Abstract
The regulation of gene expression plays an important part in cell cycle controls. We describe the molecular machinery that co-ordinates gene transcription at the M–G1 interval during the fission yeast mitotic cell cycle. A sequence is identified in the cdc15+ promoter that we call a PCB (pombe cell cycle box), which confers M–G1-specific transcription. Sequences similar to the PCB are present in the promoters of seven other genes, spo12+, cdc19+, fin1+, sid2+, ppb1+, mid1+/dmf1+ and plo1+, which we find to be transcribed at M–G1. A transcription factor complex is identified that binds to the PCB sequence, which we name PBF, for PCB-binding factor. Finally, we show that PBF binding activity and consequent gene transcription are regulated by the Plo1p protein kinase, thus invoking a potential auto-feedback loop mechanism that regulates mitotic gene transcription and passage through septation and cytokinesis.
Keywords: cell cycle/fission yeast/plo1+/transcription
Introduction
Many forms of control have been shown to regulate the mitotic cell division cycle, such as protein kinase activity, specific proteolytic degradation and changes in intracellular location. Transcription has also been found to play an important role in controlling cell cycle progress, and the cell cycle-specific regulation of gene expression is widespread. Microarray analysis has revealed in the budding yeast, Saccharomyces cerevisiae, that the transcript profile of ∼700 genes, out of a total of ∼6000, varies throughout the mitotic cell cycle (Cho et al., 1998; Spellman et al., 1998). These genes fall into a number of groups whose transcript abundance peaks at different cell cycle times. Each group of genes is regulated co-ordinately by a common DNA sequence present in their promoters, which are bound by a transcription factor complex. Examples of such groupings are the MCB–MBF group of genes at G1–S, and Mcm1p/Fkhp genes during mitosis (Futcher, 2000).
Cell cycle-regulated transcription has also been studied in the fission yeast, Schizosaccharomyces pombe. A group of genes, including cdc22+, cdc18+, cig2+, cdt1+ and mik1+, are transiently expressed at the beginning of S phase, and their products are required, either directly or indirectly, for DNA synthesis (Fernandez-Sarabia et al., 1993; Kelly et al., 1993; Connolly and Beach, 1994; Hofmann and Beach, 1994; Ng et al., 2001). The molecular components that control G1–S transcription in fission yeast comprise a transcription factor complex named DSC1 (DNA synthesis control; also called MBF), that contains the products of the cdc10+, res1+, res2+, rep1+ and rep2+ genes (Lowndes et al., 1992; Tanaka et al., 1992; Caligiuri and Beach, 1993; Miyamoto et al., 1994; Sugiyama et al., 1994; Zhu et al., 1994; Nakashima et al., 1995). DSC1 binds to MCB sequences (MluI cell cycle box; ACGCGT) that are present in the promoters of cdc22+, cdc18+, cdt1+ and cig2+, all of which are expressed maximally at the G1–S boundary during the mitotic cell cycle.
We are interested in identifying other groups of genes that are regulated co-ordinately in fission yeast at different cell cycle times, with the aim of characterizing the molecular components that control their transcription. To this end, we have studied cdc15+, a gene that is transcribed specifically during the M–G1 phase of mitotic division (Fankhauser et al., 1995; Utzig et al., 2000). We report the identification of seven other genes that are expressed coincidentally with cdc15+, and describe a promoter sequence and a transcription factor complex that regulates their cell cycle transcription. Finally, we show that Plo1p regulates the expression of these genes. We propose that Plo1p controls M–G1 cell cycle transcription during mitosis in fission yeast, as part of an auto-activatory loop that results in cytokinesis and septation.
Results
Mitotic cell cycle transcription of cdc15+
cdc15+ was first identified by mutation in the original fission yeast cell cycle screens (Nurse et al., 1976), and ascribed a function late in the cell cycle between nuclear division and early cell plate formation. Subsequent characterization of the gene product (Fankhauser et al., 1995) showed that cdc15+ is a key element in the control of the re-organization of the F-actin ring at cytokinesis (Marks et al., 1986). This ring later constricts to effect cytokinesis, once genomic segregation has occurred (Demeter and Sazer, 1998). Fankhauser et al. (1995) also showed that cdc15+ transcript abundance varied during the cell cycle, with a peak at metaphase during vegetative growth, which suggested that control of cdc15+ transcription may play a role in promoting cytokinesis.
We confirmed and extended the observation concerning cdc15+ mRNA abundance during the mitotic cell cycle, by two different synchronization methods. In synchronously dividing cells, which were either wild type (972h–) size selected by centrifugal elutriation (Figure 1A), or synchronized by transient arrest at the G2–M boundary by reversible temperature shifts of a cdc25-22 mutant (Figure 1B), cdc15+ mRNA varied in abundance in a cell cycle manner. By directly comparing the peak level of cdc15+ transcript with that of cdc22+, we confirmed that cdc15+ peaks slightly earlier in the cell cycle. This was especially apparent in the cdc25-22 mutant experiment (Figure 1B), where we took samples every 10 min after temperature release for northern analysis. As cdc22+ mRNA peaks at the G1–S boundary (White et al., 2001), these two experiments suggested that cdc15+ mRNA was present at M–G1.
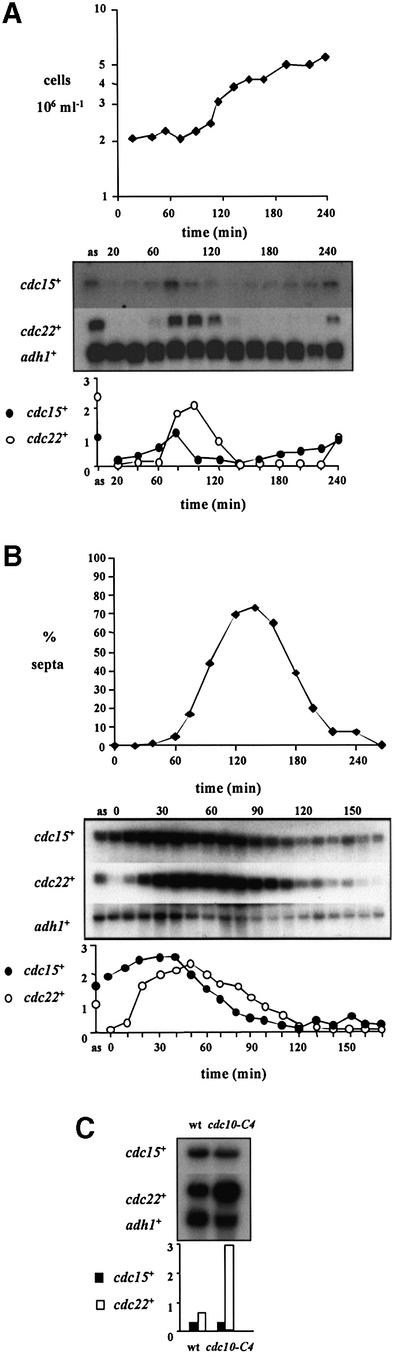
Fig. 1. cdc15+ is expressed before MCB-regulated genes in the fission yeast cell cycle, and is not controlled by DSC1. (A) A population of wild-type cells (972h–), synchronous for division, were size selected by centrifugal elutriation at 32°C, and cell samples taken every 20 min for northern blot analysis of RNA. ‘as’ indicates RNA prepared from asynchronously dividing cells prior to elutriation. The blot was hybridized consecutively with cdc15+, cdc22+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. (B) cdc25-22 cells were synchronized for cell division by transient temperature shifts. Northern blot analysis was performed on RNA samples prepared from cell samples taken at 10 min intervals following release from restrictive temperature. The blot was hybridized consecutively with cdc15+, cdc22+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. The degree of synchrony is indicated by the septation index. (C) RNA was prepared from cultures of wild-type (972h–) and cdc10-C4 cells grown at 25°C, and subjected to northern blot analysis. The membrane was hybridized consecutively with cdc15+, cdc22+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown.
cdc22+ is a member of a group of genes whose transcription is regulated co-ordinately at the G1–S boundary by the combination of a common promoter sequence present in all the genes’ promoters which is bound by a transcription factor complex. The promoter sequence is called an MCB, and the transcription factor complex, DSC1, contains the cdc10+ gene product (Lowndes et al., 1992).
cdc15+ is unlikely to be under DSC1–MCB control, because it is not transcribed at the G1–S interval. Furthermore, cdc15+ does not contain MCB sequences in its promoter region. Finally, a mutant of a component of DSC1, cdc10-C4, which results in overexpression of all known MCB-regulated genes compared with wild type (McInerny et al., 1995; Ng et al., 2001), did not affect cdc15+ transcription (Figure 1C).
