Abstract
Genetic linkage studies indicate that germline variations in a gene or genes on chromosome Xq27–28 are implicated in prostate carcinogenesis. The linkage peak of prostate cancer overlies a region of ∼750 kb containing five SPANX genes (SPANX-A1, -A2, -B, -C, and -D) encoding sperm proteins associated with the nucleus; their expression was also detected in a variety of cancers. SPANX genes are >95% identical and reside within large segmental duplications (SDs) with a high level of similarity, which confounds mutational analysis of this gene family by routine PCR methods. In this work, we applied transformation-associated recombination cloning (TAR) in yeast to characterize individual SPANX genes from prostate cancer patients showing linkage to Xq27–28 and unaffected controls. Analysis of genomic TAR clones revealed a dynamic nature of the replicated region of linkage. Both frequent gene deletion/duplication and homology-based sequence transfer events were identified within the region and were presumably caused by recombinational interactions between SDs harboring the SPANX genes. These interactions contribute to diversity of the SPANX coding regions in humans. We speculate that the predisposition to prostate cancer in X-linked families is an example of a genomic disease caused by a specific architecture of the SPANX gene cluster.
Prostate cancer is the most commonly diagnosed cancer in the United States, occurring in as many as 15% of men. Over the years, there has been an accumulation of genetic epidemiological evidence in favor of a significant hereditary component in prostate cancer susceptibility. Genetic linkage studies have been used to show that multiple chromosomal regions harbor prostate cancer susceptibility genes, including HPC1 at 1q24–25, PCAP at 1q42–43, CAPB at 1q36, hereditary prostate cancer locus (HPC20) at 20q13, a locus at 16q, and a locus at 8p22–23 (Verhage and Kiemeney 2003; Montironi et al. 2004; Rubin and De Marzo 2004; Schaid 2004). At least five candidate prostate cancer susceptibility genes have been reported so far: ELAC2, RNASEL, MSR1, CHEK2, and BRCA2 (Verhage and Kiemeney 2003; Montironi et al. 2004; Rubin and De Marzo 2004; Schaid 2004). Two of them, RNASEL and MRS1, encode proteins with critical functions in the host response to infection, suggesting that defects in pathways involving innate immunity may have a particularly important role in the initiation of prostate cancer. However, even if this is confirmed, these genes are likely to account for only a small fraction of the observed genetic predisposition to prostate cancer.
The first evidence for a hereditary prostate cancer susceptibility gene at Xq27–28 (HPCX) was provided by a genetic linkage analysis of 360 hereditary prostate cancer families (Xu et al. 1998). In this study, family members from prostate cancer pedigrees in the United States, Finland, and Sweden were genotyped. Later, the prostate cancer linkage at this region was confirmed by independent replication studies (Lange et al. 1999; Xu et al. 2003; Brown et al. 2004; Gillanders et al. 2004; Farnham et al. 2005) and by the analysis of 104 German prostate cancer families (Bochum et al. 2002).
A recent genome-wide scan for prostate cancer susceptibility genes in Finnish X-linked families provided the strongest evidence for linkage between DXS1227 and DXS297 (Gillanders et al. 2004; Baffoe-Bonnie et al. 2005). Up to that point, no candidate tumor-related genes were reported to reside within this region. The mapped region spans ∼750 kb and contains the cluster of five SPANX genes (SPANX-A1,-A2,-B,-C, and -D) and LDOC1, which is down-regulated in some cancers (Nagasaki et al. 2003) (Fig. 1). The SPANX (Sperm Protein Associated with the Nucleus on the X chromosome) multigene family encodes proteins whose expression is restricted to the normal testis, a few nongametogenic tissues, and certain tumors (Zendman et al. 1999, 2003; Westbrook et al. 2000, 2004; Goydos et al. 2002; Wang et al. 2003; Kouprina et al. 2004). SPANX genes encode small unfolded proteins (∼100 amino acid residues), to some extent resembling the high mobility group A (HMGA) proteins involved in the formation of various nucleoprotein complexes. Similar to these proteins, SPANXs can form dimers and complexes with other proteins (Westbrook et al. 2000, 2001). In transformed mammalian cells, SPANX proteins are associated with the nuclear envelope, a location similar to the one in human spermatozoa (Zendman et al. 1999; Westbrook et al. 2001, 2004).
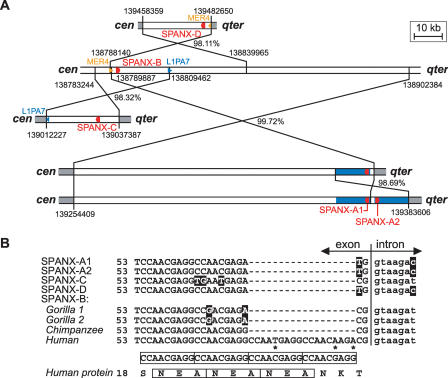
Figure 1.
Schematic structure of the SPANX-A/D locus. The scheme shows the positions of SPANX (red) and LDOC1 (green) genes. Boxes mark the positions of segmental duplications, and the box orientation corresponds to the direction of transcription of the embedded SPANX gene. The segment is drawn to scale and corresponds to a 750 kb-long genomic segment (chrX:138750001–139500000). Fragments isolated by TAR cloning are shown as well. The positions relative to the centromere (cen) and telomere (qter) are marked.
A recent analysis of SPANX gene homologs in nonhuman primates revealed that SPANX-A/D genes arose ∼7 million years ago and that their expansion is an ongoing process in hominids (Kouprina et al. 2004). In addition, this study led to the discovery of a new subfamily of SPANX genes (SPANX-N1-N5) in the human genome. The SPANX-N subfamily present in all mammals is the ancestral subfamily that gave rise to the SPANX-A/D subfamilies in the hominoid lineage. Four SPANX-N genes (N1, -N2, -N3, and -N4) were mapped ∼1.3 Mb away from the SPANX-A/D gene cluster. The fifth member of the SPANX-N subfamily, SPANX-N5, is located on the short arm of the chromosome at Xp11. Similar to other genes involved in reproduction, SPANX genes appear to have evolved under strong positive selection (Kouprina et al. 2004).
Given the possible role of SPANX-A/D genes in hereditary prostate cancer, we set out to study the entire SPANX gene cluster in families affected by prostate cancer. Because these genes are very similar and reside within large chromosomal duplications with a high level of sequence similarity, a routine PCR analysis could not be used to study individual SPANX genes. Therefore, we took advantage of transformation-associated recombination (TAR) cloning technology that allows direct isolation of full-size genes and gene clusters up to 250 kb from complex genomes (Kouprina and Larionov 2003). A comprehensive analysis of SPANX-C, SPANX-B, and SPANX-D loci is presented here.
Our analysis revealed an extensively complex and dynamic organization of the SPANX genes. A variable number of SPANX gene copies and frequent homology-based sequence transfer events between SPANX gene members were both detected. Although further study is needed, we propose that the X-linked susceptibility to prostate cancer is a genomic disorder caused by recombinational interactions between segmental duplications (SDs) representing more than 35% of the entire SPANX-A/D region.
