Abstract
A classical nuclear localization signal (NLS)-containing protein is transported into the nucleus via the formation of a NLS-substrate/importin α/β complex. In this study, we found that importin α migrated into the nucleus without the addition of importin β, Ran or any other soluble factors in an in vitro transport assay. A mutant importin α lacking the importin β-binding domain efficiently entered the nucleus. Competition experiments showed that this import pathway for importin α is distinct from that of importin β. These results indicate that importin α alone can enter the nucleus via a novel pathway in an importin β- and Ran-independent manner. Furthermore, this process is evolutionarily conserved as similar results were obtained in Saccharomyces cerevisiae. Moreover, the import rate of importin α differed among individual nuclei of permeabilized cells, as demonstrated by time-lapse experiments. This heterogeneous nuclear accumulation of importin α was affected by the addition of ATP, but not ATPγS. These results suggest that the nuclear import machinery for importin α at individual nuclear pore complexes may be regulated by reaction(s) that require ATP hydrolysis.
Keywords: ATP/importin α/nuclear pore complex/nuclear transport/time-lapse experiment
Introduction
Molecular migration between the nucleus and cytoplasm occurs through the nuclear pore complex (NPC) present in the double membrane of the nuclear envelope. The NPC, a huge proteinaceous structure composed of 50–100 different species of proteins called nucleoporins, is estimated to have a total mass of ∼125 MDa in higher eukaryotes and 66 MDa in Saccharomyces cerevisiae. Whereas molecules <20–40 kDa are able to passively diffuse through the NPC into the nucleus, the nuclear import of larger molecules is generally dependent on the presence of a specific signal sequence, the nuclear localization signal (NLS), and is often associated with a requirement for metabolic energy (Görlich and Kutay, 1999). The first identified NLS was that of the SV40 large T-antigen, which consists of a short stretch of basic amino acids, designated as the basic type NLS. This type of NLS is divided into two groups, monopartite and bipartite, based on the number of basic amino acid clusters (Dingwall and Laskey, 1991).
The nuclear import of basic type NLS-containing proteins is mediated by specific soluble factors that form a stable complex, the nuclear pore-targeting complex, in the cytoplasm (Imamoto et al., 1995a). The complex is composed of two essential components that are referred to as importin α and β (Görlich and Mattaj, 1996). In addition to these molecules, several factors participate in this transport system, including a small GTPase Ran (Moore and Blobel, 1993) and Ran-binding proteins (Paschal and Gerace, 1995). Ran has a low intrinsic activity with respect to GDP/GTP exchange and GTP hydrolysis. RCC1, a guanine nucleotide exchange factor of Ran, accelerates the dissociation of the guanine nucleotide from Ran, thereby converting RanGDP to RanGTP in the cells (Bischoff and Ponstingl, 1991). Since RCC1 is a chromatin protein and is located in the nucleus, it is generally thought that the generation of the GTP-bound form of Ran occurs in the nucleus. NTF2 facilitates the uptake of RanGDP from the cytoplasm into the nucleus in order to maintain the RanGTP gradient. In the cytoplasm, importin α interacts with a classical basic type NLS-bearing karyophile and importin β. The resultant importin α/β/NLS-substrate ternary complex migrates into the nucleoplasm through the NPC in an importin β-dependent manner. Once the importin α/β/NLS-substrate complex reaches the nucleoplasm, importin β interacts directly with RanGTP, resulting in the release of importin α and the NLS-bearing karyophile.
Structural analysis of importin α has revealed three functional domains. One is an importin β binding (IBB) domain, which is located in the region comprised of residues 10–55 in the N-terminus (Görlich et al., 1996; Weis et al., 1996). The second is a hydrophobic central domain known as the armadillo (arm) repeat domain (Yano et al., 1994). The crystal structure of yeast importin α (KAP60) or mouse importin α2 (importin αP1) showed that this central domain is composed of a tandem array of 10 arm repeats, organized in a right-handed superhelix of helices. Each arm repeat consists of three α-helices that are connected by loops. Moreover, it has been shown that a classical NLS binds to two sites within a helical surface groove of the arm repeat domain in importin α (Conti et al., 1998; Kobe, 1999; Fontes et al., 2000). The third is a short acidic domain in the C-terminus. This region binds to the cellular apoptosis susceptibility gene product (CAS), the function of which is to export importin α from the nucleoplasm (Kutay et al., 1997; Herold et al., 1998).
