 0.1 | 7.7 ± 0.2 | 4.8 ± 0.1 | 2.73 | 1.61 | 3.74 | 2.31 | 3.58 | −9.31 |
| Glyceric acid | 1.51 ± 0.03 | 0.84 ± 0.03 | 0.88 ± 0.04 | 2.27 ± 0.06 | 6.4 ± 0.2 | 0.55 | 0.58 | 1.51 | 4.24 | −6.02 | −14.02 |
| Glycerol | 26 ± 3 | 20 ± 2 | 20 ± 2 | 160 ± 30 | 18 ± 2 | 0.78 | 0.77 | 6.23 | 0.70 | 8.09 | 3.13 |
| Gly | 5.1 ± 0.3 | 2.33 ± 0.1 | 1.79 ± 0.08 | 2.97 ± 0.1 | 4.28 ± 0.08 | 0.46 | 0.35 | 0.58 | 0.84 | −11.42 | 1.04 |
| Glycolic acid | 0.9 ± 0.03 | 0.64 ± 0.01 | 0.4 ± 0.02 | 0.83 ± 0.06 | 1.3 ± 0.08 | 0.71 | 0.44 | 0.92 | 1.44 | −10.08 | −8.37 |
| His | 8 ± 1 | 1.7 ± 0.6 | 4 ± 1 | 1.4 ± 0.5 | 3.2 ± 0.5 | 0.21 | 0.55 | 0.17 | 0.40 | −5.46 | 5.29 |
| Hydroxylamine | 8 ± 2 | 8.9 ± 0.9 | 10.4 ± 0.7 | 10 ± 1 | 8.2 ± 0.7 | 1.18 | 1.39 | 1.31 | 1.10 | −0.35 | −1.61 |
| Iminodiacetic acid | 0.6 ± 0.01 | 0.168 ± 0.007 | 0.329 ± 0.005 | 0.427 ± 0.008 | 1.71 ± 0.09 | 0.28 | 0.55 | 0.71 | 2.86 | −7.27 | −12.56 |
| Inositol-β-galactoside | 24.1 ± 0.8 | 8.6 ± 0.1 | 2.44 ± 0.06 | 3.62 ± 0.08 | 8.8 ± 0.4 | 0.36 | 0.10 | 0.15 | 0.37 | −10.47 | 9.23 |
| Ile | 19.1 ± 0.4 | 9.7 ± 0.2 | 48 ± 2 | 82 ± 3 | 17.1 ± 0.8 | 0.51 | 2.50 | 4.28 | 0.90 | 10.83 | 0.25 |
| Leu | 1.53 ± 0.02 | 0.54 ± 0.01 | 0.91 ± 0.03 | 1.65 ± 0.06 | 0.63 ± 0.02 | 0.35 | 0.59 | 1.08 | 0.41 | −1.57 | 12.20 |
| Lys | 10.6 ± 0.2 | 8.8 ± 0.2 | 4.45 ± 0.08 | 5.4 ± 0.1 | 10.7 ± 0.6 | 0.83 | 0.42 | 0.51 | 1.00 | −12.69 | −1.48 |
| Malic acid | 8.32 ± 0.09 | 8.9 ± 0.1 | 2.14 ± 0.05 | 0.62 ± 0.02 | 8.9 ± 0.2 | 1.07 | 0.26 | 0.07 | 1.07 | −12.05 | −1.58 |
| Maltose | 0.51 ± 0.05 | 0.26 ± 0.02 | 0.32 ± 0.03 | 0.37 ± 0.01 | 1.4 ± 0.08 | 0.51 | 0.62 | 0.72 | 2.73 | −7.53 | −12.49 |
| Met | 0.67 ± 0.02 | 0.61 ± 0.06 | 0.123 ± 0.007 | LOD | 1.4 ± 0.2 | 0.91 | 0.18 | NA | 2.09 | −10.17 | −8.73 |
| myo-Inositol | 19.6 ± 0.6 | 6.2 ± 0.2 | 2.49 ± 0.06 | 2.53 ± 0.04 | 23.4 ± 0.4 | 0.32 | 0.13 | 0.13 | 1.19 | −11.59 | −3.99 |
| Myristic acid | 0.47 ± 0.08 | 0.51 ± 0.06 | 0.39 ± 0.03 | 0.55 ± 0.05 | 0.5 ± 0.05 | 1.10 | 0.84 | 1.18 | 1.07 | −7.31 | 1.31 |
| N-Acetyl-galactosamine | 0.041 ± 0.003 | 0.035 ± 0.003 | 0.024 ± 0.002 | 0.024 ± 0.004 | 0.077 ± 0.006 | 0.86 | 0.59 | 0.60 | 1.89 | −9.42 | −10.81 |
| N-Acetyl-l-Glu | 3.4 ± 0.2 | 1.4 ± 0.2 | 0.34 ± 0.01 | 0.4 ± 0.1 | 0.6 ± 0.05 | 0.40 | 0.10 | 0.13 | 0.18 | −8.88 | 11.41 |
| N-Acetyl-Orn | 0.48 ± 0.05 | 0.43 ± 0.03 | LOD | 0.24 ± 0.02 | 0.125 ± 0.009 | 0.90 | NA | 0.50 | 0.26 | −7.82 | 11.62 |
| Nicotinamide | 5.1 ± 0.3 | 2.4 ± 0.4 | 0.85 ± 0.08 | 1 ± 0.2 | 2.3 ± 0.3 | 0.48 | 0.17 | 0.20 | 0.46 | −10.65 | 7.29 |
| Orn | 11.5 ± 0.5 | 10.2 ± 0.2 | 0.7 ± 0.1 | 1.8 ± 0.1 | 9 ± 0.5 | 0.88 | 0.06 | 0.15 | 0.78 | −12.03 | 2.51 |
| Oxalic acid | 6.1 ± 0.6 | 5.9 ± 0.7 | 5.7 ± 0.3 | 7.4 ± 1 | 6.7 ± 0.5 | 0.97 | 0.94 | 1.21 | 1.09 | −5.43 | −4.60 |
| Phe | 13.4 ± 0.2 | 5 ± 0.1 | 5.9 ± 0.1 | 20.1 ± 0.7 | 5.2 ± 0.2 | 0.37 | 0.44 | 1.50 | 0.39 | 0.77 | 12.09 |