Results
Structure and evolution of the SPANX locus
Our previous analysis had shown that the SPANX-A/D genes reside within SDs (Stephan et al. 2002). However, neither the size nor the homology of the duplicated regions was determined because of the lack of the complete sequence of Xq27–28 at that time. The completion of the human genome, along with a new information on the organization of the SPANX genes in nonhuman primates (Kouprina et al. 2004; Ross et al. 2005), allowed us to analyze SDs within the gene cluster. Figure 1 illustrates organization of the SPANX-A/D gene cluster. As seen, the size of duplications varies from 16 kb (for SPANX-A1/A2) to 119 kb (for SPANX-B). Pairwise comparison of SDs revealed a high level of similarity (between 91% and 99%), indicating their recent evolutionary origin (Supplemental Table 3S). This comparison also identified traces of gene conversion events between the duplications (Supplemental Fig. 2S).
In our recent paper, we demonstrated that the SPANX-A/D genes appeared after a divergence between the orangutan and hominoid lineages (Kouprina et al. 2004). The expansion of the genes seems to occur by duplication of the chromosomal segment. Sequence analysis of SDs allowed us to reconstruct the evolutionary history of SPANX-A/D genes (Fig. 2A). The most likely scenario is that SPANX-A1,-A2,-C, and -D originated from the progenitor SPANX-B sequence. All duplications are found in the inverted orientations relative to a parental gene copy. The proposed scheme is supported by a comparative analysis of SPANX-C syntenic regions in the human, chimpanzee, bonobo, and gorilla. (Genomic regions from bonobo and gorilla were isolated by TAR cloning in our laboratory; GenBank accession numbers AY459387 and AY459388.) African Great Apes lack the SPANX-C specific segmental duplication (Kouprina et al. 2004). Thus, alignment of syntenic human, chimpanzee, gorilla and bonobo sequences (data not shown) allowed an accurate determination of the break points of the SPANX-C-containing SD in humans. The analysis revealed that SPANX-C is derived from SPANX-B by gene conversion-associated additional transfer of adjacent sequences (Fig. 3), as has been proposed previously for other regions by Richardson and co-authors (Richardson et al. 1998; Richardson and Jasin 2000). A sequence comparison also demonstrated a role of interspersed repeats (such as LINE1 and Alu) in generating SDs (Figs. 2A and 3).
Figure 2.
Evolution of the SPANX-A/D genes. (A) The probable scheme of evolution of the SPANX-A/D gene family by segmental duplications. The genes are marked in red. Positions relative to the centromere (cen) and telomere (qter) are marked. The numbers near duplication breakpoints indicate positions of the duplicated regions in the human genome (hg16; UCSC July 2003 genome version). SPANX-B seems to be the original locus. SPANX-B and its duplications are shown as white boxes, the sequences flanking the duplications are gray. The number next to the crossed lines shows the nucleotide identity between the duplicated regions, after excluding gaps. SPANX-D is derived from an inverted duplication of SPANX-B. The SPANX-D telomeric breakpoint colocalizes with the terminus of a MER4 element (orange), whereas the corresponding SPANX-B centromeric breakpoint contains a longer MER4 element that spans further from the duplicated region. The origin of the SPANX-C locus from SPANX-B by LINE1-mediated recombination was confirmed by a comparison with other primates (see Fig. 3). The SPANX-A1/A2 locus was probably created by a two-step process that first duplicated a large (∼115-kb) segment from the SPANX-B region and subsequently amplified an ∼17-kb-long segment containing the SPANX-A1 gene (blue boxes). (B) The origin of the human-specific 18-bp-long insertion in SPANX-B exon 1. The terminal part of the first exon contains a duplication of the 9-bp-long motif CCAACGAGG also detectable at the protein level (boxed). Whereas all other human SPANX-A/D genes and African Great Ape SPANX-B genes contain only two copies of the 9-bp repeat, the human SPANX-B gene exhibits two additional copies of the repeat. Asterisks mark deviations from the repeat consensus.
Figure 3.
Origin of the SPANX-C locus in humans. The figure shows the proposed scenario for the origin of the SPANX-C locus in humans. The scheme is based on the replication-based model of recombination with long-tract gene conversion (Richardson et al. 1998; Richardson and Jasin 2000). For simplicity, both loci are drawn in the same orientation from left to right, but, in fact, this duplication is accompanied by an inversion. The initial stimulus for recombination was a DNA break in an L1PA7 copy (dark blue, contains an AluSx insert) in the pre-C locus (preserved in the chimpanzee, bonobo, and gorilla), followed by an invasion of one end in the 44-bp-long stretch of identity (black) in the L1PA4 copy (light blue) of the SPANX-B locus. Repair synthesis continues further down past the end of L1PA4 until a random interruption in the LTR1B copy (green). The repair is then completed by nonhomologous end joining (NHEJ) of the invading strand with the original break. This model thus combines homologous recombination in the first step with nonhomologous recombination in the terminal stage. The new SPANX-C locus contains (1) a chimeric LINE1 element composed of an original L1PA7 fragment and a tail derived from L1PA4 (light blue), (2) a duplicated SPANX-B locus that is terminated with a LTR1B fragment, and (3) the terminal part of the L1PA7 element. The net result is duplication of the SPANX-C locus, while the second copy (SPANX-B) remains intact.
It is notable that the human SPANX-B gene contains a specific 18-bp insertion in the exon 1 sequence. We found that SPANX-B of African Great Apes lacks a 18-bp insertion (Fig. 2B; Supplemental Fig. 6S). This finding indicates that acquisition of the 18-bp insertion in exon 1 probably occurred after expansion of the SPANX-A/D genes.
It is well documented that SDs mediate ectopic interaction of loci that can result in chromosomal rearrangements such as duplications, deletions, and inversions (Bailey et al. 2002; Sebat et al. 2004). A high density of SDs within the HPCX mapped region at Xq27–28 suggests that the predisposition to prostate cancer in some HPCX families may be a result of genomic rearrangements mediated by SDs.
Analysis of the SPANX-C locus in X-linked prostate cancer families revealed a high rate of ectopic recombination
The inability to distinguish SPANX genes by conventional PCR methods forced us to apply a TAR cloning strategy for their mutational analysis (Supplemental Fig. 1S).
