Abstract
TWEAK and APRIL are two recently identified tumour necrosis factor (TNF) ligand family members, implicated in angiogenesis and immune regulation, respectively. TWEAK is a transmembrane protein expressed on the cell surface, whereas APRIL acts solely as a secreted factor. In this report, using RACE, RT–PCR, cDNA library screening and an RNase protection assay, we characterize a hybrid transcript between TWEAK and APRIL mRNAs. The encoded TWE-PRIL protein is composed of TWEAK cytoplasmic and transmembrane domains fused to the APRIL C-terminal domain. TWE-PRIL mRNA is expressed and translated in human primary T cells and monocytes, and endogenous TWE-PRIL protein was detected in primary human T lymphocytes and monocytic cell lines. TWE-PRIL is membrane anchored and presents the APRIL receptor-binding domain at the cell surface. It is a biologically active ligand, as it stimulates cycling in T- and B-lymphoma cell lines. Much like membrane-bound and secreted TNF-α, the different cellular localizations of TWE-PRIL and APRIL suggest that they exert distinct biological roles.
Keywords: APRIL/cell membrane anchoring/hybrid mRNA/TNF family/TWEAK
Introduction
A family of structurally related molecules termed tumour necrosis factors (TNFs) is connected intimately to the regulation of cell differentiation and death pathways. This family can be divided into two groups, ligands and receptors. TNF ligands are synthesized as transmembrane type II proteins, i.e. the N-terminal domain is oriented towards the cytoplasm, whereas the C-terminal domain is expressed on the extracellular surface. Another characteristic of TNF ligands is that their extracellular region contains a trimerization domain responsible for receptor binding. TNF ligands can induce various biological responses such as differentiation, cell death, survival or proliferation; some TNF family members induce pleiotropic responses (Smith et al., 1994).
Several members of the TNF ligand family have been described recently, including TWEAK (TNFSF12) (Chicheportiche et al., 1997) and APRIL (a proliferation-inducing ligand; TNFSF13) (Hahne et al., 1998). TWEAK appears to induce proliferation in endothelial cells, and to be implicated in angiogenesis and monocyte-mediated cytotoxicity (Lynch et al., 1999; Nakayama et al., 2000). The biological role of APRIL seems to be manifold: growth-stimulating activity in tumour cell lines (Hahne et al., 1998; Yu et al., 2000), a pro-apoptotic effect (Kelly et al., 2000) and a protective effect against death ligand-induced apoptosis (Roth et al., 2001) have been reported. Previous studies described the potential of recombinant APRIL to act as a co-stimulator of primary B and T cells in vitro (Yu et al., 2000). We recently reported the phenotype of APRIL transgenic mice, which showed that APRIL modulates B- and T-cell immunity (Stein et al., 2002). Furthermore, the elevated APRIL RNA expression levels found in a variety of tumours, as well as the increased tumour growth rate of APRIL-transfected NIH 3T3 cells in nude mice, strongly support a role for APRIL in tumour growth (Hahne et al., 1998; Kelly et al., 2000; Rennert et al., 2000).
Among TNF ligand family members, APRIL shows a unique maturation pathway. It is processed intracellularly prior to secretion and thus acts solely as a secreted factor (Lopez-Fraga et al., 2001). APRIL cleavage is mediated by furin, a ubiquitously expressed pro-protein convertase that processes many inactive precursors including hormones, growth factors and receptors (Molloy et al., 1999). Analysis of the molecular regulation of APRIL gene expression led us to identify an endogenous hybrid transcript between TWEAK and APRIL mRNAs in human primary T cells and monocytes. This transcript is translated into a functional membrane-anchored fusion protein that we termed TWE-PRIL.
Results
Two distinct APRIL transcripts are detected in human primary T cells, as well as in monocytic and tumour cell lines
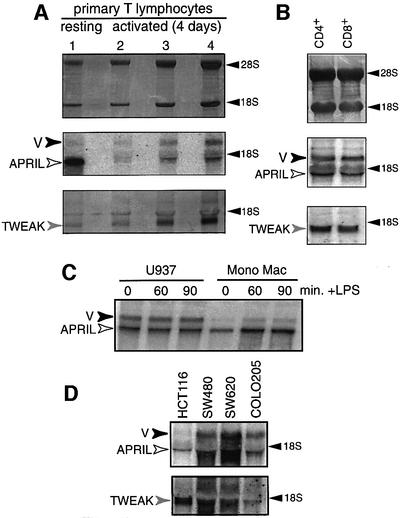
Cytoplasmic RNA from human primary T cells, resting or activated with a combination of anti-CD3 and anti-CD28 monoclonal antibodies (mAbs), was hybridized in northern blot with a probe encompassing the hAPRIL mRNA open reading frame (ORF). This showed one signal of ∼1.9 kb and a slower migrating band of ∼2.5 kb (Figure 1A), independently of the donor (Figure 1A and B; data not shown). Alignment of the DDBJ/EMBL/GenBank sequences for hAPRIL allows reconstitution of an mRNA of 1.8 kb (DDBJ/EMBL/GenBank accession No. AY081050), which, after polyadenylation, would correspond to the 1.9 kb signal. The 2.5 kb band is unlikely to represent immature APRIL mRNA, as it is found in the cytoplasmic fraction (which contains only mature mRNAs), according to the RNA isolation method used.
Fig. 1. Northern blots from T-cell, monocytic and tumour cell line RNAs revealed the presence, in addition to the canonical APRIL mRNA (white arrow), of a variant transcript (black arrow). (A) Northern blot with cytoplasmic RNAs from resting (lane 1) and 4 day-activated human primary T cells (lanes 2–4: 5, 10 and 20 µg). (B) Northern blot from total RNAs of CD4+ and CD8+ cells, purified from a population of 4 day-activated T cells. In (A) and (B), the upper panel shows methylene blue staining of the membrane; the middle panel shows two signals detected following hybridization with an RNA probe specific for the hAPRIL ORF; and the lower panel shows hybridization of the same filter with an RNA probe specific for the hTWEAK ORF. In addition to the TWEAK mRNA signal (grey arrow), a lower mobility signal was detected following longer exposure. (C) Northern blot analysis of cytoplasmic RNAs from human monocytic cell lines U937 and Mono Mac 1 activated with LPS (10 µg/ml) for 0, 60 or 90 min. Hybridization with the hAPRIL probe reveals signals for APRIL (white arrow) and the variant transcript (black arrow). (D) Northern blot of cytoplasmic RNAs from various colon carcinoma cell lines, as indicated above. Upper panel: hybridization with the hAPRIL probe shows APRIL (white arrow) and the variant (black arrow) transcripts. Lower panel: hybridization of the same filter with the hTWEAK probe reveals, in addition to TWEAK mRNA (grey arrow), a lower mobility signal upon longer exposure. 28S and 18S rRNAs are indicated.
