Abstract
Lipocalins are a group of proteins that have been characterized in bacteria, invertebrate, and vertebrate animals. However, very little is known about plant lipocalins. We have previously reported the cloning of the first true plant lipocalins. Here we report the identification and characterization of plant lipocalins and lipocalin-like proteins using an integrated approach of data mining, expression studies, cellular localization, and phylogenetic analyses. Plant lipocalins can be classified into two groups, temperature-induced lipocalins (TILs) and chloroplastic lipocalins (CHLs). In addition, violaxanthin de-epoxidases (VDEs) and zeaxanthin epoxidases (ZEPs) can be classified as lipocalin-like proteins. CHLs, VDEs, and ZEPs possess transit peptides that target them to the chloroplast. On the other hand, TILs do not show any targeting peptide, but localization studies revealed that the proteins are found at the plasma membrane. Expression analyses by quantitative real-time PCR showed that expression of the wheat (Triticum aestivum) lipocalins and lipocalin-like proteins is associated with abiotic stress response and is correlated with the plant's capacity to develop freezing tolerance. In support of this correlation, data mining revealed that lipocalins are present in the desiccation-tolerant red algae Porphyra yezoensis and the cryotolerant marine yeast Debaryomyces hansenii, suggesting a possible association with stress-tolerant organisms. Considering the plant lipocalin properties, tissue specificity, response to temperature stress, and their association with chloroplasts and plasma membranes of green leaves, we hypothesize a protective function of the photosynthetic system against temperature stress. Phylogenetic analyses suggest that TIL lipocalin members in higher plants were probably inherited from a bacterial gene present in a primitive unicellular eukaryote. On the other hand, CHLs, VDEs, and ZEPs may have evolved from a cyanobacterial ancestral gene after the formation of the cyanobacterial endosymbiont from which the chloroplast originated.
Lipocalins are an ancient and functionally diverse family of mostly extracellular proteins found in bacteria, protoctists, plants, arthropods, and chordates (Suzuki et al., 2004). They have been implicated in many important functions, such as modulation of cell growth and metabolism, binding of cell-surface receptors, nerve growth and regeneration, regulation of the immune response, smell reception, cryptic coloration, membrane biogenesis and repair, induction of apoptosis, animal behavior, and environmental stress response (Akerstrom et al., 2000; Bishop, 2000; Frenette Charron et al., 2002).
The lipocalin fold is a highly symmetrical all β-structure dominated by a single eight-stranded antiparallel β-sheet closed back on itself to form a continuously hydrogen-bonded β-barrel. This β-barrel encloses a ligand-binding site composed of both an internal cavity and an external loop scaffold (Flower et al., 2000). The structural diversity of cavity and scaffold gave rise to a variety of different binding specificities, each capable of accommodating ligands of different size, shape, and chemical character (Flower et al., 2000). Lipocalins generally bind small hydrophobic ligands such as retinoids, fatty acids, steroids, odorants, and pheromones, and interact with cell surface receptors (Flower, 2000; Flower et al., 2000).
Phylogenetic analyses of lipocalins are possible due to their highly conserved three-dimensional structure (Ganfornina et al., 2000). Three structurally conserved regions (SCRs) related to features of the β-barrel are conserved: SCR1 (strand A and the 310-like helix preceding it), SCR2 (portions of strands F and G, and the loop linking them), and SCR3 (portion of strand H, the beginning of the following helix and the loop in between). It has been suggested that bacterial lipocalins were inherited by unicellular eukaryotes and then passed on to both plants and metazoans (Bishop, 2000). According to this hypothesis, primitive metazoans spread a low number of ancient lipocalins into some of their successors, the arthropods and chordates. These primordial lipocalins were likely similar to the Lazarillo and apolipoprotein D (ApoD) proteins. Alongside the chordate radiation, the ApoD-like ancestral lipocalin suffered duplications. On one hand, it gave rise to the ancestor of retinol-binding proteins and, on the other hand, to one or more ancestors of all other paralogous groups of lipocalins that diverged into current chordate lipocalins (Sánchez et al., 2003).
Although the evolution of metazoan lipocalins is well documented (Ganfornina et al., 2000; Gutiérrez et al., 2000; Salier, 2000; Sánchez et al., 2003), very little is known of the evolution of their plant counterparts. The first evidence of the presence of putative plant lipocalins was reported by Bugos et al. (1998). These are violaxanthin de-epoxidases (VDEs) and zeaxanthin epoxidases (ZEPs), key enzymes involved in the biosynthesis of the xanthophyll pigments required for photoprotection of the photosynthetic apparatus. They share the common substrate antheraxanthin and are therefore believed to exhibit similar tertiary structure. However, the peculiar architecture of these two proteins raised doubt as to whether they truly belong to the lipocalin family (Ganfornina et al., 2000; Salier, 2000).
We have recently reported the identification of the first true plant lipocalins from wheat (Triticum aestivum) and Arabidopsis (Arabidopsis thaliana; Frenette Charron et al., 2002). The two cDNAs designated TaTIL (T. aestivum temperature-induced lipocalin) and AtTIL (Arabidopsis temperature-induced lipocalin) encode polypeptides of 190 and 186 amino acids, respectively. Structure analyses indicated the presence of the three typical SCRs that characterize lipocalins. Sequence analyses revealed that these first true plant lipocalins share similarity with three evolutionarily related lipocalins: the mammalian ApoD, the bacterial lipocalin (Blc), and the insect Lazarillo protein. The comparison of the putative tertiary structures of the human ApoD and the wheat TaTIL-1 suggests that the two proteins differ in membrane attachment and ligand interaction.
To further identify and characterize other plant lipocalins and study their putative functions, we used an integrated approach of data mining of expressed sequence tag (EST) databases, bioinformatic predictions, and structural features, as well as cellular localization, expression, phylogenetic, and comparative genomics analyses. These analyses revealed that plants possess proteins that can be classified as true lipocalins (temperature-induced lipocalins [TILs] and chloroplastic lipocalins [CHLs]) and lipocalin-like proteins (VDEs and ZEPs). The features and evolutionary origin of these proteins in plants are discussed.
RESULTS
Identification of TaTIL Homologs
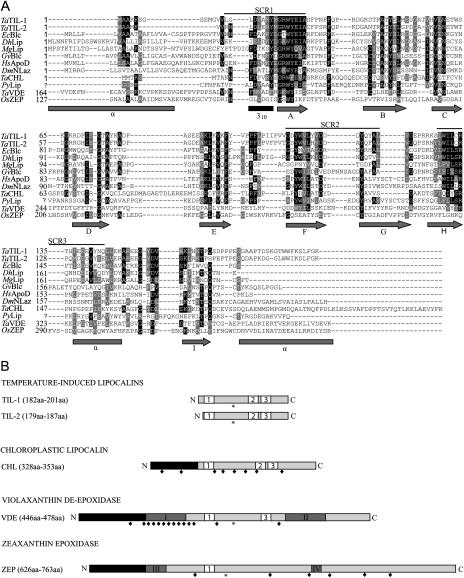
The recently identified wheat lipocalin TaTIL-1 was used to search GenBank databases. The search revealed that plants possess several homologs of this protein. A combination of EST sequencing and in silico reconstruction allowed the generation of 45 complete TaTIL-related protein sequences from plants (Table I; Supplemental Table IV). Based on size, structure, the presence of the three SCRs, and sequence similarity, these proteins were clustered into two distinct groups, TILs and CHLs. Thirty-seven TIL members sharing over 57% identity and 70% overall similarity with TaTIL-1 are found in 25 different species. Wheat possesses two different TIL members, TIL-1 and TIL-2, which share 67% identity and 79% similarity. A short region at the N terminus differentiates these two members in monocot species, but is absent in dicots. Data mining of the rice (Oryza sativa) genome revealed the existence of genes encoding TIL-1 and TIL-2 members (on chromosomes 2 and 8, respectively), whereas the Arabidopsis genome only has TIL-1 (on chromosome 5). In total, 12 plant species contain two TIL members. Sequence analyses revealed that TIL genes encode proteins ranging from 179 to 201 amino acids with a calculated molecular mass of 19 to 23 kD. All TIL homologs show a conserved putative N-glycosylation site. DGPI, PSORT, and SignalP predict a putative C-terminal cleavage site in eight out of the 37 proteins (Fig. 1; Supplemental Fig. 1). Considering this cleavage site, the calculated molecular mass of the mature TIL proteins would be 2 kD shorter than the corresponding precursor.