| Phosphoenolpyruvic acid | 1.48 ± 0.04 | 0.95 ± 0.05 | 0.279 ± 0.005 | 0.42 ± 0.01 | 0.39 ± 0.02 | 0.65 | 0.19 | 0.29 | 0.26 | −9.11 | 12.17 |
| Prephenic acid | 0.034 ± 0.003 | 0.078 ± 0.002 | 0.032 ± 0.001 | LOD | 0.04 ± 0.01 | 2.27 | 0.94 | NA | 1.16 | −6.16 | −0.56 |
| Pro | 86 ± 4 | 61 ± 4 | 29.5 ± 0.6 | 180 ± 10 | 35 ± 3 | 0.71 | 0.34 | 2.06 | 0.41 | 3.37 | 10.29 |
| Putrescine | 18.7 ± 0.5 | 13.2 ± 0.4 | 1.78 ± 0.05 | 0.129 ± 0.008 | 5.5 ± 0.3 | 0.71 | 0.09 | 0.01 | 0.29 | −10.27 | 9.86 |
| Pyruvic acid | 4.1 ± 0.3 | 2.9 ± 0.3 | 1.05 ± 0.06 | 0.53 ± 0.02 | 1.28 ± 0.08 | 0.70 | 0.25 | 0.13 | 0.31 | −9.49 | 10.47 |
| Rib | 0.46 ± 0.03 | 0.32 ± 0.03 | 1.1 ± 0.1 | 1.8 ± 0.09 | 2.2 ± 0.2 | 0.68 | 2.36 | 3.89 | 4.85 | −0.43 | −15.10 |
| Sedoheptulose-1,7-bisphosphate | LOD | LOD | LOD | 0.033 ± 0.006 | 0.015 ± 0.003 | NA | NA | NA | NA | 3.82 | −8.53 |
| Ser | 2 ± 0.03 | 0.71 ± 0.02 | 0.277 ± 0.007 | 3.28 ± 0.07 | 1.71 ± 0.06 | 0.35 | 0.14 | 1.64 | 0.85 | −3.44 | 1.82 |
| Shikimic acid | 94.9 ± 1 | 37.9 ± 0.6 | LOD | LOD | LOD | 0.40 | NA | NA | NA | −8.29 | 13.35 |
| sn-Glycerol-3-phosphate | 2.2 ± 0.2 | 0.8 ± 0.1 | 0.79 ± 0.07 | 7 ± 0.5 | 1.1 ± 0.2 | 0.37 | 0.36 | 3.25 | 0.49 | 5.88 | 5.48 |
| Succinic acid | 2.97 ± 0.09 | 7.4 ± 0.1 | 2.41 ± 0.06 | 2.72 ± 0.06 | 2.84 ± 0.05 | 2.48 | 0.81 | 0.92 | 0.96 | −4.26 | 2.44 |
| Threonic acid | 1.42 ± 0.04 | 3.5 ± 0.1 | 0.56 ± 0.01 | 3.3 ± 0.1 | 12.8 ± 0.5 | 2.47 | 0.39 | 2.33 | 9.04 | −6.10 | −14.52 |
| Thr | 7.3 ± 0.3 | 5.5 ± 0.3 | 17.9 ± 0.7 | 21.7 ± 0.5 | 11.5 ± 0.9 | 0.75 | 2.46 | 2.99 | 1.58 | 7.80 | −6.09 |
| Trehalose | 0.15 ± 0.01 | 0.093 ± 0.003 | 0.036 ± 0.002 | 0.051 ± 0.003 | 0.043 ± 0.003 | 0.63 | 0.24 | 0.35 | 0.29 | −8.83 | 12.32 |
| Trp | 0.6 ± 0.1 | 0.33 ± 0.07 | 2.9 ± 0.7 | 1.2 ± 0.3 | 0.43 ± 0.07 | 0.55 | 4.95 | 2.11 | 0.73 | 5.62 | −1.10 |
| Tyr | 8.1 ± 0.2 | 4.16 ± 0.05 | 9 ± 0.1 | 36.5 ± 0.5 | 5.8 ± 0.2 | 0.51 | 1.11 | 4.51 | 0.71 | 9.78 | 3.18 |
| Uracil | 6.86 ± 0.09 | 1.3 ± 0.1 | 0.68 ± 0.07 | 1.4 ± 0.4 | 1.44 ± 0.06 | 0.19 | 0.10 | 0.20 | 0.21 | −8.56 | 11.09 |
| Val | 9.3 ± 0.1 | 3.9 ± 0.1 | 3.1 ± 0.1 | 11.6 ± 0.4 | 4.6 ± 0.3 | 0.42 | 0.33 | 1.25 | 0.50 | −3.54 | 10.73 |
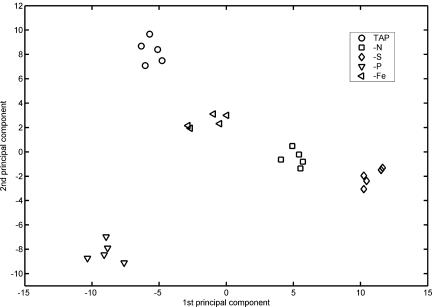
Apart from investigating differences in the levels of individual metabolites, factor analytic techniques can be used to quantitatively examine the sources of variation between different experimental conditions. Here, we used principal component analysis (PCA) to reduce the number of variables, classify metabolites, and obtain a multivariate measure of the variability of the metabolite profiles between different treatments and the relative homogeneity among replicate samples of each group. A data matrix of 170 metabolites, including both known compounds and unidentified analyte peaks, was manually inspected for false negatives and subsequently used for PCA. Projection of the resulting sample scores for the first and