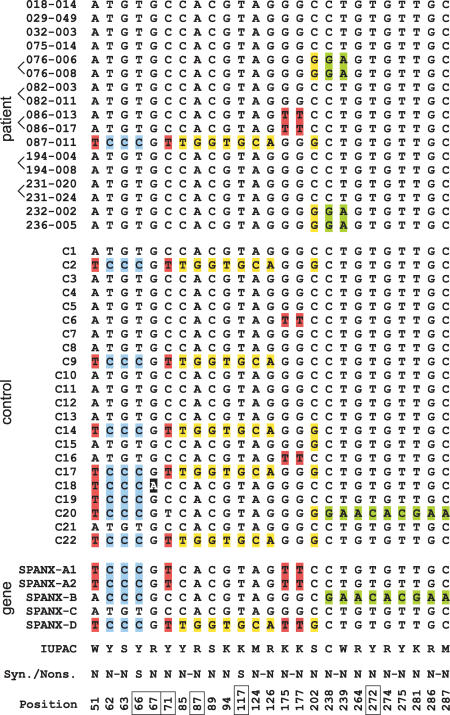
For the direct isolation of the SPANX-C genomic segment, a TAR vector was designed containing targeting sequences chosen from the unique SPANX-C flanking sequences, ∼17 kb upstream and ∼66 kb downstream from the SPANX-C gene. Thus, this TAR vector allows an 83-kb genomic DNA fragment carrying the SPANX-C gene to be cloned. The SPANX-C locus was TAR-cloned from 17 individuals (12 families) affected with prostate cancer showing linkage to Xq27 and from 22 unaffected controls (see Methods). To check for the presence of mutations in SPANX-C, exon and noncoding sequences were PCR amplified from each yeast artificial chromosome (YAC) TAR isolate using a pair of specific primers (Supplemental Table 1S). A sequence analysis of the SPANX-C gene from the patients identified 50 single nucleotide polymorphisms (SNPs) in 1515-bp-long compared segments. Eighteen SNPs were present in exon sequences, and all of them, with two exceptions, resulted in amino acid changes (Fig. 4). Alignments of SPANX-C complete gene variants are shown in Supplemental Figure 3S. As seen, there are four variants of the SPANX-C coding region in the patients. One of the variants, present in patient 087–011, contains 12 nucleotide changes that result in eight different amino acid substitutions within the SPANX protein. However, because the same nucleotide substitutions were found in unaffected individuals (Fig. 4), these changes seem to represent normal polymorphisms in SPANX-C. Still, it is notable that TCCC changes are only seen in one case, 087–011 (at positions 51, 62–63, and 66), compared with eight cases in the control group (Fig. 4). This may indicate that the “ATGT” SPANX-C allele may be a prevalent allele in patients. Comparison of these polymorphic variants with the sequences of other members of the SPANX gene family reveals that almost all nucleotide changes are caused by homology-based sequence transfer (Fig. 4). For example, a SPANX-C variant in patient 087–011 is identical to SPANX-D, which is located ∼500 kb away from the SPANX-C locus. We isolated the SPANX-D locus from the same patient by TAR cloning (see below). An analysis revealed the identity of SPANX-C and SPANX-D sequences in the same patient. This observation may be explained by gene conversion between the two genes. Thus, all nucleotide changes detected in the SPANX-C coding regions of the patients only lead to the shuffling of SPANX sequences.
Figure 4.
Analysis of SPANX-C coding sequences in X-linked families. The alignment of the SPANX-C coding sequences obtained from prostate cancer patients (top), healthy controls (middle), and genomic SPANX-A/D genes (bottom). The figure shows the results of analysis of 12 families, some of them are represented by two brothers (brackets show brothers). Note that position 67 (black shading) is a single point mutation without gene conversion; all other mutation positions are recombinations/gene conversions. IUPAC codes of the variants, synonymous (S) and nonsynonymous (N) characters of changes, and positions in the CDS alignment are shown at the bottom; positions highlighted by boxes correspond to CpG sites and their TpG/CpA variants. If several mutations were in the same codon, they were marked by a dash. In all such cases, the resulting effect was an amino acid replacement, and therefore, all such changes were marked as nonsynonymous.
We also checked for the occurrence of genomic rearrangements in the SPANX-C locus in X-linked prostate cancer families. For this purpose, we carried out a physical analysis of SPANX-C TAR YAC clones isolated from the affected families and unaffected controls. The analysis confirmed the predicted size of YACs (83 kb) with identical Alu profiles in all samples (Supplemental Fig. 7S). Thus, we conclude that there are no detectable rearrangements in the SPANX-C locus of X-linked prostate cancer patients analyzed so far.
Analysis of LDOC1 did not reveal any mutations in the coding region
SPANX-C TAR clones also contain another gene, LDOC1, which has been shown to be down-regulated in some cancers (Nagasaki et al. 1999; Inoue et al. 2005). To investigate whether X-linked patients have mutational changes in this gene, ∼1.2-kb promoter and coding sequences were PCR amplified from TAR isolates (Supplemental Table 1S) and sequenced. Sequence analysis did not reveal nucleotide differences in LDOC1 between 17 prostate cancer patients and 22 unaffected controls, except for a polymorphism within a (TG)n array present in the promoter region 720 bp upstream of the start codon. The most frequent alleles contain 14 and 15 TG (94%). Several alleles with 16 and 17 TG were also detected both in normal individuals and in patients. Based on these results, we conclude that the predisposition to prostate cancer is unlikely caused by mutations in the LDOC1 gene.
Analysis of the SPANX-D locus in X-linked families
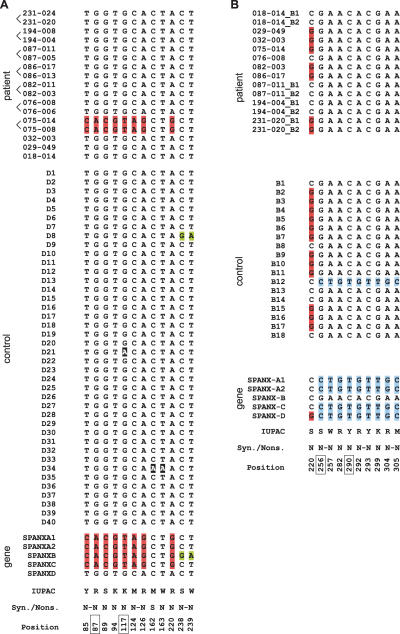
The SPANX-D locus was TAR-cloned as a 107-kb fragment from 17 patients (10 X-linked families) and from 40 unaffected controls (see Methods). To check for the presence of mutations in SPANX-D, exon and noncoding sequences were PCR amplified from each YAC TAR isolate by a pair of specific primers (Supplemental Table 1S). A sequence analysis of the SPANX-D gene from patients identified 52 SNPs in 1112-bp-long compared segments. Twelve SNPs were present in exon sequences. Alignments of the complete SPANX-D gene variants are shown in Supplemental Figure 4S. There are only two variants of the SPANX-D coding region in the patients (Fig. 5A). One of the variants, present in patients 075–014 and 075–008 (one family), contains eight nucleotide changes. Comparison of this polymorphic variant with the sequences of other members of the SPANX gene family revealed that all nucleotide changes are caused by homology-based sequence transfer from SPANX-A, which is located 75 kb away from the SPANX-D locus. In the control D8, a trace of the conversion to SPANX-B is also seen.
Figure 5.
Analysis of SPANX-D and SPANX-B coding sequences in X-linked families. The alignment of SPANX-D (A) and SPANX-B (B) coding sequences obtained from prostate cancer patients (top), healthy controls (middle), and genomic SPANX-A/D genes (bottom). The figure shows the results of analysis of 10 families, some of them are represented by two brothers (shown by brackets). When two SPANX-B copies were present in the same individual, they were marked by _B1 and _B2 suffices. IUPAC codes of the variants, synonymous (S) and nonsynonymous (N) characters of changes, and positions in the CDS alignment are shown at the bottom; positions highlighted by boxes correspond to CpG sites and their TpG/CpA variants. If several mutations occurred in the same codon, they were marked by a dash. In all such cases, the resulting effect was an amino acid replacement, and therefore, all such changes were marked as nonsynonymous.
The physical characterization of the SPANX-D TAR isolates did not reveal any detectable rearrangements. All YACs had the predicted size (107 kb) and identical Alu profiles (data not shown). Thus, we conclude that the SPANX-D locus is not rearranged in the X-linked prostate cancer patients analyzed so far.