Table I.
Nomenclature and characteristics of plant lipocalins and lipocalin-like proteins
TaTIL homologs were identified using the TaTIL-1 protein sequence as query using TBLASTN against the GenBank EST database. Overlapping ESTs were assembled using the CAP3 assembly software and a consensus cDNA sequence was deduced when three or more identical sequences could be aligned. The degree of sequence identity was determined using ALIGN on the Biology Workbench and the National Center for Biotechnology Information BLAST 2 sequences. Chl, Chloroplastic; ND, not determined; PM, plasma membrane associated.
| Name | Class | Species | Accession No. | Precursor Protein | Mature Protein | Subcellular Location | Identity/Similaritya |
|---|---|---|---|---|---|---|---|
| kD (Amino Acid) | kD (Amino Acid) | ||||||
| TILs | |||||||
| PpTIL | Mosses | Physcomitrella patens | DQ222991 | 22 (189) | 22 (189) | PM | 57%/72% |
| TrTIL | Mosses | Tortula ruralis | DQ223011 | 21 (186) | 21 (186) | PM | 59%/70% |
| PtTIL | Conifers | Pinus taeda | DQ222992 | 21 (182) | 21 (182) | PM | 62%/77% |
| HvTIL-1 | Monocots | Hordeum vulgare | DQ222974 | 22 (190) | 22 (190) | PM | 96%/97% |
| HvTIL-2 | Monocots | H. vulgare | DQ222978 | 21 (182) | 21 (182) | ND | 66%/78% |
| OsTIL-1 | Monocots | Oryza sativa | XP_466697 | 22 (195) | 22 (195) | PM | 86%/91% |
| OsTIL-2 | Monocots | O. sativa | XP_482610 | 21 (179) | 21 (179) | ND | 69%/80% |
| SoTIL | Monocots | Saccharum officinarum | DQ222989 | 22 (193) | 22 (193) | PM | 88%/95% |
| SbTIL-1 | Monocots | Sorghum bicolor | DQ222976 | 23 (201) | 23 (201) | PM | 86%/92% |
| SbTIL-2 | Monocots | S. bicolor | DQ222980 | 21 (187) | 21 (187) | ND | 64%/77% |
| TaTIL-1 | Monocots | Triticum aestivum | AAL75812 | 22 (190) | 22 (190) | PM | 100% |
| TaTIL-2 | Monocots | T. aestivum | DQ222977 | 21 (182) | 21 (182) | ND | 67%/79% |
| ZmTIL-1 | Monocots | Zea mays | DQ222975 | 23 (198) | 23 (198) | PM | 87%/95% |
| ZmTIL-2 | Monocots | Z. mays | DQ222979 | 21 (181) | 21 (181) | ND | 66%/79% |
| AtTIL | Dicots | Arabidopsis | BAB10998 | 21 (186) | 21 (186) | PM | 75%/83% |
| BnTIL | Dicots | Brassica napus | DQ222996 | 21 (187) | 21 (187) | PM | 70%/81% |
| CsTIL | Dicots | Citrus sinensis | DQ223001 | 22 (186) | 22 (186) | PM | 73%/84% |
| GmTIL | Dicots | Glycine max | DQ222990 | 21 (184) | 21 (184) | PM | 67%/81% |
| GmTIL′ | Dicots | G. max | DQ222982 | 21 (184) | 21 (184) | ND | 67%/81% |
| GaTIL | Dicots | Gossypium arboreum | DQ223000 | 21 (185) | 21 (185) | PM | 73%/85% |
| GaTIL′ | Dicots | G. arboreum | DQ222986 | 21 (179) | 21 (179) | ND | 70%/81% |
| LsTIL | Dicots | Lactuca sativa | BQ852119 | 21 (185) | 21 (185) | PM | 74%/86% |
| LeTIL | Dicots | Lycopersicon esculentum | DQ222988 | 21 (185) | 21 (185) | PM | 74%/86% |
| LeTIL′ | Dicots | L. esculentum | DQ222981 | 21 (185) | 21 (185) | ND | 74%/86% |
| MtTIL | Dicots | Medicago truncatula | DQ222994 | 21 (184) | 21 (184) | PM | 70%/82% |
| MtTIL′ | Dicots | M. truncatula | DQ222983 | 19 (168) | 19 (168) | ND | 74%/86% |
| McTIL | Dicots | Mesembryanthemum crystallinum | DQ222999 | 22 (187) | 22 (187) | PM | 72%/83% |
| McTIL′ | Dicots | M. crystallinum | DQ222985 | 22 (187) | 22 (187) | ND | 75%/84% |
| PbTIL | Dicots | Populus balsamifera | DQ223002 | 21 (185) | 21 (185) | PM | 73%/85% |
| PbTIL′ | Dicots | P. balsamifera | DQ222987 | 21 (185) | 21 (185) | ND | 74%/85% |
| PotTIL | Dicots | Populus tremula | DQ223003 | 21 (185) | 21 (185) | PM | 73%/84% |
| Pot × PotrTIL | Dicots | P. tremula × Populus tremuloides | DQ223004 | 21 (185) | 21 (185) | PM | 75%/86% |
| PaTIL | Dicots | Prunus armeniaca | DQ222998 | 21 (185) | 21 (185) | PM | 70%/84% |
| PrpTIL | Dicots | Prunus persica | DQ222997 | 21 (185) | 21 (185) | PM | 70%/84% |
| StTIL | Dicots | Solanum tuberosum | DQ222995 | 21 (186) | 21 (186) | PM | 73%/85% |
| StTIL′ | Dicots | S. tuberosum | DQ222984 | 21 (185) | 21 (185) | ND | 74%/85% |
| VvTIL | Dicots | Vitis vinifera | DQ222994 | 22 (185) | 22 (185) | PM | 72%/85% |
| CHLs | |||||||
| HvCHL | Monocots | H. vulgare | DQ223006 | 37 (336) | 26 (230) | Chl | 25%/35% |
| OsCHL | Monocots | O. sativa | XP_473969 | 37 (342) | 26 (231) | Chl | 26%/36% |
| SbCHL | Monocots | S. bicolor | DQ223005 | 38 (340) | 26 (231) | Chl | 25%/34% |
| TaCHL | Monocots | T. aestivum | DQ223009 | 37 (339) | 26 (230) | Chl | 26%/36% |
| AtCHL | Dicots | Arabidopsis | CAB41869 | 39 (353) | 26 (230) | Chl | 25%/40% |
| GmCHL | Dicots | G. max | DQ223010 | 36 (328) | 26 (231) | Chl | 26%/36% |
| InCHL | Dicots | Ipomoea nil | DQ223007 | 37 (334) | 26 (231) | Chl | 29%/41% |
| StCHL | Dicots | S. tuberosum | DQ223008 | 38 (333) | 26 (231) | Chl | 27%/39% |
| VDEs | |||||||
| OsVDE_jap | Monocots | O. sativa var. japonica | AAL09678 | 50 (446) | 40 (351) | Chl | 14% |
| OsVDE_ind | Monocots | O. sativa var. indica | AAF97601 | 50 (446) | 40 (351) | Chl | 14% |
| SoVDE | Monocots | S. officinarum | CAB59211 | 54 (472) | 40 (351) | Chl | 13% |
| TaVDE | Monocots | T. aestivum | AAK38177 | 51 (454) | 40 (351) | Chl | 13% |
| AtVDE | Dicots | Arabidopsis | NP_172331 | 52 (462) | 40 (352) | Chl | 14% |
| CsVDE | Dicots | C. sinensis | AAL67858 | 54 (473) | 39 (344) | Chl | 13% |
| NtVDE | Dicots | Nicotiana tabacum | AAC50031 | 55 (478) | 40 (347) | Chl | 14% |
| LsVDE | Dicots | L. sativa | AAC49373 | 54 (473) | 40 (351) | Chl | 14% |
| ZEPs | |||||||
| CrZEP | Algae | C. reinhardtii | AAO34404 | 81 (763) | 72 (676) | Chl | 10% |
| CspZEP | Algae | Chlamydomonas sp. W80 | AAO48941 | 78 (727) | 68 (627) | Chl | 9% |