In resting T cells, the relative expression level of the canonical APRIL transcript (1.9 kb) is ∼6-fold higher than that of the variant (v, 2.5 kb). In activated T cells, however, the transcripts are expressed at similar levels (Figure 1A). This ratio is maintained in T cells activated with anti-CD3/CD28 or phytohaemagglutinin (PHA) for 2–8 days (not shown), coinciding with activation-induced proliferation. To analyse the two T-cell subpopulations, CD4+ and CD8+ cells were purified from T cells activated with anti-CD3/CD28 mAb for 4 days. Northern blot analysis of CD4+ and CD8+ cell RNA revealed a similar expression pattern for APRIL and the variant transcript in both cell types (Figure 1B). mRNA expression levels of another TNF ligand superfamily member, TWEAK, showed no significant variation following T-cell activation or in the two T-cell subpopulations (Figure 1A and B).
As APRIL RNA expression is described in monocytes (Shu et al., 1999), APRIL transcript expression was analysed in two human monocytic cell lines (Figure 1C). In Mono Mac 1 and U937 cells, APRIL and the variant mRNAs were both detected, and their relative expression level did not vary following lipopolysaccharide (LPS) activation: 6-fold higher for APRIL in Mono Mac 1 cells, and 2-fold higher in U937 cells.
High APRIL mRNA levels are reported in tumour cell lines and in primary tumour tissues (Hahne et al., 1998; Kelly et al., 2000; Rennert et al., 2000). Analysis of several colorectal carcinoma cell lines shows differential but clear expression of APRIL and the variant mRNAs (Figure 1D). In the same cell lines, TWEAK mRNA expression levels also vary. As for APRIL, variant mRNA expression in distinct tumour cell lines may indicate a role during tumour formation.
APRIL and the variant transcripts are engaged in protein synthesis in human primary T cells and monocytes
Protein synthesis is regulated by mRNA expression on the one hand, and by translational control on the other. Indeed, mRNA expression levels do not necessarily reflect the synthesis rate of the protein encoded (Pradet-Balade et al., 2001). To analyse the translational regulation of APRIL and the variant mRNAs, primary T cell and monocyte cytoplasmic extracts were fractionated on sucrose gradients. This technique allows separation of translated mRNA (contained in monosome and polysome fractions) from untranslated mRNA (in fractions containing free ribonucleo-particles). The distribution of a given mRNA species across these fractions reflects the extent to which the mRNA is actually engaged in protein synthesis.
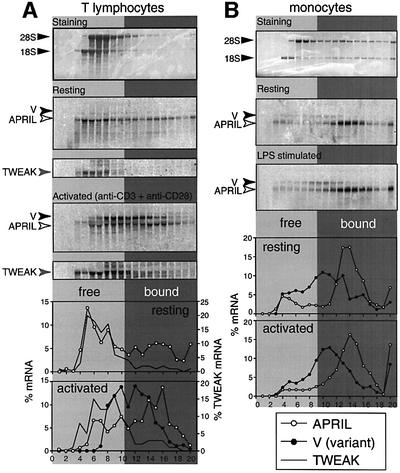
In resting T cells, the canonical APRIL transcript is found predominantly in ribosome-free fractions, with a smaller part found in polysome-bound fractions, suggesting low translation efficiency for APRIL mRNA in these cells (Figure 2A). In T lymphocytes activated by a combination of anti-CD3/CD28 mAb for 4 days, there is a larger proportion of polysome-bound APRIL transcript, so that translation efficiency of APRIL mRNA increases following T-cell activation (Figure 2A). Since different RNA quantities are loaded on each sucrose gradient, the total signals for APRIL mRNA do not reflect the variation in mRNA expression upon T-cell activation. In activated T cells, the majority of the variant transcript is engaged in translation. It is also detected, although at a lower intensity, in gradients from resting T cells, principally in the ribosome-free fractions (Figure 2A).
Fig. 2. APRIL and the variant transcripts are engaged in protein synthesis in human primary T cells and monocytes. Cytoplasmic extracts from human primary T lymphocytes (A) and human primary monocytes (B) were fractionated on a sucrose gradient to separate mRNAs according to ribosome loading. Upper panels show rRNA distribution in gradients following methylene blue staining of the membranes. (A) Resting T lymphocytes were compared with lymphocytes activated for 4 days with anti-CD3 and anti-CD28. Middle (resting) and lower (activated) panels show the distribution of APRIL (white arrow) and variant (V, black arrow) transcripts in the sucrose gradient fractions from resting and activated cells, visualized by hybridization with the hAPRIL RNA probe. The same membranes, hybridized with the hTWEAK probe, show the distribution of TWEAK mRNA (grey arrow) and of a lower mobility mRNA. The quantification of signals for APRIL, variant and TWEAK mRNAs in the sucrose gradients is plotted below. (B) The middle (resting) and lower (LPS-stimulated) panels show the distribution of APRIL (white arrow) and variant (V, black arrow) transcripts in sucrose gradient fractions from human primary monocytes, resting or LPS stimulated for 1.5 h, as visualized following hybridization with the hAPRIL RNA probe. The quantification of signals in the sucrose gradients is plotted below.
Hybridization of the same filters with the hTWEAK probe shows that this mRNA is present mainly in the ribosome-free fractions, without any significant alteration in translation efficiency following T-cell activation (Figure 2A). This may explain why a previous study failed to detect TWEAK protein on human T cells.
Human primary monocytes were LPS treated for 1.5 h, a time at which maximum TNF-α secretion is observed (Espel et al., 1996). Resting and activated monocytes express both APRIL and the variant mRNAs (Figure 2B). The majority of APRIL transcript and a significant fraction of the variant mRNA are involved in translation, as they are found mostly in the polysomal fractions (Figure 2B). There is no significant change in the translation efficiency of either of these transcripts following LPS stimulation, nor in their relative expression levels, 6-fold higher for APRIL (Figure 2B).
Finally, analysis of the distribution of the variant transcript in the sucrose gradients demonstrates that it is a mature mRNA that gives rise to protein synthesis in primary T cells and monocytes.
Identification of the variant transcript as a TWEAK–APRIL hybrid mRNA
To identify the nature of vAPRIL mRNA, cDNAs from resting and activated human primary T cells were amplified by RT–PCR using primers that anneal throughout the APRIL coding region. This invariably resulted in specific amplification of single bands fully compatible with the APRIL sequence, suggesting that the variant transcript differs from APRIL RNA in either its 5′ or 3′ regions. Several 3′ RACE experiments using T cell poly(A)+ RNA and various kits (see Materials and methods) resulted in amplification of a single band, which was identical in resting and activated T cells, and corresponded to the 3′ region of the APRIL sequence (DDBJ/EMBL/GenBank accession No. AY081051). 5′ RACE experiments using T-cell cDNA allowed amplification of a fragment from both resting and activated T cells, whose sequence corresponded to the 5′ region of APRIL mRNA. An additional band was amplified from activated T-cell cDNA, which contained APRIL nucleotides 528–835 (DDBJ/EMBL/GenBank accession No. AY081050) preceded by a sequence unrelated to APRIL. The latter region was 100% identical to hTWEAK nucleotides 215–555 (DDBJ/EMBL/GenBank accession No. AF00558372), suggesting the expression of a TWEAK–APRIL hybrid mRNA.
