Abstract
Translation initiation factor 3 (eIF3) of Saccharo myces cerevisiae forms a multifactor complex (MFC) with eIFs 1, 2, 5 and Met-tRNAiMet. We previously constructed a subunit interaction model for the MFC. Here we incorporated affinity tags into the three largest eIF3 subunits (eIF3a/TIF32, eIF3b/PRT1 and eIF3c/NIP1) and deleted predicted binding domains in each tagged protein. By characterizing the mutant subcomplexes, we confirmed all key predictions of our model and uncovered new interactions of NIP1 with PRT1 and of TIF32 with eIF1. In addition to the contact between eIF2 and the N-terminal domain (NTD) of NIP1 bridged by eIF5, the C-terminal domain (CTD) of TIF32 binds eIF2 directly and is required for eIF2–eIF3 association in vivo. Overexpressing a CTD-less form of TIF32 exacerbated the initiation defect of an eIF5 mutation that weakens the NIP1–eIF5–eIF2 connection. Thus, the two independent eIF2–eIF3 contacts have additive effects on translation in vivo. Overexpressing the NIP1-NTD sequestered eIF1–eIF5–eIF2 in a defective subcomplex that derepressed GCN4 translation, providing the first in vivo evidence that association with eIF3 promotes binding of eIF2 and Met-tRNAiMet to 40S ribosomes.
Keywords: eukaryotic translation initiation factor/multifactor complex/protein synthesis/ternary complex/translational control
Introduction
Assembly of the 80S translation initiation complex is a multiple step process involving a large number of soluble eukaryotic initiation factors (eIFs). According to current models, a ternary complex (TC) comprised of eIF2, GTP and Met-tRNAiMet binds to the 40S ribosome with the help of eIFs 1, 1A and 3. The 43S pre-initiation complex thus formed interacts with mRNA in a manner stimulated by eIF4F, poly(A)-binding protein and eIF3, and the resulting 48S complex scans the mRNA until the Met-tRNAiMet base-pairs with the AUG start codon. On AUG recognition, the eIF5 stimulates GTP hydrolysis by eIF2, the eIFs are ejected and the 60S subunit finally joins with the 40S–Met-tRNAiMet–mRNA complex. For a new round of initiation, the ejected eIF2–GDP complex must be recycled to eIF2–GTP by the exchange factor eIF2B (reviewed in Hershey and Merrick, 2000; Hinnebusch, 2000).
The eIF3 is intriguing because it can bind directly to the 40S ribosome and stimulates the binding of both TC and mRNA to 40S subunits in vitro (Hershey and Merrick, 2000; Hinnebusch, 2000). While mammalian eIF3 contains 11 different subunits, the yeast factor has only five core subunits (eIF3a/TIF32, eIF3b/PRT1, eIF3c/NIP1, eIF3i/TIF34 and eIF3g/TIF35) and one substoichiometric component (eIF3j/HCR1). The five-subunit complex purified from yeast can restore binding of Met-tRNAiMet (Danaie et al., 1995; Phan et al., 1998) and mRNA (Phan et al., 2001) to 40S ribosomes in heat-inactivated prt1-1 (eIF3b) mutant extracts. Thus, yeast eIF3 possesses two critical functions ascribed to the more complex mammalian factor. The role of mammalian eIF3 in promoting mRNA binding to the ribosome often is attributed to its interaction with the largest subunit of the cap-binding complex eIF4F (Hershey and Merrick, 2000); however, this interaction has not been shown for the same factors in yeast. The ability of eIF3 to promote TC binding to the 40S ribosome is not understood at the molecular level in either system.
Interactions among the yeast eIF3 subunits have been studied extensively by yeast two-hybrid analysis and in vitro binding assays (Asano et al., 1998; Phan et al., 1998; Valášek et al., 2001). The results of these studies (summarized in Figure 1) suggest that TIF34 and TIF35 bind to the PRT1 C-terminal domain (CTD), while TIF32 binds to the RNA recognition motif (RRM) in the PRT1 N-terminal domain (NTD). TIF32 binds to PRT1 through an internal domain related in sequence to eIF3j/HCR1, and HCR1 binds to both the RRM in PRT1 and TIF32. NIP1 interacts with the N-terminal half of TIF32 but not with PRT1 (Figure 1). In accordance with this model, PRT1 can form distinct subcomplexes in vivo, one containing PRT1, TIF32 and NIP1, and the other comprised of PRT1, TIF34 and TIF35. While the PRT1–TIF32–NIP1 subcomplex could restore 40S binding of Met-tRNAiMet and mRNA in the prt1-1 extract, the PRT1–TIF34–TIF35 subcomplex was relatively inert (Phan et al., 2001). Consistently, expression of an N-terminally truncated form of PRT1 sequestered TIF34 and TIF35 in an inactive subcomplex that could not associate with ribosomes, and had a dominant-negative effect on cell growth (Evans et al., 1995; Valášek et al., 2001).
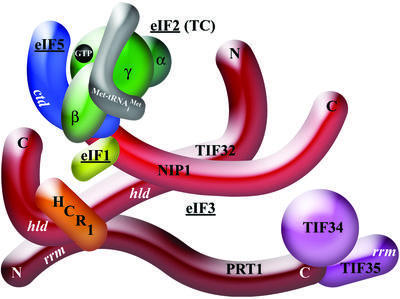
Fig. 1. A three-dimensional model of the MFC based largely on binary interactions between isolated subunits. The labeled protein subunits are shown roughly in proportion to their molecular weights. ctd, C-terminal domain; hld, HCR1-like domain; rrm, RNA recognition motif. See text for further details.
eIF3 is physically associated with other eIFs in yeast. It co-purified with eIF1 (Naranda et al., 1996; Phan et al., 1998) and contained nearly stoichiometric amounts of eIF5 when purified by affinity chromatography (Phan et al., 1998). In vitro, eIF1 and the eIF5-CTD can bind simultaneously to the NIP1-NTD (Phan et al., 1998; Asano et al., 1999, 2000). Consistently, eIF1 and eIF5 co-purified with the PRT1–TIF32–NIP1 subcomplex, but not with the PRT1–TIF34–TIF35 subcomplex (Phan et al., 2001; Figure 1). Both eIF5 and eIF1 were implicated in selecting AUG as the start codon (Huang et al., 1997), and their related functions in scanning may be coordinated by mutual association with NIP1 (Asano et al., 2000). Interestingly, the eIF5-CTD can interact simultaneously with NIP1 and the β-subunit of eIF2 in vitro (Asano et al., 1999, 2000), suggesting that the eIF5-CTD bridges an interaction between eIFs 2 and 3 (Figure 1).
Consistent with the model shown in Figure 1, we showed previously that a multifactor complex (MFC) containing eIFs 1, 2, 3 and 5 and Met-tRNAiMet could be affinity purified from yeast and can exist free of ribosomes in vivo. A cluster of alanine substitutions in the eIF5-CTD (tif5-7A) that disrupted eIF5 interactions with NIP1 and eIF2β in vitro also destabilized the MFC in vivo. The tif5-7A mutant has a temperature-sensitive phenotype (Ts–) and a diminished rate of translation initiation that is partially suppressed by overexpressing the TC, which restores high levels of the MFC in the mutant cells. Hence, we proposed that the MFC is an important initiation intermediate in vivo (Asano et al., 1999, 2000).
The presence of eIF2 and eIF3 in the MFC could provide an explanation for the ability of eIF3 to stimulate TC binding to 40S ribosomes in yeast extracts (Hinnebusch, 2000), with eIF5 serving as an adaptor between eIF3 bound to the 40S subunit and the TC. Consistent with this idea, the tif5-7A mutation reduced binding of TC to 40S subunits in the mutant extract in a manner that was rescued by wild-type eIF5. In tif5-7A mutant cells, however, we observed an accumulation of 48S complexes containing eIFs 1, 2 and 3 but lacking eIF5. Hence, we concluded that stable association of eIF5 with the MFC is more critically required in vivo for a step subsequent to TC binding, either scanning, AUG recognition or GTP hydrolysis by eIF2 (Asano et al., 2001).
Since the adaptor function of the eIF5-CTD is not essential for TC binding in vivo, we sought to identify other contacts between eIF3 and eIF2 that could facilitate 43S complex formation. It was also important to test the in vivo relevance of our subunit interaction model shown in Figure 1. To accomplish these goals in the current study, we made deletions of predicted binding domains for various MFC components in affinity-tagged forms of the three largest eIF3 subunits, and determined the compositions of the affinity-purified complexes. The results confirmed the subunit interactions depicted in Figure 1 and revealed new contacts between NIP1 and PRT1 and between TIF32 and eIF1. We also uncovered the first direct contact between eIF2 and eIF3 and obtained genetic evidence of its importance for translation initiation in vivo. Finally, we showed that sequestering eIF2 in a subcomplex with the NIP1-NTD, eIF5 and eIF1 derepresses GCN4 translation. This last result provides the first in vivo evidence that eIF3 promotes TC binding to the 40S ribosome.