second principal components, which together account for 54% of the total sample variance, clearly separated the five experimental groups (Fig. 3), indicating a high suitability of metabolic readouts to study environmental effects in Chlamydomonas. It was apparent that the replicate analysis of samples of each individual treatment gave very similar profiles, which validates high reproducibility of the experimental procedure from cell harvest to data analysis. Distances between the groups of samples give a measure of the overall difference between the metabolite profiles of different treatments. Interestingly, score values for the first and second principal components for the −Fe, −N, and −S samples correlate. This suggests that the first two principal components, rather than explaining specific effects of adaptation to Fe, N, or S deprivation, measure parameters of the general response to nutrient deficiency with common effects in all three treatments with the most severe impact observed under S-depleted conditions. P deprivation, however, is transduced mainly as difference in score value for principal component two, suggesting that adjustment to P deficiency may share the general response developing in the other nutrient stress situations during 24-h treatment only to a limited extent. This notion is supported by the varying growth rates (controls: 1.6 doublings per day; −Fe: 1.6; −N: 0.7; −S: 0.7; and −P: 1.3) during the 24-h period of treatment. Besides cessation of growth, the regulation of photosynthetic electron transport has been characterized as a general response to adjust metabolism and sustain viability when nutrient levels fall (Grossman and Takahashi, 2001). While the decline of photosynthetic electron transport in N-deprived Chlamydomonas cells is due to the loss of the cytochrome b6f complex (Peltier and Schmidt, 1991), during P and S limitation cells undergo a transition of the photosynthetic apparatus to state 2, which allows them to more effectively dissipate excess absorbed excitation energy. Both conditions result in a 75% decrease of maximal O2 evolution that correlates with the reduction of electron flow through PSII, although with P deprivation a longer treatment is needed to observe the effect (Wykoff et al., 1998). The delayed response of P-deprived cells may be due to the mobilization of internal phosphate reserves. Chlamydomonas cells possess electron-dense vacuoles containing large amounts of pyrophosphate and short and long chain polyphosphate alongside with magnesium, calcium, and zinc (Ruiz et al., 2001). They presumably serve as Pi storage and buffer a critical Pi concentration in the cytoplasma, although also P-starved cells contain polyphosphate bodies of the normal size (Siderius et al., 1996) and most of the inorganic phosphate taken up by Pi-starved Chlamydomonas cells is still accumulated as polyphosphate (Hebeler et al., 