These experiments showed that cdc15+ is representative of a new type of mitotic cell cycle-expressed gene in fission yeast, which is transcribed at the M–G1 boundary.
Regulation of cdc15+ transcription
To see whether variation in cdc15+ mRNA levels during the cell cycle was due to transcriptional regulation, we tested the ability of DNA fragments from the promoter region of cdc15+ to confer M–G1 transcription to a heterologous gene. A number of cdc15+ promoter fragments were cloned into the reporter plasmid pSPΔ178 (Lowndes et al., 1992), and the cell cycle-dependent transcription of lacZ analysed by northern blot analysis. One fragment, corresponding to bases –157 to –53 relative to the cdc15+ ATG, conferred M–G1 transcription to lacZ, coincident with endogenous cdc15+ cell cycle expression in synchronized wild-type cells size selected by elutriation (Figure 2A). Similar results were obtained in cdc25-22 cells synchronized by transient temperature arrest (data not shown). These experiments showed that the cyclic behaviour of cdc15+ mRNA abundance was due to transcriptional regulation and not mRNA stability, and that this fragment of DNA contained the cis-acting DNA element that controlled this expression.
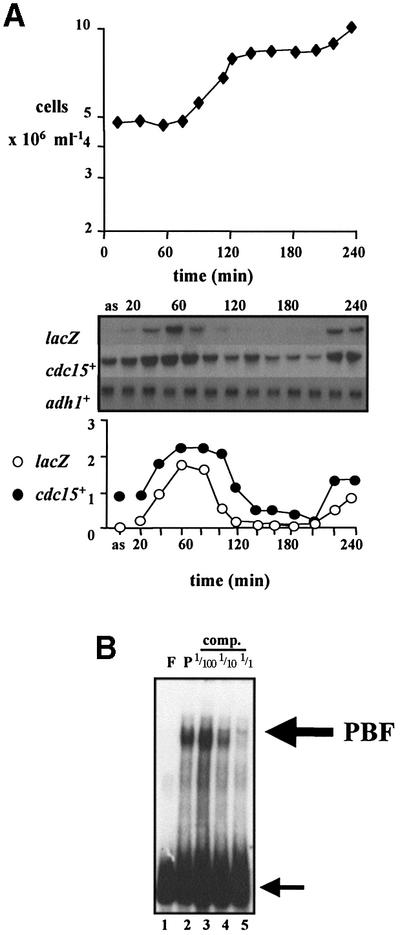
Fig. 2. Characterization of a region of the cdc15+ promoter that confers M–G1 transcription, and identification of a transcription factor complex, PBF, that binds to it. (A) A fragment from the cdc15+ promoter was inserted into pSPΔ178 (Lowndes et al., 1992) to create pSPΔ178. 15UAS, transformed into wild-type cells, and cells synchronous for division were size selected by centrifugal elutriation at 32°C, with samples taken every 20 min for northern blot analysis of RNA. ‘as’ indicates RNA prepared from asynchronously dividing cells prior to elutriation. The blot was hybridized consecutively with lacZ, cdc15+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. (B) The same cdc15+ promoter fragment as in (A) was used as labelled probe in gel retardation analysis with total fission yeast protein extracts. Lane F, free probe; lane P, 20 µg of protein with probe. Competition reactions were performed with the same unlabelled cdc15+ promoter DNA with 1/100, 1/10 and 1 M excess. The large arrow indicates PBF, and the small arrow indicates free probe.
This same cdc15+ promoter fragment was then labelled and used as the probe in gel retardation experiments with protein extracts from wild-type fission yeast cells to identify the protein complex that bound to this sequence (Figure 2B). Because this complex potentially regulates cdc15+ cell cycle transcription, we named it PBF for pombe cell cycle box (PCB)-binding factor (see later).
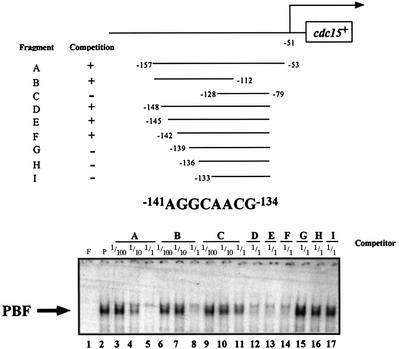
We mapped the region of contact between PBF and the cdc15+ promoter by competition experiments using fragments of DNA from the cdc15+ promoter region (Figure 3). Significantly, whereas fragment F was able to compete, fragment G could not (compare lanes 14 and 15), with the difference between these two DNAs being only 3 bp. This observation implicated this region of DNA as being important for interaction with PBF. The sequence around this region, which corresponded to –141 to –134 relative to the ATG of cdc15+, is AGGCAACG.
Fig. 3. Mapping of the interaction site between PBF and the cdc15+ promoter. Gel retardation analysis was performed using the cdc15+ promoter fragment as labelled probe. Lane F, free probe; lane P, 20 µg of protein with probe. Competition reactions were performed with unlabelled DNAs corresponding to different fragments of the cdc15+ promoter, designated with the letters A–I, with 1/100, 1/10 and 1 M excess, as indicated.
A family of M–G1-regulated genes
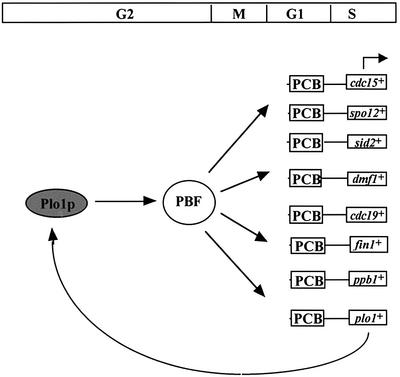
Using the sequence defined by gel retardation analysis in the cdc15+ promoter, we searched the fission yeast genome database (Wood et al., 2002) for other genes that contained similar sequences in their promoter regions. We took two approaches to this, both examining gene promoters randomly and looking specifically in the promoter regions of genes that are known to be cell cycle regulated in budding yeast at M–G1. Having identified potential genes (Figure 4A), we analysed their transcription profile during a mitotic cell cycle. These data are shown in Figure 4B. Interestingly, spo12+, cdc19+, fin1+, mid1+/dmf1+, sid2+, ppb1+ and plo1+ all showed a similar transcription profile to that of cdc15+, suggesting that the same molecular processes may regulate transcription of all of these genes. spo12+ previously has been reported to be cell cycle expressed, coincident with cdc15+ (Samuel et al., 2000). It may be significant that most of these genes form a group that is implicated in the execution or regulation of cytokinesis and septation. Outside this group is cdc19+, a member of the MCM class of protein required for DNA replication that have been shown to be loaded onto chromosomes during metaphase (Kearsey et al., 2000).