Analysis of the SPANX-B locus revealed a variable number of gene copies
The SPANX-B locus is located 238 kb away from SPANX-C (Fig. 1). To analyze this locus, a TAR cloning vector was constructed that allowed SPANX-B to be isolated as a 63-kb genomic segment from 14 prostate cancer patients (10 X-linked families) and 18 unaffected controls (see Methods). Similarly to the SPANX-C and SPANX-D analysis, promoter, exon, and intron sequences were PCR amplified from each YAC TAR isolate using a pair of specific primers and sequenced. Forty-one SNPs were identified in 1112-bp-long compared segments. Most of them (29 SNPs) were in the noncoding region. The only change in the coding region that resulted in amino acid replacement in the patients was a C/G transversion at position 230 (Fig. 5B). The same substitution was found in the unaffected controls. Similar to other SPANX loci, the identified variants of SPANX-B result from homology-based sequence transfer events between SPANX-B and other gene family members (Fig. 5B; Supplemental Fig. 5S). The lower frequency of SPANX-B sequence variants compared with SPANX-C may be due to the fact that SPANX-B is the most proximal locus and/or that it contains multiple copies of the gene (see below). Also, the presence of an 18-bp insertion in exon 1 caused by the expansion of a minisatellite may suppress gene conversion of the SPANX-B gene.
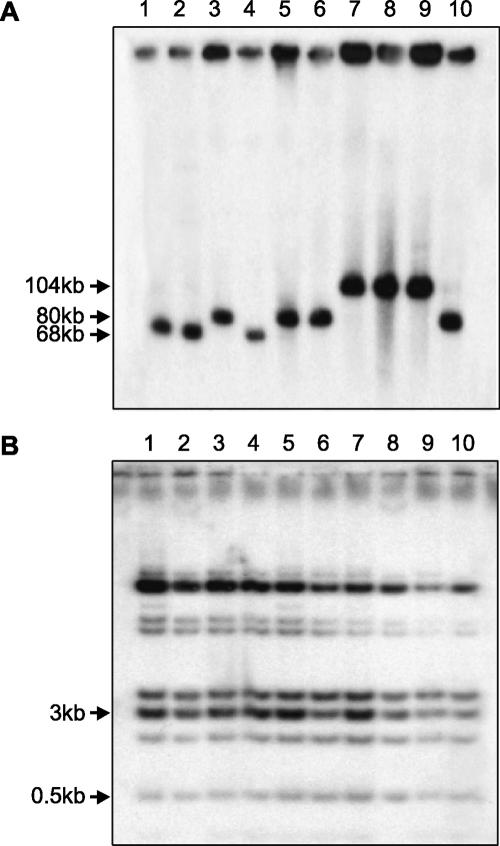
Unexpectedly, sequencing of the cloned PCR fragments revealed several polymorphic SPANX-B gene copies in some patients (e.g., 087–011, 194–004, and 231–020) (Supplemental Fig. 5S). A further physical analysis of the SPANX-B TAR isolates from the patients showed variability in the size of inserts, ranging from 63 kb to >100 kb (Fig. 6A). At the same time, the Alu profiles of the clones were identical (Fig. 6B). We propose that these results can be explained by the presence of a different number of tandem duplications carrying the SPANX-B gene within the inserts. The partial sequencing of several SPANX-B TAR isolates confirmed this suggestion and showed that a greater length of the insert was indeed due to the tandem duplication of a 12-kb DNA segment carrying the SPANX-B gene.
Figure 6.
Physical characterization of the SPANX-B genomic TAR isolates. (A) Chromosomal-size DNA was isolated from yeast transformants containing a SPANX-B locus, digested with NotI, CHEF gel electrophoresis-separated, and blot-hybridized with an exon 2 probe. The SPANX-B locus was analyzed in 14 patients and 18 controls; the figure illustrates only 10 randomly chosen individuals. Lanes 1–5 correspond to four unaffected controls (C1, C2, C3, C4 and C3); lanes 6–10 correspond to five patients (236–005, 029–049, 032–003, 075–014 and 236–005). For control C3 and patient 236–005, two independent TAR isolates were included into the analysis to exclude internal deletions during isolation. The leading bands from 60 kb to 90 kb correspond to the linearized molecules of SPANX-B, correspondingly. (B) Alu profile characterization of TAR YACs containing SPANX-B. Total yeast DNA was isolated from the transformants and digested to completion with TaqI. Fragments were separated by gel electrophoresis, transferred to a nylon membrane, and hybridized with an Alu probe.
Formally, different sizes of inserts in the TAR YACs may result from the recombinational interaction of multiple tandem repeats during cloning in yeast. Therefore, it was necessary to confirm the presence of such tandem duplications in TAR clones by a direct analysis of genomic DNA from patients. The genomic DNA was digested with BamHI and hybridized to a unique probe corresponding to a region outside of a 119-kb SPANX-B containing SD (see Methods for details). As predicted, BamHI digestion produced fragments of sizes 28 kb, 40 kb, 52 kb, and 64 kb, corresponding to one, two, three, and four copies of SPANX-B, respectively (Fig. 7A,B). The BamHI fragments from the genomic DNA and TAR isolate from the same patient were identical in size, indicating a high accuracy of SPANX-B isolation by TAR. Similarly, we determined the number of SPANX-B copies in all TAR clones isolated from patients and unaffected controls. This analysis showed that some controls also contain more than one copy of SPANX-B (Fig. 7B).
Figure 7.
Copy number of the SPANX-B gene. (A) The scheme of the 12-kb tandem repeats in individuals containing one, two, or three copies of the SPANX-B gene. (B) Genomic DNA from 28 individuals (11 cases and 18 controls) was digested with BamHI, CHEF gel electrophoresis-separated, and blot-hybridized with a specific probe to reveal the presence of a 12-kb SPANX-B tandem repeat. The figure shows only 13 individuals. Lanes 1–4 correspond to four controls, C3, C2, C6, and C4. Lanes 5–13 correspond to nine patients (029–049, 018–014, 075–014, 086–013, 087–011, 194–004, 082–003, 076–008, 236–005). The fragments of the predicted size of 28 kb, 40 kb, 52 kb, and 64 kb correspond to one, two, three, and four copies of SPANX-B, respectively. (C) The scheme represents the proportion of affected (patients, top) and unaffected (controls, bottom) individuals with one, two, three, and more than four copies of the SPANX-B gene. Presented are the results from 66 unaffected controls and 84 patients with the hereditary prostate cancer. Note that prostate cancer pedigrees analysis did not reveal linkage of the cancer predisposition to HPCX in these patients. Eleven patients (086–017, 076–008, 087–011, 194–004, 236–005, 029–049, 075–014, 239–019, 082–003, 032–003, and 231–020) were checked both by real-time PCR and by blot hybridization. The numbers in parentheses correspond to the number of individuals with the given number of SPANX-B copies.
To check whether the copy number of SPANX-B is different in cases and controls, additional samples were needed. However, analysis of clinical materials using Southern blot hybridization is limited because of the requirement for a large amount of high-molecular-weight DNA. Therefore, we applied a quantitative real-time PCR for analysis of additional samples (see Methods). Specific amplification of the SPANX-B sequence from genomic DNA was achieved using specific primers developed for the exon 1 sequence containing a 18-bp SPANX-B-specific insertion (Supplemental Table 1S). The use of the control DNA samples containing a known number of SPANX-B copies (i.e., determined by Southern blot hybridization) allowed the reaction to be calibrated and reproducible results to be obtained. The use of real-time PCR increased the number of screened individuals to 50. Figure 7C and Supplemental Table 4S summarize our results on the determination of the SPANX-B gene copy number. As seen, this number is variable both in cases and in controls. The maximum copy number of the SPANX-B gene (12 and 14 copies) was detected in two patients, 084–009 and 051–014. However, this SPANX-B amplification does not seem to be linked to prostate cancer predisposition because the difference in copy numbers between the cases and controls is not statistically significant.