To confirm this result, an unamplified, activated human T-cell cDNA library was screened with a probe corresponding to the hAPRIL ORF. Of 48 clones isolated, 23 corresponded to APRIL, another 23 to the TWEAK–APRIL hybrid (Figure 3A) and the remaining two clones encoded unknown cDNAs. All clones lacked the ATG initiation site. To determine whether the TWEAK portion of the TWEAK–APRIL hybrid mRNA encompasses the initiation codon of the TWEAK ORF, RT–PCR experiments were performed using a 5′ primer annealing 30 nucleotides upstream of the TWEAK initiation ATG and a 3′ primer annealing downstream of the hybrid junction, in the APRIL segment (Figure 3A). Fragments of predicted lengths were amplified from cDNA of primary activated T cells and primary monocytes; sequencing showed that the TWEAK–APRIL hybrid mRNA encompasses the TWEAK mRNA 5′ region, including its initiation codon. As the two ORFs are fused in-frame, translation of the hybrid mRNA, starting at the TWEAK initiation codon, would produce a TWEAK–APRIL fusion protein with an estimated mol. wt of 38 kDa. Translation in the two other reading frames shows a succession of stop codons, giving rise to ORFs shorter than 300 nucleotides. We thus conclude that the TWEAK–APRIL hybrid mRNA is translated into a fusion protein, which we term TWE-PRIL. This fusion protein consists of the intracellular, transmembrane and stalk region of TWEAK fused to the extracellular receptor-binding part of APRIL (Figure 3A).
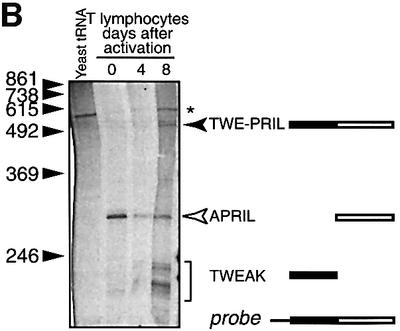
Fig. 3. The variant transcript is a TWEAK–APRIL hybrid mRNA. (A) Sequence of hTWE-PRIL mRNA and the protein encoded. Primers used for PCR experiments are indicated by an arrow. The putative transmembrane region (deduced from that of hTWEAK) is shown as a black box, and the putative furin cleavage site (deduced from that of hAPRIL) as a dark grey box with white letters. The white box shows the portion of the sequence identical to that of hTWEAK mRNA; the grey box shows that corresponding to hAPRIL mRNA. (B) RNase protection assay using a probe encompassing the TWEAK–APRIL junction of the TWE-PRIL hybrid mRNA shows four specific protected fragments. One corresponds to TWE-PRIL mRNA, one to APRIL, and the doublet to TWEAK mRNA. The asterisk indicates a non-specific band, visible in yeast tRNA; molecular weights are shown on the left.
To exclude that the identified TWEAK–APRIL hybrid 5′ RACE products and cDNA clones were due to a cDNA synthesis artefact, and to confirm expression of the hybrid mRNA, RNase protection assays were performed on human T-cell poly(A)+ RNAs using a cDNA overlapping the TWEAK–APRIL junction as probe template (Figure 3B). We detected a protected fragment of ∼290 nucleotides corresponding to the APRIL portion of the probe, common to resting and activated T cells (with less intensity in the latter, as predicted). In activated T cells, additional protected fragments were detected, one of ∼530 nucleotides corresponding to the protected TWEAK– APRIL probe, and a faster migrating doublet of ∼240 nucleotides, corresponding to the TWEAK fragment. These data demonstrate the presence of a hybrid TWEAK–APRIL mRNA in T lymphocytes, which corresponds to the slower migrating vAPRIL detected in the northern blot.
Genomic structure of the TWEAK and APRIL loci
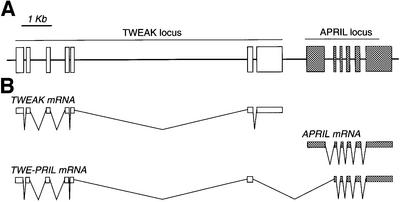
The APRIL gene has been mapped to human chromosome 17 (17p13.1). To address the mechanism of TWE-PRIL mRNA generation, the genomic organization of the APRIL locus was analysed by screening genomic bacterial artificial chromosome (BAC) clones. As a result, BAC clone L144-L13 was identified as encompassing the APRIL gene and its flanking regions. APRIL contains six exons and five introns, which are transcribed into canonical APRIL mRNA. Further sequencing of the flanking regions showed that the TWEAK gene lies 878 bp upstream of the putative APRIL transcription start site, and that both genes are in tandem, with the same orientation (Figure 4A). These data were confirmed recently by the GenBank human genome database, which also provided the full sequence of the TWEAK gene, composed of seven exons and six introns. TWE-PRIL mRNA encompasses TWEAK exons 1–6, fused to APRIL exons 2–6 using the splice donor/acceptor sites of TWEAK and APRIL mRNAs (Figure 4B). The genomic organization of the loci, together with the structure of TWE-PRIL mRNA, are compatible with intergenic splicing between TWEAK and APRIL genes, giving rise to the TWE-PRIL transcript.
Fig. 4. Organization of the human TWEAK and APRIL gene loci. (A) TWEAK and APRIL genes are in the same orientation and separated by <1 kb. (B) TWEAK mRNA is composed of TWEAK exons 1–7, APRIL mRNA of six APRIL exons, whereas the TWE-PRIL transcript comprises TWEAK exons 1–6 fused to APRIL exons 2–6.
TWE-PRIL is expressed as a membrane-anchored protein
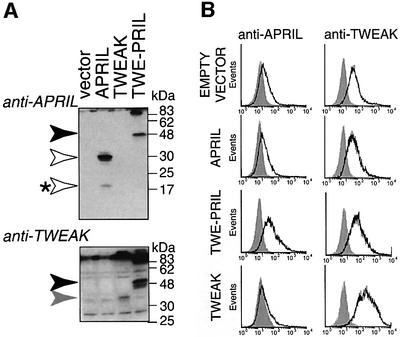
TWEAK was described as a type II cell surface protein (Nakayama et al., 2000), whereas APRIL is processed intracellularly, acting as a secreted factor (Lopez-Fraga et al., 2001). Its structure suggests that, in TWE-PRIL, the APRIL extracellular region is presented at the cell surface. To determine this, human embryonic kidney (HEK) 293T cells were transiently transfected for 24–36 h with constructs of the full ORFs encoding TWEAK, APRIL and TWE-PRIL, or empty vector as a control. Protein expression was verified by western blot analysis of cell lysates using a polyclonal anti-APRIL antibody affinity purified on a peptide encompassing amino acids 125–146 of APRIL (Lopez-Fraga et al., 2001), and a polyclonal anti-human TWEAK antibody (Figure 5A). The anti-APRIL antibody recognized unprocessed (30 kDa) and processed (17 kDa) forms in APRIL transfectants. The anti-TWEAK antibody did not react with lysates of APRIL transfectants, whereas TWEAK transfectants showed a specific signal using anti-TWEAK, but not anti-APRIL antibody (Figure 5A). Lysates of cells transfected with a TWE-PRIL-coding expression vector showed specific bands, at a mol. wt of 48 kDa, when probed with anti-APRIL and anti-TWEAK antibodies (Figure 5A). The difference from the molecular weight estimated for the protein encoded by the TWE-PRIL ORF, 38 kDa, may result from post-translational modifications, as described for APRIL (Hahne et al., 1998). TWE-PRIL also appears to form SDS-stable aggregates, probably trimers, which are detected near the top of the gel. Soluble APRIL was detected in culture supernatant of APRIL-transfected cells and, in lower amounts, in the supernatants of TWE-PRIL transfectants, suggesting that TWE-PRIL may be processed at the cell surface (not shown).