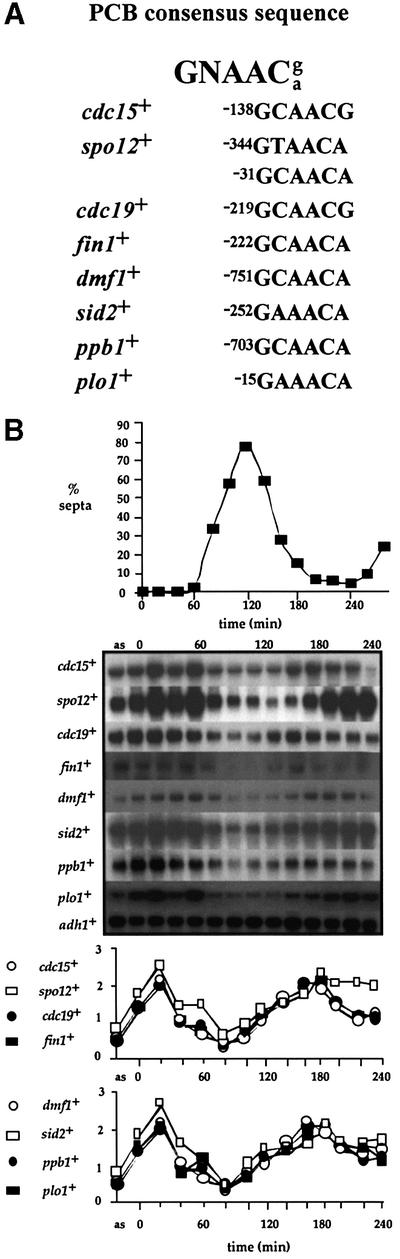
Fig. 4. A group of seven genes are expressed during mitosis in fission yeast coincident with cdc15+. (A) Predicted consensus sequence for the PCB element in fission yeast. Sequences related to the PCB consensus present in M–G1-expressed gene promoters are listed, with numbers referring to their position relative to each gene’s ATG. (B) cdc25-22 cells were synchronized for cell division by transient temperature shifts. Northern blot analysis was performed on RNA from cell samples taken at 20 min intervals following release from the restrictive temperature. ‘as’ indicates RNA prepared from asynchronously dividing cells prior to temperature shifts. The blot was hybridized consecutively with cdc15+, spo12+, cdc19+, mid1+/dmf1+, fin1+, sid2+, ppb1+, plo1+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. The degree of synchrony achieved is indicated by the septation index.
Defining the PCB sequence
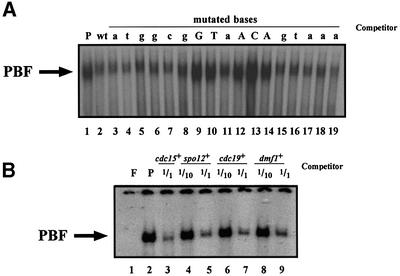
To define more precisely the consensus sequence required for M–G1 transcription in fission yeast, we adopted two approaches. First, we carried out a series of competitive gel retardation experiments using a fragment of DNA from the spo12+ promoter that contained a sequence related to the sequence defined in the cdc15+ promoter. A 20 bp fragment from the spo12+ promoter containing the putative upstream sequence (position –344; Figure 4A) successfully competed with PBF (Figure 5A, lane 2), demonstrating that both promoters recognize the same transcription factor complex. Individual base pairs required for PBF to bind spo12+ DNA were determined by assaying the effect of mutating single base pairs on competitive binding activity. A series of DNAs were synthesized containing consecutive single base pair mutations (A/T to G, or C/G to T), and their ability to bind PBF assayed. As shown in Figure 5A, the central GT and ACA (lanes 9, 10 and 12–14) were all critical for binding, as their individual mutation resulted in loss of PBF binding activity. The fact that mutating the T in the central GT had such an effect was surprising, as this base pair is not conserved amongst other genes (Figure 4A); possibly this base pair is important in the spo12+ promoter in combination with some other base(s).
Fig. 5. Defining the PCB sequence. (A) Base pairs required in spo12+ PCB to bind PBF. Gel retardation analysis was performed using the cdc15+ promoter fragment as labelled probe. Lane P, 20 µg of protein with probe. Competition reactions were performed using cold DNA fragments corresponding to the spo12+ PCB, where single consecutive bases were mutated in separate DNAs, with A/T to G, or C/G to T, with 1 M excess. The central GT and ACA are required for successful competition with PBF. (B) PBF binds promoter fragments containing PCB sequences from other genes transcribed at M–G1 during mitosis. Gel retardation analysis was performed using the cdc15+ promoter fragment as labelled probe. Lane F, free probe; lane P, 20 µg of protein with probe. In alternate lanes, 1 and 1/10 M excess unlabelled competitor promoter DNA fragments from various fission yeast mitotic M–G1-expressed genes was added to the reaction mixture prior to electrophoresis.
In the second approach, we tested whether PBF bound DNA from the promoters of other M–G1-transcribed genes containing sequences related to the PCB sequence (Figure 5B). We performed competitive gel retardation experiments using fragments from the cdc19+ and mid1+/dmf1+ promoters, and demonstrated that these DNAs also bound PBF.
These experiments allowed us to predict GNAACg/a as a putative consensus sequence for the fission yeast M–G1 cis-acting promoter element (Figure 4A). We suggest naming this sequence PCB for pombe cell cycle box.
Cell cycle-dependent behaviour of PBF binding to PCBs
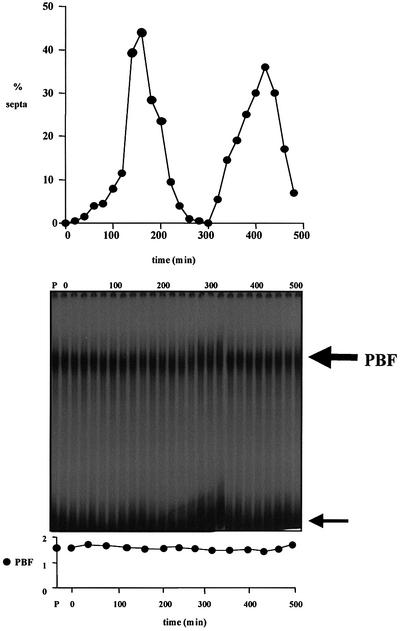
A possible mechanism for PBF and PCB regulation of cell cycle transcription of cdc15+ is by periodic binding of the transcription factor to the promoter sequence during the cell cycle. We tested this hypothesis in three different experiments. In the first experiment, we size selected small wild-type cells (972h–) by centrifugal elutriation, and followed a synchronous population during two divisions. Gel retardation analysis of PBF in such synchronized cells revealed constitutive binding of PBF to PCBs during the cell cycle (Figure 6). Similar results were obtained with synchronized cells made by transient temperature arrest using the cdc10-M17 mutation (data not shown; Kim and Huberman, 2001).
Fig. 6. Cell cycle binding of PBF to PCBs. A population of wild-type cells (972h–) synchronous for division were size selected by centrifugal elutriation at 25°C, and cell samples taken every 20 min for gel retardation analysis. PBF was detected using the cdc15+ promoter fragment as labelled probe with 20 µg of protein in each sample. Lane P, 20 µg of protein from asynchronous cells with probe. The large arrow indicates PBF, and the small arrow indicates free probe. The ratio of PBF to free probe is plotted.
In the third experiment, we examined PBF binding to PCBs in protein extracts from fission yeast cells arrested at discrete stages of the cell cycle (G1, S, G2 and M), using a variety of cdc– mutants. In agreement with the previous experiments, PBF bound to PCBs in cells arrested at all cell arrest points (data not shown). These three experiments suggested that PBF binds to PCBs throughout the cell cycle.
plo1-ts35 affects PBF binding activity in vitro and PCB-regulated gene transcription in vivo
We were interested in identifying genes that regulated cell cycle expression of the cdc15+ group of genes through the PBF transcription factor complex. One potential candidate is the plo1+ polo-like kinase, which is known to have a central role in regulating late mitotic events in fission yeast (Ohkura et al., 1995; Bahler et al., 1998; Tanaka et al., 2001).