Discussion
The recent expansion of SPANX genes has led to a potentially unstable region at Xq27
Analysis of the genomic region carrying evolutionarily young SPANX-A/D genes revealed its dynamic structure. Both frequent gene deletion/duplication and homology-based sequence transfer events were identified within this region and are presumably due to the presence of large SDs with a high level of sequence similarity representing approximately one third of the region.
The comparison of SD sequences showed that SPANX-A1, -A2, -C, and -D genes arose as a result of duplications of the SPANX-B locus during the last 5–7 million years. This estimate is in agreement with our previous results based on comparison of the syntenic regions in primates (Kouprina et al. 2004). All the duplication events were accompanied by inversions. Many X- and Y-linked genes expressed in the testis are located in similar large inverted repeats (Skaletsky et al. 2003; Warburton et al. 2004). Such an organization seems to be related to male germline expression and/or to maintenance of sequence integrity by gene conversion repair. The results of a detailed analysis of the SPANX-C locus are consistent with LINE1-mediated duplication via long-tract gene conversion with the additional transfer of adjacent sequences (Richardson et al. 1998; Richardson and Jasin 2000). Direct experimental evidence demonstrates that double-stranded breaks artificially induced in LINE1 constructs can be repaired by gene conversion with various genomic LINE1 elements (Tremblay et al. 2000). Long-track gene conversion is also implicated in an Alu-mediated interchromosomal duplication (Babcock et al. 2003). This mechanism is particularly attractive for gene duplications, as it prevents chromosomal translocations and the net result is one duplication, with the second participating locus remaining intact (Richardson and Jasin 2000).
The discovery of a variable number of a 12-kb tandem duplication containing the SPANX-B gene indicates that the expansion of SPANX genes is still an ongoing process. This process is most likely specific to the human lineage because both the chimpanzee and the gorilla contain a single copy of SPANX-B(V. Larionov, unpubl.). It is notable that the human SPANX-B gene contains a specific 18-bp insertion that is absent in African Great Apes. This insertion seems to have a functional significance, as it is retained in all copies of SPANX-B. It is interesting that this insertion presumably encodes a site of protein glycosylation. The presence of this site seems to be the reason that this protein is localized in the cytoplasm in contrast with other SPANXs (Westbrook et al. 2004). The fact that the expansion of SPANX genes in recent evolution was initiated twice by the SPANX-B locus may be related to a high density of interspersed repeats in this region.
Roughly two thirds of the SPANX sequence variants identified in this study can be explained by shuffling variants among alleles and paralogs, indicating a high frequency of ectopic recombination between the genes. A high frequency of converted SPANX alleles together with diversifying selection may provide a material for accelerated evolution of the SPANX genes (Birkhead and Pizzari 2002; Ball and Parker 2003; Kouprina et al. 2004). Although additional studies are required to make a final conclusion on the mechanism of recombination, the most likely mechanism is gene conversion. There are several reports on gene conversion in the human genome (e. g., Chen and Ferec 2000; Newman and Trask 2003); in the majority of cases, a conclusion concerning gene conversion was made based on the comparison of DNA sequences obtained from different individuals. In the case of SPANX genes (given the advantage of TAR cloning), we can isolate donor and target sequences from the same individual. Such data may provide a unique opportunity to analyze the mechanism of gene conversion in humans.
The presence of multiple copies of SPANX genes with similar sequences in the human genome raises the question of whether all of them have the same function. According to the classical model of gene family evolution (Ohno 1970), the absence of inactivating mutations in duplicated paralogs is an indication of functional diversification, whereby duplicated genes diverge in functional properties. Our results showed that expansion of the SPANX-A/D family was not accompanied by inactivation of gene family members. All of the genes have identical 5′,3′, and intron sequences, and none of them have stop codons in the open reading frame (ORF). This is quite unexpected because gene conversion should accelerate the process of disseminating mutations in ORFs (Chen and Ferec 2000; Newman and Trask 2003). The absence of inactivating mutations in the SPANX genes suggests that they encode proteins with important distinctive functions in reproduction. For example, different SPANX sequence variants may be important for cryptic female choice between spermatozoa. Additional studies are required to elucidate the role of individual SPANX proteins in spermatogenesis and in fertilization.
How the dynamic structure of the SPANX gene cluster may be related to prostate cancer susceptibility
There may be two explanations of how SPANX genes contribute to prostate malignancy. First, if these genes are transcribed (at least transiently) in prostate, then mutations in any of them may result in the production of an abnormal protein (or protein complex) that functions as an oncogene. The fact that we did not find any specific alterations in three of the five SPANX genes in X-linked families does not contradict this mutational hypothesis. Mutations may occur in the SPANX-A1 or SPANX-A2 genes, the analysis of which is now in progress. A mutant phenotype may also be caused by a specific combination of “normal” SPANX-A/D alleles. The identified frequent “ATGT” allele (with changes at positions 51, 62, 63, and 66) in the SPANX-C locus in patients may be an indication of such a preferential haplotype.
Alternatively, if SPANX genes are similar to other cancer/testis-specific genes, they would be exclusively expressed in testis (Zendman et al. 1999; Westbrook et al. 2000; Wang et al. 2003; Zendman et al. 2003; Kouprina et al. 2004; Salemi et al. 2004; Scanlan et al. 2004; Westbrook et al. 2004). Thus, activation of any of these genes may result in malignancy. This hypothesis is supported by recent experimental data. We found that ectopic expression of the prevalent SPANX-C allele is sufficient to induce a transformed phenotype in mammalian cells, as manifested by foci formation (V. Larionov, in prep.). If other SPANX alleles have a similar phenotype, hereditary prostate cancer may result from a genomic rearrangement(s) that releases the SPANX genes from a stringent transcriptional control. Such a genomic rearrangement may be common in X-linked families, and it may result from a high density of SDs within the SPANX gene cluster. Based on the different orientations of SDs at Xq27, their recombinational interactions may result in deletions/duplications and inversions of the SPANX-containing genomic regions. Indirect evidence of large deletions within this region has been obtained recently from the analysis of prostate carcinomas (Kibel et al. 2003). An illegitimate recombination between non-allelic homologous sequences may result in an increase of the SPANX gene copy number (as we observed for the SPANX-B gene) and thereby lead to up-regulation of gene expression. It is also possible that recombinational interactions between SPANX-associated duplications affect the expression of genes in the regions flanking the SPANX-A/D gene cluster.
There has been a rapid growth of literature describing pathological rearrangements caused by interactions between SDs (Shaw and Lupski 2004). A common name for such abnormalities is “genomic disorders.” A dynamic structure of the HPCX candidate region at Xq27 suggests that prostate malignancy may be a novel genomic disorder. To clarify the types of rearrangements that lead to prostate cancer susceptibility, further analyses are needed, including immunodetection of SPANX proteins in prostate samples, mutational analysis of SPANX-A1 and SPANX-A2 genes, and searches for deletions/duplications and inversions at SPANX intergenic regions in X-linked families, all of which are now in progress in our laboratory.