1992). Therefore, it has been suggested that apart from Pi storage these organelles may serve also as internal calcium stores and may have a role in inositol-1,4,5-trisphosphate/calcium signaling (Siderius et al., 1996). The positional shift of the metabolite profiles for P-deprived cells after projection onto the plane defined by principal components 1 and 2 as compared to the other treatments may reflect the divergent alteration of metabolism due to the mobilization of internal Pi reserves and alleviation of external phosphate deficiency during the first 24 h of treatment. The contribution of individual metabolites to the differences observed is quantified via the loading of the metabolite for the respective principal component (Fig. 4). Among the group of metabolites with high absolute loading values for the first principal component are Glc-6-phosphate, Tyr, Trp, and yCR610 (high positive loadings), and glyceraldehyde-3-phosphate, Lys, and yCR535 (small negative loadings). A high positive loading value usually indicates abundance of these metabolites in samples with high score value for the corresponding principal component, whereas for small negative loading metabolite levels and score value are inversely correlated. This is confirmed for Tyr (PC1 loading = 0.098) with pool ratios increasing from 0.51 (−Fe) to 1.11 (−N) and 4.51 (−S), and for glyceraldehyde-3-phosphate (PC1 loading = −0.0829) with pool ratios decreasing from 1.22 (−Fe) to 0.98 (−N) and 0.50 (−S). TAP-cultured cells and −P cells, which are not very well discriminated by the first PC, nevertheless can be clearly separated using the linear combination of metabolite responses that constitutes the second principal component. Here, phosphoenolpyruvate, shikimate, and analyte yCR635 have among the highest loadings, whereas yCR758, N-acetyl-Orn, and maltotriose have very small loadings, indicating high discriminative power of these metabolites for TAP versus −P cultures in this set of five experimental groups.
Figure 3.
Sample scores for the first and second principal components extracted from the normalized peak response data of the five experimental groups. Each group is represented by five samples. The first and second principal components account for 35.2% and 19.1% of the total sample variance, respectively.
Figure 4.
Loading plots of the first (A) and second (B) principal components. For each principal component, the metabolites with the five largest absolute loading values are indicated. Peaks that could not be identified by library comparison are marked by “yCR” plus ID number. High positive loadings point to a more intense response of the corresponding metabolites in samples that have a high score value for this principal component. Samples with a small negative score value contain comparatively less of the metabolites with large positive loadings and more of those with small negative loadings.