| OsZEP | Monocots | O. sativa | BAB39765 | 68 (626) | 60 (548) | Chl | 12% |
| AtZEP_col | Dicots | Arabidopsis | AAM13144 | 74 (667) | 65 (586) | Chl | 11% |
| AtZEP_ler | Dicots | Arabidopsis | AAG17703 | 74 (667) | 65 (586) | Chl | 11% |
| AtZEP_? | Dicots | Arabidopsis | AAF82390 | 74 (667) | 65 (586) | Chl | 11% |
| CaZEP | Dicots | Capsicum annuum | Q96375 | 72 (660) | 63 (582) | Chl | 12% |
| CuZEP | Dicots | Citrus unshiu | BAB78733 | 73 (664) | 64 (586) | Chl | 11% |
| LeZEP | Dicots | L. esculentum | P93236 | 73 (669) | 64 (583) | Chl | 11% |
| NpZEP | Dicots | Nicotiana plumbaginifolia | CAA65048 | 73 (663) | 64 (583) | Chl | 11% |
| NtZEP | Dicots | N. tabacum | S69548 | 73 (663) | 64 (583) | Chl | 11% |
| PaZEP | Dicots | P. armeniaca | O81360 | 72 (661) | 63 (580) | Chl | 12% |
With respect to the wheat TaTIL-1; for VDEs and ZEPs, only the similarity is indicated.
Figure 1.
Structure of plant lipocalins and lipocalin-like proteins. A, Alignment of the deduced amino acid sequences of wheat lipocalins with a select set of related lipocalins. Identical residues are in black and similar residues are in gray. The three SCRs that provide a signature for lipocalins are indicated. The secondary structure predicted from already published models (Bishop et al., 1995; Peitsch and Boguski, 1990) is presented under the alignment. Arrows, gray rectangles, and black rectangle represent β-strands, α-helix, and the 310 α-helix, respectively. B, Schematic representation of plant lipocalins. Plant lipocalins and lipocalin-like proteins can be classified in four groups. White boxes labeled 1, 2, and 3 represent the three SCRs (SCR1, SCR2, and SCR3) that characterize lipocalins. Black rectangles represent chloroplast transit peptides. Dark-gray rectangles with roman numerals represent, respectively: I, Cys-rich region; II, Glu-rich region; III, ADP-binding site; and IV, FAD-binding site. Stars and lozenges represent conserved N-glycosylation sites and conserved Cys residues, respectively.
The second group, CHLs, was found in eight species and shares 25% identity and 35% similarity with TaTIL-1 (Table I). CHL homologs encode proteins ranging from 328 to 340 amino acids with a calculated molecular mass of 36 to 39 kD. TargetP and ChloroP predict N-terminal chloroplastic transit peptides with high scores (over 0.800; Fig. 1; Supplemental Fig. 2). However, those transit peptides do not show any conservation in cleavage site position nor in length (17–68 amino acids). Pairwise sequence alignments (Supplemental Fig. 2) predict chloroplast transit peptide cleavage sites near the beginning of SCR1 in both monocot and dicot sequences. CHL homologs also possess eight conserved Cys residues probably involved in the three-dimensional structure of the protein by forming disulfide bridges (Fig. 1B; Supplemental Fig. 2).
Lipocalin-Like Proteins: VDEs and ZEPs
It has been suggested that xanthophyll cycle enzymes are lipocalin members (Bugos et al., 1998; Hieber et al., 2000). However, no ESTs corresponding to VDEs or ZEPs were found using TaTIL-1 as query. VDE and ZEP protein sequences from Arabidopsis were thus used to search GenBank databases. This search identified eight and 12 complete sequences corresponding to VDEs and ZEPs, respectively (Fig. 1; Supplemental Figs. 3 and 4). VDE and ZEP sequences share less that 15% similarity with TaTIL-1. VDE homologs encode proteins ranging from 446 to 478 amino acids with a calculated molecular mass of 50 to 55 kD (Table I). Each of the eight VDE homologs possesses an N-terminal transit peptide that targets the protein to the chloroplast (Fig. 1B; Supplemental Fig. 3). Considering the cleavage of the transit peptide, the calculated molecular mass of the mature VDE proteins would be in the range of 39 to 40 kD. VDE homologs show a conserved putative N-glycosylation site and 14 conserved Cys residues. Of those 14 Cys residues, 11 form a Cys-rich region in the N-terminal portion. The C-terminal portion contains 47% charged residues, most of which being Glu residues forming a Glu-rich region. All VDE proteins possess the first lipocalin signature SCR1 next to the Cys-rich region. In six of the eight VDE sequences, SCR1 fits the consensus, while three VDE sequences show discrepancies according to the Prosite database. All VDEs exhibit the two invariant amino acids G and W that are key features of SCR1 (Flower et al., 2000). SCR3 is also found in the Glu-rich C-terminal region, and the conserved R residue that characterizes this fingerprint is conserved. SCR2 is not present in the VDE sequences.
ZEP homologs encode proteins ranging from 626 to 763 amino acids with a calculated molecular mass of 68 to 80 kD (Table I). As for the VDE proteins, they possess an N-terminal transit peptide that targets the protein to the chloroplast (Fig. 1; Supplemental Fig. 4). After the cleavage of the transit peptide, the calculated molecular mass of the mature ZEP proteins ranges from 60 to 72 kD. ZEP homologs possess a conserved putative N-glycosylation and two conserved Cys residues. In addition, ZEP proteins contain an ADP-binding domain in their N-terminal portion and a FAD-binding domain in their C-terminal portion (Marin et al., 1996). The two invariant amino acids G and W that are key features of SCR1 are also present. ZEPs differ from TILs and CHLs in that they do not possess SCR2 and SCR3. ZEP proteins show 28% identity and 44% similarity with monooxygenases and oxidases that contain ADP-binding and FAD-binding domains found in bacteria and cyanobacteria (Supplemental Fig. 5).