Saccharomyces cerevisiae contains a single importin α, referred to as Srp1p, which is an essential gene originally identified as a suppressor of the temperature-sensitive RNA polymerase I mutation (Yano et al., 1992). In contrast to yeast, mammals, such as the human and mouse, possess at least six importin α isoforms [importin α1, αS1, NPI1; importin α2, αP1, PTAC58, Rch1; importin α3, αQ1, Qip1; importin α4, αQ2; importin α6 (identified only in human); and importin αS2, NPI2] (Cortes et al., 1994; Cuomo et al., 1994; Imamoto et al., 1995b; Köhler et al., 1997; Tsuji et al., 1997). These importin α proteins can be classified into three subfamilies. One subfamily consists of importin α1, α6 and αS2; the second, importin α2; and the third includes importin α3 and α4. The amino acid sequence identity among these three subfamilies is ∼50%, and the identity between members in a subfamily is ∼80–85%. These known importin α molecules interact specifically with importin β, resulting in high-affinity binding to classical NLSs. They are also exported by CAS from the nucleoplasm. The results of several investigations suggest that these importin α molecules show different expression patterns in adult tissues or cell lines, and display specific recognition for distinct classical NLSs or karyophiles (Prieve et al., 1996; Miyamoto et al., 1997; Nadler et al., 1997; Sekimoto et al., 1997; Tsuji et al., 1997). The existence of distinct importin α isoforms in mammals implies that different isoforms could recognize distinct target proteins.
The dynamic movement of importin β has been extensively analyzed and a large body of data has accumulated to show that importin β by itself translocates through the NPC bi-directionally by binding directly to nucleoporins. However, the intracellular behavior of importin α has not been extensively examined. The goal of this study was to develop a better understanding of the intracellular dynamics of importin α. As a result, we found that importin α alone can migrate into the nucleus in an importin β- and Ran-independent manner. In addition, our findings show that the pathway of importin α import is distinct from that of importin β import. Furthermore, this novel import is conserved from mammals to yeast, suggesting that the importin α import may have physiological significance.
Results
The IBB domain-lacking importin α mutant is localized in the nucleus when transiently expressed in mammalian cells
Importin α is known to be a classical NLS receptor and to function as an adapter molecule between an NLS-bearing karyophile and importin β. Our previous studies showed, by indirect immunofluorescence with anti-importin α antibodies, that importin α is localized throughout the cytoplasm and nucleus, and that a portion of cytoplasmically injected anti-importin α antibodies migrated into the nucleus in a piggy-back fashion (Imamoto et al., 1995b), indicating that endogenous importin α traverses the nuclear envelope in living mammalian cells.
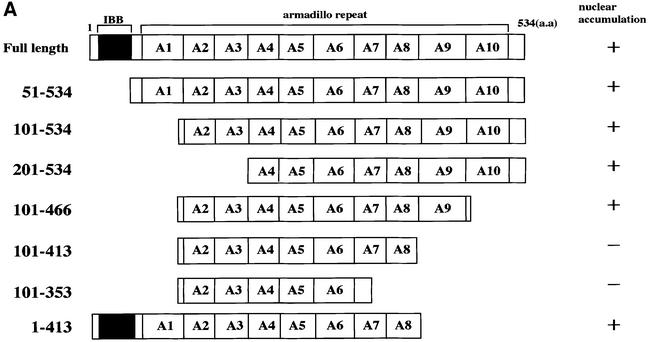
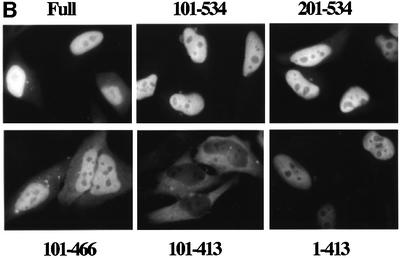
In this study, to analyze the dynamic behavior of importin α more precisely in living cells, we first transiently expressed a variety of GFP–importin α constructs in HeLa cells. As shown in Figure 1, full-length importin α was predominantly localized in the nucleus, which is consistent with the current model in which importin α enters the nucleus via complex formation with a karyophile and importin β. However, we found that an importin α mutant (101–534) lacking the IBB domain, which is unable to bind to importin β, also accumulated in the nucleus, indicating that importin α is able to migrate into the nucleus in an IBB domain-independent manner. In addition, it was found that this migration was dependent on the C-terminal arm repeat domain containing arm repeat 9 and a portion of arm repeat 10 (Figure 1). Furthermore, this IBB domain-independent nuclear import of importin α is distinguished from the import through complex formation with importin β, because a mutant (1–413) lacking arm 9 and a portion of arm 10 but containing the IBB domain could accumulate in the nucleus. These findings suggest two possibilities: (i) importin α has the ability to migrate into the nucleus by itself; or (ii) importin α is carried into the nucleus in a piggy-back fashion by binding to multimeric karyophiles complexed with importin α/β.