Results
Mapping interactions between TIF32 and NIP1 in vitro
In previous in vitro binding experiments (GST pull-downs), we mapped the binding domains in PRT1, TIF34 and TIF35 for the other eIF3 subunits, but had not completed a similar analysis for TIF32 and NIP1 (Asano et al., 1998; Valášek et al., 2001). Using the same in vitro approach, we determined that residues 200–600 in TIF32 are necessary and sufficient for interaction with GST– NIP1 in vitro and that the principal TIF32-binding domain in NIP1 lies just C-terminal to that for eIFs 1 and 5, between residues 157 and 370. A detailed description of these results is provided in the Supplementary data available at The EMBO Journal Online. The new findings on TIF32–NIP1 interactions have been incorporated into the model shown in Figure 1.
In vivo confirmation that TIF32 and eIFs 1 and 5 bind to the N-terminal half of NIP1 and evidence for a novel NIP1–PRT1 contact
We showed previously that a His8-tagged form of PRT1 (PRT1-His) expressed in wild-type cells competes with endogenous PRT1 for association with the MFC components, all of which co-purified with PRT1-His on nickel affinity resin. Deleting the RRM domain of PRT1-His eliminated all MFC components except TIF34 and TIF35 from the purified preparation, providing in vivo evidence that the RRM is required to tether the PRT1–TIF34–TIF35 subcomplex to the rest of the MFC (Valášek et al., 2001; Figure 1; Table I). Here we employed a similar approach to test the prediction that the NIP1-NTD is required to tether eIFs 1, 5 and 2 to eIF3, but is dispensable for eIF3 integrity. We inserted a His8 tag at the C-terminus of NIP1 and introduced the tagged allele (NIP1-His) into a wild-type strain. NIP1-His was purified from the whole-cell extract (WCE) on nickel–agarose resin and the co-purifying proteins detected by western blotting. Probing the eluate with antibodies against the His8 tag showed that full-length NIP1-His was purified quantitatively from the WCE. As expected, the eluate also contained the core eIF3 subunits, HCR1, and eIFs 1, 5 and 2, indicating that a proportion of the entire MFC had co-purified with NIP1-His (Figure 2C, lanes 3 and 4). Importantly, little or none of these proteins were purified from the parental strain containing only untagged NIP1 (Figure 2C, lanes 1 and 2). The purified complex was free of ribosomes, as no 40S ribosomal protein S22 was detected in the eluate from the NIP1-His strain (lanes 3 and 4).
Table I. Phenotypes of mutations analyzed in this studya.
| Alleleb | Mutationc | Binding domain affectedd | Co-purifying factorse | Complementing functionf | Dom. Slg–g/suppressed byh | Dom. Gcd–i/suppressed by hc eIF2-IMT4j |
|---|---|---|---|---|---|---|
| TIF32 mutations | ||||||
| lc empty vector | NA | NA | None | – | – | – |
| hc TIF32-His | None | None | All | + | – | – |
| lc TIF32-Δ8-His | [Δ1–199] | None | All | Partial | – | ND |
| hc TIF32-Δ8-His | [Δ1–199] | None | ND | Partial | + | – |
| lc TIF32-Δ86-His | [Δ1–199; 791–964] | PRT1k, HCR1, eIF2 | All | – | – | ND |
| hc TIF32-Δ86-His | [Δ1–199; 791–964] | PRT1k, HCR1, eIF2 | ND | – | – | – |
| hc TIF32-Δ4-His | [Δ1–790] | PRT1k, NIP1, eIF1 | eIF2 | – | – | – |
| lc TIF32-Δ5-His | [Δ642–964] | PRT1, HCR1, eIFs 1, 2 | NIP1, eIF5 | – | – | ND |
| hc TIF32-Δ5- His | [Δ642–964] | PRT1, HCR1, eIFs 1, 2 | ND | – | +/hc NIP1 | – |
| +/hc eIF2 + IMT4 (partial) | ||||||
| lc TIF32-Δ6-His | [Δ791–964] | PRT1k, HCR1, eIF2 | NIP1, PRT1, TIF34, TIF35, eIFs 1, 5 | – | – | ND |
| hc TIF32-Δ6-His | [Δ791–964] | PRT1k, HCR1, eIF2 | ND | – | +/hc eIF2 + IMT4 | – |
| |
||||||
| NIP1 mutations | ||||||
| sc empty vector | NA | NA | None | – | – | – |
| hc NIP1-His | None | None | All | + | – | –/ND |
| sc NIP1-N′-His | [Δ206–812] | TIF32, PRT1 | eIFs 1, 2, 5 | – | – | – |
| hc NIP1-N′-His | [Δ206–812] | TIF32, PRT1 | eIFs 1, 2, 5 | – | – | ++/Yes |
| sc NIP1-C-His | [Δ1–156] | eIF1, eIF5 | PRT1, TIFs 32, 34, 35 | – | – | – |
| hc NIP1-C-His | [Δ1–156] | eIF1, eIF5 | PRT1, TIFs 32, 34, 35 | – | – | – |
| hc NIP1-ΔJ-His | [Δ157–371] | TIF32 | eIFs 1, 2, 5 | – | – | – |
| hc NIP1-ΔA-His | [Δ371–812] | PRT1 | TIF32, eIFs 1, 2, 5 | – | – | – |
| hc NIP1-ΔB′-His | [Δ571–812] | PRT1 | All | – | – | – |
| |
||||||
| PRT1 mutations | ||||||
| sc empty vector | NA | NA | None | – | – | – |
| hc PRT1-His | None | None | All | + | – | – |
| hc PRT1-Δ5′-His | [Δ551–724] | NIP1, TIFs 34, 35 | TIF32 | – | – | – |
| sc PRT1-Δ7′-His | [Δ641–724] | TIFs 34, 35 | TIF32, NIP1, eIF5 | – | – | ND |
| hc PRT1-Δ7′-His | [Δ641–724] | TIFs 34, 35 | ND | – | +/hc NIP1 + TIF32 | – |
| sc PRT1-ΔRRM-Hisl | [Δ35–136] | TIF32, HCR1 | TIFs 34, 35 | – | +/hc TIF34 + TIF35 | ND |
aThe LEU2 or URA3 plasmids containing TIF32-His, NIP1-His, PRT1-His, or their mutant derivatives, were introduced into strains YLV314U [MATa ura3::URA3::rpg1-1 trp1-1::TRP1::rpg1-Δ2 ade2-1 can1-100 leu2-3 112 his3-11,15], B8302 [MATa cyc1-NLS cyc7-67 ura3-52 lys5-10 nip1-1] or H1676 [MATa prt1-1 leu2-3 leu2-112 ura3-52], respectively, to analyze complementation of the Ts– phenotypes of these strains. All of the plasmids were also introduced into W303 [MATa ade2-1 trp1-1 can1-100 leu2-3 112 his3-11,15 ura3] and H2881 [MATa trp1-1 leu2-3 112 ura3-52 gcn2Δ] to test for dominant slow growth (Slg–) or Gcd– phenotypes, respectively. NA, not applicable; ND, not determined.
bSingle-copy (sc) vectors YCplac33 for NIP1 and YCplac11 for PRT1; low-copy (lc) vector pRS315 for TIF32; high-copy (hc) vector YEplac181 for TIF32, NIP1 and PRT1.
cNumbers in parentheses indicate the amino acids of the particular protein that were deleted.
dComplete or partial deletion of the predicted binding domain(s) for the listed protein(s) in the His8-tagged protein under study.
eList of protein(s) that co-purified with the His8-tagged protein under study in Ni2+ chelation chromatography, summarized from Figures 2–4.
fComplementation of the growth defect at 37°C in transformants of the rpg1-1, nip1-1 or prt1-1 Ts– strains YLV314U, B8302 and H1676, respectively, after 3 days of incubation; –, no complementation; +, complementation similar to the wild-type allele.
gDom. Slg–, dominant slow growth phenotype at 30°C in the presence of a wild-type chromosomal copy of the corresponding gene in W303; –, no dominant Slg– phenotype; +, dominant Slg– phenotype evident.
hPlasmids YEpNIP1T carrying NIP1; p1780-IMT containing SUI2, SUI3, SUI4 and IMT4 (encoding the three subunits of eIF2 and tRNAiMet, respectively), and pLPY-NIP1-TIF32 bearing NIP1 and TIF32 were used to complement the dominant Slg– phenotype conferred by the relevant mutant alleles in W303 at 30°C.
iGcd– phenotypes were recognized by suppression of the 3-AT-sensitive phenotype of the gcn2Δ allele in transformants of H2881 evaluated on SC-Leu-His medium containing 10 mM 3-AT for the TIF32-His and PRT1-His alleles and on SC-Leu-His with 20 mM 3-AT for the NIP1-His alleles. Growth of isogenic GCN2 strain H2880 [MATa trp1-1 leu2-3,112 ura3-52] and the gcn2Δ strain H2881 transformed with an empty vector on these media were scored as +++++ and –, respectively.
jStrains containing particular NIP1-His alleles were transformed with p1780-IMT and tested for suppression of the Gcd– phenotype under the same conditions as in i, except that SC-Leu-Ura-His with 20 mM 3-AT was used; Yes, complete suppression; No, no suppression.
kOnly one of the two PRT1-binding domains in TIF32 affected.
lDetermined in Valášek et al. (2001).