Fig. 5. TWE-PRIL is expressed at the cell surface of HEK 293T transfectants. (A) Protein expression by HEK 293T cells transfected with expression vector, empty or encoding hAPRIL, hTWEAK or TWE-PRIL, was analysed by western blotting of NP-40 cell lysates separated by 15% SDS–PAGE under non-reducing conditions, using polyclonal anti-APRIL and anti-TWEAK antibodies. White arrows indicate signals corresponding to APRIL; the asterisk shows intracellularly processed APRIL. Black and grey arrows indicate TWE-PRIL- and TWEAK-specific signals, respectively. (B) FACS analysis of HEK 293T transfected cells as in (A), using anti-APRIL and -TWEAK antibodies. In each panel, staining with secondary antibody alone is shown in grey as a reference. One representative staining of three is shown.
Cell surface expression levels were determined in the same transfectant samples by fluorescence-activated cell sorting (FACS) analysis, by comparison with cells treated with secondary antibody alone (Figure 5B). No specific staining by the anti-APRIL antibody was observed at the surface of APRIL, TWEAK or control transfectants. In contrast, TWE-PRIL-transfected cells were stained by the anti-APRIL antibody. Conversely, anti-TWEAK antibody showed specific staining in TWEAK-transfected cells, no staining for APRIL transfectants and marginal staining for TWE-PRIL-transfected cells (Figure 5B). The fact that anti-TWEAK antibody detects TWE-PRIL in western blot, but produces low surface staining in FACS analysis, suggests steric inaccessibility of native TWE-PRIL epitopes.
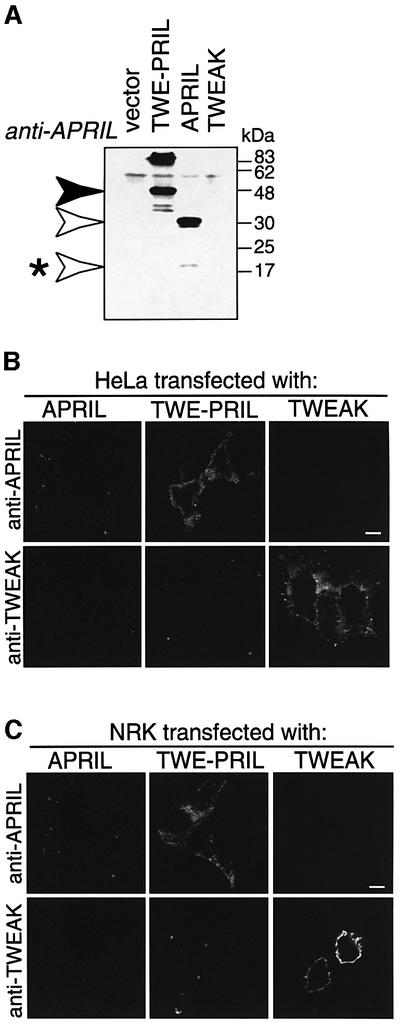
Surface TWE-PRIL expression was confirmed further by surface immunofluorescence staining of non-permeabilized HeLa and NRK transfectants incubated with specific anti-APRIL and anti-TWEAK antibodies prior to fixation. Western blot analysis of cell lysates confirmed expression of the transfected plasmids while showing no detectable signal in cells transfected with the empty vector (Figure 6A; data not shown). In both cell types, fluorescence microscopy demonstrated specific surface staining of TWE-PRIL-expressing cells after incubation with anti-APRIL, but not anti-TWEAK antibody. TWEAK-expressing cells were positive after staining with anti-TWEAK, but not anti-APRIL antibody. As predicted, APRIL-expressing cells showed no staining with either anti-TWEAK or anti-APRIL antibody (Figure 6B and C). These results concur with the FACS analysis and confirm cell surface expression of TWE-PRIL.
Fig. 6. TWE-PRIL is expressed at the cell surface of HeLa and NRK transfectants. (A) Protein expression of HeLa cells transfected with expression vector, either empty (control) or encoding hAPRIL, hTWEAK or hTWE-PRIL, was analysed by western blotting of cell lysates, using polyclonal anti-APRIL antibody, as for Figure 5A. White arrows indicate signals corresponding to hAPRIL; the asterisk shows intracellularly processed APRIL. The black arrow indicates the TWE-PRIL-specific signal. (B and C) Immunofluorescence analysis of HeLa (B) or NRK (C) cells transfected with APRIL, TWE-PRIL or TWEAK. Cells were exposed to the indicated antibodies prior to fixing, revealing cell surface expression of the corresponding antigens. Bar = 200 µm.
Since recombinant TWE-PRIL is expressed as a transmembrane protein, membrane-enriched lysates (containing plasma membrane and organelles) were prepared from human primary T cells and monocytic cell lines, to detect endogenous TWE-PRIL. The lysates were analysed by western blotting with anti-APRIL antibody, in comparison with lysates of 293T cells transfected with vector encoding APRIL, TWE-PRIL or empty vector. In human T cells, both resting and activated for 4 days with anti-CD3/CD28 antibody, two APRIL forms were recognized specifically, corresponding to full-length (30 kDa) and processed (17 kDa) APRIL, in agreement with its processing in the Golgi apparatus prior to secretion (Lopez-Fraga et al., 2001). Both APRIL forms were more abundant in activated than in resting T cells, possibly because of the increased translation efficiency of its mRNA (Figure 2A). In the activated T cells, a third protein was also detected whose mobility was identical to that of recombinant TWE-PRIL (Figure 7). TWE-PRIL is less abundant than APRIL, reflecting the differences in the translation efficiency of their mRNAs, while the absence of signal for TWE-PRIL in the resting T cells may be explained by the low translation efficiency of its mRNA in those cells (Figure 2A). In addition, U937 and Mono Mac 1 cells showed signals for full-length and mature APRIL, as well as TWE-PRIL proteins (Figure 7; data not shown). Importantly, an identical profile was obtained when a different anti-APRIL antibody was used that recognizes a distinct region in APRIL (amino acids 215–236) and TWE-PRIL, excluding the possibility that the protein detected is due to aspecific cross-reactivity (not shown). These data thus indicate that the endogenous TWE-PRIL protein is expressed at detectable levels in activated primary T lymphocytes and in monocytic cell lines.