Fig. 1. Nuclear accumulation of importin α lacking an IBB domain. (A) Summary and the cellular localization of the importin α (mNPI2) deletion mutants used in this transfection analysis. These mutants were constructed into pEGFP-C2 transfection vectors (see Materials and methods). (B) Cellular localization of these mutants in transiently transfected HeLa cells. After transfection (12 h), the cells were fixed with 3.7% formaldehyde in PBS and the subcellular localization of EGFP-fused proteins was observed.
Importin α migrates into the nucleus in the absence of cytosol in vitro
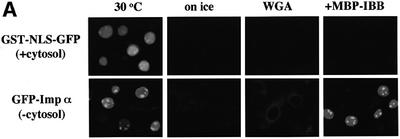
In order to characterize the IBB domain-independent nuclear import of importin α, we next used digitonin-permeabilized cells. Consistent with the previous results, the nuclear import of the SV40 T-antigen NLS fused with GST and GFP (GST–NLS–GFP) was observed only in the presence of cytosolic extract and an ATP regeneration system (Figure 2A). Surprisingly, we found that recombinant GFP–importin α accumulated in the nucleus, even in the absence of exogenous cytosolic extracts. The nuclear migration of importin α was sensitive to temperature and was inhibited by wheatgerm agglutinin (WGA), which is known to bind to glycosylated nucleoporins. These results indicate that the nuclear migration of importin α does not result from passive diffusion, but that this protein migrates into the nucleus through the gated channels of the NPC.
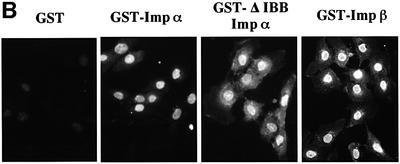
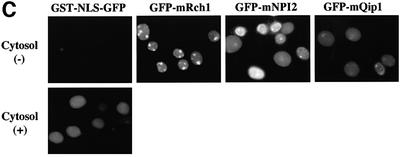
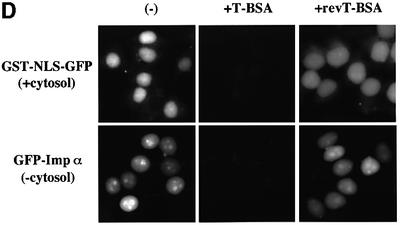
Fig. 2. Importin α is able to migrate into the nucleus in an importin β-independent manner in an in vitro assay. (A–D) Cells were treated with 40 µg/ml digitonin in TB (see Materials and methods) for 5 min on ice, and after washing with PBS twice, the cells were incubated with 10 µl of testing solution. (A) Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–importin α (mRch1) with TB alone or 2.5 µM GST–NLS–GFP with cytosolic extracts prepared from mouse Ehrlich ascites tumor cells and an ATP regeneration system for 20 min at 30°C or on ice. The other import reactions of GFP–importin α or GST–NLS–GFP were performed for 20 min at 30°C after pretreatment with 0.4 mg/ml WGA for 10 min at 30°C, or in the presence of 25 µM MBP–IBB for 20 min at 30°C. (B) Digitonin-permeabilized MDBK cells were incubated with 1 µM wild-type GST–importin α (NPI1) or 1 µM GST–ΔIBB importin α (NPI1; 78–534 amino acids) for 20 min at 30°C. As a control, 1 µM GST alone or 1 µM GST–importin β was used. To detect the GST portion, anti-GST–antibody (B-14; a mouse monoclonal IgG; Santa Cruz Biotechnology, Inc.) (2 µg/ml) was used and detected with RITC-conjugated goat anti-mouse IgG. (C) Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–mRch1, 2.5 µM GFP–mNPI2 and 2.5 µM GFP–mQip1 alone or 2.5 µM GST–NLS–GFP in the presence or absence of Ehrlich cytosolic extracts and an ATP regeneration system for 20 min at 30°C. (D) Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–importin α (mRch1) alone or 2.5 µM GST–NLS–GFP with Ehrlich ascites tumor cells cytosolic extracts and an ATP regeneration system in the presence of 25 µM T-BSA or 25 µM revT-BSA for 20 min at 30°C.
To rule out the possibility that this cytosol-independent nuclear import of importin α is dependent upon endogenous importin β remaining in the digitonin-permeabilized cells, maltose binding protein (MBP)-fused IBB domain-containing protein (MBP–IBB) was added to the permeabilized cells. As shown in Figure 2A, we found that the addition of the IBB domain had no effect on the nuclear accumulation of importin α, indicating that the remaining importin β in permeabilized cells is not involved in the nuclear migration of importin α and that importin α is not transported into the nucleus in a piggy-back fashion by binding to another karyophile/importin α/β complex.
In order to confirm that the cytosol-independent import of importin α is independent of the IBB domain, we employed a GST–importin α mutant lacking the IBB domain (GST–ΔIBB importin α) in the in vitro assay. As shown in Figure 2B, the IBB domain-lacking mutant of importin α efficiently migrated into the nucleus. In addition, three isoforms of importin α from the mouse, importin α2/mRch1, importin αS2/mNPI2 and importin α3/mQip1, entered the nucleus in a similar manner (Figure 2C), indicating that the three distinct importin α subfamilies have the same activity with respect to their ability to enter the nucleus.