Localization of the TIL-1 Lipocalin
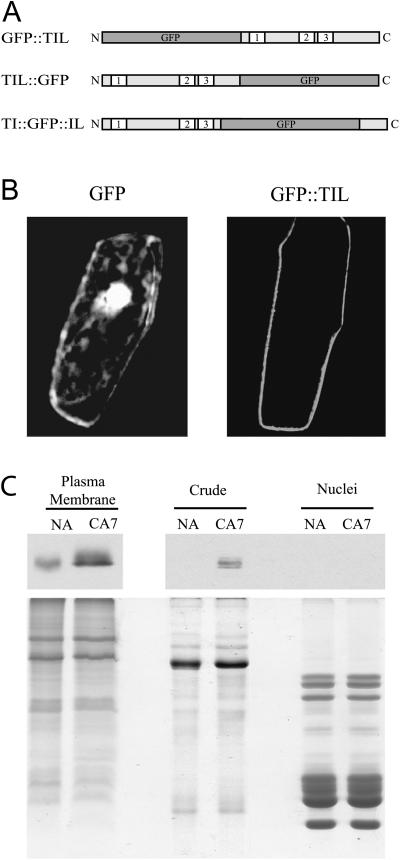
No targeting peptide was found in TIL-1. However, Blc and Lazarillo are known to be anchored to the plasma membrane (PM; Bishop, 2000). We therefore performed transient expression analysis of green fluorescent protein (GFP) fusion proteins in onion (Allium cepa) epidermal cells to establish the subcellular location of the AtTIL-1 protein. To assess for a possible effect of the GFP moiety on subcellular localization, three constructs were generated (Fig. 2A). The results show that the GFP∷TIL fusion accumulates specifically at the PM (Fig. 2B). The two other constructs showed the same localization pattern (data not shown). In contrast, the fluorescence is visible throughout the cell when the GFP protein is in its native state (negative control; Fig. 2B). These data show that TIL proteins accumulate at the PM.
Figure 2.
Cellular localization of the plant TIL lipocalins. A, Schematic representation of GFP fusions used in the transient expression experiments. N and C are the amino and carboxy termini of the proteins, respectively; 1, 2, and 3 indicate the three SCRs. B, Transient expression assays of GFP-TIL fusions. Plasmids carrying the fusions were transformed into onion epidermal cells by microprojectile bombardment. Confocal images of GFP fluorescence were captured 20 h after transformation. Only the GFP∷AtTIL data are shown since the three constructs gave the same fluorescence pattern. The color figure is shown in Supplemental Figure 6. C, Biochemical fractionation analysis. Wheat protein extracts were prepared and subjected to SDS-PAGE and western-blot analyses. Top, Western-blot results obtained using the anti-TaTIL antibody (1/25,000, 10-s exposure for the PM fractions; 1/2,500, 5-min exposure for the other fractions). Bottom, Coomassie Brilliant Blue-stained gel showing the quality of the preparations. Typical protein patterns are observed for each fraction. NA, Nonacclimated plants grown for 7 d; CA7, plants grown for 7 d at 24°C, then cold acclimated at 4°C for 7 d.
The AtTIL localization result obtained by transient expression in onion cells was confirmed by biochemical fractionation in wheat. The immunoblot results in Figure 2C show that cold acclimation (CA) induces a high accumulation of TaTIL-1 in an enriched PM fraction of cold-acclimated wheat but not in nuclei. The protein is also detected in a total soluble extract, but at a lower level.
Expression Studies
Induction by Abiotic Stresses
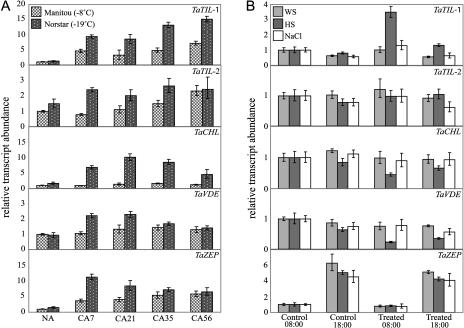
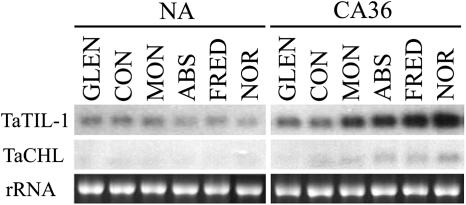
Expression analyses of the wheat lipocalin genes were carried out using quantitative real-time PCR. The data show that a low-temperature (LT) treatment induces the accumulation of the TaTIL-1, TaTIL-2, and TaZEP transcripts in both less tolerant (Manitou) and hardy wheat (Norstar; Fig. 3A). This increase is greater in the hardy winter cultivar. TaCHL and TaVDE transcripts also accumulate during CA, but only in the tolerant wheat. To determine whether plant lipocalin genes are regulated by other stresses, plants were subjected to different treatments. Results in Figure 3B show that a heat shock induces TaTIL-1 expression while it represses TaCHL and TaVDE. There is no significant change in response to water or salt stress. The TaTIL-1 and TaCHL transcripts accumulate differentially in various wheat cultivars showing different levels of freezing tolerance, indicating that their expression is associated with the plant's capacity to develop freezing tolerance (Fig. 4).
Figure 3.
Expression analysis of wheat lipocalins in response to abiotic stresses. Plants were treated, then total RNA was isolated from leaves, reverse transcribed, and subjected to quantitative real-time PCR. Relative transcript abundance was calculated and normalized with respect to the 18S rRNA transcript level. Data shown represent mean values obtained from four independent amplification reactions (n = 4), and the error bars indicate the range of possible RQ values defined by the se of the delta threshold cycles (Cts). This experiment was repeated three times with similar results. A, Expression during CA of wheat seedlings. Tissues were sampled at 18:00. Spring wheat (T. aestivum L. cv Manitou; LT50 of −8°C) and winter wheat (T. aestivum L. cv Norstar; LT50 −19°C) were cold acclimated at 4°C for the indicated number of days. NA, Nonacclimated plants grown for 7 d; CA7 to CA56, plants cold acclimated for 7 to 56 d. B, Expression following exposure of wheat seedlings to abiotic stresses. Winter wheat (T. aestivum L. cv Norstar; LT50 −19°C) plants were treated as follows: WS, dehydrated in a 30% polyethylene glycol solution (water stress); HS, exposed to 40°C for 1 h (heat shock); and NaCl, treated with 300 mm NaCl for 12 and 22 h (salt stress). The sampling time (08:00 and 18:00) indicates treatment periods of 12 and 22 h, respectively.
Figure 4.
Expression analysis of wheat TaTIL and TaCHL lipocalins in various wheat cultivars showing varying levels of freezing tolerance. Nonacclimated (NA) control plants were maintained at 20°C for 7 d, while other plants were cold acclimated at 4°C for 36 d (CA36) after the 7-d germination period. Tissues were sampled at 18:00. RNA was extracted and analyzed on a denaturing RNA gel blot. Spring wheat genotypes (T. aestivum L. cv Glenlea [GLEN; LT50 −8°C] and cv Concorde [CON; LT50 −8°C]); winter wheat genotypes (T. aestivum L. cv Monopole [MON; LT50 −15°C], cv Absolvent [ABS; LT50 −16°C], cv Fredrick [FRED; LT50 −16°C], and cv Norstar [NOR; LT50 −19°C]); rRNA, ethidium bromide-stained 28S ribosomal RNA included to show RNA loads (7.5 μg).
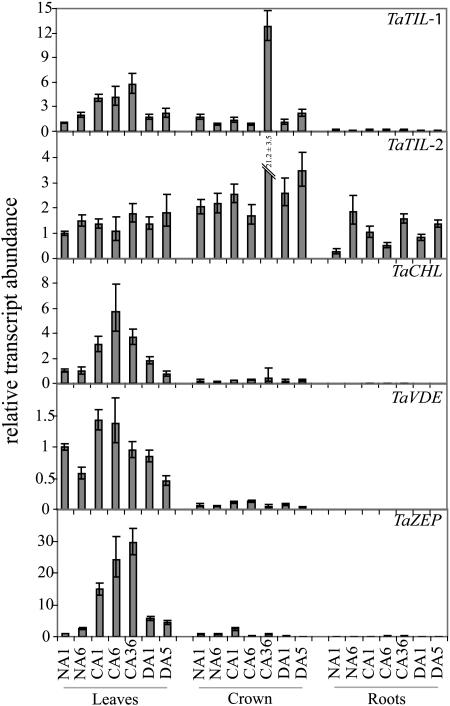
TaTIL-1, TaTIL-2, TaCHL, TaVDE, and TaZEP transcripts all accumulate in response to CA in green leaves (Fig. 5). The maximal accumulation is seen after 6 d of CA for TaCHL and 36 d for TaTIL-1 and TaZEP. When the plants are deacclimated at 24°C for 1 and 5 d, all transcripts decline to the nonacclimated control levels. TaTIL-1 and TaTIL-2 transcripts accumulate to a higher level in crown compared to leaves after 36 d of CA. The results also show that TaTIL-2 is the only wheat lipocalin expressed in roots.