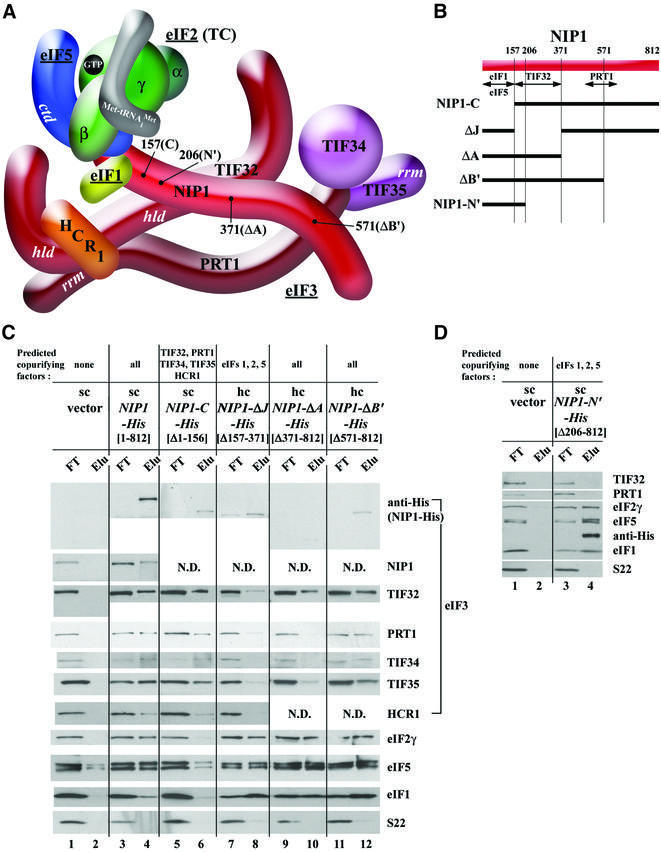
Fig. 2. Analysis of purified subcomplexes formed in vivo by truncated forms of NIP1-His. (A) Three-dimensional model of the MFC modified from Figure 1 to incorporate the results of in vitro binding assays in Supplementary figure S1 and data from (C) and (D) regarding the deduced interaction between C-terminal segments of NIP1 and PRT1. The boundaries of the relevant truncations [summarized in (B)] are indicated at the appropriate positions in NIP1. (B) Schematic of NIP1 with arrows delimiting the minimal binding domains for the indicated proteins identified by in vitro binding assays (summarized in Figure 1) or deduced from results shown below. The lines beneath the schematic depict various truncated NIP1-His proteins (designated on the left) that were expressed in yeast and purified by Ni2+ chelation chromatography. (C) WCEs prepared from transformants of yeast strain W303 bearing empty vector (lanes 1 and 2), single copy (sc) plasmid YCpNIP1-His (lanes 3 and 4), sc YCpNIP1-C-His (lanes 5 and 6), high copy (hc) YEpNIP1-ΔJ-His (lanes 7 and 8), hc YEpNIP1-A-His (lanes 9 and 10) and hc YEpNIP1-ΔB′-His (lanes 11 and 12) were incubated overnight with Ni2+-NTA–silica resin and the bound proteins were eluted and subjected to western blot analysis. Odd-numbered lanes each contained 3% of the flow-through fractions (FT); even-numbered lanes contained 33% of the total volume of the corresponding eluted fractions (Elu). For each construct, the encoded NIP1 amino acids are indicated in brackets beneath the construct name. The proteins predicted to co-purify with each NIP1-His protein, based on the model shown in (A), are listed at the very top of the panel. (D) The same as (C) except that sc YCpNIP1-N′-His was used for lanes 3 and 4.
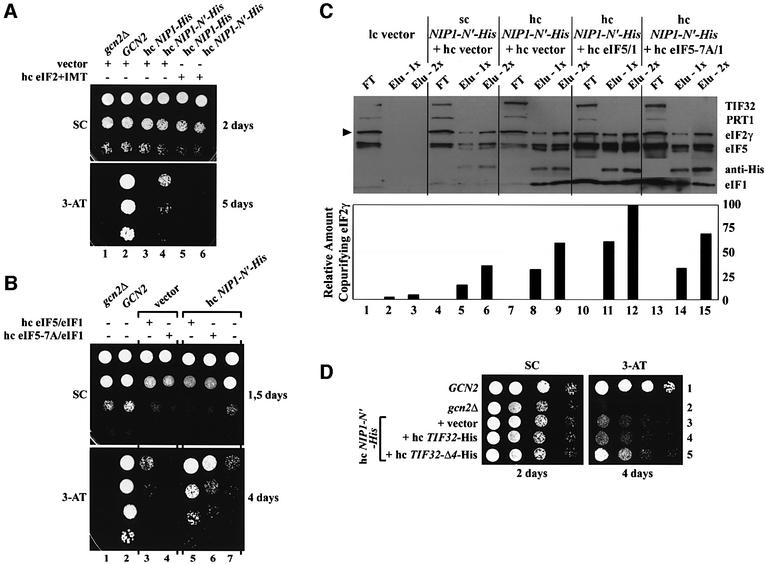
We then conducted the same experiment using a His8-tagged NIP1 allele lacking the N-terminal 156 residues (NIP1-C-His) lacking the predicted binding domain for eIFs 1 and 5 (Figure 2A and B). The eIF3 subunits co-purified with NIP1-C-His to nearly the same extent observed for full-length NIP1-His, whereas eIFs 1, 2 and 5 were recovered at the low background levels observed with untagged NIP1 (Figure 2C, lanes 5 and 6 versus 1–4). Very different results were obtained for the His8-tagged protein encoded by NIP1-ΔJ-His, which lacks the internal residues 157–371 encompassing the predicted TIF32-binding domain (Figure 2A and B). This protein failed to co-purify with other core eIF3 subunits but retained strong association with eIFs 1, 5 and 2 (Figure 2C, lanes 7 and 8). These and all subsequent affinity purifications of mutant proteins were carried out repeatedly with very reproducible results. These data confirm our prediction that the N-terminus of NIP1 is required for eIF3 association with eIFs 1, 5 and 2, but is dispensable for integrity of the eIF3 complex in vivo. They also show that the adjacent domain in NIP1 is required for incorporation into the eIF3 complex, but is dispensable for its association with eIFs 1, 5 and 2. We extended the last conclusion by showing that a His8-tagged segment containing only the N-terminal 205 residues of NIP1 (encoded by NIP1-N′-His; Figure 2A and B) co-purified with eIFs 1, 2 and 5, but not with the eIF3 subunits (Figure 2D).
According to our model, truncating NIP1-His after residue 370 should not affect the composition of the MFC, because the truncated protein would retain the binding sites for eIFs 1, 5 and 2 in the NIP1-NTD and the newly identified binding site for TIF32 between residues 157 and 370 (see Supplementary figure S1; Figure 2B). Con sistently, the NIP1-His protein lacking residues 371–812 (NIP1-ΔA-His; Figure 2A and B) co-purified with eIFs 1, 5 and 2, and also TIF32, but showed greatly diminished association with PRT1, TIF34 and TIF35 (Figure 2C, lanes 9 and 10 versus 3 and 4). This last result suggests that deletion of NIP1 residues 371–812 eliminated PRT1, which in turn removed TIF34 and TIF35, from the MFC. The loss of PRT1–TIF34–TIF35 was much less pronounced for a smaller truncation (NIP1-ΔB′-His; Figure 2B), which removed all residues C-terminal to position 570 (Figure 2C, lanes 11 and 12). Together, these results suggest that residues 371–570 of NIP1 contain a binding site for PRT1 that is required for tight association of the PRT1–TIF34–TIF35 subcomplex with the MFC (Figure 2A and B).
In an effort to confirm this last conclusion, we tested a GST fusion containing the C-terminal portion of NIP1 (residues 157–812, GST–NIP1-C) for in vitro interaction with recombinant 35S-labeled fragments of PRT1. The C-terminal PRT1 fragments Δ3 and Δ5 containing the predicted NIP1-binding domain (Figure 3B), bound to GST–NIP1-C at higher levels than to GST alone (Figure 3D, lane 3 versus 2). Although the interaction was weak, it appeared to be specific because a shorter [35S]PRT1 fragment (Δ7) did not bind to GST–NIP1-C, even though the latter bound strongly to GST–TIF34 and GST–TIF35 in previous experiments (Valášek et al., 2001). Only weak binding was detected between GST–NIP1-C and a larger segment of PRT1 (Δ2; Figure 3B and D), and previously no binding was demonstrated between full-length NIP1 and PRT1 (Asano et al., 1998). To account for these last results, we suggest that the NIP1-binding domain in [35S]PRT1-Δ2 is obscured by a non-physiological interaction with the N-terminal portion of PRT1, which in native eIF3 would interact with TIF32. Additional evidence for an interaction between the C-terminus of both NIP1 and PRT1 is presented below.