We next examined the effect of basic type NLS-containing substrates on the import of importin α. When an excess amount of unlabeled T-BSA, which is the SV40 T-antigen NLS conjugated to BSA, was added to the assay, the cytosol-independent nuclear import of importin α was strongly inhibited, while it was not affected at all by the addition of unlabeled reverse T-BSA (revT-BSA) (Figure 2D). In addition, the presence of an excess amount of T-antigen NLS peptide or of GST–NLS–GFP recombinant protein also dramatically inhibited the nuclear import of importin α (data not shown). These results indicate that importin α has the ability to migrate into the nucleus by itself in an importin β-independent manner when it is not bound to basic type NLS-containing proteins, and that importin α itself possesses the necessary and sufficient information to be translocated via the NPC.
Nuclear import of importin α occurs without the GTP hydrolysis of Ran
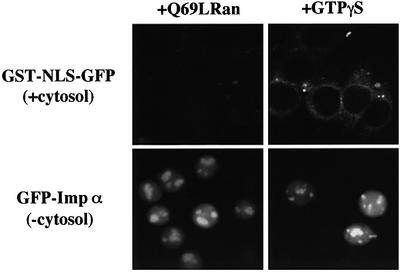
To further verify that the migration of importin α into the nucleus is not dependent on Ran, we examined the effect of a dominant-negative Ran mutant that is defective in GTP hydrolysis, Q69LRanGTP, which has been demonstrated to strongly inhibit the Ran-dependent import of known substrates (Palacios et al., 1996), on the cytosol-independent nuclear migration of importin α. As shown in Figure 3, Q69LRanGTP had no effect on importin α import, while the cytosol-dependent nuclear import of GST–NLS–GFP was significantly inhibited. Furthermore, it was found that a non-hydrolyzable GTP analog, GTPγS, did not inhibit the nuclear migration of importin α. These data strongly suggest that Ran does not play a role in the nuclear import of importin α.
Fig. 3. Imporin β-independent nuclear import of importin α occurs without the support of GTP hydrolysis of Ran. Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–importin α (mRch1) alone, or 2.5 µM GST–NLS–GFP with Ehrlich cytosolic extracts and an ATP regeneration system, in the presence of 25 µM Q69LRanGTP or 1 mM GTPγS for 20 min at 30°C.
Importin α import does not require ATP hydrolysis
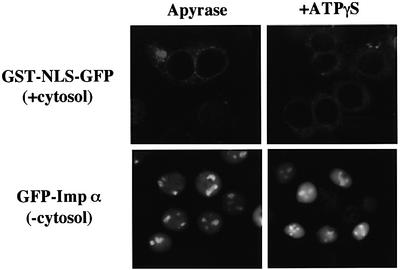
As discussed above, importin α migrated into the nucleus without the exogenous addition of ATP. In order to determine the energy requirement for importin α import, digitonin-permeabilized cells were treated with apyrase. As shown in Figure 4, the pre-incubation of the cells with apyrase had no effect on importin α import, whereas the import of GST–NLS–GFP was completely abolished in the apyrase-pretreated permeabilized cells. In addition, a non-hydrolyzable ATP analog, ATPγS, had no effect on the nuclear import of importin α. These data suggest that the requirement for ATP and its hydrolysis is much less for the import of importin α, compared with conventional NLS-mediated nuclear import.
Fig. 4. Nuclear import of importin α is distinct from that of a conventional NLS-containing karyophile in terms of its requirement for ATP hydrolysis. Digitonin-permeabilized MDBK cells were incubated with TB containing 0.1 U/ml apyrase (Sigma) and 2% BSA for 5 min at 30°C. After rinsing the cells with TB, they were incubated with import mixtures containing 2.5 µM GFP–importin α (mRch1) alone or 2.5 µM GST–NLS–GFP with Ehrlich ascites tumor cell cytosolic extracts and ATP regeneration system for 20 min at 30°C. The permeabilized cells were also incubated with the import mixtures in the presence of 1 mM ATPγS for 20 min at 30°C.