Methods
Analysis of the SPANX SDs
The positions of SPANX-A/D SDs were determined using local BLAT (Kent 2002) searches of the SPANX-A/D genes with flanking sequences against the human genome (hg16; UCSC July 2003 genome version). The exact positions of duplication breakpoints were determined by visual inspection. The origin of duplication was estimated as follows: For each SPANX-A/D locus, we first identified the duplicated counterpart with the longest alignment. To estimate which of the two loci was the parent locus (substrate) for duplication and which one was the result (product) of duplication, we analyzed the repetitive elements in the breakpoints of both duplications. If the terminus of one duplication breakpoint corresponded with the end of a repeat sequence, but the same repeat extended further down into the flanking regions in the second duplication, we considered the first duplication as a product and the second duplication as a substrate of duplication. Repetitive elements were detected by CENSOR (Jurka et al. 1996; http://www.girinst.org/Censor_Server-Data_Entry_Forms.html) with Repbase Update libraries (Jurka 2000; http://www.girinst.org/Repbase_Update.html). Gene conversion regions were predicted using GENECONV (Sawyer 1989; http://www.math.wustl.edu/∼sawyer/geneconv/).
Subjects
A detailed description of the study samples is presented elsewhere (Xu et al. 2003; Gillanders et al. 2004; Baffoe-Bonnie et al. 2005). Briefly, transformed lymphoblast cell lines were developed from 21 patients (12 families) linked to the HPCX region. DNA from these cell lines prepared in agarose plugs was used for TAR cloning experiments and Southern blot hybridization. In addition, DNA from 84 patients with the hereditary prostate cancer was extracted from peripheral blood using a QIAamp DNA Blood Kit (Qiagen, http://www.qiagen.com). This DNA was used to determine the SPANX-B gene copy number using a quantitative real-time PCR. Genomic DNA from 66 normal individuals used as eligible controls (Caucasians) was purchased from Coriell Institute for Medical Research.
TAR cloning of SPANX-C, SPANX-D, and SPANX-B genomic regions
The scheme of TAR cloning is described in Supplemental Figure 1S. TAR cloning experiments were carried out as described previously (Leem et al. 2003). Three vectors, TAR-C, TAR-D, and TAR-B, were constructed using a basic vector pVC604. The vector TAR-C contained 5′ 164-bp and 3′ 187-bp targeting hooks, specific to the unique sequences flanking SPANX-C. The vector TAR-D contained 5′ 203-bp and 3′ 227-bp targeting hooks, specific to the unique sequences flanking SPANX-D. The TAR-B vector contained 5′ 257-bp and 3′ 284-bp targeting hooks, specific to the unique sequences flanking SPANX-B. The 5′ and 3′ targeting sequences correspond to positions 39,708–39,872 and 122,818–123,004 in the BAC AL109699 (SPANX-C), positions 71,638–71,887 and 134,177–134,457 in the BAC AL451048 (SPANX-B), and positions 43,511–43,713 and 150,550–150,775 in the BAC AL121881 (SPANX-D), respectively. The targeting sequences were amplified from human genomic DNA using specific primers (Supplemental Table 1S). The vectors were linearized with either SphI or SalI before being used in the TAR cloning experiments. Genomic DNA in agarose plugs was prepared from cell lines as described previously (Leem et al. 2003). Commercially available human male DNA, chimpanzee (Pan troglodytes) DNA, and gorilla (Gorilla gorilla) DNA (Coriell Institute for Medical Research) were also used for TAR cloning. It is notable that gorilla and chimpanzee SPANX-B genomic regions were TAR isolated using human-specific targeting hooks. To identify the clones positive for SPANX-C, SPANX-D, and SPANX-B, yeast transformants were examined with PCR using pairs of specific diagnostic primers (Supplemental Table 1S). The yield of SPANX-positive clones was approximately the same (1%–5%) with genomic DNA prepared in agarose plugs and commercially available DNA. It is worth noting that all three SPANX loci were successfully TAR cloned from all X-linked patients analyzed in this study that suggests that there are no genomic rearrangements within the targeted regions.
Physical characterization of the TAR isolates
The sizes and Alu profiles of TAR isolates were determined as described previously (Leem et al. 2003). Chromosomal-size DNA was isolated from yeast transformants containing the SPANX gene, digested with NotI, Clamped Homogeneous Electrical Field gel Electrophoresis (CHEF) gel electrophoresis-separated, and blot-hybridized with the exon 2 probe (Supplemental Table 1S). For Alu profile characterization of TAR YACs, total yeast DNA was isolated from the transformants and digested to completion with TaqI. Fragments were separated by gel electrophoresis, transferred to a nylon membrane, and hybridized to an Alu probe.
Sequencing of SPANX-C, SPANX-D, SPANX-B, and LDOC1 genes
The promoter, coding and intronic regions, and 3′ noncoding sequences of the SPANX-C, SPANX-D, and SPANX-B genes were PCR amplified from the TAR YAC isolates using a set of SPANX gene-specific primers (Supplemental Table 1S). Two pairs of primers were designed to amplify the coding and promoter regions of the LDOC1 gene as 683- and 497-bp fragments, correspondingly, using specific primers (Supplemental Table 1S). The PCR fragments were cloned into a TA vector. Sequence forward and reverse reactions were run on a PE-Applied Biosystem 3100 Automated Capillary DNA Sequencer. In addition, several SPANX YAC TAR isolates were partially sequenced after their conversion into BACs. Complete sequences of SPANX-C, SPANX-D, and SPANX-B alleles were named and numbered according to the clone/accession identifier (Supplemental Table 2S). To sequence the inserts in TAR clones, YACs were retrofitted into BAC/YACs using the BRV1 retrofitting vector and then transformed into a DH10B Escherichia coli strain (Kouprina et al. 1998). Before sequencing, the integrity of the inserts in BACs was confirmed by comparing their Alu profiles with those of the original TAR yeast isolates. Sequences were aligned with MAVID (Bray and Pachter 2004; http://baboon.math.berkeley.edu/mavid/). Pictures of alignments and alignment schemes were prepared in WAViS (Zika et al. 2004; http://wavis.img.cas.cz). Database searches were performed using versions of the BLAST program appropriate for different types of sequence comparisons: BLASTN for nucleotide sequences, BLASTP for protein sequences, and TBLASTN for searching a nucleotide database translated in 6 frames with a protein query (Altschul et al. 1990).
Determination of the copy number of the SPANX-B-containing repeat by Southern blot hybridization
The presence of 12-kb tandem duplications containing the SPANX-B gene was first detected by SfiI digestion of TAR YAC isolates. There are two SfiI sites in the exon 1 sequence of SPANX-B. Thus, when the SPANX-B gene is tandemly repeated, the exon 2-specific probe (positions 111,320–111,659 in AL451048 BAC) visualizes an extra 12-kb band (Supplemental Table 1S). The presence of a 12-kb tandem duplication was further confirmed by partial sequencing of several SPANX-B TAR isolates (Supplemental Table 2S). Based on the available genomic sequence, BamHI digestion produces a 28-kb SPANX-B-containing fragment. So, to determine the number of SPANX-B tandem repeats, genomic DNA from patients and normal individuals was digested by BamHI, CHEF-separated, and blot-hybridized with a probe specific to a unique sequence upstream of SPANX-B (Supplemental Table 1S). The hybridization visualizes a 28-kb fragment when there is one copy of the SPANX-B gene. Each additional copy of the tandem duplication containing the SPANX-B gene increases the size of the detectable fragment by 12 kb (28 kb, 40 kb, 52 kb, 64 kb).