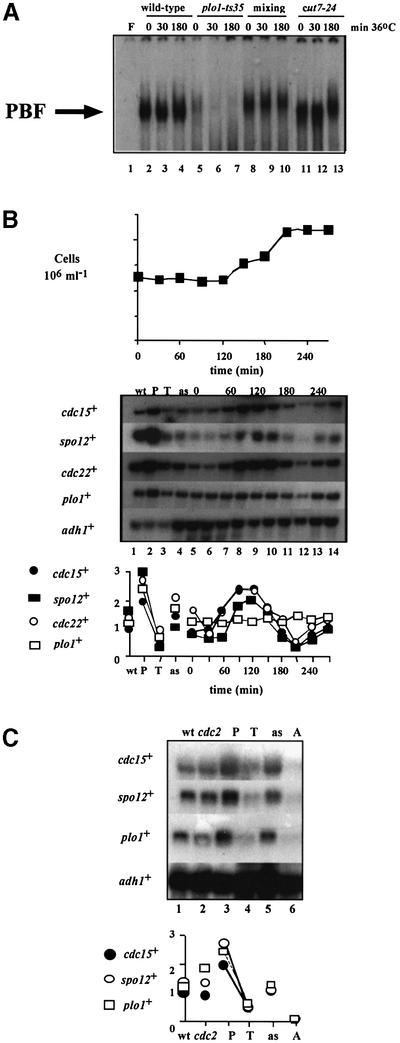
If Plo1p controls PCB-regulated genes, then compromising Plo1p function might affect both PBF activity and the periodic transcription of these genes. We therefore initially determined PBF binding activity in vitro by gel retardation analysis in five different temperature-sensitive plo1– alleles. Interestingly, the mutant with the strongest phenotype, plo1-ts35, resulted in the loss of PBF binding at both permissive and restrictive temperatures (Figure 7A, lanes 5–7). plo1-ts35 cells block mitotic progression as cells are unable to form a mitotic spindle. We therefore asked whether the lack of PBF binding in plo1-ts35 at 36°C was an indirect consequence of the early mitotic arrest, by assessing PBF activity in cut7-24. cut7-24 mutant cells arrest at the same stage of mitosis as plo1-ts35, because of defects in a mitotic motor protein that is required to form the spindle (Hagan and Yanagida, 1990). cut7-24 cells contained PBF at 36°C, indicating that loss of PBF in plo1-ts35 was not a consequence of the cell cycle arrest in this strain (Figure 7A, lanes 11–13). Furthermore, the loss of PBF activity in plo1-ts35 was not due to dominant inhibition of binding in plo1-ts35 cells, as mixing extracts from wild-type and mutant cells had no effect on PBF (Figure 7A, lanes 8–10). plo1-ts35 is a recessive, loss-of-function, allele: haploid plo1-ts35 cells containing a single copy of plo1+ are phenotypically wild type (data not shown). Therefore, loss of PBF in plo1-ts35 was likely to be due to loss of a Plo1p function, rather than dominant interference with activation by another effector.
Fig. 7. plo1-ts35 affects PBF in vitro and PCB-regulated gene transcription in vivo. (A) Gel retardation analysis was performed using the cdc15+ promoter fragment as labelled probe with 20 µg of protein extracts from wild-type (972h–), plo1-ts35 and cut7-24 cells grown at permissive (25°C) and restrictive temperatures (36°C), for the indicated times. In the mixing experiment, protein extracts from wild-type and plo1-ts35 cells were mixed before electrophoresis. Lane F indicates free probe. (B) Cell cycle transcription of cdc15+, spo12+ and plo1+ in plo1-ts35 cells. A population of plo1-ts35 cells, synchronous for division, was size selected by centrifugal elutriation at 25°C. Cell samples were taken before elutriation (as) and at 30 min intervals after elutriation for northern blot analysis of RNA. Control RNA samples from asynchronous wild-type (972h–) cells (wt), and peak (P) and trough (T) cdc15+ mRNA cell cycle samples from the experiment shown in Figure 1A were included. The blot was hybridized consecutively with cdc15+, spo12+, cdc22+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. (C) Transcription of cdc15+, spo12+ and plo1+ in plo1-ts35 cdc2-33 arrested cells. plo1-ts35 cdc2-33 cells were cell cycle arrested by temperature shift after enriching for early G2 cells by elutriation (Tanaka et al., 2001). Cell samples were taken before elutriation (as), and at the arrest (A) after elutriation for northern blot analysis of RNA. Control RNA samples from asynchronous wild-type (972h–) cells (wt), asynchronous cdc2-33 cells (cdc2), and peak (P) and trough (T) cdc15+ mRNA cell cycle samples from the experiment shown in Figure 1B were included. The blot was hybridized consecutively with cdc15+, cdc22+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown.
To determine whether the loss of PBF activity affected the periodic transcription of its target genes, we examined the cell cycle profile of cdc15+ and spo12+ mRNA in plo1-ts35 cells synchronized by centrifugal elutriation at the permissive temperature of 25°C. The profiles of both transcripts were altered, and they were no longer expressed at the M–G1 boundary but instead were delayed and transcribed coincident with cdc22+ at G1–S (Figure 7B). Furthermore, plo1+ itself was no longer transcribed in a cell cycle-dependent manner in plo1-ts35 cells, but mRNA levels were constant at low levels throughout the cell cycle. The level of plo1+ mRNA in plo1-ts35 cells was similar to the lowest trough level of the profile seen in a wild-type cell cycle (Figure 7B; compare lane 3 with 5–14). This observation implies that plo1+ regulates its own expression, consistent with the presence of a PCB sequence in its promoter (Figure 4A). A reduction in plo1+ transcript levels at 25°C is consistent with the minor (5%) misshapen septum phenotype seen at this temperature in these cells (data not shown). The alteration in transcript periodicity in plo1-ts35 was specific to cdc15+, spo12+ and plo1+, as the cdc22+ transcript, which is under different cell cycle regulation, peaked in G1 phase (Figure 7B).
We also tested the effect of a plo1-ts35 temperature arrest on PCB-regulated gene transcription. A plo1-ts35 cdc2-33 culture was incubated at 36°C for 150 min after enrichment for early G2 cells by elutriation. Cells taken for northern blot analysis contained very low levels of cdc15+, spo12+ and plo1+ transcript (Figure 7C, lane 6), consistent with the suggestion that plo1+ is regulating the expression of these genes. These transcripts were present at higher levels in similarly arrested cdc2-33 cells (data not shown), and in cdc25-22 cells arrested at the same cell cycle stage (Figure 1B).
To summarize these three experiments, plo1-ts35 affected PBF binding in vitro, and cdc15+, spo12+ and plo1+ transcription in vivo, suggesting that plo1+ controls M–G1 PCB-regulated gene expression through PBF.
Overexpression of plo1+ causes overexpression of PCB-regulated genes
Previous experiments have demonstrated the critical role of plo1+ in controlling late cell mitotic events, as its overexpression in interphase cells resulted in premature septation (Ohkura et al., 1995). This control of septation by plo1+ is thought to be, in part, through it activating the regulatory septation initiation network (SIN; Tanaka et al., 2001). However, the active SIN needs to have the relevant target molecules in the F-actin ring in order to stimulate contraction of this ring and concomitant septation. Given that Plo1p is required for PBF activity and cell cycle transcription of PCB-regulated genes, we next asked whether overexpression of plo1+ would also provide the target molecules for the SIN by inducing transcription of PCB-regulated genes in interphase cells.
plo1+ transcription was induced in elutriated cdc10-129 cells, immediately transferred to 36°C and so arrested in G1 (Ohkura et al., 1995), and cdc15+ and spo12+ mRNA levels were monitored. plo1+ overexpression caused multiple rounds of septation and resulted in abnormally high levels of both transcripts (Figure 8A; compare lanes 4–9 with 10–15). Significantly, the level of cdc15+ and spo12+ mRNA in these cells was similar to the levels seen at the peak of cell cycle expression of either gene in wild-type cells at M–G1 (Figure 8A; compare lane 2 with 10–15). We observed similar levels of overexpression for other PCB-regulated genes in G1-arrested cells overexpressing plo1+ (data not shown).