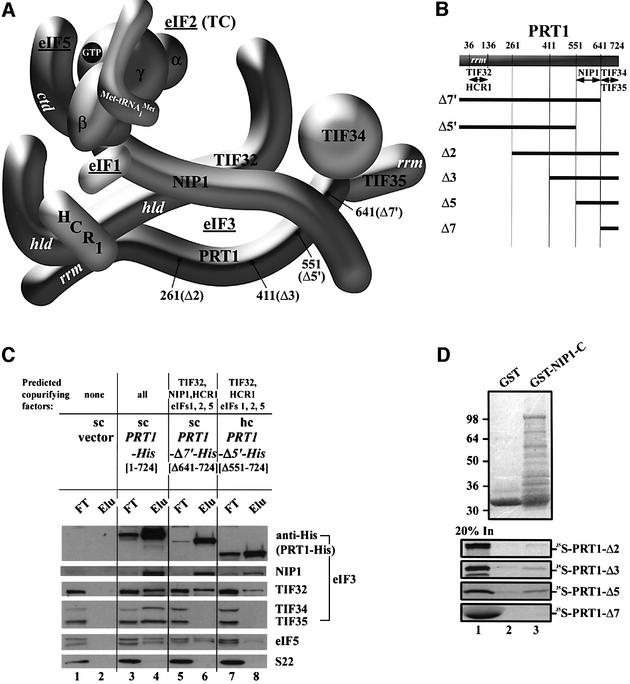
Fig. 3. Analysis of purified subcomplexes formed in vivo by truncated forms of PRT1-His. (A and B) Same as Figure 2A and B except that truncations in PRT1-His are under study; (B) also includes schematics of [35S]PRT1 segments used for the in vitro binding assays shown in (D). (C) Nickel chelation purification of protein complexes containing His-tagged PRT1 proteins conducted as described in Figure 2C, except that WCEs were prepared from W303 transformants bearing empty vector (lanes 1 and 2), sc YCpPRT1-His (lanes 3 and 4), sc YCpPRT1-Δ7′-His (lanes 5 and 6) and hc YEpPRT1-Δ5′-His (lanes 7 and 8). (D) The [35S]PRT1 fragments designated in (B) were synthesized in vitro and labeled with [35S]methionine (lane 1) then used in pull-down assays with the GST fusion containing the NIP1-C fragment described in Figure 2B (lane 3) or with GST alone (lane 2). The bound proteins were detected by autoradiography (lower panel) or Coomassie Blue staining (upper panel).
In vivo evidence that the PRT1-CTD has independent binding sites for NIP1 and TIF34/TIF35
To demonstrate that the extreme C-terminus of PRT1 is required for association of TIF34 and TIF35 in vivo, we purified His-tagged PRT1 proteins with C-terminal truncations. Deletion of the last 83 residues from the tagged PRT1 protein (PRT1-Δ7′-His; Figure 3A and B) eliminated TIF34 and TIF35 without affecting the levels of TIF32 and NIP1 in the purified complex (Figure 3C, lanes 5 and 6). Thus, the extreme C-terminus of PRT1 is required only for association of TIF34 and TIF35 with eIF3 in vivo. A more extensive truncation after residue 551 was made that eliminates the NIP1-binding domain predicted from the in vitro binding data discussed above (PRT1-Δ5′-His; Figure 3A and B). The PRT1-Δ5′-His protein showed diminished association with TIF32, NIP1 and eIF5 compared with that seen for the longer PRT1-Δ7′-His protein (Figure 3C, lanes 7 and 8 versus 5 and 6). These findings support the idea that a binding site for NIP1 is located just upstream from the TIF34–TIF35-binding site at the C-terminus of PRT1.
Evidence that the TIF32-CTD directly interacts with eIF2 in addition to the PRT1-NTD and HCR1
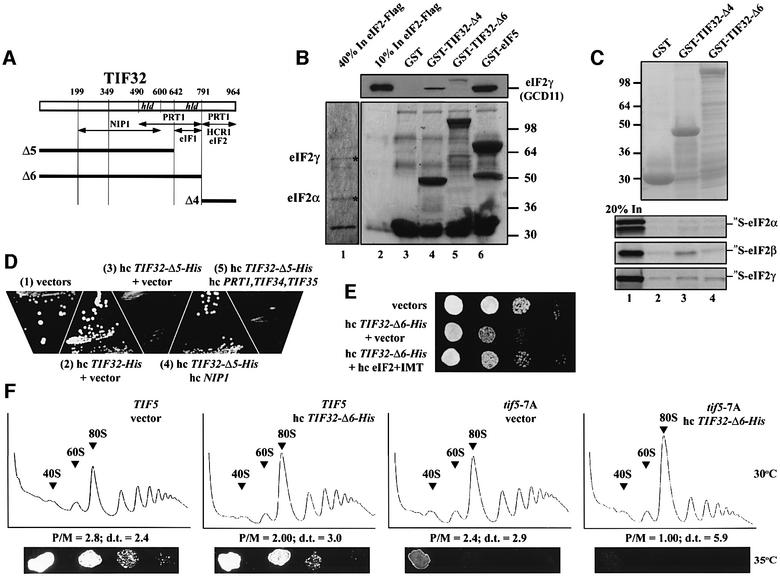
We proceeded next to identify the domains in TIF32 that mediate its interactions with other eIF3 subunits in vivo by expressing full-length or truncated versions of His-tagged TIF32. The C-terminal half of TIF32 can be divided into two domains, the HCR1-like domain (HLD) and the extreme C-terminus (Figure 4A and B), and both were shown to interact specifically with PRT1 and HCR1 in vitro (Valášek et al., 2001). To test the validity of these results, we deleted the C-terminal 174 residues of TIF32-His (TIF32-Δ6-His), removing one of two PRT1-binding domains and the HCR1-binding domain (Figure 4A and B). As predicted, this deletion reduced, but did not abolish the association of PRT1, TIF34 and TIF35 with the tagged protein, and completely eliminated interaction of HCR1 with TIF32-His (Figure 4C, lanes 7 and 8 versus 3 and 4). Unexpectedly, this deletion nearly eliminated association of eIF2 with the MFC, without substantially affecting the content of NIP1, eIF5 or eIF1 in the preparation. We also analyzed a more extensive C-terminal truncation after residue 642 (TIF32-Δ5-His) that encroached on the HLD domain and removed part of the second PRT1-binding domain (Figure 4A and B). This deletion led to complete loss of PRT1, TIF34, TIF35 and HCR1 from the purified complex (Figure 4C, lanes 5 and 6) without any reduction in the NIP1 or eIF5 content of the preparation. As observed for the smaller C-terminal truncation (Δ6), eIF2 was nearly absent from the TIF32-Δ5-His preparation; in addition, eIF1 was also missing. These results demonstrate that the extreme C-terminus and adjacent HLD in TIF32 both contribute to the interactions with PRT1 that tether the PRT1–TIF34–TIF35 subcomplex to TIF32 and the rest of the MFC. However, they also revealed an unexpected requirement for the extreme C-terminus of TIF32 for stable binding of eIF2 to the MFC that is independent of the bridging function of eIF5. Moreover, the loss of eIF1 that occurred without a reduction in NIP1 association with TIF32-Δ5-His, coupled with the wild-type level of eIF1 associated with TIF32-Δ6-His, implies that TIF32 residues 642–791 contribute to eIF1 binding to the MFC. This conclusion was supported further by our finding that TIF32 residues 491–791 fused to GST specifically interacted with eIF1 in an in vitro binding assay. A detailed description of this last result is provided in the Supplementary data.
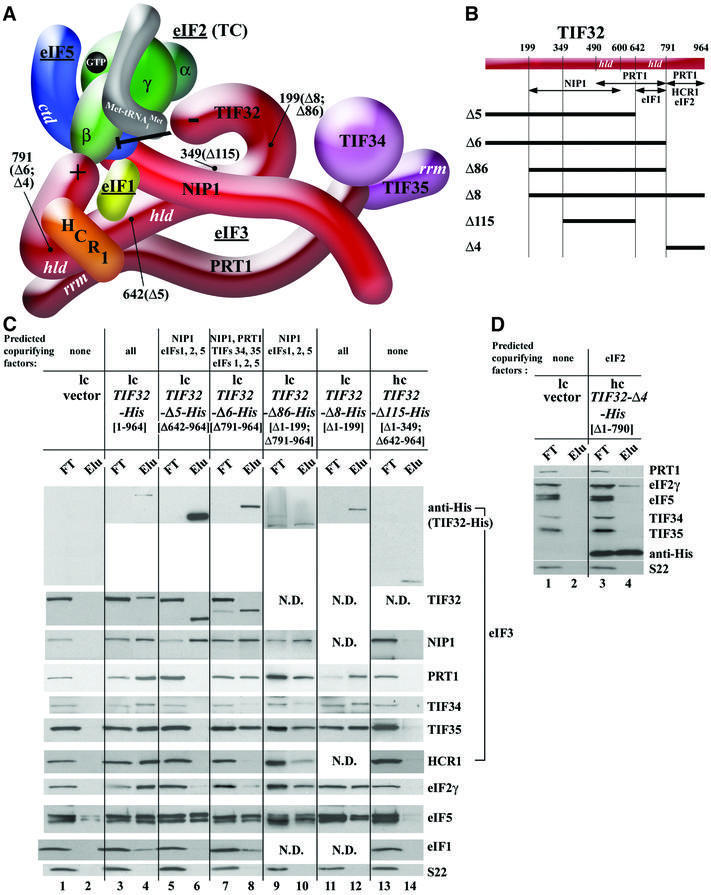
Fig. 4. Analysis of purified subcomplexes formed in vivo by truncated forms of TIF32-His. (A and B) Same as Figure 2A and B except that truncations in TIF32-His are under study. The model in (A) was modified to incorporate novel interactions of the TIF32-CTD with eIFs 1 and 2, and the proposed inhibitory effect of the TIF32-NTD on the interaction of NIP1-NTD–eIF5 with eIF2, deduced from results described in (C) and (D) and Figure 5. (C) Nickel chelation purification of protein complexes containing His-tagged TIF32 proteins conducted as described in Figure 2C, except that WCEs were prepared from W303 transformants bearing empty vector (lanes 1 and 2), low copy (lc) plasmid pRSTIF32-His (lanes 3 and 4), lc pRSTIF32-Δ5-His (lanes 5 and 6), lc pRSTIF32-Δ6-His (lanes 7 and 8), lc pRSTIF32-Δ86-His (lanes 9 and 10), lc pRSTIF32-Δ8-His (lanes 11 and 12) and hc YEpTIF32-Δ115-His (lanes 13 and 14). (D) The same as (C) except that hc YEpTIF32-Δ4-His was used for lanes 3 and 4.