Determination of the SPANX-B gene copy number by quantitative real-time PCR
The TaqMan probe and primers were designed using the primer Express software (Applied Biosystems), following the criteria indicated in the program. We designed the specific TaqMan SPANX-B probe to be complementary to exon 1 (Supplemental Table 1S). The SPANX-B probe contains a fluorophore 5′ FAM as a reporter. SPANX-B forward and reverse primers were created from exon 1. Notably, the reverse primer corresponds to a unique 18-bp insert present only in the SPANX-B gene (Supplemental Table 1S), providing a specific amplification of the SPANX-B gene sequence. The size of the amplicon is 78 bp. We used an RNAaseP kit as an internal reference (Applied Biosystems). This kit contains 20xRNAaseP mix with a VIC-labeled probe and specific primers for the RNAaseP gene. We performed separate and multiplex (i.e., the amplification of SPANX-B and RNAaseP was performed in the same tube) pre-runs, varying the concentrations of primers and probe to obtain the highest intensity and specificity of the reporter fluorescent signal. In the control experiments, we obtained amplification efficiency close to 100% for the two genes, signifying that both reactions proceeded with very high efficiencies. PCR was carried out using an ABI prism 7700 (Applied Biosystems) in a 96-well optical plate with a final reaction volume of 50 μL. All reactions in each plate were prepared from a single PCR Mastermix consisting of 2×TaqMan Universal PCR Master Mix, 900 nM SPANX-B forward primer, 900 nM SPANX-B reverse primer, 250 nM SPANX-B probe, 20×RNAaseP Mix, and HPLC pure water. A total of 50 ng of DNA template (5 μL) was dispensed into each of the three sample wells for triplicate reactions. Each sample was run in triplicates to quantify the SPANX-B gene compared with the internal RNAaseP control gene. Thermal cycling conditions included a pre-run of 2 min at 50°C and 10 min at 95°C. Cycle conditions were 40 cycles at 95°C for 15 sec and 60°C for 1 min, according to the TaqMan Universal PCR Protocol (ABI). Each plate run was monitored with three control male samples containing known SPANX-B copy numbers (1 copy, 2 copies, and 4 copies). No-template control (background) was also included in each assay. The relative copy number of SPANX-B was calculated for each sample using the comparative CT method. Patient DNA was extracted from the peripheral blood using a QIAamp DNA Blood Kit recommended for obtaining a high-quality DNA template, which is very important for the reliability of the experiment. DNA was diluted in HPLC pure water to a concentration of ∼ 20 ng/μL and stored at 4°C.
Acknowledgments
We thank Ms. Nina Kouprina for professional editing of this manuscript and we also thank the anonymous referees for suggestions that led to substantial improvements in this paper. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY787057–AY787130 and AY920931–AY920987.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4212705. Freely available online through the Genome Research Immediate Open Access option.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Babcock, M., Pavlicek, A., Spiteri, E., Kashork, C.D., Ioshikhes, I., Shaffer, L.G., Jurka, J., and Morrow, B.E. 2003. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 13: 2519–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffoe-Bonnie, A.B., Smith, J.R., Stephan, D.A., Schleutker, J., Carpten, J.D., Kainu, T., Gillanders, E.M., Matikainen, M.P., Teslovich, T.M., Tammela, T.L.J., et al. 2005. A major locus for hereditary prostate cancer in Finland: Localization by linkage disequilibrium of a haplotype in the HPCX region. Hum. Genet. 117: 307–316. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Ball, M.A. and Parker, G.A. 2003. Sperm competition games: Sperm selection by females. J. Theor. Biol. 224: 27–42. [DOI] [PubMed] [Google Scholar]
- Birkhead, T.R. and Pizzari, T. 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3: 262–273. [DOI] [PubMed] [Google Scholar]
- Bochum, S., Paiss, T., Vogel, W., Herkommer, K., Hautmann, R., and Haeussler, J. 2002. Confirmation of the prostate cancer susceptibility locus HPCX in a set of 104 German prostate cancer families. Prostate 52: 12–19. [DOI] [PubMed] [Google Scholar]
- Bray, N. and Pachter, L. 2004. MAVID: Constrained ancestral alignment of multiple sequences. Genome Res. 14: 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W.M., Lange, E.M., Chen, H., Zheng, S.L., Chang, B., Wiley, K.E., Isaacs, S.D., Walsh, P.C., Isaacs, W.B., Xu, J., et al. 2004. Hereditary prostate cancer in African American families: Linkage analysis using markers that map to five candidate susceptibility loci. Br. J. Cancer 90: 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.M. and Ferec, C. 2000. Gene conversion-like missense mutations in the human cationic trypsinogen gene and insights into the molecular evolution of the human trypsinogen family. Mol. Genet. Metab. 7: 463–469. [DOI] [PubMed] [Google Scholar]
- Farnham, J.M., Camp, N.J., Swensen, J., Tavtigian, S.V., and Albright, L.A. 2005. Confirmation of the HPCX prostate cancer predisposition locus in large Utah prostate cancer pedigrees. Hum. Genet. 116: 179–185. [DOI] [PubMed] [Google Scholar]
- Gillanders, E.M., Xu, J., Chang, B.L., Lange, E.M., Wiklund, F., Bailey-Wilson, J.E., Baffoe-Bonnie, A., Jones, M., Gildea, D., Riedesel, E., et al. 2004. Combined genome-wide scan for prostate cancer susceptibility genes. J. Natl. Cancer Inst. 96: 1240–1247. [DOI] [PubMed] [Google Scholar]
- Goydos, J.S., Patel, M., and Shih, W. 2002. NY-ESO-1 and CTp11 expression may correlate with stage of progression in melanoma. J. Surg. Res. 98: 76–80. [DOI] [PubMed] [Google Scholar]
- Inoue, M., Takahashi, K., Niide, O., Shibata, M., Fukuzawa, M., and Ra, C. 2005. LDOC1, a novel MZF-1-interacting protein, induces apoptosis. FEBS Lett. 579: 604–608. [DOI] [PubMed] [Google Scholar]
- Jurka, J. 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet. 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Jurka, J., Klonowski, P., Dagman, V., and Pelton, P. 1996. CENSOR—A program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem 20: 119–121. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibel, A.S., Faith, D.A., Bova, G.S., and Isaacs, W.B. 2003. Xq27–28 deletions in prostate carcinoma. Genes Chrom. Cancer 37: 381–388. [DOI] [PubMed] [Google Scholar]
- Kouprina, N. and Larionov, V. 2003. Exploiting the yeast Saccharomyces cerevisiae for the study of the organization of complex genomes. FEMS Microbiol. Rev. 27: 629–649. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., Annab, L., Graves, J., Afshari, C., Barrett, J.C., Resnick, M.A., and Larionov, V. 1998. Functional copies of a human gene can be directly isolated by TAR cloning with a small 3′ end target sequence. Proc. Natl. Acad. Sci. 95: 4469–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina, N., Mullokandov, M., Rogozin, I., Collins, K., Solomon, G., Risinger, J., Koonin, E., Barrett, J.C., and Larionov, V. 2004. The SPANX gene family of cancer-testis specific antigens: Rapid evolution, an unusual case of positive selection and amplification in African Great Apes and hominids. Proc. Natl. Acad. Sci. 101: 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, E.M., Chen, H., Brierley, K., Perrone, E.E., Bock, C.H., Gillanders, E., Ray, M.E., and Cooney, K.A. 1999. Linkage analysis of 153 prostate cancer families over a 30-cM region containing the putative susceptibility locus HPCX. Clin. Cancer Res. 5: 4013–4020. [PubMed] [Google Scholar]
- Leem, S.H., Noskov, V.N., Park, J.E., Kim, S.I., Larionov, V., and Kouprina, N. 2003. Optimum conditions for selective isolation of genes from complex genomes by transformation-associated recombination cloning. Nucleic Acids Res. 31: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montironi, R., Scarpelli, M., and Lopez Beltran, A. 2004. Carcinoma of the prostate: Inherited susceptibility, somatic gene defects and androgen receptors. Virchows Arch. 444: 503–508. [DOI] [PubMed] [Google Scholar]
- Nagasaki, K., Manabe, T., Hanzawa, H., Maass, N., Tsukada, T., and Yamaguchi, K. 1999. Identification of a novel gene, LDOC1, down-regulated in cancer cell lines. Cancer Lett. 140: 227–234. [DOI] [PubMed] [Google Scholar]
- Nagasaki, K., Schem, C., von Kaisenberg, C., Biallek, M., Rosel, F., Jonat, W., and Maass, N. 2003. Leucine-zipper protein, LDOC1, inhibits NF-kappaB activation and sensitizes pancreatic cancer cells to apoptosis. Int. J. Cancer 105: 454–458. [DOI] [PubMed] [Google Scholar]
- Newman, T. and Trask, B.J. 2003. Complex evolution of 7E olfactory receptor genes in segmental duplications. Genome Res. 5: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer, Berlin, Germany.