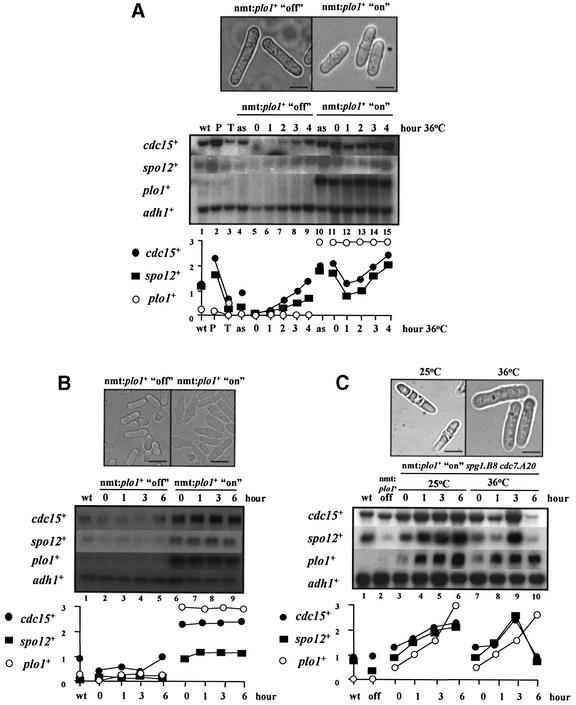
Fig. 8. Effect of overexpressing plo1+ on PCB-regulated gene transcription. (A) Time course of cdc15+ and spo12+ mRNA levels during the overexpression of plo1+ in cdc10-129 arrested cells, following enrichment of G2 cells by centrifugal elutriation (Mulvihill et al., 1999). Two cultures of asynchronous cdc10-129 cells containing pREP1:plo1+ were grown for 15 h in EMM at 25°C, in one case in the presence of thiamine (nmt:plo1+ off), and in the other in the absence of thiamine (nmt:plo1+ on). G2 cells were size selected by centrifugal elutriation, and immediately transferred to 36°C. Cell samples were taken before elutriation (as) and at hourly time points after elutriation (0–4), for northern blot analysis of RNA. Control RNA samples from asynchronous wild-type (972h–) cells (wt), and peak (P) and trough (T) cdc15+ mRNA cell cycle samples from the experiment shown in Figure 1B were included. The blot was hybridized consecutively with cdc15+, spo12+, plo1+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. Representative phase contrast micrographs of cells in both cultures are shown. Bar = 10 µm. (B) Time course of cdc15+ and spo12+ mRNA levels during the overexpression of plo1+ in wild-type cells. A culture of asynchronous wild-type cells containing pREP1:plo1+ was grown in the presence of thiamine (nmt:plo1+ off) to early exponential stage of growth at 25°C. The culture was then split in two: one half grown in EMM in the absence of thiamine (nmt: plo1+ on), the other half in EMM in the presence of thiamine (nmt:plo1+ off), both for 15 h. Cell samples were taken for northern blot analysis of RNA at the times indicated. A control RNA sample from asynchronous wild-type (972h–) cells (wt) was included. The blot was hybridized consecutively with cdc15+, spo12+, plo1+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. Representative phase contrast micrographs of cells in both cultures are shown. Bar = 10 µm. (C) Time course of cdc15+ and spo12+ mRNA levels during the overexpression of plo1+ in cdc7.A20 spg1.B8 arrested cells (Tanaka et al., 2001). A culture of asynchronous cdc7.A20 spg1.B8 cells containing pREP1:plo1+ were grown for 15 h in EMM at 25°C, in the absence of thiamine (nmt: plo1+ on). The culture was then split in two: one half grown at 25°C, and the other half transferred to 36°C. Cell samples were taken after temperature shift (0–6) for northern blot analysis of RNA. Control RNA samples from asynchronous wild-type (972h–) cells (wt), and cdc7.A20 spg1.B8 cells containing pREP1:plo1+ grown in the presence of thiamine (nmt:plo1+ off) were included. The blot was hybridized consecutively with cdc15+, spo12+, plo1+ and adh1+ probes, the latter as a loading control. Quantification of each transcript against adh1+ is shown. Representative phase contrast micrographs of cells in both cultures are shown. Bar = 10 µm.
To confirm that inappropriate activation of PCB-regulated genes by plo1+ overexpression was not due to an artefact of the cdc10-129 arrest, we performed a similar experiment in wild-type cells. plo1+ was overexpressed in a culture of asynchronous wild-type cells, and cdc15+ and spo12+ transcript levels were monitored. Transcripts of both genes were present at higher levels in these cells relative to cells where plo1+ was not overexpressed (Figure 8B).
Plo1p control of PCB-regulated genes does not require the active SIN
Plo1p activates the SIN to control the timing of septation. It was possible that Plo1p controls the transcription of PCB-regulated genes indirectly by activating the SIN. To test this possibility, we asked whether overexpression of plo1+ promoted cdc15+ and spo12+ transcription in cells where SIN function was inactivated by temperature-sensitive mutation. plo1+ was overexpressed in cdc7.A20 spg1.B8 cells at 36°C. As both the cdc7.A20 and spg1.B8 gene products are inactive at this temperature, plo1+ overproduction cannot activate the SIN and so is unable to promote septation (Schmidt et al., 1997; Tanaka et al., 2001). cdc15+ and spo12+ were transiently overexpressed in these cells (Figure 8C; compare lanes 5 and 9). This result suggests that induction of cdc15+ and spo12+ transcription by plo1+ is not dependent on the SIN, although SIN activity is required to maintain PBF activity.
Discussion
plo1+ regulates M–G1 transcription
In fission yeast, the formation of the septum is co-ordinated with mitotic progression (Le Goff et al., 1999). After commitment to mitosis, an F-actin ring forms in the middle of the cell. This ring contains the products of the mid1+/dmf1+ and cdc15+ genes. Later in mitosis, after the nuclei have separated, constriction of the F-actin ring and subsequent formation of the primary septum are induced by the activation of a protein kinase cascade called the septum initiation network (SIN). The activity of the small G protein, Spg1p, controls the activity of this cascade: septation is inhibited if Spg1p is maintained in an inactive state by the bipartite GAP complex Cdc16p–Byr4p.
Plo1p has already been shown to regulate more than one aspect of septation. It physically binds to Mid1p/Dmf1p and is required for the recruitment of this protein to the F-actin ring (Bahler et al., 1998). Loss of either Plo1p or Mid1p/Dmf1p function results in malformed or inappropriately positioned septa (Chang et al., 1996; Sohrmann et al., 1996; Bahler et al., 1998). Plo1p also acts later in mitosis when it activates the SIN (Tanaka et al., 2001). Furthermore, a screen for mutations that are dependent upon elevated levels of Plo1p for survival has identified links with genes encoding modulators of the actin cytoskeleton, sce3+, cdc8+ and rho1+ (Cullen et al., 2000). Thus, plo1+ controls septation through regulating actin ring formation, the co-ordinated constriction of this ring and septum deposition.
We describe an additional mechanism by which plo1+ regulates late mitotic events. We show that Plo1p controls expression of a group of genes late in the cell cycle (Figure 9). These genes include cdc15+, ppb1+ and sid2+, which have important roles in different aspects of septation. cdc15+ controls deposition of the actin ring (Fankhauser et al., 1995; Utzig et al., 2000), ppb1+ regulates the degradation of the primary septum (Yoshida et al., 1994), and sid2+ is a component of the SIN that determines the timing of actin ring constriction (Balasubramanian et al., 1998; Sparks et al., 1999).