Nuclear import of importin α is saturable but does not compete with importin β
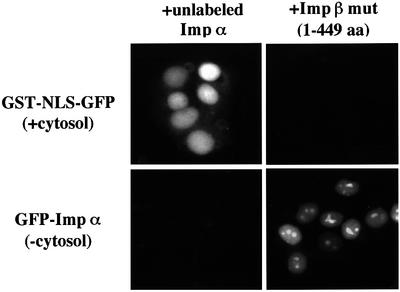
To further characterize importin α import, we examined the saturability of the import process. For this experiment, an excess amount of untagged importin α was added, in the presence of GFP–importin α, to the in vitro assay system. Under these conditions, the nuclear accumulation of GFP–importin α was greatly diminished, while no effect was observed for the nuclear import of GST–NLS–GFP (Figure 5). In contrast, an importin β mutant (1–449 amino acids), which is known to inhibit the nuclear import of both the importin α/β/NLS-substrate complex and importin β alone through a specific interaction with nucleoporins (Kose et al., 1997), had no effect on importin α import. Collectively, these results clearly demonstrate that importin α import is saturable and involves specific interactions, probably with the NPC, that are distinct from that of both importin α/β/NLS-substrate complex and importin β alone.
Fig. 5. Nuclear import of importin α is saturable but does not compete with importin β. Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–importin α (mRch1) alone, or 2.5 µM GST–NLS–GFP with Ehrlich ascites tumor cells cytosolic extracts and ATP regeneration system, in the presence of an excess (∼10×) amount of untagged importin α (mRch1), or 25 µM importin β mutant (1–449 amino acids) for 20 min at 30°C.
Importin β-independent nuclear import activity of importin α is conserved in yeast
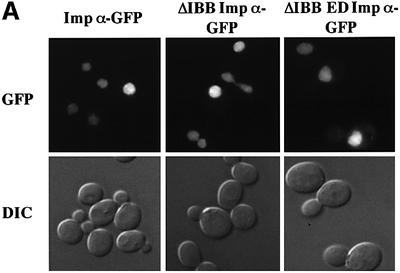
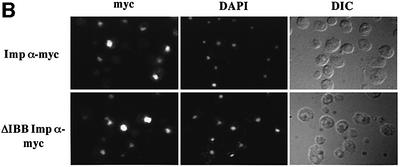
To further examine the process of importin β-independent nuclear import of importin α, we examined whether it also occurred in yeast. Results shown in Figure 6 indicate that yeast importin α that lacks the IBB domain (ΔIBB importin α) is efficiently localized to the nucleus and this localization is indistinguishable from that of wild-type importin α. Similar results are obtained whether importin α proteins are visualized as C-terminal GFP fusion proteins (Figure 6A) or epitope tagged and detected with an anti-myc antibody (Figure 6B). To confirm that import of ΔIBB importin α is not mediated by binding to NLS cargo that contains multiple NLSs, we examined whether a mutant of importin α that cannot bind NLS cargo (Gruss et al., 2001), ΔIBB ED importin α, could still enter the nucleus. Results shown in Figure 6A indicate that ΔIBB ED importin α is localized to the nucleus as efficiently as either wild-type or ΔIBB importin α. This suggests that yeast ΔIBB importin α does not enter the nucleus by binding to cargo that contains more than one nuclear targeting signal and piggy-backing into the nucleus via another import receptor that recognizes that cargo. Taken together, these results support the finding that there is an importin β-independent mechanism for nuclear import of importin α and suggest that this mechanism has been conserved through evolution.
Fig. 6. Saccharomyces cerevisiae importin α enters the nucleus in an importin β-independent manner. (A) Wild-type yeast cells were transformed with importin α–GFP, ΔIBB importin α–GFP or ΔIBB ED importin α–GFP. GFP fusion proteins were visualized by direct fluorescence microscopy. Corresponding differential interference contrast (DIC) images are shown. (B) Myc-tagged importin α proteins were detected by indirect immunofluorescence using an anti-myc antibody. Cells were also stained with DAPI to show the position of the nucleus. Corresponding DIC images are shown.