To determine whether the isolated TIF32-CTD can interact with eIF2 in vivo, we expressed in yeast cells the C-terminal 174 residues of the protein tagged with His8 (TIF32-Δ4-His; Figure 4A and B). As shown in Figure 4D, a small fraction of eIF2, but no detectable eIF5 or eIF3 subunits, co-purified with TIF32-Δ4-His. We also tested the TIF32-CTD for binding to purified eIF2 in vitro. The eIF2 bearing a FLAG epitope tag on the β-subunit was purified from yeast (Figure 5B, lane 1) and incubated with GST fusions containing different segments of TIF32, GST–eIF5 or GST alone. Consistent with previous results (Asano et al., 1999), eIF2-FLAG interacted strongly with GST–eIF5 (Figure 5B, lane 6). Importantly, eIF2-FLAG bound to the C-terminus of TIF32 in construct GST– TIF32-Δ4 (Figure 5B, lane 4) but not to the GST–TIF32 fusion lacking only the CTD (Δ6, lane 5). Thus, the TIF32-CTD can interact directly with eIF2 in vivo and in vitro. To investigate which subunit of eIF2 interacts with the TIF32-CTD, all three subunits were synthesized in vitro and tested for binding to GST fusions containing the TIF32-CTD. As shown in Figure 5C, only the β-subunit interacted specifically with GST–TIF32-Δ4 (lane 3 versus 2 and 4), as observed for the eIF2 complex. Below, we present genetic data supporting the importance of the TIF32-CTD interaction with eIF2 in vivo.
Fig. 5. The TIF32-CTD interacts with eIF2 in vitro and this contact is important for efficient translation initiation in vivo. (A) The same as in Figure 4B. (B) Flag-tagged eIF2 was purified from WCEs using anti-FLAG affinity resin and used in pull-down assays with GST alone (lane 3) or GST fusions to TIF32-Δ4, TIF32-Δ6 or TIF5 (eIF5) (lanes 4–6). Binding of eIF2 was detected by western blotting with antibodies against eIF2γ (GCD11). Lane 2 contained 10% of the eIF2 added to each reaction. (C) Binding of 35S-labeled eIF2 subunits to GST–TIF32-Δ4, GST–TIF32-Δ6 (lanes 3 and 4) or GST alone (lane 2), as described in Figure 3D. (D) Transformants of strain W303 containing empty vector (1), hc YEpTIF32-His (2) or hc YEpTIF32-Δ5-His-U (3–5) were co-transformed with empty vector (1–3), hc YEpNIP1-His (4) and hc pLPY-PRT1-His-TIF34-HA-TIF35-Flag (5). The resulting strains were streaked on SD medium and incubated at 30°C for 2 days. (E) The dominant-negative Slg– phenotype of hc TIF32-Δ6-His is suppressed by high-copy eIF2 and tRNAiMet. Transformants of strain W303 containing empty vectors; hc YEpTIF32-Δ6-His and empty vector (YEp24); and hc YEpTIF32-Δ6-His and p1780-IMT were spotted in four serial dilutions on SD medium and incubated at 30°C for 2 days. (F) Overexpressing the CTD-less TIF32-Δ6-His protein severely exacerbates the translation initiation defect in tif5-7A Ts– cells. Upper panels: strains H2898 (TIF5) and H2899 (tif5-7A) were transformed with empty vector or hc YEpTIF32-Δ6-His-U, grown in SD medium to an OD600 of ∼1.5, and 50 µg/ml cycloheximide was added 5 min prior to harvesting. WCEs were prepared and resolved on 5–45% sucrose gradients. The positions of 40S and 60S subunits and 80S ribosomes are indicated. P/M, ratio of the A254 in the combined polysome fractions to that in the 80S peak; d.t., cell doubling time in hours. Bottom panels: the latter strains were spotted in four serial dilutions on SD medium and incubated at 35°C for 2 days.
Evidence that the N- and C-termini of TIF32 have opposing effects on interactions of TIF32 and NIP1 with eIF2
We wished to provide in vivo evidence that the region immediately N-terminal to the HLD in TIF32 contains the binding site for NIP1, as predicted by our model (Figure 4A and B). Towards this end, we created internal deletions removing part or all of the predicted NIP1-binding site between residues 200 and 600 in TIF32-His; however, all of these proteins were very unstable. Accordingly, we determined the effects of truncating the protein after residue 350, removing ∼40% of the predicted NIP1-binding domain, in the context of TIF32-Δ5-His, to produce TIF32-Δ115-His (Figure 4A and B). Whereas TIF32-Δ5-His co-purified with NIP1 and eIF5, the TIF32-Δ115-His protein did not (Figure 4C, lanes 5 and 6 versus 13 and 14). Note that the Δ115 protein was detected in the eluate at levels comparable with that of full-length TIF32-His (Figure 4C, lanes 4 and 14). Thus, residues N-terminal to residue 350 in TIF32 are required for interaction with NIP1 and, by extension, with eIF5.
We also constructed a smaller truncation of the TIF32-NTD in the context of TIF32-Δ6-His that leaves the predicted NIP1-binding domain intact (TIF32-Δ86-His; Figure 4A and B). Unexpectedly, this protein co-purified with all MFC components, including eIF2 (Figure 4C, lanes 9 and 10). This result was surprising because eIF2 did not co-purify with the parental construct TIF32-Δ6-His (Figure 4C, lanes 7 and 8). Hence, removing the NTD of TIF32 partially restored association of eIF2 with the rest of the mutant MFC, which lacks the eIF2-binding site in the TIF32-CTD. Removal of the TIF32-NTD from full-length TIF32 (TIF32-Δ8-His; Figure 4A and B) had no effect on MFC composition (Figure 4C, lanes 11 and 12 versus 3 and 4). Thus, the TIF32-NTD seems to inhibit association of eIF2 with the MFC, imposing a requirement for the TIF32-CTD–eIF2 interaction for retention of eIF2 in the MFC. In the absence of the inhibitory TIF32-NTD, the NIP1-NTD is sufficient for a strong interaction with the eIF5–eIF1–eIF2 subcomplex (Figure 2D).
Effects of truncating NIP1, TIF32 or PRT1 on eIF3 function in vivo
Each of the alleles encoding a truncated eIF3 subunit was tested for its ability to complement the Ts– phenotypes of tif32, nip1 or prt1 mutants. Except for TIF32-Δ8-His, none did so, whether present on high-copy (hc) or low-copy (lc) plasmids (summarized in Table I). TIF32-Δ8-His in lc or hc partially complemented the rpg1-1 Ts– allele of TIF32. The lack of complementing function for certain alleles possibly could arise from their reduced expression. Indeed, western blot analysis with anti-His6 antibodies suggested that NIP1-His mutant proteins were expressed at lower levels than full-length NIP1-His (Figure 2C). However, these last results may not reflect the true levels of the tagged proteins, as suggested by the disparate western signals obtained using anti-His6 versus anti-TIF32 antibodies for TIF32-His, TIF32-Δ5-His and TIF32-Δ6-His (Figure 4C, lanes 4, 6 and 8). If we compare the amounts of MFC components that co-purified with each mutant protein with those obtained for the corresponding full-length proteins instead, it appeared that most of mutant proteins were expressed at levels comparable with that of the full-length versions (Figures 2–4). Moreover, many alleles were expressed from lc plasmids in the analysis of Figures 2–4 (NIP1-C-His, NIP1-N′-His, PRT1-Δ7′-His, TIF32-Δ5, -Δ6, -Δ86 and -Δ8-His) but had no complementing function even when expressed from hc plasmids (Table I). For these latter alleles, it seems clear that the deletions impair the essential functions of the corresponding eIF3 subunits.