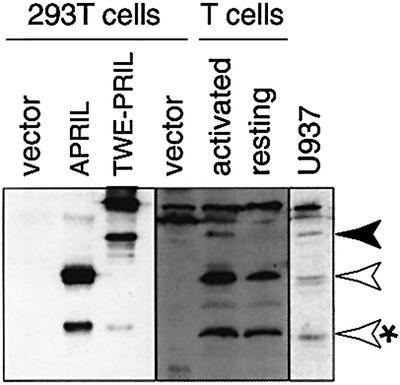
Fig. 7. Endogenous APRIL and TWE-PRIL are detected in human primary T cells and monocytic cells. Membrane-enriched lysates were prepared from resting and activated primary T cells, the U937 monocytic cell line and, as controls, 293T cells transfected with empty vector or vector encoding APRIL and TWE-PRIL, as indicated. Western blot analysis of these extracts using an anti-APRIL antibody shows endogenous expression of full-length (white arrow) and processed APRIL (white arrow, *) in all cells tested. A 48 kDa protein corresponding to TWE-PRIL was also detected in activated T cells and U937 (black arrow) cells. Note that although a longer exposure was required to visualize TWE-PRIL in T cells, no signal corresponding to APRIL or TWE-PRIL was observed in empty vector cell lysates.
TWE-PRIL is a biologically active protein
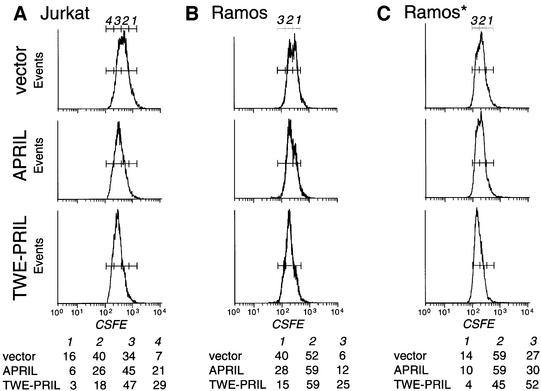
To test the biological activity of TWE-PRIL, transfected 293T cells were irradiated and then co-cultured with Jurkat (a human T-lymphoma cell line) or Ramos (a human B-lymphoma cell line) cells as target. Previously, it has been reported that soluble, recombinant APRIL is capable of stimulating proliferation as well as apoptosis in tumour cell lines (Hahne et al., 1998; Kelly et al., 2000; Yu et al., 2000). We therefore analysed these effects in the co-culture setting. Neither recombinant APRIL nor APRIL-transfected cells induced apoptosis in our hands. Similarly, TWE-PRIL-transfected cells failed to elicit apoptosis in the Jurkat or Ramos target cells (not shown). We therefore focused on the proliferation-stimulating effect of APRIL. To this end, target cells were labelled with carboxyfluorescein succinimidyl ester (CSFE) prior to co-culturing, to monitor cell divisions of the target cells co-cultured with the different transfectants. Each division decreases the CSFE intensity by 2-fold and this can be detected easily by FACS (Figure 8). Jurkat cells cultured with APRIL transfectants showed an increase in cell division, as compared with those cultured with control transfectants, whereas Jurkat cells cultured with TWE-PRIL transfectants showed an even greater increase (Figure 8A). Such an effect was also observed using Ramos cells as targets, after 2–5 days co-culture (Figure 8B and C). In similar experiments, the supernatant of the transfected cells was aspirated prior to target cell addition, to remove any secreted molecules, as the rates of synthesis of transfected proteins are significantly decreased after cell irradiation. Under these conditions, Jurkat and Ramos cells showed little response to APRIL transfectants, as this depends on APRIL secreted into the medium. In contrast, these cells remained fully responsive to TWE-PRIL transfectants (Figure 8C; data not shown). Taken together, these data indicate that TWE-PRIL is a membrane-bound protein that is functional as it can induce proliferation, in a manner comparable with APRIL.
Fig. 8. TWE-PRIL is a biologically active protein. CSFE-labelled Jurkat (A) and Ramos (B and C) cells were cultured on monolayers of 293T cells transfected with empty expression vector or vector encoding APRIL or TWE-PRIL, as indicated. Cell distribution in 2-fold decreasing peaks (as indicated by gates) analysed by FACS after 4 (A) or 3 (B and C) days of co-culture, are indicated below the FACS profiles. In (C), the culture supernatant of the 293T cells was replaced by fresh medium before addition of Ramos cells, to remove any fibroblast-secreted protein. Data shown are representative of five independent experiments.
Discussion
Northern blotting of human primary T lymphocytes, monocytes and tumour cell lines revealed the presence, in addition to the canonical APRIL mRNA, of another as yet undescribed hybrid TWE–PRIL transcript. Hybridization of the filters with a TWEAK probe shows a signal corresponding to the hybrid mRNA (especially evident in sucrose gradients from T lymphocytes) after longer exposure because this probe recognizes a smaller part of the TWE-PRIL transcript than the APRIL probe. RT–PCR and 3′ RACE experiments showed that the APRIL and variant transcripts shared their 3′-untranslated regions (3′-UTRs) and part of the coding sequence. 5′ RACE experiments indicated that the variant transcript comprises, 5′ to the APRIL mRNA fragment, part of another TNF ligand mRNA, namely TWEAK. Expression of this hybrid mRNA was corroborated by screening an unamplified cDNA library derived from activated T lymphocytes. Of the clones isolated, one half corresponded to APRIL cDNA and the other half to TWE-PRIL, in agreement with the relative expression of these transcripts in activated T cells. Finally, RNase protection assay confirmed expression of the hybrid mRNA at levels comparable with those of APRIL mRNA. RT–PCR assays further indicated that TWE-PRIL transcripts encompass TWEAK exons 1–6, including the AUG initiation codon, fused in-frame to APRIL exons 2–6. The splice donor and acceptor sites used for the hybrid transcript are the same as those used for splicing between exons 6 and 7 of TWEAK (donor) and exons 1 and 2 of APRIL (acceptor). TWEAK and APRIL genes map to human chromosome 17 (17.p13.1), are located in tandem and are separated by <1 kb. This organization of the APRIL and TWEAK locus is conserved in the mouse genome (DDBJ/EMBL/GenBank accession No. Mm11 WIFeb01 222). Although alteration of this locus by genomic recombination in T cells and monocytes cannot be formally ruled out, intergenic splicing between TWEAK and APRIL appears to be the simplest explanation for the production of this hybrid mRNA.
A few cases of intergenic splicing between neighbouring genes have been described in eukaryotes (Fears et al., 1996; Magrangeas et al., 1998; Moore et al., 1999; Finta and Zaphiropoulos, 2000; Millar et al., 2000; Communi et al., 2001), although no function could be postulated for the resulting fusion protein. TWE-PRIL would be the first example of intergenic splicing affecting two genes of the same family, resulting in a fusion protein that shares the features typical of TNF ligands. In contrast to previously reported intergenic splicings, TWE-PRIL mRNA expression levels are comparable with that of APRIL (1:6 to 1:1 under different conditions). The TWEAK/TWE-PRIL transcript ratio appears to vary in tumour cell lines, and it is important to note that the TWEAK polyadenylation signal is predictably strong, even stronger than that of APRIL, according to the Polyadq software (http://argon.cshl.org/tabasha/polyadq_form.htlm). These characteristics indicate that TWE-PRIL mRNA is not a by-product of inefficient termination in TWEAK mRNA, but that its expression is tightly regulated, and involves as yet unidentified factors. Simultaneous expression of distinct alternatively spliced transcripts has already been described (Black, 1998). Several models could be envisaged, in which promoters or splice factors would influence the choice of the TWEAK polyadenylation site or of the acceptor splice site (from APRIL exon 2 versus TWEAK exon 7) (for reviews see Smith and Valcarcel, 2000; Proudfoot et al., 2002). Finally, the endogenous protein is detected in primary and tumour cells, and was shown to be functional. Thus, the expression of TWE-PRIL mRNA appears to have functional implications, rather than merely generating genetic diversity, as previously hypothesized for other hybrid RNAs (Magrangeas et al., 1998).