In conclusion, metabolite profiling appears to be a well suited method to detect numerous changes of metabolite levels in response to environmental stimuli. The method introduced here will enable more refined experimental setups to detect more subtle changes in metabolite levels. This will be useful to derive specific hypotheses of how metabolic activity adjusts in response to external stimuli and also for investigation of Chlamydomonas mutants. The level of accuracy observed in this study suggests that Chlamydomonas may be highly suitable as a model for metabolomic research, especially in the context of photosynthetic processes. The possibility to control growth conditions precisely and to obtain large amounts of homogeneous sample material is a clear advantage in the attempt to study complex metabolic changes in fine-scaled gradients of conditions or with high temporal resolution.
MATERIALS AND METHODS
Cell Culture
Chlamydomonas reinhardtii strains CC-125 wt mt+ and CC-400 cw15 mt+ were acquired from the Chlamydomonas Genetics Center.
Strain CC-125 was cultured in TAP medium (Harris, 1989) at 25°C under constant illumination with cool-white fluorescent bulbs at a fluence rate of 90 μmol m−2 s−1 and with continuous shaking. Single colonies were used to inoculate a starter culture, which was harvested at late log-phase and used to inoculate a new culture at a starting density of 5 × 105 cells/mL. After 48 h, cells were harvested by centrifugation, washed twice with sterile 20 mm TRIS pH 7.0, supplied with 300 μm CaCl2, 400 μm MgCl2, and 7 mm KCl, and resuspended at a starting density of 1 × 106 cells/mL in TRIS-buffered media depleted of N (Farr et al., 1994), S (Davies et al., 1994), P (Quisel et al., 1996), or Fe (Herbik et al., 2002) prepared as described earlier, grown for 24 h, and harvested at final densities of 5 to 8 × 106 cells/mL.
Cell Number and Chlorophyll Determination
Correlation between turbidity and cell density was verified in advance by hemocytometer cell count with an optical density (γ = 750 nm) of one corresponding to 1.5 × 107 cells/mL. Chlorophyll a and b levels were determined spectrophotometrically (Wintermans and Demots, 1965; Harris, 1989).
Control of Metabolite Leakage during Quenching
Strains CC-125 and CC-400 were grown for 22 h from a starting density of 1 × 106 cells/mL in unlabeled TAP medium to a density of 6.8 × 106 cells/mL.
Ten microliters of [U-14C]acetate (Amersham; 200 μCi mL−1) were added to 10 mL of the cell suspension, which was put back to the same culture conditions. After 3 h of incubation, cells were harvested by centrifugation for 5 min at 2,000g at 4°C. The culture supernatant was carefully removed and residual medium extracted with a pipette. Cells were gently resuspended in 10 mL of ice-cold (unlabeled) TAP medium to wash off all remaining activity from medium residuals and interstitial spaces. Resuspended cells were collected by centrifugation for 5 min at 2,000g at 4°C, and the supernatant was carefully removed. The pellet was resuspended in 10 mL of room-temperature (unlabeled) TAP and gently agitated on a shaker for 15 min in the light at room temperature to allow for equilibration of the cells after two rounds of centrifugation.
Aliquots of 170 μL of the cell suspension were subjected to the quenching procedure described below. The quenched samples were prepared in six replicates: Cells of three of the samples were harvested after 3 min by centrifugation, whereas the other three were kept for 30 min at −25°C before pelleting. A total of 625 μL of the supernatant, corresponding to 125 μL of cell suspension, was used for scintillation. Measurements of the total amount of incorporated radioactivity were made by using 125 μL of the washed and equilibrated cell suspension with an addition of 500 μL of quench solution directly for scintillation.
Control samples for leakage inflicted by the centrifugation itself were prepared by harvesting an aliquot of the washed and equilibrated cell suspension at 2,500g for 5 min at 4°C and subjecting the supernatant to quenching in parallel with the cell suspension.
Using 10 mL of scintillation cocktail (Ready Safe; Beckman Coulter) per sample, scintillation for 14C was performed on an LS 6500 scintillation counter (Beckman Coulter).
Solvents and Chemicals
HPLC-grade methanol and chloroform were supplied by Merck. MSTFA was supplied by Macherey-Nagel. [U-13C]Sorbitol was purchased from Isotec. All other chemicals, in the highest grade available, were supplied by Sigma-Aldrich.