It was not surprising that deletions which disrupted the core eIF3 complex lacked complementing function in vivo, including ΔJ and ΔA in NIP1-His, Δ7′ and Δ5′ in PRT1-His, and Δ5 and Δ115 in TIF32-His (Table I). The deletions of the NTD of NIP1-His (NIP1-C-His) and the extreme C-terminus of TIF32 (TIF32-Δ6-His) left the core eIF3 complex intact and only severed the interactions between eIF3 and other eIFs in the MFC. The fact that these two well-expressed mutant proteins had no complementing function (Table I) supports the notion that interactions between eIF3 and eIFs 1, 2 and 5 mediated by the NIP1-NTD, and between eIF2 and the TIF32-CTD, are crucial for translation initiation in vivo. Several deletions did not impair formation of the MFC complex detectably, including NIP1-ΔB′, TIF32-Δ8 and TIF32-Δ86. It remains to be seen what aspect of eIF3 function is impaired by these mutations in vivo.
Truncated TIF32 and PRT1 alleles with dominant-negative effects on growth
The biochemical results presented in Figures 2–4 indicate that the truncated eIF3 subunits reside in defective subcomplexes lacking one or more components of the MFC. If these subcomplexes exist in vivo, then we might expect to observe dominant Slg– phenotypes when the mutant alleles are introduced on hc plasmids. In accordance with this prediction, we found that TIF32-Δ5-His conferred a strong Slg– phenotype, and that TIF32-Δ6-His and PRT1-Δ7′-His produced moderate Slg– phenotypes, when present on hc plasmids (Table I; Figure 5D and E).
As shown above, TIF32-Δ5-His formed incomplete complexes with NIP1 and eIF5 that were devoid of other MFC components (Figure 4C, lanes 5 and 6); thus, the Slg– phenotype of hc TIF32-Δ5-His probably results from sequestering NIP1 in non-functional subcomplexes with eIF5. Consistently, the Slg– phenotype of hc TIF32-Δ5-His was suppressed by a hc NIP1 plasmid (Figure 5D, sector 3 versus 4). (Presumably, eIF5 normally is present in excess of eIF3 so that providing more NIP1 is sufficient to restore high levels of intact MFC.) A hc plasmid with PRT1, TIF34 and TIF35 did not suppress the Slg– phenotype of hc TIF32-Δ5-His (Figure 5D, sector 3 versus 5), in agreement with the fact that the PRT1–TIF34–TIF35 subcomplex does not bind to TIF32-Δ5-His. Similarly, PRT1-Δ7′-His formed incomplete complexes lacking TIF34 and TIF35 (Figure 3C), so that its dominant Slg– phenotype could result from sequestering NIP1 and TIF32 in non-functional complexes. Consistently, the Slg– phenotype of hc PRT1-Δ7′-His was suppressed by a hc plasmid bearing NIP1 and TIF32 (data not shown).
The TIF32-His-Δ6 product forms a MFC that lacks only eIF2 (Figure 4C, lanes 7 and 8), and the dominant Slg– phenotype of this allele in high copy (Table I) supports the idea that interaction of eIF2 with the TIF32-CTD (lacking in this allele) is an important aspect of eIF3 function. In support of this interpretation, the Slg– phenotype of hc TIF32-Δ6-His was suppressed by hc plasmid p1780-IMT encoding all three subunits of eIF2 and tRNAiMet, components of the TC (Figure 5E; Table I). Moreover, hc TIF32-Δ6-His exacerbated the Ts– phenotype conferred by tif5-7A (Figure 5F, bottom panels), the eIF5-CTD mutation that destabilizes interaction between eIFs 2 and 3 that is bridged by eIF5. Combining these mutations leads to a marked reduction in polysomes at the expense of monosomes relative to that seen in the single mutants [compare the polysome:monosome ratios (P/M) in Figure 5F]. As shown for hc TIF32-Δ6-His in Figure 5E, the Ts– phenotype of a tif5-7A mutant also was partially suppressed by overexpressing the TC from p1780-IMT (Asano et al., 1999). Together, these findings suggest that the independent contacts between eIF2 and eIF3 involving the TIF32-CTD and eIF5-CTD have additive stimulatory effects on translation initiation in vivo. In this view, overexpressing the TC can restore its stable association with other components of MFC when one or the other contact with eIF3 is impaired in the tif5-7A or hc TIF32-His-Δ6 mutants.
The fact that hc PRT1-Δ5-His did not produce a dominant Slg– phenotype is consistent with our finding that PRT1-Δ5-His interacts weakly with TIF32 and NIP1, and not at all with TIF34 and TIF35 (Figure 3C). Most of the NIP1-His mutant alleles even in high copy seem to co-purify with a smaller proportion of eIF3 than do TIF32-His mutants (Figure 2C versus Figure 4C), possibly due to reduced expression compared with the wild type. Hence, the lack of dominant Slg– phenotypes for NIP1-His mutant proteins (Table I) can be explained by proposing that a relatively larger proportion of the MFC remains intact in these cells. The same explanation may apply to TIF32-Δ86-His.
Evidence that sequestering the TC by NIP1-N′-His, eIF5 and eIF1 reduces its binding to 40S subunits in vivo
NIP1-N′-His was expressed at relatively high levels and it bound strongly to eIFs 1, 2 and 5, but not to any other eIF3 subunits (Figure 2D). Presumably, overexpression of NIP1-N′-His does not sequester a large enough proportion of these factors to reduce MFC formation substantially and confer a dominant Slg– phenotype. However, it did produce a dominant Gcd– phenotype in high copy (Figure 6A; Table I), indicating constitutive derepression of GCN4 mRNA translation. GCN4 encodes a transcriptional activator of amino acid biosynthetic genes, and efficient translation of its mRNA is restricted to amino acid starvation conditions where the α subunit of eIF2 is phosphorylated by GCN2 (Hinnebusch, 1997). Phos phorylated eIF2 competitively inhibits eIF2B, reducing formation of eIF2–GTP from eIF2–GDP and lowering TC formation. The eIF2 phosphorylation and attendant TC depletion produced by GCN2 in starved cells is sufficient to induce GCN4 translation without impairing cell growth. Because gcn2Δ mutants cannot induce GCN4, they fail to grow on medium containing the inhibitor of histidine biosynthesis, 3-aminotriazole (3-AT). Mutations in eIF2 or eIF2B subunits that reduce TC formation derepress GCN4 translation constitutively and render gcn2Δ cells 3-AT-resistant (3-ATR). Similarly, hc NIP1-N′-His allowed gcn2Δ cells to grow on medium containing 20 mM 3-AT, a Gcd– phenotype, whereas cells overexpressing full-length NIP1-His showed no growth on 3-AT plates (Figure 6A, lanes 1, 3 and 4).
Fig. 6. Evidence that sequestration of the TC by NIP1-N′-His, eIF5 and eIF1 impedes its binding to 40S subunits in vivo. (A) Overexpression of the NIP1-NTD confers a Gcd– phenotype that is suppressed by simultaneous overexpression of eIF2 and tRNAiMet. Isogenic strains H2881 (gcn2Δ), H2880 (GCN2) and H2881 carrying either hc NIP1-His (YEpNIP1-His) or hc NIP1-N′-His (YEpNIP1-N′-His) were transformed with either empty vector (YEp24) or hc eIF2 + IMT (p1780-IMT). The resulting strains were spotted in three serial dilutions on SD medium containing 20 mM 3-AT (lower panel), and incubated at 33°C for 2 or 5 days. (B) High-copy eIF5 and eIF1 (encoded by TIF5 and SUI1), but not eIF5-7A and eIF1, exacerbate the Gcd– phenotype of hc NIP1-N′-His. The same as (A) except that H2881 carrying empty vector (YEplac181) or hc NIP1-N′-His (YEpNIP1-N′-His) was co-transformed with hc eIF5–eIF1 (YEpTIF5 + SUI1) or hc eIF5-7A–eIF1 (YEpTIF5-7A + SUI1). (C) Nickel chelation affinity purification of protein complexes containing His-tagged NIP1-N′ protein conducted as described in Figure 2C, except that WCEs were prepared from W303 transformants bearing empty vectors (lanes 1–3), sc YCpNIP1-N′-His and empty vector (lanes 4–6), hc YEpNIP1-N′-His and empty vector (lanes 7–9), hc YEpNIP1-N′-His and hc YEpTIF5 + SUI1 (lanes 10–12), or hc YEpNIP1-N′-His and hc YEpTIF5-7A + SUI1 (lanes 13–15). Lanes 1, 4, 7, 10 and 13 each contained 3.3% of the flow-through fractions from Ni2+-NTA–silica binding (FT); lanes 2, 5, 8, 11 and 14 contained 16.5% (Elu-1x); and lanes 3, 6, 9, 12 and 15 contained 33% (Elu-2x) of the total eluates. The relative amount of eIF2γ (marked with the arrowhead) in each eluate was quantified using the NIH Image program (version beta 3b) and plotted in the histogram below. (D) The same as (A) except that H2881 carrying hc NIP1-N′-His (YEpNIP1-N′-His-U) was co-transformed with empty vector (YEplac181), hc TIF32-His (YEpTIF32-His) or hc TIF32-Δ4-His (YEpTIF32-Δ4).