TWEAK, like most TNF ligand family members described so far, and despite the presence of a furin cleavage site in its stalk region, was detected as a membrane-anchored protein in interferon-γ-activated primary human monocytes (Nakayama et al., 2000). Conversely, APRIL is processed intracellularly prior to secretion (Lopez-Fraga et al., 2001). TWE-PRIL, which combines the TWEAK cytoplasmic domain and a portion of the stalk region with the APRIL extracellular trimerization domain, was detected on the surface of intact transfected cells by FACS and immunofluorescence analysis. It is thus likely that the TWEAK cytoplasmic tail, shared by TWE-PRIL, anchors this protein to the plasma membrane, exposing the APRIL-like domain to the extracellular milieu.
TNF ligands are characterized by their C-terminal domain, which mediates their binding to the cognate receptor. TWE-PRIL and APRIL share their receptor-binding domain, and may thus bind the same receptors. Transmodulin activator and cAML interactor (TACI) and B-cell maturation antigen (BCMA) are APRIL receptors, which are shared with B lymphocyte stimulator (BLyS), another TNF ligand family member (Yu et al., 2000). RT–PCR experiments showed that both receptors are expressed on Ramos cells (not shown), which were APRIL sensitive in co-culture experiments. In addition, there is growing evidence for the existence of another, as yet unidentified receptor that binds to APRIL but not to BLyS (Rennert et al., 2000). This unknown APRIL receptor is expressed on Jurkat cells, which lack TACI and BCMA (Rennert et al., 2000) but responded to APRIL secreted by 293T transfected cells. Furthermore, both Ramos and Jurkat cells were responsive to transmembrane TWE-PRIL. These data indicate that TWE-PRIL may recognize the same receptors as APRIL. Consistently with this hypothesis, a BLyS–APRIL chimeric protein, in which the APRIL receptor-binding domain is anchored at the cell membrane by the BLyS transmembrane domain, which results in a structure similar to that of TWE-PRIL, binds BCMA (Rennert et al., 2000). This demonstrates that membrane anchoring does not alter APRIL binding specificity. By comparison with other ligand–receptor interactions within the TNF family, APRIL and TWE-PRIL may have different affinities for their cognate receptors. It has been reported that soluble TNF-α can be a more potent activator of TNF receptor 2 than the membrane-bound form (Grell et al., 1995). Furthermore, soluble and membrane-bound TNF-α have been demonstrated to trigger distinct signals (Alexopoulou et al., 1997; Kusters et al., 1997; Ruuls et al., 2001). It was reported recently that membrane-bound TNF-α supports many features of lymphoid organ structure, whereas secreted TNF-α is needed for optimal proinflammatory functions (Ruuls et al., 2001). Several other members of the TNF ligand family are expressed as membrane-anchored proteins that can be cleaved into a soluble form, including TWEAK and BLyS. In those cases, both forms are biologically active, yet no distinct function can be attributed to either form (Nakayama et al., 2000; Nardelli et al., 2001). Further experiments are necessary to decipher the physiological role of TWE-PRIL; maybe some of the functions ascribed to APRIL may in fact be exerted by TWE-PRIL, as recombinant soluble APRIL proteins are very hydrophobic and tend to form aggregates in solution that could mimic membrane-anchored TWE-PRIL. Nevertheless, it is clear that the functions of APRIL and TWE-PRIL differ in that APRIL can act at a distance as a secreted cytokine, while TWE-PRIL expectedly acts in cell–cell contact. In resting and activated T lymphocytes, TWEAK mRNA distribution in sucrose gradients did not vary, so that translation efficiency, in particular, of APRIL mRNA appears to be increased specifically following T-lymphocyte activation (Figure 1A). TNF-α is the only known member of the TNF ligand family described as translationally controlled, involving trans-acting factors that bind to its 3′-UTR AU-rich (ARE) motifs (Mijatovic et al., 2000). APRIL mRNA lacks any such motif, but comparison of the human and mouse mRNA sequences (DDBJ/EMBL/GenBank accession No. AF294825) suggests that a conserved 170 nucleotide motif 5′ to the AUG initiation codon may be involved in its translational regulation.
In conclusion, this report constitutes the first description of an endogenous fusion protein, TWE-PRIL, which is translated from a hybrid transcript between TWEAK and APRIL mRNAs, two members of the TNF ligand family. This protein allows membrane anchoring of the APRIL receptor-binding domain, which otherwise is secreted. Generation of appropriate knockout mice will allow definition of the specific roles of TWE-PRIL and APRIL.
Materials and methods
Cells and culture conditions
Human primary T cells were isolated by Ficoll-Paque (Pharmacia, Uppsala, Sweden) from buffy coats from healthy donors, followed by Percoll (Pharmacia) gradient centrifugation, then activated with a combination of anti-CD3 (clone 3Tb) and soluble anti-CD28 mAb (kindly provided by Dr D.Olive, Marseille, France) as described (Mikulits et al., 2000). CD4+ and CD8+ T-lymphocyte populations were positively selected using mouse anti-hCD4 and anti-hCD8 antibodies, respectively (BD PharMingen), and recovered with goat anti-mouse Ig antibody coupled to magnetic beads (Dynabead, Sunnyvale, CA).
Human primary monocytes were isolated from peripheral blood mononuclear cells by density centrifugation on Ficoll gradients and adherence on gelatin (Merck, Darmstadt, Germany)-coated flasks (Espel et al., 1996). After 3 days culture, a >95% pure monocyte population was recovered and allowed to rest for 3 h (37°C, 5% CO2) prior to LPS stimulation (Sigma, St Louis, MO).
Cell lines used (Mono Mac 1, U937, colon carcinoma cells, NRK, HeLa, Ramos, Jurkat and 293T HEK cells; all from the American Type Culture Collection, Manassas, VA) were cultured at 1 × 106 cells/ml on Petri dishes in 10% endotoxin-free fetal calf serum (FCS)/Dulbecco’s modified Eagle’s medium supplemented with 2 mM l-glutamine.
RNA preparation, poly(A)+ purification, RNase protection assay, northern blot analysis and sucrose gradient fractionation
Cytoplasmic RNA was prepared using NP-40 and total RNA by the guanidinium–thiocyanate–acid phenol method (Müllner and Garcia-Sanz, 1997c). When indicated, RNA populations were poly(A)+ enriched by two rounds of purification using the Oligotex-mini kit (Qiagen, Hilden, Germany), and yields were monitored by northern blotting.
Hybrid TWE-PRIL mRNA was analysed in poly(A)+ RNAs by RNase protection assay using a specific SP6 antisense transcript as probe (Garcia-Sanz and Müllner, 1997). After separation of protected fragments on a 6% sequencing gel, signals were quantified by phosphorimaging (Molecular Dynamics, Sunnyvale, CA).