The rate of import of importin α differs among individual nuclei of permeabilized cells
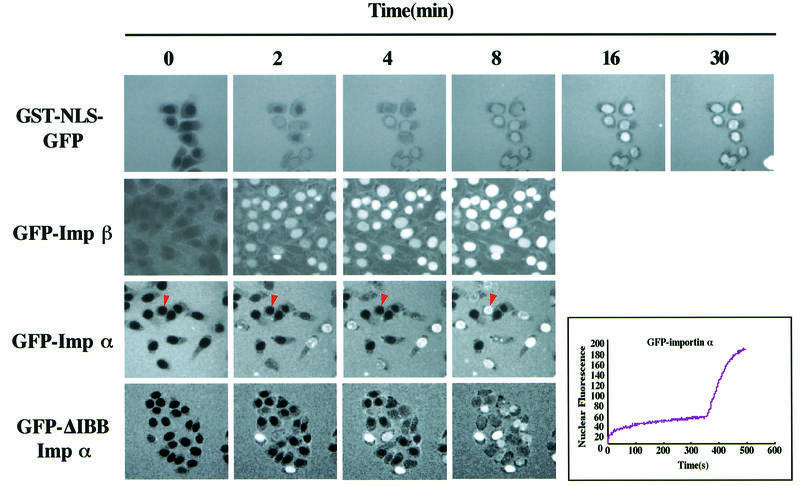
To estimate the rate of entry of importin α into the nucleus in permeabilized cells, we monitored its import into the nucleus by time-lapse photography using confocal microscopy. For control experiments, the rate of entry of GST–NLS–GFP and GFP–importin β was examined. The nuclear import of GST–NLS–GFP was monitored at intervals of 3 s in the presence of importin α/β, Ran, NTF2 and an ATP regenerating system, and GFP–importin β, GFP–importin α and GFP–ΔIBB importin α were monitored at intervals of 1 s in the absence of soluble factors and ATP. As shown in Figure 7, we observed a steady increase in nuclear fluorescence of GST–NLS–GFP that continued for ∼25 min and found that the fluorescence was concentrated in almost all of the nuclei at nearly the same rate. Nuclear import of GFP–importin β occurred more rapidly than that of GST–NLS–GFP and reached a plateau within ∼4 min and, like GST–NLS–GFP, the nuclear accumulation of GFP–importin β occurred at almost the same rate in almost all of the nuclei. The nuclear import of importin α, however, was very unique and significantly different from that of GST–NLS–GFP and importin β. That is, GFP–importin α accumulated in individual nuclei at distinctly different rates, although the nuclear accumulation of importin α was observed to be as rapid as that of importin β in a few cells. For the most unique example, we observed a nucleus (shown by an arrowhead in Figure 7) where GFP–importin α was rapidly concentrated in the nucleus after an incubation of ∼6 min. Importantly, GFP–ΔIBB importin α also accumulated in individual nuclei in a similar manner to the wild-type importin α, which clearly eliminates the possibility that endogenous residual importin β remaining in the individual permeabilized cells may be recycled and thus affect the influx kinetics of GFP–importin α. These results suggest that each nucleus may be functionally different with respect to the import of importin α. Furthermore, more careful observation indicated that GFP–importin α transiently concentrated at the nuclear periphery, suggesting that importin α may interact with nucleoporins at the nuclear pore before translocation into the nucleoplasm and that the functional heterogeneity of each nucleus for importin α import may be the result of structural and/or functional heterogeneity of the NPCs of individual nuclei.
Fig. 7. Time-lapse analysis of importin α nuclear import. Four micromolar GST–NLS–GFP, 4 µM GFP–importin β, 4 µM GFP–importin α (mRch1) and 4 µM GFP–ΔIBB importin α (hRch1) were added to the digitonin-permeabilized MDBK cells and their accumulation into nuclei was recorded in real time by confocal microscopy. The nuclear import of GST–NLS–GFP was monitored in the presence of recombinant importin α (mRch1), importin β, RanGDP, p10/NTF2 and an ATP regeneration system, and that of GFP–importin β, GFP–importin α and GFP–ΔIBB importin α was monitored in the absence of any soluble factors and exogenous ATP. The image of GST–NLS–GFP was captured 500 times at intervals of 3 s. The images of GFP–importin β, GFP–importin α and GFP–ΔIBB importin α were captured 500 times at intervals of 1 s. A nucleus indicated by an arrowhead showed that GFP–importin α was rapidly concentrated in the nucleus just after an incubation of ∼6 min. The change in the mean fluorescence intensity of the nucleus with time was plotted.
The heterogeneity of the nuclear import rate of importin α in individual permeabilized cells can be recovered by the addition of ATP
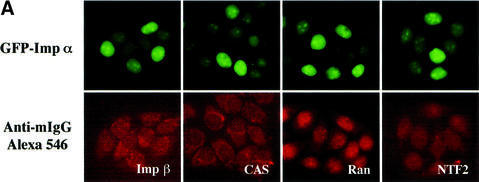
To determine whether the heterogeneity of the import rate of importin α for individual nuclei is affected by residual amounts of endogenous soluble transport factors such as importin β, CAS, Ran and NTF2 remaining in the permeabilized cells, we performed indirect immunofluorescence on permeabilized cells using specific antibodies. These factors remained in each permeabilized cell to almost the same extent, not only just after the permeabilization (data not shown) but also after the import reaction (Figure 8A), suggesting that the heterogeneity was not due to these residual, soluble factors.