Fig. 9. Model outlining the regulation of M–G1-specific transcription in fission yeast by Plo1p. A group of eight genes which are expressed periodically during the fission yeast cell cycle, with a peak of expression at M–G1, are required for cytokinesis and septation. This co- ordinate expression is controlled by a combination of a promoter sequence present in the gene promoters called a PCB (pombe cell cycle box), bound by a transcription factor complex PBF (PCB-binding factor). PBF activity is regulated by Plo1p; thus, potentially, plo1+ regulates its own expression in a positive feedback loop.
Our evidence for the role of plo1+ in controlling the PCB-regulated genes comes partly from using a temperature-sensitive mutant of plo1+, plo1-ts35. plo1-ts35 is a recessive, loss-of-function, mutant allele, which arrests mitotic progression due to an inability to form the mitotic spindle at the restrictive temperature. The septa in plo1-ts35 cells at 36°C resemble those in mid1–/dmf1– cells in being misshapen and positioned inappropriately. The plo1-ts35 mutant not only abolished PBF binding to the cdc15+ promoter DNA in vitro (Figure 7A), but delayed cell cycle transcription of cdc15+ and spo12+ in vivo (Figures 7B). This latter phenotype was specific to the PCB-regulated genes, as cdc22+, a G1–S cell cycle-regulated gene under DSC1–MCB control, was not affected. Importantly, these effects were seen at the permissive temperature for plo1-ts35, when mitotic progression was essentially normal. Therefore, they were not the secondary consequence of a mitotic defect in plo1-ts35, but were a direct consequence of compromised Plo1p function. A minority (5%) of plo1-ts35 septa are misplaced or misshapen, which suggests that the periodicity of transcription that is missing in plo1-ts35 may be important to ensure fidelity of septation.
The consequence of ectopic overexpression of plo1+ confirmed the role of Plo1p in regulating PBF function, as expression in both G1-arrested and wild-type cells elevated transcription of cdc15+ and spo12+ (Figure 8A and B). Overexpression of plo1+ in G1 and G2 has been shown previously to cause premature SIN activation and septation (Ohkura et al., 1995; Mulvihill et al., 1999; Tanaka et al., 2001). We have established that overexpression of plo1+ promoted transcription of the PCB-regulated genes in cells in which the SIN had been inactivated (Figure 8C). This indicates that plo1+ induces PCB-mediated transcription independently of its function in regulating the SIN. It is interesting, however, to note that PBF activation by plo1+ overproduction was only transient when SIN function was absent. This suggests that the SIN is required to maintain PBF-mediated transcription, once it has been activated by Plo1p. The transient nature of PBF activation is reminiscent of the transient induction of the EMTOC upon plo1+ induction in SIN-defective cells (Heitz et al., 2001). We suggest that overexpression of plo1+ results in septation in G1 and G2 cells in part because it activates the SIN, but also because it results in expression of PCB-regulated genes, some of which are either components or substrates of the SIN.
To summarize these experiments, we have shown that manipulating plo1+ resulted in changes in the expression of PCB-regulated genes. Overexpressing plo1+ elevated PCB-regulated gene transcription, whereas compromising plo1+ function by mutation (implying reduced plo1+ activity) delayed or abolished cdc15+ expression. Com bining these observations with the fact that plo1-ts35 also affected PBF binding in vitro, we suggest that plo1+ regulates septation by both SIN activation and through controlling expression of the cdc15+ group of genes through the PCB and PBF transcription machinery.
Feedback model
plo1+ itself was also cell cycle regulated at M–G1, raising the possibility that there is a positive feedback loop regulating late mitotic events (Figure 9). Potentially, plo1+ could regulate its own expression through controlling the activity of PBF. plo1-ts35 affected plo1+ transcription (Figure 7B and C), and plo1+ contains a putative PCB sequence in its promoter (Figure 4A). plo1+ transcription was affected differently from other PCB-regulated genes in plo1-ts35, with mRNA being present at low levels throughout the cell cycle (Figure 7B). A possible explanation for this difference is that plo1-ts35 initially results in reduced levels of plo1+ transcription and Plo1p protein, which subsequently results in delayed cdc15+ and spo12+ transcription. At the very least, this result suggests that other levels of control of the plo1+ gene are likely, which is consistent with the observation that Plo1p protein levels are not cell cycle regulated (Mulvihill et al., 1999).
The fact that Plo1p is a protein kinase suggests that Plo1p may regulate PBF through phosphorylation. Modulation of transcription factor activity by phosphorylation is widespread and well studied (Whitmarsh and Davis, 2000).
Components of PBF
What are the molecular components of PBF? In budding yeast, the transcription factor complex that operates late in the cell cycle, and controls the homologue to plo1+, CDC5, is composed of Mcm1p, Fkh1p and Fkh2p (Koranda et al., 2000; Kumar et al., 2000; Pic et al., 2000; Zhu et al., 2000). Fkh1p and Fkh2p are both forkhead-type transcription factors. A gene containing a forkhead motif, named sep1+, has been identified in fission yeast in a mutant screen for genes required for septation (Ribar et al., 1997) and, provocatively, a sep1Δ mutant has been shown to affect cdc15+ transcription during the cell cycle (Zilahi et al., 2000). Unfortunately, we have been unable to show that Sep1p is part of PBF in gel retardation studies, with either epitope-tagged or deletion versions of the gene (S.S.Ng, M.Anderson, C.J.McInerny and V.Simanis, data not shown).
Meiotic transcription
The PCB-regulated genes are also transcribed specifically during meiosis, suggesting that these genes have a role in regulating the sexual life cycle in fission yeast (Mata et al., 2002; our unpublished data). The induction of cdc15+ occurred after pre-meiotic S phase, at a time corresponding to the first and second meiotic divisions, implying that these genes may be important for these events. While little is known about the function of the PCB-regulated genes during meiosis, it is clear that Plo1p displays discrete localization to the SPB and spindle during this alternative life cycle (Mulvihill, 1999).
The meiotic regulation of cdc15+ may occur through PBF and the PCB elements. There is precedence in fission yeast for groups of genes being regulated by a similar transcription factor complex in both mitosis and meiosis: DSC1 and MCB sequences control cdc22+ and the rec+ genes during the two life cycles (L.Cunliffe, S.White and C.J.McInerny, unpublished data).
Conservation of controls
Homologues to a number of the genes that we have identified as being regulated at the M–G1 interval in fission yeast are also cell cycle regulated in budding yeast. These include CDC5 (plo1+), DBF2 (sid2+), SPO12 (spo12+) and MCM2 (cdc19+), and are transcribed slightly earlier in the cell cycle during M phase (Cho et al., 1998; Spellman et al., 1998). This difference may arise from the alternative approaches to cytokinesis that have been adopted by the two organisms. Budding yeast generate the F-actin ring used for cytokinesis when they commit to the cell cycle at START. This ring persists until after anaphase when the SIN equivalent, the MEN, is activated and promotes both mitotic exit and cytokinesis (Bardin and Amon, 2001). Given that MEN activation follows so rapidly after commitment to anaphase, transcription of the CDC5 group of genes at the metaphase–anaphase boundary would prepare the cell for rapid and efficient utilization of the ring during cytokinesis.
In the case of MCM genes, these have also been shown to be expressed during late mitosis in budding yeast (Cho et al., 1998; Spellman et al., 1998). This control is probably relevant to the fact that these proteins are loaded onto chromosomes during late mitosis at anaphase, as an important part of regulating initiation of the subsequent S phase (Kearsey et al., 2000).
The fact that genes with similar functions are transcribed specifically late in the cell cycle in both organisms suggests that such regulation of expression is significant. It will be important to establish if related mechanisms are conserved in other eukaryotes.
Materials and methods
Media and general techniques
General molecular procedures were performed as described by Sambrook et al. (1989), while the media used for the propagation of S.pombe were as described by Moreno et al. (1991). The standard genetic procedures of Gutz et al. (1974) and Kohli et al. (1977) were followed.