For northern blot analysis, RNA samples were separated by electrophoresis through denaturing 1.2% formaldehyde–agarose gels and transferred to nylon membranes (GeneScreen, NEN, Boston, MA). rRNA distribution was visualized by methylene blue staining. Northern blots were hybridized sequentially with cRNA probes specific for human APRIL and human TWEAK, after [α-32P]CTP labelling using SP6 polymerase. Hybridization was performed for 2 days at 65°C, followed by washes, with a final wash at 74°C for 45 min (Müllner and Garcia-Sanz, 1997a). Filters subsequently were exposed and quantified by phosphorimaging.
Sucrose gradient fractionation was performed essentially as described (Müllner and Garcia-Sanz, 1997b). Cell extracts were prepared by lysis at 4°C in extraction buffer (10 mM Tris–HCl pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 20 mM dithiothreitol and 500 U/ml RNAsin), and nuclei and mitochondria removed by centrifugation (12 000 g, 1 min, 4°C). The supernatant was supplemented with 150 µg/ml cycloheximide, 665 mg/ml heparin and 1 mM phenylmethylsulfonyl fluoride (PMSF), layered onto a 10 ml linear sucrose gradient and centrifuged in an SW41Ti rotor (Beckman, Palo Alto, CA; 180 000 g 120 min, 4°C) without braking. Fractions (550 µl) were collected and digested with 100 µg of proteinase K (30 min, 37°C). RNA was recovered by ethanol precipitation and analysed by northern blotting.
RT–PCR and RACE experiments
5′ and 3′ RACE experiments were performed using the Marathon cDNA Amplification kit or the SMART kit (Clontech) and the 5′/3′ Race kit (Roche, Switzerland). Amplifications were performed with Advantaq 2 or Advantaq-GC 2 (both from Clontech), according to the manufacturer’s instructions for reaction mix and cycling conditions.
TWE-PRIL cloning and genomic sequencing
TWE-PRIL clones were obtained from an unamplified cDNA library (Edge BioSystems, Gaithersburg, MD) derived from T cells of healthy blood donors, which had been stimulated with 1 µg/ml PHA for 1 day and expanded in the presence of interleukin-2 (100 U/ml) for an additional 7–10 days. For screening, a probe comprising the extracellular domain of hAPRIL was used (for experimental details, see the manufacturer’s protocol; Edge BioSystems, MD).
For genomic sequencing, a BAC clone (L144-L13) was identified from a human BAC DNA pool as described by the supplier (Genome Systems, St Louis, MO). Clone L144-L13 was analysed by subsequent sequencing using various primers designed from the published APRIL cDNA sequence (Hahne et al., 1998).
Cell transfections and co-culture conditions
Constructs containing the APRIL, TWEAK or TWE-PRIL ORFs were cloned into the pCcDNA3.1/Hygro(+) expression vector (Invitrogen, Gaithersburg, MD) and transfected into HEK 293T, HeLa and RNK cells using the Lipofectamine method (Invitrogen). Cell populations were analysed 24–36 h after transfection. For co-culture experiments, 293T cells were irradiated (30 Gy) 16 h after transfection, before target cell addition.
Polyclonal antibody production
Generation of polyclonal anti-APRIL antibodies was described recently (Lopez-Fraga et al., 2001). Polyclonal anti-TWEAK antibodies were generated in rabbits after immunization with a recombinant soluble form of TWEAK spanning extracellular amino acids 106–249. Purified IgG fractions were used for western blot analysis or FACS staining.
Subcellular fractionation and western blot analysis
For subcellular fractionation, cells were incubated (5 min, on ice) with TES buffer (10 mM Tris pH 7.5, 1 mM EDTA, 0.25 M sucrose), lysed by 10 passages through a 25 gauge needle and depleted of nuclei (800 g). Nucleus-depleted lysates were enriched for the membrane fraction by ultracentrifugation (100 000 g, 1 h). The pelleted fraction containing mitochondria, microsomes, plasma membrane and organelles was then resuspended in TES containing 1% NP-40 to solubilize the proteins. This fraction is referred to as membrane-enriched lysates.
Cytoplasmic lysates were prepared in NP-40 buffer (1% NP-40, 1% leupeptin, 1% aprotinin, 1 mM PMSF in 50 mM Tris pH 7.4). After protein quantification to ensure equal sample loading, lysates were separated by SDS–PAGE and transferred to nitrocellulose. Equal loading and transfer were verified in each experiment by Ponceau S staining of the membrane (Sigma). Immunoblots were probed using 2 µg/ml affinity-purified anti-APRIL or 20 µg/ml anti-TWEAK antibody and developed using the ECL system (Amersham Pharmacia Biotech).
Cytofluorometric analysis and CSFE labelling
Transfected cells were incubated with primary antibody (20 min, on ice) in phosphate-buffered saline (PBS) supplemented with 2% FCS and sodium azide. After primary antibody staining, cells were washed and incubated with the second antibody following the same protocol. For CSFE labelling, cells were incubated with 2.5 µM CSFE in PBS (3 min, room temperature), followed by incubation (1 min) with 1:5 (v/v) FCS. Cells were washed twice, incubated (1 h, 37°C), washed again and added to the transfectants. Cell surface protein expression and CSFE staining were monitored by flow cytometry (XL, Coulter, Miami, FL) and analysed with the manufacturer’s software package.
Immunofluorescence and image acquisition
Cells were cultured overnight in chamber slides, washed with PBS, treated with 5% bovine serum albumin, 10% goat serum in PBS (≥30 min, 4°C), and incubated for 1 h with 2 µg/ml anti-APRIL or 20 µg/ml anti-TWEAK antibody, and for 1 h with Cy2- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch). Cells were washed again, then fixed in 4% paraformaldehyde (15 min, room temperature). Images were obtained using an Ar–Kr laser and a TCS-NT Leica confocal imaging system (Leica Microsystems, Heidelberg, Germany).
Acknowledgments
Acknowledgements
We are grateful to Dr M.Campanero for critically reviewing the manuscript, C.Mark for editorial assistance, M.Obrero for plasmid preparation, M.C.Moreno for help with FACS, T.Santos for help with cell fractionation, and P.Lucas, G.del Real and M.Fresno for providing buffy coats. This work was supported in part by an EU-TMR Network grant (contract number ERBFMRXCT980197, to J.A.G.-S.). M.L.F. is supported by the Consejería de Educación y Cultura de la Comunidad de Madrid, financed by the Fondo Social Europeo. The Department of Immunology and Oncology was founded and is supported by the Spanish Council for Scientific Research (CSIC) and by the Pharmacia Corporation.