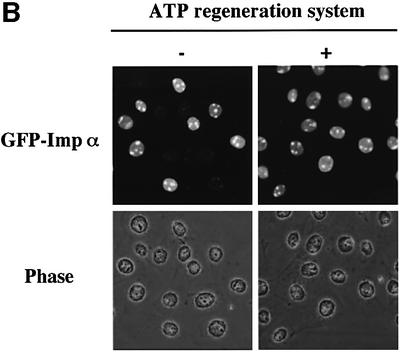
Fig. 8. Incubation of permeabilized cells with ATP affects the nuclear import efficiency of importin α. (A) After digitonin-permeabilized HeLa cells were fixed with 3.7% formaldehyde in PBS, endogenous importin β, CAS, Ran and NTF2 were stained with respective specific antibodies (mouse monoclonal antibodies; Transduction Laboratories). These antibodies were detected by Alexa 546-conjugated goat anti-mouse IgG (Molecular Probes). (B) Digitonin-permeabilized MDBK cells were incubated with 2.5 µM GFP–importin α (mRch1) in the absence or presence of an ATP regeneration system for 20 min at 30°C.
Although we demonstrated that the nuclear import of importin α does not require ATP or its hydrolysis, we noted that the addition of ATP accelerated the rate of importin α import. Therefore, we added an ATP regeneration system to the in vitro assay. As a result, we found that the addition of an ATP regeneration system, but not ATPγS, dramatically increased the nuclear accumulation of importin α and diminished the heterogeneity (Figure 8B). These results suggest that reaction(s) requiring ATP hydrolysis may alter the function of the NPC, thus affecting the efficiency of the importin β-independent nuclear import of importin α.
Discussion
This study provides a clear demonstration that all subgroups of mouse importin α (GFP–mRch1, GFP– mNPI2 and GFP–mQip1) are able to migrate into the nucleus without the addition of exogenous cytosol or importin β in an in vitro assay (Figure 2C). The reaction has no requirement for Ran or its GTP hydrolysis (Figure 3). In addition, the import is sensitive to WGA and temperature, suggesting that importin α migrates into the nucleus through the gated channels of the NPC (Figure 2A). These results show that importin α is able to behave in two different ways in cells. One is to function as an adapter molecule between NLS-containing proteins and importin β to support NLS-mediated import, and the other is to migrate into the nucleus by itself without the aid of other soluble factors.
It has been reported that some proteins are able to migrate into the nucleus in the absence of other soluble factors and in a Ran-independent manner. Importin β was first demonstrated to migrate rapidly into the nucleus by itself in a Ran-independent manner (Kose et al., 1997). Unlike importin α, the nuclear import of importin β is temperature independent. Moreover, although the import of importin α was saturable, importin β did not compete with the importin α import (Figure 5). These results indicate that the import pathway for importin α is clearly distinct from that of importin β.
Yokoya et al. (1999) demonstrated that β-catenin, which is known to contain the characteristic arm repeat motifs, is imported into the nucleus in a manner similar to importin β in a Ran-independent manner. Unlike importin α, the import of β-catenin is competitively inhibited by importin β-family molecules. In addition, the import of two uracil-rich small nuclear ribonucloprotein (U snRNP) components U1A and U2B” into the nucleus has also been reported to occur in a Ran-independent manner and in the absence of soluble cytosolic factors (Hetzer and Mattaj, 2000). The import of these two proteins is essentially different from that of importin α with respect to their ATP requirement, because the import of importin α does not require ATP and its hydrolysis (Figure 4), while U1A import requires ATP hydrolysis. Furthermore, it was recently demonstrated that the nuclear import of RCC1 can proceed by at least two distinct mechanisms (Nemergut and Macara, 2000). The first is a classical importin α/β-dependent import pathway mediated by the N-terminal domain (NTD) of RCC1, which contains basic amino acid residues. The second pathway is not dependent on the NTD and does not require importin α, importin β or the addition of any other soluble factors. Although the NTD-independent import of RCC1 is saturable, sensitive to temperature and occurs in energy-depleted cells like the importin α import, the mutant of importin β (45–462 amino acids) competitively inhibits the import of RCC1. Moreover, the NTD-independent pathway of RCC1 is not inhibited by WGA treatment, which differs from the import of importin α. From these results, we assume that importin α is transported into the nucleus by a quite novel process.
How then is importin α transported into the nucleus? Importin α possesses the ability to be constitutively translocated through the NPC by itself like importin β and β-catenin. Importin β possesses tandem repeating motifs called HEAT motifs and is translocated through the NPC via specific direct interaction with saturable site(s) of NPC components. β-catenin possesses 12 tandem repeating motifs called arm, and the arm repeats of β-catenin have been reported to be necessary and sufficient for its nuclear accumulation (Funayama et al., 1995). In addition, it has been demonstrated that importin β competes with the nuclear import of β-catenin (Yokoya et al., 1999). Furthermore, it has been shown that the structures of arm motifs and HEAT motifs are fundamentally similar (Malik et al., 1997), suggesting that these repeating motifs may share a functional similarity. From these findings, it has been proposed that β-catenin migrates into the nucleus via an interaction with the same site of the NPC as importin β. As shown in the Results, it was consistently found that arm repeat 9 and a portion of arm repeat 10 of importin α are required for its nuclear import (Figure 1). However, importin β did not inhibit the nuclear import of importin α, although importin α competitively inhibited its own import (Figure 5). Therefore, the possibility that importin α may require a specific carrier protein, which is tightly retained in the permeabilized cells and does not compete with importin β for binding to NPC, cannot be excluded at this time. Alternatively, importin α may interact with site(s) of the NPC other than those that bind importin β. In fact, certain nucleoporins containing FXFG repeats, Nup153 (Moroianu et al., 1997) and Nup2p (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000), have been reported to interact with importin α. Further studies will be required in order to determine whether importin α is translocated into the nucleus via direct interactions with specific site(s) on the NPC, and if so, which nucleoporins are involved in the import of importin α.