- Richardson, C. and Jasin, M. 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 20: 9068–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C., Moynahan, M.E., and Jasin, M. 1998. Double-strand break repair by interchromosomal recombination: Suppression of chromosomal translocations. Genes & Dev.. 12: 3831–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, M.T., Grafham, D.V., Coffey, A.J., Scherer, S., McLay, K., Muzny, D., Platzer, M., Howell, G.R., Burrows, C., Bird, C.P., et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, M.A. and De Marzo, A.M. 2004. Molecular genetics of human prostate cancer. Mod. Pathol. 17: 380–388. [DOI] [PubMed] [Google Scholar]
- Salemi, M., Calogero, A.E., Di Benedetto, D., Cosentino, A., Barone, N., Rappazzo, G., and Vicari, E. 2004. Expression of SPANX proteins in human-ejaculated spermatozoa and sperm precursors. Int. J. Androl. 27: 134–139. [DOI] [PubMed] [Google Scholar]
- Sawyer, S.A. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526–538. [DOI] [PubMed] [Google Scholar]
- Scanlan, M.J., Simpson, A.J., and Old, L.J. 2004. The cancer/testis genes: Review, standardization, and commentary. Cancer Immun. 4: 1–15. [PubMed] [Google Scholar]
- Schaid, D.J. 2004. The complex genetic epidemiology of prostate cancer. Mol. Genet. 13: R103–R121. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525–528. [DOI] [PubMed] [Google Scholar]
- Shaw, C.J. and Lupski, J.R. 2004. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 13: R57–R64. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. [DOI] [PubMed] [Google Scholar]
- Stephan, D.A., Howell, G.R., Teslovich, T.M., Coffey, A.J., Smith, L., Bailey-Wilson, J.E., Malechek, L., Gildea, D., Smith, J.R., Gillanders, E.M., et al. 2002. Physical and transcript map of the hereditary prostate cancer region at Xq27. Genomics 79: 41–50. [DOI] [PubMed] [Google Scholar]
- Tremblay, A., Jasin, M., and Chartrand, P. 2000. A double-strand break in a chromosomal LINE element can be repaired by gene conversion with various endogenous LINE elements in mouse cells. Mol. Cell. Biol. 20: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage, B.A. and Kiemeney, L.A. 2003. Inherited predisposition to prostate cancer. Eur. J. Epidemiol. 18: 1027–1036. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Zhang, Y., Liu, H., Salati, E., Chiriva-Internati, M., and Lim, S.H. 2003. Gene expression and immunologic consequence of SPAN-Xb in myeloma and other hematologic malignancies. Blood 101: 955–960. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E., Giordano, J., Cheung, F., Gelfand, Y., and Benson, G. 2004. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14: 1861–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook, V.A., Diekman, A.B., Klotz, K.L., Khole, V.V., von Kap-Herr, C., Golden, W.L., Eddy, R.L., Shows, T.B., Stoler, M.H., Lee, C.Y., et al. 2000. Spermatid-specific expression of the novel X-linked gene product SPAN-X localized to the nucleus of human spermatozoa. Biol. Reprod. 63: 469–481. [DOI] [PubMed] [Google Scholar]
- Westbrook, V.A., Diekman, A.B., Naaby-Hansen, S., Coonrod, S.A., Klotz, K.L., Thomas, T.S., Norton, E.J., Flickinger, C.J., and Herr, J.C. 2001. Differential nuclear localization of the cancer/testis-associated protein, SPAN-X/CTp11, in transfected cells and in 50% of human spermatozoa. Biol. Reprod. 64: 345–358. [DOI] [PubMed] [Google Scholar]
- Westbrook, V.A., Schoppee, P.D., Diekman, A.B., Klotz, K.L., Allietta, M., Hogan, K.T., Slingluff, C.L., Patterson, J.W., Frierson, H.F., Irvin Jr., W.P., et al. 2004. Genomic organization, incidence, and localization of the SPAN-X family of cancer-testis antigens in melanoma tumors and cell lines. Clin. Cancer. Res. 10: 101–112. [DOI] [PubMed] [Google Scholar]
- Xu, J., Meyers, D., Freije, D., Isaacs, S., Wiley, K., Nusskern, D., Ewing, C., Wilkens, E., Bujnovszky, P., Bova, G.S., et al. 1998. Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat. Genet. 20: 175–179. [DOI] [PubMed] [Google Scholar]
- Xu, J., Gillanders, E.M., Isaacs, S.D., Chang, B.L., Wiley, K.E., Zheng, S.L., Jones, M., Gilde, D., Riedesel, E., Albertus, J., et al. 2003. Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate 57: 320–325. [DOI] [PubMed] [Google Scholar]
- Zendman, A.J., Cornelissen, I.M., Weidle, U.H., Ruiter, D.J., and van Muijen, G.N. 1999. CTp11, a novel member of the family of human cancer/testis antigens. Cancer Res. 59: 6223–6229. [PubMed] [Google Scholar]
- Zendman, A.J., Zschocke, J., van Kraats, A.A., de Wit, N.J., Kurpisz, M., Weidle, U.H., Ruiter, D.J., Weiss, E.H., and van Muijen, G.N. 2003. The human SPANX multigene family: Genomic organization, alignment and expression in male germ cells and tumor cell lines. Gene 309: 125–133. [DOI] [PubMed] [Google Scholar]
- Zika, R., Paces, J., Pavlicek, A., and Paces, V. 2004. WAViS server for handling, visualization and presentation of multiple alignments of nucleotide or amino acids sequences. Nucleic Acids Res. 32: W48–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Web site references
- http://www.girinst.org/Censor_Server-Data_Entry_Forms.html; Censor
- http://www.girinst.org/Repbase_Update.html; Repbase update.
- http://www.math.wustl.edu/∼sawyer/geneconv/; GENECONV
- http://www.qiagen.com; Qiagen
- http://baboon.math.berkeley.edu/mavid/; MAVID
- http://wavis.img.cas.cz/; WAViS