The strains used in this study are shown in Table I. plo1-ts35 was identified in a screen for diploidizing mutants (Broek et al., 1991), which had a spindle formation defect, and is mutated in the kinase domain (F.H.MacIver, F.E.Stevens, D.M.Glover and I.M.Hagan, unpublished data).
Table I. Strain list.
| Collection No. | Genotype | Origin |
|---|---|---|
| GG1 | 972h– (wild type) | Laboratory stock |
| GG 194 | h– cdc2-33 | Laboratory stock |
| GG 202 | h– cdc10-M17 | Laboratory stock |
| GG 249 | h+ cdc10-C4 | Laboratory stock |
| GG 394 | h– cdc25-22 ura-D18 pSPΔ178.15UAS | This study |
| GG 479 | h– cdc10 129 leu1-32 pREP1:plo1+ | Laboratory stock |
| GG 481 | h– plo1-ts35 cdc2-33 | This study |
| GG 603 | h– ura4-D18 pSPΔ178.15UAS | This study |
| GG 615 | h- leu1-32 pREP:plo1+ | This study |
| IH 136 | h– cut7-24 leu1-32 | Laboratory stock |
| IH 1751 | h– cdc7.A20 spg1.B8 leu1-32 ura4-D18 pREP1:plo1+ | Laboratory stock |
| IH 1758 | h– plo1-ts35 ura-D18 leu1-32 | This study |
For physiological experiments, cells were grown routinely in minimal medium (EMM) with shaking, at 25 or 32°C. Temperature-sensitive mutants were incubated at the restrictive temperature of 36°C in order to display their mutant phenotype.
Populations of synchronously dividing fission yeast cells were prepared by use of a Beckman elutriator rotor (Creanor and Mitchison, 1982). Synchronization of cells by transient temperature shifts in the cdc25-22 mutant was achieved by growing cells to mid-exponential growth at 25°C, before shifting to 36°C; cells were then shifted back to 25°C after 3.5 h to enter the mitotic cell cycle in synchrony. plo1-ts35 cdc2-33 and cdc2-33 cells, grown in YE, were elutriated to produce early G2 cells, before transfer to 36°C for 2.5 h. Samples subsequently were removed for RNA extraction, and to measure septation indices by microscopic examination.
To overexpress plo1+ using the pREP1 vector (Maundrell, 1993), cells were grown in EMM with 5 µg/µl thiamine (nmt1+ promoter ‘off’) to the early exponential stage of growth. Cells were washed three times in thiamine-free EMM, and then grown for 15 h in thiamine-free EMM (nmt1+ promoter ‘on’), at the same temperature. For cdc arrest experiments, the next day cells were synchronized by elutriation, and immediately transferred to 36°C (Ohkura et al., 1995).
Cell number per ml of liquid culture was determined from a sample fixed in a 0.1% formaldehyde/0.1% sodium chloride solution. Following sonication, cells were counted electronically with a Z2 Coulter Counter. Flow cytometry analysis (fluorescence-activated cell sorting; FACS) was performed as previously described (McInerny et al., 1995), using the FACScan system and the Cell Quest analysis program (Becton Dickinson, USA); 10 000 cells were analysed per time point.
DNA constructs
The PCB-containing fragment DNA from the cdc15+ promoter was amplified by PCR with XhoI restrictions sites with oligos GCGCTCGAGTATTGTGCACTCAGATAGGCA and GCGCTCGAGAAATTCCTGGTAGTCGATTTC, and cloned into pSPΔ178 (Lowndes et al., 1992) to create pSPD178.15UAS (GB 194). pREP1:plo1+ (pHN204; Ohkura et al., 1995) was transformed into leu1-32, cdc10-129 and cdc7.A20 spg1.B8 cells.
RNA manipulations
Schizosaccharomyces pombe total RNA was prepared (McInerny et al., 1995) using a Ribolyser (Hybaid Ltd, UK), and northern blot analysis carried out using GeneScreen membrane (NEN, Life Science Products Inc., USA), following the manufacturer’s suggested protocol. Northern blots were hybridized with the DNA probes made by PCR corresponding in each case to ∼1 kb of each gene’s open reading frame. DNA probes were labelled with [α-32P]dCTP using the random hexanucleotide labelling procedure of Feinberg and Volgelstein (1983). Equal loading of RNA in each lane was confirmed by hybridization with an adh1+ probe. Transcripts were quantified using NIH software, and the ratios, relative to the invariant adh1+ mRNA, estimated and plotted.
Gel retardation analysis (band shift)
Whole-cell protein extracts were generated from wild-type (972h–) cells, and gel retardation analysis was performed as previously described (Ng et al., 2001), using a labelled cdc15+ promoter fragment made with oligos GCGGAATTCTATTGTGCACTCAGATAGGCA and GCGCTCGAGAAATTCCTGGTAGTCGATTTC. This fragment corresponds to ‘A’ in Figure 3. To generate other cdc15+ promoter fragments for competition experiments shown in Figure 3, the following oligos were used: ‘B’, GCGGTCGAGGTGACAACCGTCCCTAGCAAG and GCGGAATTGTATTGTGCACTCAGATAGGCA; ‘C’, TAGGGACGGTTGTCACCG; ‘D’, CTCAGATAGGCAACGGTTG; ‘E’, AGATAGGCAACGGTTGCTA; ‘F’, TAGGCAACGGTTGATAGG; ‘G’, GCAACGGTTGCTAGGGAC; ‘H’, ACGGTTGCTAGGGACGGTTG; and ‘I’, GTTGCTAGG GACGGTTGTC were each amplified with the oligo GCGGTCGAGAGAGTAAACATGTTTGTTTAG.
For the spo12+ promoter analysis, two 20mer oligos containing the spo12+ PCB, ATGGCGGTAACAGTAAAAGT and ACTTTTACTGTTACCGAACT, were annealed and used in competition reactions. Individual base pair requirements for PBF binding were established by creating 20mers as before, but with consecutive base pair substitutions A/T to G, or C/G to T.
Competition reactions were completed using promoter fragments amplified with oligos from the gene promoters mid1+/dmf1+, CGTTGCTATCAACAAACTTC and TTGTATTTTTGACTGATAGC; and cdc19+, GGAAAATGTAGTGATACCTG and CAACTAAACGTTGGCAAATA.
Acknowledgments
Acknowledgements
We would like to thank Viesturs Simanis and Matthias Sipiczki for strains, Anne Graham for technical support, Lesley Cunliffe and Rebecca Thomson for unpublished data, and other members of the Glasgow and Manchester labs for suggestions and encouragement during the course of this work. The work was supported by grants from the Wellcome Trust, the BBSRC and The Royal Society to C.J.M., and Cancer Research UK and the BBSRC to I.M.H and D.M.G.