Figure 5.
Expression analysis of wheat lipocalins in different tissues. Plants were grown for 7 d at 20°C. Nonacclimated (NA) control plants were maintained at 20°C for 1 and 6 d. Cold-acclimated (CA) plants were transferred at 4°C for 1, 6, and 36 d. After CA, some plants were transferred at 20°C for 1 and 5 d for deacclimation (DA). Leaf, crown, and root tissues were sampled at 18:00 and RNA was extracted and analyzed as described in Figure 3.
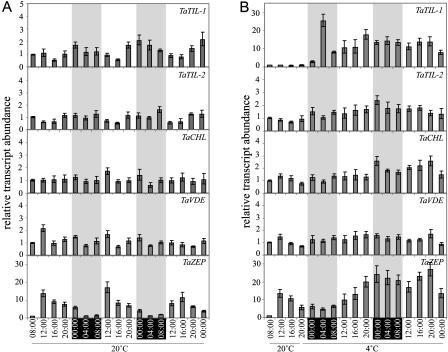
Regulation during the Diurnal Cycle
Recent evidence has emerged on the regulation of stress-regulated gene expression by the circadian clock (Fowler et al., 2005). In addition, Thompson et al. (2000) reported that the expression of ZEP genes in tomato (Lycopersicon esculentum) is under the control of circadian regulation. Based on the classification of ZEP proteins as lipocalin-like proteins, we thus performed expression analyses to determine whether oscillation in transcript accumulation of these genes occurs during a 16-h-light/8-h-dark regime at 20°C (Fig. 6A). TaZEP transcripts accumulate to lower levels during dark periods while they accumulate up to 15-fold in the presence of light, reaching a maximum after 4 h near diurnal time 12:00. This oscillation was observed over three cycles. TaTIL-1 also demonstrated a diurnal oscillation over three cycles. However, the oscillation is less pronounced and reaches a maximal accumulation level at diurnal time 00:00 and a minimal accumulation level at diurnal time 16:00. TaTIL-2, TaCHL, and TaVDE transcript accumulation is not under the control of diurnal regulation. To evaluate the effect of LT on diurnal oscillation, plants were exposed to 4°C under the same light/dark regime (Fig. 6B). Upon exposure to LT, the diurnal oscillation of the TaZEP transcript accumulation is deregulated.
Figure 6.
Expression analysis of wheat lipocalins in response to diurnal cycles. Plants were germinated for 8 d at 20°C under a 16-h-day/8-h-night photoperiod. Beginning on day 8 at 8:00, plants were grown at 20°C for 12 h (from 08:00–20:00) then kept at 20°C for 52 h (A, controls) or transferred at 4°C for 52 h (B, cold acclimated). Leaf samples were harvested at each time point, and RNA was extracted and analyzed as described in Figure 3. Gray areas represent the dark periods.
Evolution of Lipocalins
To investigate the evolutionary origin of plant lipocalins, we searched for homologs in ancient plants and algae. Data mining of nonredundant sequence databases, EST databases, and other genome projects showed that sequences encoding homologs of TILs and CHLs are found in ancient plants like mosses, coniferals, gnetales, and cycads (Table I). No entries encoding TIL or CHL homologs were found for the green algae Chlamydomonas reinhardtii or the red algae Cyanidioschyzon merolae. However, three plant lipocalin-related ESTs were identified in the red algae Porphyra yezoensis. A survey of 14 cyanobacterial genome project databases revealed that the cyanobacterium Gloeobacter violaceus PCC7421 is the only cyanobacterial strain that possesses a lipocalin gene. A search of 31 fungi genomes revealed lipocalin homologs in two different fungi, the yeast Debaryomyces hansenii CBS767 and the foliar plant pathogen Magnaporthe grisea strain 70-15.
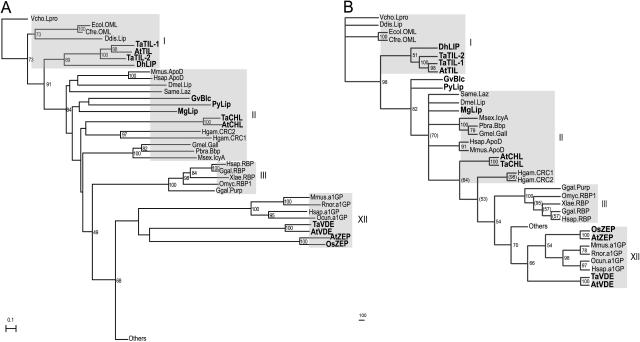
The relationships between plant lipocalins, ancient lipocalins, and other family members were determined by building phylogenetic trees (Fig. 7). Our goal was not to redesign the evolution scheme of the lipocalin family, but to trace the origin of plant lipocalins. We therefore used the dataset from Ganfornina et al. (2000) and appended the plants, algae, cyanobacteria, and fungi sequences (this study) and the newly identified epidydimal lipocalin sequence (Suzuki et al., 2004). To reduce the complexity, we removed closely related sequences from the original alignment of Ganfornina et al. (2000). However, each of the 14 clades was represented. We thus aligned 84 lipocalin sequences and reconstructed phylogenetic trees using the neighbor-joining (NJ) method (Fig. 7A) and the maximum likelihood (ML)-based method (Fig. 7B). For the latter, we first computed a global tree (Supplemental Fig. 9) and then refined the part of the tree containing the new sequences using the corresponding subset of the initial alignment and the same phylogenetic reconstruction methodology.
Figure 7.
Phylogenetic analyses of selected lipocalins. A, The NJ tree was built from the alignment presented in Supplemental Figure 7 and rooted with the VchoLpro taxon. Only part of the tree is shown. The global tree is presented in Supplemental Figure 8. The scale bar represents the branch length (number of amino acid substitutions/site). B, The ML tree was built from the alignment presented in Supplemental Figure 7 and rooted with the VchoLpro taxon. Bootstrap values in parentheses indicate nodes that were refined for better resolution. Only part of the tree is shown. The global tree is presented in Supplemental Figure 9. The scale bar represents the branch length (number of amino acid substitutions/100 residues). The gray boxes, identified by roman numerals, represent the clade nomenclature (Gutiérrez et al., 2000).
In comparison with the original alignment by Ganfornina et al. (2000), our alignment contains long gaps due to the presence of ZEP and VDE sequences and the lower conservation of the SCR2 and SCR3 signatures. Both trees obtained from this alignment are supported by strong bootstrap values and agree well with the 14 major lipocalin clades already identified (Supplemental Figs. 8 and 9; Ganfornina et al., 2000; Sánchez et al., 2003). The branching pattern suggests that the plant TILs, the yeast DhLIP, the cyanobacterium GvBlc, and the red algae PyLIP diverged early from Blcs (ML bootstrap values of 819 and 700). This is in agreement with a previous phylogenetic study that included a plant lipocalin (Sánchez et al., 2003). The fungus MgLIP and plant CHL lipocalins are incorporated into clade II along with the insect Lazarillo and the mammalian ApoD. The two lipocalin-like groups, VDEs and ZEPs, are in clade XII with a1GP lipocalins. The a1GP protein has been described as an outlier lipocalin due to the lower conservation of motifs SCR2 and SCR3 (Ganfornina et al., 2000). As these motifs are not well conserved in VDEs and ZEPs, it is not surprising to see these three proteins in the same clade. However, the branching pattern inside this clade is not supported by high bootstrap values in both trees. It is worth noting that exclusion of the VDE and ZEP sequences from the phylogenetic analysis results in the positioning of clade XII near clade XIII in the trees, as reported by Ganfornina et al. (2000). Apart from this, the branching pattern of the trees is not affected.