References
- Alexopoulou L., Pasparakis,M. and Kollias,G. (1997) A murine transmembrane tumor necrosis factor (TNF) transgene induces arthritis by cooperative p55/p75 TNF receptor signaling. Eur. J. Immunol., 27, 2588–2592. [DOI] [PubMed] [Google Scholar]
- Black D.L. (1998) Splicing in the inner ear: a familiar tune, but what are the instruments? Neuron, 20, 165–168. [DOI] [PubMed] [Google Scholar]
- Chicheportiche Y., Bourdon,P.R., Xu,H., Hsu,Y.M., Scott,H., Hession,C., Garcia,I. and Browning,J.L. (1997) TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J. Biol. Chem., 272, 32401–32410. [DOI] [PubMed] [Google Scholar]
- Communi D., Suarez-Huerta,N., Dussossoy,D., Savi,P. and Boeynaems,J.M. (2001) Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. J. Biol. Chem., 276, 16561–16566. [DOI] [PubMed] [Google Scholar]
- Espel E., Garcia-Sanz,J.A., Aubert,V., Menoud,V., Sperisen,P., Fernandez,N. and Spertini,F. (1996) Transcriptional and translational control of TNF-α gene expression in human monocytes by major histocompatibility complex class II ligands. Eur. J. Immunol., 26, 2417–2424. [DOI] [PubMed] [Google Scholar]
- Fears S., Mathieu,C., Zeleznik-Le,N., Huang,S., Rowley,J.D. and Nucifora,G. (1996) Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl Acad. Sci. USA, 93, 1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finta C. and Zaphiropoulos,P.G. (2000) The human CYP2C locus: a prototype for intergenic and exon repetition splicing events. Genomics, 63, 433–438. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanz J.A. and Müllner,E.W. (1997) RNase protection and primer extension. In Lefkovits,I. (ed.), Immunology Methods Manual. Vol. 1. Academic Press, London, UK, pp. 425–438.
- Grell M. et al. (1995) The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell, 83, 793–802. [DOI] [PubMed] [Google Scholar]
- Hahne M. et al. (1998) APRIL, a new ligand of the tumor necrosis factor family, stimulates tumor cell growth. J. Exp. Med., 188, 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K., Manos,E., Jensen,G., Nadauld,L. and Jones,D.A. (2000) APRIL/TRDL-1, a tumor necrosis factor-like ligand, stimulates cell death. Cancer Res., 60, 1021–1027. [PubMed] [Google Scholar]
- Kusters S. et al. (1997) In vivo evidence for a functional role of both tumor necrosis factor (TNF) receptors and transmembrane TNF in experimental hepatitis. Eur. J. Immunol., 27, 2870–2875. [DOI] [PubMed] [Google Scholar]
- Lopez-Fraga M., Fernandez,R., Albar,J.P. and Hahne,M. (2001) Biologically active APRIL is secreted following intracellular processing in the Golgi apparatus by furin convertase. EMBO rep., 2, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C.N., Wang,Y.C., Lund,J.K., Chen,Y.W., Leal,J.A. and Wiley,S.R. (1999) TWEAK induces angiogenesis and proliferation of endothelial cells. J. Biol. Chem., 274, 8455–8459. [DOI] [PubMed] [Google Scholar]
- Magrangeas F. et al. (1998) Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor α-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J. Biol. Chem., 273, 16005–16010. [DOI] [PubMed] [Google Scholar]
- Mijatovic T., Houzet,L., Defrance,P., Droogmans,L., Huez,G. and Kruys,V. (2000) Tumor necrosis factor-α mRNA remains unstable and hypoadenylated upon stimulation of macrophages by lipopolysaccharides. Eur. J. Biochem., 267, 6004–6012. [DOI] [PubMed] [Google Scholar]
- Mikulits W., Pradet-Balade,B., Habermann,B., Beug,H., Garcia-Sanz,J.A. and Mullner,E.W. (2000) Isolation of translationally controlled mRNAs by differential screening. FASEB J., 14, 1641–1652. [DOI] [PubMed] [Google Scholar]
- Millar J.K., Christie,S., Semple,C.A. and Porteous,D.J. (2000) Chromosomal location and genomic structure of the human translin-associated factor X gene (TRAX; TSNAX) revealed by intergenic splicing to DISC1, a gene disrupted by a translocation segregating with schizophrenia. Genomics, 67, 69–77. [DOI] [PubMed] [Google Scholar]
- Molloy S.S., Anderson,E.D., Jean,F. and Thomas,G. (1999) Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol., 9, 28–35. [DOI] [PubMed] [Google Scholar]
- Moore R.C. et al. (1999) Ataxia in prion protein (PrP)-deficient mice is associated with upregulation of the novel PrP-like protein doppel. J. Mol. Biol., 292, 797–817. [DOI] [PubMed] [Google Scholar]
- Müllner E.W. and Garcia-Sanz,J.A. (1997a) Analysis of RNA expression by Northern blotting. In Lefkovits,I. (ed.), Immunology Methods Manual. Vol. 1. Academic Press, London, UK, pp. 407–424.
- Müllner E.W. and Garcia-Sanz,J.A. (1997b) Polysome gradients. In Lefkovits,I. (ed.), Immunology Methods Manual. Vol. 1. Academic Press, London, UK, pp. 457–462.
- Müllner E.W. and Garcia-Sanz,J.A. (1997c) Preparation of RNA. In Lefkovits,I. (ed.), Immunology Methods Manual. Vol. 1. Academic Press, London, UK, pp. 389–406.
- Nakayama M., Kayagaki,N., Yamaguchi,N., Okumura,K. and Yagita,H. (2000) Involvement of TWEAK in interferon γ-stimulated monocyte cytotoxicity. J. Exp. Med., 192, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli B. et al. (2001) Synthesis and release of B-lymphocyte stimulator from myeloid cells. Blood, 97, 198–204. [DOI] [PubMed] [Google Scholar]
- Pradet-Balade B., Boulme,F., Beug,H., Mullner,E.W. and Garcia-Sanz,J.A. (2001) Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci., 26, 225–229. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger,A. and Dye,M.J. (2002) Integrating mRNA processing with transcription. Cell, 108, 501–512. [DOI] [PubMed] [Google Scholar]
- Rennert P. et al. (2000) A soluble form of B cell maturation antigen, a receptor for the tumor necrosis factor family member APRIL, inhibits tumor cell growth. J. Exp. Med., 192, 1677–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth W., Wagenknecht,B., Klumpp,A., Hahne,M., Tschopp,J. and Weller,M. (2001) Protection from death ligand-induced apoptosis by APRIL, a new ligand of the tumor necrosis factor family. Cell Death Differ., 8, 403–410. [DOI] [PubMed] [Google Scholar]
- Ruuls S.R. et al. (2001) Membrane-bound TNF supports secondary lymphoid organ structure but is subservient to secreted TNF in driving autoimmune inflammation. Immunity, 15, 533–543. [DOI] [PubMed] [Google Scholar]
- Shu H.-B., Hu,W.-H. and Johnson,H. (1999) TALL-1 is a novel member of the TNF family that is down-regulated by mitogens. J. Leukoc. Biol., 65, 680–683. [PubMed] [Google Scholar]
- Smith C.A., Farrah,T. and Goodwin,R.G. (1994) The TNF receptor superfamily of cellular and viral proteins: activation, costimulation and death. Cell, 76, 959–962. [DOI] [PubMed] [Google Scholar]
- Smith C.W. and Valcarcel,J. (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci., 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Stein J.V. et al. (2002) APRIL modulates B and T cell immunity. J. Clin. Invest., 109, 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. (2000) APRIL and TALL-I and receptors BCMA and TACI: system for regulating humoral immunity. Nat. Immunol., 1, 252–256. [DOI] [PubMed] [Google Scholar]