Harvest, Quenching, and Extraction
At the incubation site, the cell suspension was injected into a stirred solution composed of 32.5% methanol in water supplied with 300 μm CaCl2, 400 μm MgCl2, and 7 mm KCl, which corresponds to the molarity of macro salts in standard TAP medium. The solution was prechilled at −25°C and used at a ratio of 4:1 (v/v) of quenching solution to cell suspension for rapid cooling of cells. During injection, the quenching solution was stirred for rapid dilution of injected cells. Centrifuge tubes containing the solution during harvest were cooled in an ethanol/dry ice bath to keep sample temperature below −20°C. Cells were collected by centrifugation at 2,500g for 5 min with the centrifuge and rotor precooled at −20°C. Supernatant was decanted and residual liquid carefully removed. The pellet was flash-frozen in liquid nitrogen. Throughout homogenization of the recovered material with pestle and mortar, the preparation was kept in a bath of liquid N2. Cells were extracted by adding 8.33 mL per 109 cells of MCW (10:3:1, v/v/v) prechilled at −20°C.
GC-TOF Measurement
Further processing of the extract, including removal of particulate material, phase separation by addition of 0.4 volume parts water, centrifugation, solvent evaporation to complete dryness, and methoxyamine/trimethylsilylation derivatization, was carried out as described earlier (Roessner et al., 2001). GC-TOF analysis was performed on an HP5890 gas chromatograph with splitless injection of 1 μL at 230°C and 15°C/min GC temperature ramping from 85°C (2 min isocratic) to 360°C, using a 30-m MDN35 column (0.32-mm i.d., 0.25-μm film; Macherey-Nagel) at a constant flow rate of 2 mL/min. The Pegasus II TOF (Leco) mass spectrometer ion source operated at −70 kV filament voltage with ion source and transfer line temperature set at 250°C. Data were acquired at a rate of 20 spectra per second in the mass range of m/z 85 to 500.
Data Analysis
Chromatographic files were processed with ChromaTOF software (Leco). Samples were compared to reference chromatograms containing equal amounts of extract from all experimental groups involved and peaks were reported at a signal to noise ratio of 20. Analyte spectra were searched against custom spectrum libraries and identified based on retention index and spectrum similarity match. Peak lists were checked for false negatives and artifact peaks removed. Peak area was normalized to cell number and internal standard [U-13C]sorbitol. PCA was carried out after standardization of the data using the functions provided in Matlab 6.5 (Mathworks).
Acknowledgments
We thank the Chlamydomonas Genetics Center for providing the C. reinhardtii wild-type and mutant strains. We also would like to thank Aenne Eckardt for excellent technical assistance, and Prof. Lothar Willmitzer for valuable and helpful discussions.
This work was supported by the Max Planck Society.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christian Bölling (boelling@mpimp-golm.mpg.de).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.071589.
References
- Abadia J, Lopez-Millan AF, Rombola A, Abadia A (2002) Organic acids and Fe deficiency: a review. Plant Soil 241: 75–86 [Google Scholar]
- Barsch A, Patschkowski T, Niehaus K (2004) Comprehensive metabolite profiling of Sinorhizobium meliloti using gas chromatography-mass spectrometry. Funct Integr Genomics 4: 219–230 [DOI] [PubMed] [Google Scholar]
- Behrenfeld MJ, Kolber ZS (1999) Widespread iron limitation of phytoplankton in the south pacific ocean. Science 283: 840–843 [DOI] [PubMed] [Google Scholar]
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9: 418–425 [DOI] [PubMed] [Google Scholar]
- Buchholz A, Hurlebaus J, Wandrey C, Takors R (2002) Metabolomics: quantification of intracellular metabolite dynamics. Biomol Eng 19: 5–15 [DOI] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz F, Grossman AR (1994) Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning W, van Dam K (1992) A method for the determination of changes of glycolytic metabolites in yeast on a subsecond time scale using extraction at neutral pH. Anal Biochem 204: 118–123 [DOI] [PubMed] [Google Scholar]
- Eckhardt U, Buckhout TJ (1998) Iron assimilation in Chlamydomonas reinhardtii involves ferric reduction and is similar to Strategy I higher plants. J Exp Bot 49: 1219–1226 [Google Scholar]