Another small difference between our analyses and those of others (Ganfornina et al., 2000; Gutiérrez et al., 2000; Sánchez et al., 2003; Suzuki et al., 2004) is that in the ML tree, the two lipocalins Hsap.Lcn5 and Ggal.QS-21 are relocated to the miscellaneous clade that already contains Mmus.Lcn11, Hsap.Lcn9, and Lviv.ESP. In the NJ tree, only Ggal.QS-21 is relocated to this clade.
DISCUSSION
We recently reported the identification of the first true lipocalins from plants, TaTIL-1 and AtTIL, which possess the three SCRs that characterize lipocalins (Frenette Charron et al., 2002). Data mining of various databases using the TaTIL-1 sequence as query resulted in the identification of all available full-length plant lipocalins. Protein sequence alignments revealed that these proteins can be classified into four groups based on structural features conserved among typical lipocalins. Two of these groups, TILs and CHLs, are bonafide lipocalins. Monocotyledonous species possess genes encoding two different members of the TIL group, TIL-1 and TIL-2, which are regulated by abiotic stresses. On the other hand, there is no conclusive evidence of the existence of these two forms in dicotyledonous plants. Members of the CHL group are expressed specifically in photosynthetic tissues of higher plants in response to LT exposure. The presence of a transit peptide at their N terminus suggests that they may play a role in the chloroplast during CA.
TaTIL-1 and AtTIL proteins share similarity with three evolutionarily related lipocalins, ApoD, Blc, and Lazarillo (Frenette Charron et al., 2002). Since the latter two proteins are known to be anchored to membranes, we hypothesized that TILs were also membrane associated. Our localization studies showed that TIL-1 is indeed localized at the PM level. This result is supported by proteomic analyses of PM proteins from Arabidopsis (Kawamura and Uemura, 2003). TILs do not bear a signal peptide; therefore, bioinformatic analyses were used to determine which type of attachment is responsible for the PM localization. These analyses suggested the presence of a C-terminal cleavage site and a favorable environment (proper hydrophobic tail length and hydrophilic region length) for the addition of a glycosylphospatidylinositol (GPI) anchor in eight of the 37 reconstructed TIL proteins. Addition of a GPI anchor would result in the cleavage of the C-terminal end of TILs. To determine whether this is the case, a C-terminal TIL∷GFP fusion was tested by transient expression. The addition of a GPI anchor would result in the cleavage of the TIL∷GFP fusion in two separate proteins, TIL and GFP, and GFP would be able to move freely in the cytoplasm. Our results demonstrate that TILs are associated with the PM, but not via a GPI anchor, since the GFP fluorescence is always observed at the PM level. The fact that the N-terminal, internal, and C-terminal GFP fusions are localized at the PM suggests that TILs could be targeted to this site via the hydrophobic loop between β-strands 5 and 6, as we suggested previously (Frenette Charron et al., 2002).
Despite the presence of SCRs in members of the other two groups, VDEs and ZEPs, many questions have been raised as to whether they truly belong to the lipocalin family. The size and the exon-intron architecture of these xanthophyll cycle enzymes show no significant similarity to the genomic organization of typical lipocalin genes (Gutiérrez et al., 2000; Salier, 2000). VDEs are predicted to be lipocalin-like proteins with a central barrel structure flanked by a Cys-rich N-terminal domain and a Glu-rich C-terminal domain (Fig. 1; Hieber et al., 2002). ZEPs possess ADP and FAD-binding domains and only fit the description of lipocalins based on a low SCR1 similarity (Fig. 1). On the other hand, the 44% sequence similarity with monooxygenases would instead classify ZEP proteins in the latter family. According to our phylogenetic analyses, VDEs and ZEPs are positioned in clade XII together with a1GP. Since a1GP is found only in marsupials and placental mammals, it is unlikely that this grouping reflects a genuine evolutionary relationship. Given the features of VDEs and ZEPs, the strict definition of lipocalins, and their positioning in the phylogenetic trees, it is difficult to consider them part of the lipocalin family. Rather, they could be classified as lipocalin-like proteins. The apparent fusion of a true plant lipocalin with other proteins during evolution was proposed to explain the atypical structures of these enzymes (Ganfornina et al., 2000). The appearance of proteins with novel functions would have been an evolutionary advantage in that it would have provided plants with enhanced protection against photooxidative damage.
Important clues regarding the evolution of plant lipocalins in our study come from the finding of a lipocalin homolog in the cyanobacterium G. violaceus PCC7421. Cyanobacteria are unicellular organisms that carry a complete set of genes for oxygenic photosynthesis, the most fundamental life process on earth. The chloroplasts in higher plants are believed to have evolved from cyanobacterial ancestors who developed an endosymbiontic relationship with a eukaryotic host cell (Delwiche et al., 1995). To this day, sequence information is available for 14 complete and two partial genomes of cyanobacteria. Among these, only G. violaceous possesses a lipocalin gene. Unlike most recent cyanobacteria, this strain lacks thylakoids, and phycobilisomes are attached to the PM. Recent molecular phylogenetic analyses show that G. violaceus is a member of early branching of the cyanobacterial lineage (Delwiche et al., 1995) and could thus be the oldest known cyanobacterium. This suggests that G. violaceous or a close relative might have been the initial donor that gave rise to the chloroplast structures of higher plants. These observations reveal that certain lipocalins were associated with photosynthetic membranes early in the evolution.
No TIL homologs were found in the primitive photosynthetic green algae C. reinhardtii nor in the red algae C. merolae, two species for which extensive genomic information is available. However, a homolog was found in the red algae P. yezoensis. Red algae (Rhodophyta) are photoautotrophic eukaryotes characterized by a lack of flagella and the presence of phycobiliproteins within the plastid (Bold and Wynne, 1985; South and Whittick, 1987). Porphyra species are blade-forming red seaweeds and are among the simplest of red algae. Some are extremely tolerant to desiccation and are found in the highest, driest reaches of the littoral zone in cold temperate and boreal regions. The presence of a lipocalin in this species may be related to its desiccation tolerance. The close positioning, in the phylogenetic trees, of the cyanobacterial GvBlc with PyLip from a photosynthetic red algae supports the hypothesis of lateral transfer.
Another novel finding is the identification of lipocalins in two fungi species, D. hansenii and M. grisea. D. (Torulaspora) hansenii is a cryotolerant marine yeast that tolerates salinity levels up to 24%, whereas common yeast (Saccharomyces cerevisiae) growth is inhibited at 10% salinity. D. hansenii is the most common species found in all types of cheeses (Fleet, 1990). It is also common in other dairy products (Seiler and Busse, 1990) because of its ability to grow in the presence of high salt at LT and to metabolize lactic and citric acids. M. grisea, the causal agent of rice blast disease, is one of the most devastating threats to food security worldwide (Zeigler et al., 1994). M. grisea shows excellent adaptation abilities to a wide spectrum of stresses (Ikeda et al., 2001). The presence of lipocalins in these fungi species may explain their abiotic stress tolerance. It is possible that CHLs and lipocalin-like proteins could have arisen from a lateral transfer following infection by a fungus such as M. grisea. It has been suggested that such transfers could explain the presence of M. grisea DNA in plant genomes (Kim et al., 2000). On the other hand, gene duplication and/or fusion can also be proposed to explain the presence of the different proteins in higher plants genomes.