We reasoned that the Gcd– phenotype of hc NIP1-N′-His could result from sequestering TC in a non-functional complex that is defective for 40S binding. Supporting this prediction, overexpressing all four components of the TC from p1780-IMT fully suppressed the 3-ATR phenotype of hc NIP1-N′-His (Figure 6A, lanes 4 and 6). Given that interaction of NIP1-N′-His with eIF2 should be bridged by eIF5 and stabilized by eIF1 (Asano et al., 2000), overexpressing eIFs 1 and 5 should enhance the ability of NIP1-N′-His to sequester TC in defective complexes. Moreover, this enhancement should be diminished if we overexpress eIF5-7A instead of wild-type eIF5, as the former is impaired for interactions with eIF2β and the NIP1-NTD (Asano et al., 1999, 2000). In accordance with these predictions, the Gcd– phenotype of hc NIP-N-His was strongly enhanced by a hc plasmid encoding eIF5 and eIF1, but less so by one encoding eIF5-7A and eIF1 (Figure 6B, lanes 5–7). In agreement with previous findings (Asano et al., 1999), hc eIF5/eIF1 alone, but not hc eIF5-7A/eIF1, had a moderate Gcd– phenotype (Figure 6B, lanes 3 and 4).
We next sought biochemical evidence that co-overexpressing eIF1 and eIF5 (but not eIF5-7A) increases the amount of eIF2 that is sequestered with NIP1-N′-His. As shown in Figure 6C, ∼2-fold greater amounts of eIFs 1, 5 and 2 co-purified with NIP1-N′-His when the latter was overexpressed from a hc plasmid (lanes 7–9) versus a lc plasmid (lanes 4–6). Importantly, the amount of co-purifying eIF2 increased further (∼1.7-fold) along with that of eIF1 and wild-type eIF5 when the latter two proteins were co-overexpressed with NIP1-N′-His (lanes 10–12 versus 7–9), but increased only ∼1.2-fold when eIF5-7A replaced wild-type eIF5 (lanes 13–15 versus 7–9). Thus, the Gcd– phenotype of these strains correlates with the amount of eIF2 physically sequestered by the NIP1-NTD.
Having shown above that the TIF32-CTD fragment encoded by TIF32-Δ4-His also binds eIF2 in vivo (Figure 4D), we reasoned that combining this allele with hc NIP1-N′-His should exacerbate the effect of the latter on GCN4 expression. Consistently, the Gcd– phenotype of hc NIP1-N′-His was enhanced by hc TIF32-Δ4-His but not by hc TIF32-His or hc TIF32-Δ6-His (lacking the CTD) (Figure 6D). These findings support the idea that TC is recruited to 40S ribosomes most effectively when it is associated with intact eIF3 in the MFC.
Discussion
In vivo confirmation of subunit interactions in the MFC
Most of the previous information regarding the subunit organization of eIF3 and its interactions with eIFs 1, 5 and 2 was derived from studies of binary interactions between pairs of recombinant proteins. Here, we analyzed subunit interactions in the context of native MFC by expressing His-tagged versions of NIP1, TIF32 or PRT1 lacking various internal segments, and characterizing the mutant subcomplexes each formed in vivo. In this way, we confirmed the importance of all binary interactions predicted previously, and we uncovered novel interactions between eIF3 subunits PRT1 and NIP1, and between TIF32 and eIFs 1 and 2.
By characterizing the MFC subcomplexes formed by truncated versions of PRT1-His, we confirmed that PRT1 has binding domains for the other four core eIF3 subunits. The extreme CTD is required only to tether TIF34 and TIF35 to the rest of the complex (Figure 3C, Δ7′). Just N-terminal to that is the NIP1-binding domain, and removing this segment (Δ5′) weakened, but did not abolish, interaction of PRT1-His with the rest of the MFC. The residual interaction can be attributed to the TIF32-binding domain at the N-terminus of PRT1, and we showed previously that deleting the PRT1-NTD dissociated the PRT1-His–TIF34–TIF35 subcomplex from the rest of the MFC (Valášek et al., 2001). The latter implies that the PRT1–NIP1 interaction is not sufficient for complex formation by these two proteins in vivo.
NIP1 contains distinct binding domains for TIF32 and PRT1. Deleting the PRT1-binding domain in the C-terminal half of NIP1-His reduced the amount of PRT1–TIF34–TIF35 subcomplex associated with NIP1-His without decreasing its interaction with TIF32 (Figure 2C, ΔA). Deleting the TIF32-binding domain in the N-terminal half of the protein dissociated NIP1-His from all eIF3 subunits (ΔJ). This last result provides independent evidence that the NIP1–PRT1 interaction is too weak for complex formation by these subunits in the absence of TIF32. Importantly, the binding domains for TIF32 and PRT1 could be removed from NIP1-His without impairing its association with eIFs 1, 2 and 5, as this requires only the NIP1-NTD (Figure 2D).
Consistent with the above conclusions, we found that TIF32 also contains separate binding domains for NIP1 and PRT1. Deleting the two binding domains for PRT1 in the C-terminal portion of TIF32-His dissociated the PRT1–TIF34–TIF35 subcomplex from TIF32-His, but did not impair binding of NIP1 and eIF5 to TIF32 (Figure 4C, Δ5). This result, plus the results for NIP1-ΔA-His cited above, shows that a stable TIF32–NIP1–eIF5 subcomplex can exist in vivo. Previously, we showed that a stable TIF32–PRT1-His binary complex could be purified from yeast, whereas a PRT1-His–NIP1 complex was undetectable (Phan et al., 2001). Thus, stable association between PRT1 and NIP1 is dependent on the separate contacts each protein makes with TIF32 (Figure 4A).
Direct interaction between eIF2 and eIF3 via the TIF32-CTD
Although the isolated NIP1-NTD can bind to eIFs 1, 5 and 2 in vivo, the TIF32-CTD is required additionally to tether eIFs 1 and 2 to the intact MFC. Deleting the C-terminal 174 residues of TIF32-His (Δ6) completely eliminated eIF2 from the MFC (Figure 4C). Deleting the next 149 residues in TIF32-His (Δ5) led to complete loss of the PRT1–TIF34–TIF35 subcomplex and also removed eIF1 from the MFC. Importantly, neither deletion dissociated eIF5 or NIP1 from the truncated TIF32-His protein. This is significant because eIF5 and NIP1 previously were the only known binding partners for eIF2 and eIF1 in the MFC. We determined that residues 491–791 in TIF32 encompassing the HLD are sufficient for interaction with eIF1 in vitro (Supplementary figure S1D). We also showed that the TIF32-CTD can interact directly with eIF2 both in vivo (Figure 4D) and in vitro (Figure 5B), Consistent with this last finding, overexpression of the TIF32-CTD (Δ4) exacerbated the Gcd– phenotype conferred by overexpressing the NIP1-NTD (N′) (Figure 6D). The latter was attributed to sequestering eIF2 in non-productive complexes with NIP1-NTD (Figure 6C).
Deleting the TIF32-CTD destroyed the ability of the resulting allele (TIF32-Δ6-His) to complement the Ts– phenotype of a chromosomal tif32 mutant. Moreover, hc TIF32-Δ6-His conferred a dominant Slg– phenotype that could be suppressed by overproducing the TC (Figure 5E), and hc TIF32-Δ6-His significantly reduced the rate of translation initiation when introduced into the Ts– tif5-7A mutant (Figure 5F). The tif5-7A mutation weakens the indirect eIF2–eIF3 interaction bridged by the eIF5-CTD, and also was partially suppressed by overproducing the TC (Asano et al., 1999). Together, these results provide compelling evidence that the TIF32-CTD and the NIP1-NTD have overlapping functions in linking together eIFs 2 and 3 in the MFC.
It is intriguing that loss of eIF2 from the MFC caused by deleting the TIF32-CTD (Δ6) was suppressed by additionally removing the TIF32-NTD by the Δ86 mutation (Figure 4C). Our interpretation of this unexpected result is that the TIF32-CTD–eIF2 interaction can compensate for a negative effect of the TIF32-NTD on the interaction of eIF2 with the NIP1-NTD–eIF5 module. While it is clearly necessary to confirm the hypothetical inhibitory function of the TIF32-NTD with point mutations, our interpretation provides an economical way of explaining why the TIF32-CTD is required for eIF2 binding to the MFC even though the NIP1-NTD–eIF5 subcomplex is sufficient for eIF2 binding outside of the MFC (Figure 2D). The putative negative function of the TIF32-NTD might be involved in disassembly of the MFC on the 40S subunit at some point in the initiation pathway.
Does eIF3 stimulate TC recruitment to the 40S ribosome in vivo?
Overexpression of the NIP1-NTD had a Gcd– phenotype that was enhanced by simultaneously overexpressing eIFs 1 and 5. These factors formed a stable NIP1-NTD– eIF5–eIF1–eIF2 subcomplex in vivo that was dependent on the eIF5-CTD. Thus, overexpression of eIF5-7A, impaired for interactions with eIF2β and NIP1, failed to enhance NIP1-NTD–eIF2 association significantly (Figure 6C). Because overexpressing eIF5-7A did not strongly enhance the Gcd– phenotype of overexpressed NIP1-NTD (Figure 6B), this phenotype clearly results from sequestering eIF2 in a defective subcomplex with NIP1-NTD, eIF5 and eIF1.