- Farr TJ, Huppe HC, Turpin DH (1994) Coordination of chloroplastic metabolism in N-limited Chlamydomonas reinhardtii by redox modulation. 1. The activation of phosphoribulosekinase and glucose-6-phosphate dehydrogenase is relative to the photosynthetic supply of electrons. Plant Physiol 105: 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O (2002) Metabolomics: the link between genotypes and phenotypes. Plant Mol Biol 48: 155–171 [PubMed] [Google Scholar]
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Grossman A, Takahashi H (2001) Macronutrient utilization by photosynthetic eukaryotes and the fabric of interactions. Annu Rev Plant Physiol Plant Mol Biol 52: 163–210 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A (2003) Extracellular matrix and sex-inducing pheromone in Volvox. Int Rev Cytol 227: 131–182 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Sourcebook : A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego [DOI] [PubMed]
- Hebeler M, Hentrich S, Mayer A, Leibfritz D, Grimme LH (1992) Phosphate regulation and compartmentation in Chlamydomonas reinhardtii studied by in vivo P-31-NMR. Photosynth Res 34: 199 [Google Scholar]
- Herbik A, Bolling C, Buckhout TJ (2002) The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol 130: 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam SH, Buchanan MJ, Shin HC, Snell WJ (1985) The Chlamydomonas cell wall: characterization of the wall framework. J Cell Biol 101: 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2002) Systems biology: a brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Kromer JO, Sorgenfrei O, Klopprogge K, Heinzle E, Wittmann C (2004) In-depth profiling of lysine-producing Corynebacterium glutamicum by combined analysis of the transcriptome, metabolome, and fluxome. J Bacteriol 186: 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynnes JA, Derzaph TLM, Weger HG (1998) Iron limitation results in induction of ferricyanide reductase and ferric chelate reductase activities in Chlamydomonas reinhardtii. Planta 204: 360–365 [Google Scholar]
- Peltier G, Schmidt GW (1991) Chlororespiration: an adaptation to nitrogen deficiency in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 88: 4791–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel JD, Wykoff DD, Grossman AR (1996) Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol 111: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz FA, Marchesini N, Seufferheld M, Govindjee, Docampo R (2001) The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem 276: 46196–46203 [DOI] [PubMed] [Google Scholar]
- Sauer U, Schlattner U (2004) Inverse metabolic engineering with phosphagen kinase systems improves the cellular energy state. Metab Eng 6: 220–228 [DOI] [PubMed] [Google Scholar]
- Shi Y, Evans JE, Rock KL (2003) Molecular identification of a danger signal that alerts the immune system to dying cells. Nature 425: 516–521 [DOI] [PubMed] [Google Scholar]
- Siderius M, Musgrave A, van den Ende H, Koerten H, Cambier P, van der Meer P (1996) Chlamydomonas eugametos (chlorophyta) stores phosphate in polyphosphate bodies together with calcium. J Phycol 32: 402–409 [Google Scholar]
- Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T (2002) Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 74: 2233–2239 [DOI] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ III, Doyle J (2004) Robustness of cellular functions. Cell 118: 675–685 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Braby CE, Grossman AR (2001) Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol 127: 665–673 [PMC free article] [PubMed] [Google Scholar]
- Tolstikov VV, Fiehn O (2002) Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal Biochem 301: 298–307 [DOI] [PubMed] [Google Scholar]
- Voigt J, Frank R (2003) 14-3-3 proteins are constituents of the insoluble glycoprotein framework of the Chlamydomonas cell wall. Plant Cell 15: 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt J, Munzner P, Vogeler HP (1991) The cell-wall glycoproteins of Chlamydomonas reinhardtii: analysis of the in vitro translation products. Plant Sci 75: 129–142 [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O (2004) Differential metabolic networks unravel the effects of silent plant phenotypes. Proc Natl Acad Sci USA 101: 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger HG (1999) Ferric and cupric reductase activities in the green alga Chlamydomonas reinhardtii: experiments using iron-limited chemostats. Planta 207: 377–384 [Google Scholar]
- Wintermans J, Demots A (1965) Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta 109: 448–453 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell 3: 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]