References
- Bahler J., Steever,A.B., Wheatley,S., Wang,Y.l., Pringle,J.R., Gould,K.L. and McCollum,D. (1998) Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol., 143, 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum,D., Chang,L., Wong,K.C., Naqvi,N.I., He,X., Sazer,S. and Gould,K.L. (1998) Isolation and characterization of new fission yeast cytokinesis mutants. Genetics, 149, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J. and Amon,A. (2001) Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol., 2, 815–826. [DOI] [PubMed] [Google Scholar]
- Broek D., Bartlett,R., Crawford,K. and Nurse,P. (1991) Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature, 349, 388–393. [DOI] [PubMed] [Google Scholar]
- Caligiuri M. and Beach,D. (1993) Sct1 functions in partnership with Cdc10 in a transcription complex that activates cell cycle START and inhibits differentiation. Cell, 72, 607–619. [DOI] [PubMed] [Google Scholar]
- Chang F., Woollard,A. and Nurse,P. (1996) Isolation and characteriz ation of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci., 109, 131–142. [DOI] [PubMed] [Google Scholar]
- Cho R.J. et al. (1998) A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell, 2, 65–73. [DOI] [PubMed] [Google Scholar]
- Connolly T. and Beach,D. (1994) Interaction between the Cig1 and Cig2 B-type cyclins in the fission yeast cell cycle. Mol. Cell. Biol., 14, 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanor J. and Mitchison,J.M. (1982) Patterns of protein synthesis during the cell cycle of the fission yeast Schizosaccharomyces pombe. J. Cell Sci., 58, 263–285. [DOI] [PubMed] [Google Scholar]
- Cullen C.F., May,K.M., Hagan,I.M., Glover,D.M. and Ohkura,H. (2000) A new genetic method for isolating functionally interacting genes: high plo1+-dependent mutants and their suppressors define genes in mitotic and septation pathways in fission yeast. Genetics, 155, 1521–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J. and Sazer,S. (1998) imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J. Cell Biol., 143, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Reymond,A., Cerutti,L., Utzig,S., Hofmann,K. and Simanis,V. (1995) The S.pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell, 82, 435–444. [DOI] [PubMed] [Google Scholar]
- Feinberg P. and Volgelstein,B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem., 1, 6–13. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sarabia M.J., McInerny,C.J., Harris,P., Gordon,C.B. and Fantes,P.A. (1993) The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol. Gen. Genet., 238, 241–251. [DOI] [PubMed] [Google Scholar]
- Futcher B. (2000) Microarrays and cell cycle transcription in yeast. Curr. Opin. Cell Biol., 12, 710–715. [DOI] [PubMed] [Google Scholar]
- Gutz H., Heslot,H., Leupold,U. and Loprieno,N. (1974) Schizosaccharo myces pombe. In King,R.C. (ed.), Handbook of Genetics. Vol. I. Plenum, New York, NY, pp. 395–446.
- Hagan I. and Yanagida,M. (1990) Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature, 347, 563–566. [DOI] [PubMed] [Google Scholar]
- Heitz M.J., Petersen,J., Valovin,S. and Hagan,I.M. (2001) MTOC formation during mitotic exit in fission yeast. J. Cell Sci., 114, 4521–4532. [DOI] [PubMed] [Google Scholar]
- Hofmann J.F.X. and Beach,D. (1994) cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J., 13, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S.E., Montgomery,S., Labib,K. and Lindner,K. (2000) Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J., 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J., Martin,G.S., Forsburg,S.L., Stephen,R.J., Russo,A. and Nurse,P. (1993) The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell, 74, 371–382. [DOI] [PubMed] [Google Scholar]
- Kim S.M. and Huberman,J.A. (2001) Regulation of replication timing in fission yeast. EMBO J., 20, 6115–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Hottinger,H., Munz,P., Strauss,A. and Thuriaux,P. (1977) Genetic mapping in Schizosaccharomyces pombe by mitotic and meiotic analysis and induced haploidization. Genetics, 87, 423–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda M., Schleiffer,A., Endler,L. and Ammerer,G. (2000) Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature, 406, 94–98. [DOI] [PubMed] [Google Scholar]
- Kumar R., Reynolds,D.M., Shevchenko,A., Shevchenko,A., Goldstone,S.D. and Dalton,S. (2000) Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol., 10, 896–906. [DOI] [PubMed] [Google Scholar]
- Le Goff X., Utzig,S. and Simanis,V. (1999) Controlling septation in fission yeast: finding the middle and timing it right. Curr. Genet., 35, 571–584. [DOI] [PubMed] [Google Scholar]
- Lowndes N.F., McInerny,C.J., Johnson, A.L., Fantes,P.A. and Johnston,L.H. (1992) Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature, 355, 449–453. [DOI] [PubMed] [Google Scholar]
- Marks J., Hagan,I.M. and Hyams,J.S. (1986) Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl., 5, 229–241. [DOI] [PubMed] [Google Scholar]
- Mata J., Lyne,R., Burns,G. and Bahler,J. (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet., 32, 143–147. [DOI] [PubMed] [Google Scholar]
- Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- McInerny C.J., Kersey,P.J., Creanor,J. and Fantes,P.A. (1995) Positive and negative roles for cdc10 in cell cycle gene expression. Nucleic Acids Res., 23, 4761–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M., Tanaka,K. and Okayama,H. (1994) res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J., 13, 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Mulvihill D.G.P. (1999) The characterisation of the activity and localisation of the S.pombe Polo-like kinase Plo1p. PhD thesis, University of Dundee, Dundee, UK.
- Mulvihill D.P, Petersen,J., Ohkura,H., Glover,D.M. and Hagan,I.M. (1999) Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell, 10, 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N., Tanaka,K., Sturm,S. and Okayama,H. (1995) Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J., 14, 4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.S., Anderson,M., White,S. and McInerny,C.J. (2001) mik1+ G1–S transcription regulates mitotic entry in fission yeast. FEBS Lett., 503, 131–134. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux,P. and Nasmyth,K. (1976) Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 146, 167–178. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Hagan,I.M. and Glover,D.M. (1995) The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring and septum, can drive septum formation in G1 and G2 cells. Genes Dev., 9, 1059–1073. [DOI] [PubMed] [Google Scholar]
- Pic A. et al. (2000) The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J., 19, 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribar B., Banrevi,A. and Sipiczki,M. (1997) sep1+ encodes a transcription-factor homologue of the HNF-3/forkhead DNA-binding-domain family in Schizosaccharomyces pombe. Gene, 202, 1–5. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Samuel J.M., Fournier,N., Simanis,V. and Millar,J.B. (2000) spo12 is a multicopy suppressor of mcs3 that is periodically expressed in fission yeast mitosis. Mol. Gen. Genet., 264, 306–316. [DOI] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann,M., Hofmann,K., Woollard,A. and Simanis,V. (1997) The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev., 11, 1519–1534. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Fankhauser,C., Brodbeck,C. and Simanis,V. (1996) The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev., 10, 2707–2719. [DOI] [PubMed] [Google Scholar]
- Sparks C.A., Morphew,M. and McCollum,D. (1999) Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol., 146, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A., Tanaka,K., Okazaki,K., Nojima,H. and Okayama,H. (1994) A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J., 13, 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Okazaki,K., Okazaki,N., Ueda,T., Sugiyama,A., Nijima,H. and Okayama,H. (1992) A new cdc gene required for S-phase entry of Schizosaccharomyces pombe encodes a protein similar to the cdc10+ and SWI4 gene products. EMBO J., 11, 4923–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Petersen,J., MacIver,F., Mulvihill,D.P., Glover,D.M. and Hagan,I.M. (2001) The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J., 20, 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzig S., Fankhauser,C. and Simanis,V. (2000) Periodic accumulation of cdc15 mRNA is not necessary for septation in Schizosaccharomyces pombe. J. Mol. Biol., 302, 751–759. [DOI] [PubMed] [Google Scholar]
- White S., Khaliq,F., Sotiriou,S. and McInerny,C.J. (2001) The role of DSC1 components cdc10+, rep1+ and rep2+ in MCB gene transcription at the mitotic G1–S boundary in fission yeast. Curr. Genet., 40, 251–259. [DOI] [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davis,R.J. (2000) Regulation of transcription factor activity by phosphorylation. Cell. Mol. Life Sci., 57, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Toda,T. and Yanagida,M. (1994) A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci., 107, 1725–1735. [DOI] [PubMed] [Google Scholar]
- Zhu G., Spellman,P.T., Volpe,T., Brown,P.O., Botstein,D., Davis,T.N. and Futcher,B. (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature, 406, 90–94. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Takeda,T., Nasmyth,K. and Jones,N. (1994) pct1+, which encodes a new DNA-binding partner of p85cdc10, is required for meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev., 8, 885–898. [DOI] [PubMed] [Google Scholar]
- Zilahi E., Salimova,E., Simanis,V. and Sipiczki,M. (2000) The S.pombe sep1 gene encodes a nuclear protein that is required for periodic expression of the cdc15 gene. FEBS Lett., 481, 105–108. [DOI] [PubMed] [Google Scholar]