GCN4 translation is regulated by four short upstream open reading frames (uORFs). Ribosomes that have translated the first uORF and resumed scanning must rebind the TC before reaching uORF4 to be prevented from reaching the GCN4 start codon. A reduction in TC levels caused by mutations in eIF2B allows a fraction of reinitiating 40S ribosomes to bypass uORF4 and reinitiate at GCN4, even in cells lacking the eIF2α kinase GCN2 (Gcd– phenotype). The fact that the Gcd– phenotype of hc NIP1-NTD was suppressed by overexpressing the TC implies that it results from reduced TC binding to the 40S ribosomes en route to uORF4. This, in turn, suggests that the subcomplex containing eIF2, eIF1, eIF5 and the NIP1-NTD is defective in delivering TC to scanning 40S ribosomes. The same can be said for the binary complex containing eIF2 and the TIF32-CTD, as overexpression of the latter (hc TIF32-Δ4-His) exacerbated the Gcd– phenotype of hc NIP1-N′-His (Figure 6D). Recent biochemical experiments confirm that overexpression of NIP1-N′-His sequesters a fraction of eIF2 in non-ribosomal complexes (L.Valášek, B.Szamecz, K.H.Nielsen and A.G.Hinnebusch, unpublished observations). These results provide the first in vivo evidence that incorporation of TC into the MFC enhances its binding to 40S subunits. They are consistent with biochemical data obtained in the 1970s indicating that mammalian eIF3 stimulates TC binding to 40S subunits in vitro, and with more recent in vitro data from yeast showing reduced TC binding in prt1-1 and tif5-7A mutant extracts (Hershey and Merrick, 2000; Hinnebusch, 2000). Because eIF3 and eIF2 are thought to have distinct binding sites on the 40S ribosome (Hinnebusch, 2000), their physical association in the MFC may permit cooperative binding of these factors to the 40S subunit.
Although sequestering eIF2 in a subcomplex with NIP1-N′-His produced a Gcd– phenotype, the tif5-7A mutation did not (Asano et al., 1999), even though it destabilized the MFC and reduced TC binding to 40S subunits in vitro (Asano et al., 2001). In fact, tif5-7A cells showed an accumulation of eIF2 and eIF3 in 43–48S complexes that lacked only eIF5. Based on this and other findings, we concluded previously that the eIF5-CTD is more important for scanning, AUG recognition or eIF5 GAP function than for TC binding to 40S ribosomes (Asano et al., 2001). Overexpressing the CTD-less TIF32-Δ6-His exacerbated the translation initiation defect in tif5-7A cells (Figure 5F), presumably by generating a defective MFC lacking two contacts between eIF2 and eIF3; however, a Gcd– phenotype still was not observed. Thus, it appears that loss of the TIF32-CTD–eIF2 interaction produced by hc TIF32-Δ6-His exacerbates the rate-limiting defect in scanning, AUG recognition or GAP function conferred by tif5-7A, rather than abolishing TC binding to 40S subunits.
Even though TC binding can occur in the hc TIF32-Δ6-His tif5-7A mutant, we still might expect to observe a Gcd– phenotype if the rate of this reaction is reduced by loss of multiple contacts between eIF2 and eIF3, as more 40S subunits would reach uORF4 before they rebind the TC. One way to resolve this paradox is to propose that the predicted Gcd– phenotype of the hc TIF32-Δ6-His and tif5-7A alleles is suppressed by their additional effects on scanning. In this view, the reduced rate of ribosomal scanning from uORF1 to uORF4 would compensate for the decreased rate of TC binding, with no net increase in the number of 40S subunits that arrive at uORF4 without binding the TC. Alternatively, the mutations may cause a delay in GTP hydrolysis by the fraction of 40S subunits that rebind the TC before reaching uORF4, causing them to pause at the uORF4 start codon. This would hinder the movement of other 40S subunits located upstream and provide them with additional time to rebind the TC before reaching uORF4. In fact, certain mutations in eIF3 subunits prevent derepression of GCN4 translation (Gcn– phenotype) most probably because of impaired scanning or a delay in GTP hydrolysis (K.H.Nielsen and A.G.Hinnebusch, unpublished data). In contrast, overexpression of NIP-N′-His should not interfere with the proposed functions of eIF5 and eIF3 in scanning and should only reduce the amount of TC physically associated with eIF3 in the MFC. As such, the hc NIP-N′-His allele can selectively decrease the rate of TC binding to 40S subunits and thereby increase GCN4 translation. It remains to be seen whether point mutations in eIF3 subunits can be obtained that selectively impair TC binding without impairing other downstream functions of eIF3, to produce a Gcd– phenotype.
Materials and methods
Yeast strains and plasmids
Strains used in this study are listed in Table II, and details of their construction are available in the Supplementary data, as are the plasmids employed and details of their construction.
Table II. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| F556 (W303) | MATa ade2-1 trp1-1 can1-100 leu2-3, 112 his3-11, 15 ura3 | A.Hopper |
| YLV314U | MATa ura3::URA3::rpg1-1 trp1-1::TRP1::rpg1-Δ2 ade2-1 can1-100 leu2-3,112 his3-11,15 | Valášek et al. (1998) |
| B8302 | MATa cyc1-NLS cyc7-67 ura3-52 lys5-10 nip1-1 | D.Goldfarb |
| H1676 | MATa prt1-1 leu2-3 leu2-112 ura3-52 | |
| H2880 | MATa trp1-1 leu2-3,112 ura3-52 | Nielsen et al.a |
| H2881 | MATa trp1-1 leu2-3,112 ura3-52 gcn2Δ | Nielsen et al.a |
| H2924 | MATα leu2-3,112 ura3-52 ino1 gcd6Δ gcn2Δ HIS4-LacZ::ura3-52 (YDpGCD6-7A, LEU2; p1780-FL, SUI2 SUI3-FL GCD11 URA3) | |
| H2898 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif5Δ::hisG tif34Δ::hisG p[TIF5-FL TRP1] p[TIF34-HA LEU2] | |
| H2899 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ tif5Δ::hisG tif34Δ::hisG p[tif5-FL-7A TRP1] p[TIF34-HA LEU2] |
aK.H.Nielsen, L.Valášek and A.G.Hinnebusch, in preparation.
GST pull-down experiments
A description of GST pull-down experiments can be found in the Supplementary data.
Ni2+ chelation chromatography of eIF3 complexes containing His-tagged proteins
His8-tagged proteins and associated complexes were purified essentially as described previously (Phan et al., 1998; Valášek et al., 2001) with modifications described in the Supplementary data.
Polysome profile analysis
Preparation of WCEs and subsequent polysome analysis were conducted essentially as described previously (Valášek et al., 2001).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to Věra Valášková for her dedicated help with the figures, Amy A.Mathew and Nilsa Rivera del Valle for technical assistance, Thomas Dever for critical reading of the manuscript, and to the members of the Hinnebusch and Dever laboratories for helpful discussions and suggestions during the course of this work.
References
- Asano K., Phan,L., Anderson,J. and Hinnebusch,A.G. (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem., 273, 18573–18585. [DOI] [PubMed] [Google Scholar]
- Asano K., Krishnamoorthy,T., Phan,L., Pavitt,G.D. and Hinnebusch,A.G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP–GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J., 18, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5 and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Shalev,A., Phan,L., Nielsen,K., Clayton,J., Valášek,L., Donahue,T.F. and Hinnebusch,A.G. (2001) Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J., 20, 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaie P., Wittmer,B., Altmann,M. and Trachsel,H. (1995) Isolation of a protein complex containing translation initiation factor Prt1 from Saccharomyces cerevisiae. J. Biol. Chem., 270, 4288–4292. [DOI] [PubMed] [Google Scholar]
- Evans D.R.H., Rasmussen,C., Hanic-Joyce,P.J., Johnston,G.C., Singer,R.A. and Barnes,C.A. (1995) Mutational analysis of the Prt1 protein subunit of yeast translation initiation factor 3. Mol. Cell. Biol., 15, 4525–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- Hinnebusch A.G. (1997) Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem., 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 185–243.
- Huang H., Yoon,H., Hannig,E.M. and Donahue,T.F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev., 11, 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranda T., MacMillan,S.E., Donahue,T.F. and Hershey,J.W. (1996) SUI1/p16 is required for the activity of eukaryotic translation initiation factor 3 in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L., Zhang,X., Asano,K., Anderson,J., Vornlocher,H.P., Greenberg,J.R., Qin,J. and Hinnebusch,A.G. (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol., 18, 4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L., Schoenfeld,L.W., Valášek,L., Nielsen,K.H. and Hinnebusch,A.G. (2001) A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J., 20, 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Trachsel,H., Hašek,J. and Ruis,H. (1998) Rpg1, the Saccharomyces cerevisiae homologue of the largest subunit of mammlian translation initiation factor 3, is required for translational activity. J. Biol. Chem., 273, 21253–21260. [DOI] [PubMed] [Google Scholar]
- Valášek L., Phan,L., Schoenfeld,L.W., Valášková,V. and Hinnebusch,A.G. (2001) Related eIF3 subunits TIF32 and HCR1 interact with an RNA recoginition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J., 20, 891–904. [DOI] [PMC free article] [PubMed] [Google Scholar]