The plant lipocalins and lipocalin-like proteins properties, their tissue specificity, and their transcript accumulation in response to temperature stress suggest a possible protection role against stress damage. Their association with the chloroplast (CHLs, VDEs, and ZEPs) and the PM (TILs) in the green leaves supports the idea that these proteins may act as scavengers of potentially harmful molecules known to be induced by temperature stress and excess light. The lipocalin-like protein VDEs and ZEPs catalyze the interconversions between the carotenoids violaxanthin, antheraxanthin, and zeaxanthin in higher plants under stress conditions to form the zeaxanthin that protects the photosynthetic apparatus against the effect of excessive light (Havaux and Kloppstech, 2001). Our previous work demonstrated that the photosynthetic acclimation to LT mimics the photosynthetic acclimation to high light because both conditions result in a comparable reduction state of PSII (Ndong et al., 2001). Based on this comparison and the data in this study, we hypothesize that the other plant lipocalins and the CHLs in particular may protect the photosynthetic apparatus against the deleterious effect of temperature stress. The work is in progress to determine the exact function of these novel members of plant lipocalins.
MATERIALS AND METHODS
Data Mining
TaTIL homologs were identified using the TaTIL-1 protein sequence as query (accession no. AAL75812) using TBLASTN against the GenBank EST database. Overlapping ESTs were assembled using the CAP3 assembly software (http://fenice.tigem.it/bioprg/interfaces/cap3.html) and a consensus cDNA sequence was deduced when three or more identical sequences could be aligned. The degree of sequence identity was determined using ALIGN on the Biology Workbench (http://workbench.sdsc.edu) and The National Center for Biotechnology Information BLAST2 sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). Sequences were aligned and analyzed using ClustalW on the Biology Workbench. Shading of amino acids was performed with BOXSHADE (http://ulrec3.unil.ch/software/boxshade/boxshade.html). PSORT, iPSORT (http://psort.nibb.ac.jp), TargetP, version 1.01 (http://www.cbs.dtu.dk), SignalP, version 2.0 (http://www.cbs.dtu.dk), and ChloroP (http://www.cbs.dtu.dk) were used to detect specific targeting sequences. For functional domain identification, we first used ScanProsite to scan the Prosite database, then most of the software available on the ExPASy server (http://ca.expasy.org). DGPI was used for GPI-anchoring site prediction (http://129.194.186.123/GPI-anchor/index_en.html).
Plant Material and Growth Conditions
In this study, we used two spring wheat genotypes (Triticum aestivum L. cv Glenlea, LT50 [lethal temperature that kills 50% of the seedlings] −8°C; and cv Concorde, LT50 −8°C), and four winter wheat genotypes (T. aestivum L. cv Monopole, LT50 −15°C; cv Absolvent, LT50 −16°C; cv Fredrick, LT50 −16°C; and cv Norstar, LT50 −19°C). Plants were grown in a mixture of 50% black earth and 50% Pro-Mix (Premier) for 7 d under a 16-h-d photoperiod with a light intensity of 250 μmol m−2 s−1 at 20°C. Heat shock (40°C) and cold treatments (4°C) were performed by changing the temperature in the growth chamber, while salt stress (0.3 m NaCl) and osmotic stress (30% [w/v] PEG-6000) were performed by saturating the soil with these solutions. A total of eight seedlings were harvested on dry ice at different time points, as stated in the figures, and immediately frozen at −70°C.
Cellular Localization of TILs
Transient Expression of GFP Fusions
AtTIL cDNA fragments were PCR amplified using the primers described in Supplemental Table I, then cloned in the pAVA321 vector (von Arnim et al., 1998) to generate three constructs. The chimeric genes encode GFP∷TIL and TIL∷GFP fusion proteins, and another protein in which the GFP protein is inserted within the TIL sequence (TI∷GFP∷IL; Fig. 2A). Plasmid DNA was coated onto M17 tungsten particles (Bio-Rad) and delivered into onion (Allium cepa) epidermal cells by particle bombardment (Shieh et al., 1993). Images were captured on a MRC1024 confocal system with a Nikon Eclipse TE300 inverted microscope and analyzed using LaserSharp software (Bio-Rad).
Subcellular Fractionation
Organellar protein fractions were prepared from leaves of control and 7-d cold-acclimated winter wheat cv Norstar. PMs were isolated by two-phase partitioning as described by Zhou et al. (1994). Nuclei were isolated as previously described (Vazquez-Tello et al., 1998), and then nuclear proteins were extracted using the TRI-Reagent (Molecular Research Center) following the manufacturer's recommendations. Total soluble proteins were prepared as described (Vazquez-Tello et al., 1998). Samples were separated on 12% SDS-PAGE gels, and the rabbit anti-TaTIL-1 antibody was used for the immunoblot analysis. Detection was performed with a peroxidase-coupled anti-rabbit IgG secondary antibody and the western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer).
Expression Analyses by Quantitative Real-Time PCR
RNA Isolation and cDNA Synthesis
Total RNA was isolated using the RNeasy plant mini kit (Qiagen). For expression analyses in the different wheat cultivars, RNA was separated on a formaldehyde agarose gel, transferred to a positively charged nylon membrane, and then hybridized sequentially to TaTIL and TaCHL 32P-labeled probes. All other expression analyses were performed using quantitative real-time PCR. Purified RNA (2.8 μg) was reverse transcribed in a 20-μL reaction volume using the SuperScript II first-strand synthesis system for reverse transcription (RT)-PCR (Invitrogen). Parallel reactions were run for each RNA sample in the absence of SuperScript II (no RT control) to assess for genomic DNA contamination. The reactions were terminated by heat inactivation at 70°C for 15 min. Subsequently, the cDNA products were treated with 2 units of RNase H for 20 min at 37°C, then diluted in water to 20 ng μL−1, and stored at −20°C.
Design of Gene-Specific Primers
The genome of hexaploid wheat contains three genomes inherited from three diploid ancestors. Primers were specifically designed to monitor the expression of the three copies of each gene in the same RT reaction. In addition, primers for TaCHL, TaVDE, and TaZEP were designed onto exon junctions to avoid genomic DNA amplification. The gene architecture of TaTIL-1 and TaTIL-2 did not allow for the design of LUX primers on the exon-exon junction. Fluorescent LUX primers as well as nonfluorescent primers (Supplemental Table I) were designed using a combination of the LUX Designer-Desktop version (Invitrogen) and Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). BLASTN searches were performed to confirm the gene specificity of the primers. Primers were synthesized by Invitrogen.
PCR Amplification
Quantitative real-time PCR assays were performed in quadruplicate on an ABI PRISM 7000 sequence detection system (Applied Biosystems) using 18S ribosomal RNA as internal standard. From the diluted cDNA, 1 μL (20 ng) was used as template in a 50-μL PCR reaction containing 1× platinum quantitative PCR SuperMix-UDG, 0.15 μm of nonfluorescent primer, 0.3 μm of LUX fluorescent primer, and ROX reference dye. The PCR thermal-cycling parameters were 50°C for 2 min, 95°C for 2 min, followed by 50 cycles of 95°C for 20 s and 60°C for 1 min. Each experiment was replicated at least three times.
Data Analysis
All calculations and statistical analyses were performed using SDS RQ Manager 1.1 software using the 2−ΔΔCt method with a relative quantification (RQ)min/RQmax confidence set at 95% (Livak and Schmittgen, 2001). The error bars display the calculated maximum (RQmax) and minimum (RQmin) expression levels that represent se of the mean expression level (RQ value). Collectively, the upper and lower limits define the region of expression within which the true expression level value is likely to occur (SDS RQ Manager 1.1 software user manual; Applied Biosystems). Amplification efficiency (98% to 100%) for the six primer sets was determined by amplification of cDNA dilution series using 80, 20, 10, 5, 2.5, and 1.25 μg per reaction (data not shown). Specificity of the RT-PCR products was assessed by gel electrophoresis. A single product with the expected length was detected for each reaction.
Phylogenetic Analyses
Proteins used in this analysis and the FASTA files are presented in Supplemental Tables II and III. The ClustalX version 1.83 software (Thompson et al., 1997) was used to generate the sequence alignment using the following parameters: gap-opening penalty of 15.0, gap-extension penalty of 0.30, and the substitution Gonnet scoring matrix. The alignment was adjusted manually to respect the position of the three SCRs, the glycosylation sites, and the Cys residues, and then used to generate phylogenetic trees based on two methods. The NJ tree was generated using TREECON for Windows (version 1.3b) with 100 bootstrap replicates and with the distance calculation set to the Poisson correction (van de Peer and de Wachter, 1994). PHYML (Guindon and Gascuel, 2003) was used to perform ML analyses with the evolution model JTT (Jones et al., 1992) and all other parameters were set to their default value. One thousand bootstrap replicates were performed with Seqboot and a consensus tree was computed with Consense following a strict-majority rule. The latter two programs are part of the PHYLIP version 3.6 package (Felsenstein, 1993). The ML trees were rooted with the VchoLpro taxon and displayed with TreeView version 1.6.6 (Page, 1996).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers given in Table I and Supplemental Table II.
Supplementary Material
Acknowledgments
We thank Dr. Sánchez and Dr. Ganfornina (Departamento de Bioquímica y Biología Molecular y Fisiología-IBGM, Universidad de Valladolid-CSIC, Valladolid, Spain) for providing their lipocalin sequence alignment. We also thank M. Champoux, C. Plouffe, G. Brault, N.A. Kane, and D. Flipo (Département des Sciences Biologiques, Université du Québec à Montréal) for technical assistance.
This work was supported by Genome Canada/Génome Québec and Natural Sciences and Engineering Research Council of Canada.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Fathey Sarhan (sarhan.fathey@uqam.ca).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070466.
References
- Akerstrom BD, Flower R, Salier JP (2000) Lipocalins: unity in diversity. Biochim Biophys Acta 1482: 1–8 [DOI] [PubMed] [Google Scholar]
- Bishop RE (2000) The bacterial lipocalins. Biochim Biophys Acta 1482: 73–83 [DOI] [PubMed] [Google Scholar]
- Bishop RE, Penfold SS, Frost LS, Holtje JV, Weiner JH (1995) Stationary phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein D. Implications for the origin of lipocalins. J Biol Chem 270: 23097–23103 [DOI] [PubMed] [Google Scholar]
- Bold HC, Wynne MJ (1985) Introduction to the Algae. Structure and Reproduction. Prentice Hall, Englewood Cliffs, NJ
- Bugos RC, Hieber AD, Yamamoto HY (1998) Xanthophyll cycle enzymes are members of the lipocalin family, the first identified from plants. J Biol Chem 273: 15321–15324 [DOI] [PubMed] [Google Scholar]
- Delwiche CF, Kuhsel M, Palmer JD (1995) Phylogenetic analysis of tufA sequences indicates a cyanobacterial origin of all plastids. Mol Phylogenet Evol 4: 110–128 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1993) PHYLIP, Phylogeny Inference Package, Version 3.6. Distributed by the author. Department of Genetics, University of Washington, Seattle
- Fleet G (1990) Yeasts in dairy products. J Appl Bacteriol 68: 199–211 [DOI] [PubMed] [Google Scholar]
- Flower DR (2000) Beyond the superfamily: the lipocalin receptors. Biochim Biophys Acta 1482: 327–336 [DOI] [PubMed] [Google Scholar]
- Flower DR, North AC, Sansom CE (2000) The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 1482: 9–24 [DOI] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette Charron JB, Breton G, Badawi M, Sarhan F (2002) Molecular and structural analyses of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett 517: 129–132 [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Gutiérrez G, Bastiani M, Sánchez D (2000) A phylogenetic analysis of the lipocalin protein family. Mol Biol Evol 17: 114–126 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Gutiérrez G, Ganfornina MD, Sánchez D (2000) Evolution of the lipocalin family as inferred from a protein sequence phylogeny. Biochim Biophys Acta 1482: 35–45 [DOI] [PubMed] [Google Scholar]
- Havaux M, Kloppstech K (2001) The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 213: 953–966 [DOI] [PubMed] [Google Scholar]
- Hieber AD, Bugos RC, Verhoeven AS, Yamamoto HY (2002) Overexpression of violaxanthin de-epoxidase: properties of C-terminal deletions on activity and pH-dependent lipid binding. Planta 214: 476–483 [DOI] [PubMed] [Google Scholar]
- Hieber AD, Bugos RC, Yamamoto HY (2000) Plant lipocalins: violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim Biophys Acta 1482: 84–91 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nakayashiki H, Takagi M, Tosa Y, Mayama S (2001) Heat shock, copper sulfate and oxidative stress activate the retrotransposon MAGGY resident in the plant pathogenic fungus Magnaporthe grisea. Mol Gen Genet 266: 318–325 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. CABIOS 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36: 141–154 [DOI] [PubMed] [Google Scholar]
- Kim NS, Park NI, Kim SH, Kim ST, Han SS, Kang KY (2000) Isolation of TC/AG repeat microsatellite sequences for fingerprinting rice blast fungus and their possible horizontal transfer to plant species. Mol Cells 10: 127–134 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- Ndong C, Danyluk J, Huner NPA, Sarhan F (2001) Survey of gene expression in winter rye during changes in growth temperature, irradiance or excitation pressure. Plant Mol Biol 45: 691–703 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Peitsch MC, Boguski MS (1990) Is apolipoprotein D a mammalian bilin-binding protein? New Biol 2: 197–206 [PubMed] [Google Scholar]
- Salier JP (2000) Chromosomal location, exon/intron organization and evolution of lipocalin genes. Biochim Biophys Acta 1482: 25–34 [DOI] [PubMed] [Google Scholar]
- Sánchez D, Ganfornina MD, Gutiérrez G, Marín A (2003) Exon-intron structure and evolution of the lipocalin gene family. Mol Biol Evol 20: 775–783 [DOI] [PubMed] [Google Scholar]
- Seiler H, Busse M (1990) The yeasts of cheese brines. Int J Food Microbiol 11: 289–303 [DOI] [PubMed] [Google Scholar]
- Shieh MW, Wessler SR, Raikhel NV (1993) Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol 101: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South GR, Whittick A (1987) Introduction to Phycology. Blackwell Scientific Publications, London
- Suzuki K, Lareyre JJ, Sánchez D, Gutiérrez G, Araki Y, Matusik RJ, Orgebin-Crist MC (2004) Molecular evolution of epididymal lipocalin genes localized on mouse chromosome 2. Gene 339: 49–59 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peer Y, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Vazquez-Tello A, Ouellet F, Sarhan F (1998) Low temperature-stimulated phosphorylation regulates the binding of nuclear factors to the promoter of wcs120, a wheat cold-specific gene. Mol Gen Genet 257: 157–166 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Zeigler RS, Tohme J, Nelson J, Levy M, Correa F (1994) Linking blast population analysis to resistance breeding: a proposed strategy for durable resistance. In RS Ziegler, SA Leong, PS Teng, eds, Rice Blast Disease. CAB International, Wallingford, UK, pp 267–292
- Zhou BL, Arakawa K, Fujikawa S, Yoshida S (1994) Cold-induced alterations in plasma membrane proteins that are specifically related to the development of freezing tolerance in cold-hardy winter wheat. Plant Cell Physiol 35: 175–182 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.