As shown in Figure 6, we demonstrated that the nuclear import activity of importin α is conserved from yeast to mammals. Hübner et al. (1999) showed that the plant Arabidopsis thaliana importin α binds with a high affinity to a tested classical NLS and mediates targeting of NLS-containing transport substrates to the nuclear envelope in the presence of Ran and NTF2 without the aid of importin β. At present, although insufficient data are available to understand the physiological significance of the observed import of mammalian and yeast importin α, it has been reported that importin α might have an additional essential role(s) distinct from the NLS receptor in nuclear protein transport, such as roles relating to the regulation of protein degradation and mitotic onset (Tabb et al., 2000). Thus, we can speculate on some possible roles for importin β-independent importin α import. First, importin α may carry unknown cargoes into the nucleus in an importin β-independent manner, as reported for plant importin α (Hübner et al., 1999). In this case, since mouse importin α is not able to enter the nucleus in the presence of multivalent (T-BSA) or monovalent (T-peptide or GST–NLS–GFP) conventional basic type NLS-containing substrates (Figure 2D), this suggests that cargoes bearing a novel type of NLS, thereby binding to a different site of importin α, may be transported to the nucleus by importin α without the aid of importin β. Secondly, importin α solely migrates into the nucleus, where it binds to certain nuclear proteins, regulating their nuclear function. Since a basic type NLS of karyophiles is often located in their DNA binding domain, an excess amount of nuclear importin α may mask the DNA binding domain via interacting with the NLS, thus suppressing their intranuclear function. Thirdly, the nuclear import efficiency may be regulated by controlling the amount of cytoplasmic importin α. Fourthly, importin α may enter the nucleus by itself to regulate mitotic events, since it has recently been demonstrated that importin α has an inhibitory effect on spindle assembly in mitosis (Gruss et al., 2001). Further studies will be required to explore these and other possibilities. In any case, since the import activity appears to be widely conserved among species, we suggest that the importin β/Ran-independent nuclear import ability of importin α has a significant physiological role in eukaryotic cells.
By time-lapse photography, we found that although the nuclear accumulation of GFP–importin α in a few cells occurred as rapidly as that of GFP–importin β, the rate or timing of nuclear import of GFP–importin α differed among cells (Figure 7). On the other hand, GST–NLS–GFP accumulated evenly in almost all the nuclei when the reaction was performed in the presence of importin α, β, Ran, NTF2 and an ATP regeneration system, or GFP–importin β alone. Therefore, it would be interesting to assume that functional diversity exists in individual NPCs or that the transport activity of NPCs may be differentially regulated in individual cells. Moreover, it was found that the heterogeneity observed in the importin α import is apparently diminished by the addition of an ATP regeneration system to the in vitro assay (Figure 8B). The integrity of the nuclear envelope was not injured as the result of ATP treatment of permeabilized cells, because GST–NLS–GFP did not enter the nucleus in the absence of soluble factors following ATP treatment (data not shown). Although the issue of how ATP acts on NPCs is not yet resolved, speculation concerning this phenomenon can be made. First, certain ATPase(s) may function at or near the NPCs to regulate the transport through the nuclear pores in a cell-specific manner. ATPases are known to function in diverse biological events such as selective ion transport, actin-based motility, membrane traffic and nuclear envelope formation (Hetzer et al., 2001). Secondly, chaperone-like activity may control the structure and/or function of the NPCs through ATP hydrolysis. Thirdly, certain kinase(s) may phosphorylate a population of nucleoporins, thus regulating the transport efficiency of the NPCs. It has been consistently reported that the phosphorylation of nuclear transport machinery regulates the nuclear import of a karyophilic protein in vitro (Mishra and Parnaik, 1995), and it has been speculated that different kinases either positively or negatively regulate the activity of the nuclear import machinery (Kehlenbach and Gerace, 2000). In order to understand the mechanism of this ATP requirement, the identification and characterization of the regulator(s) of NPC function will be required.
Another important issue that should be explored is the generality for importin α import. It would be interesting to know whether molecules other than importin α are transported into the nucleus through this importin β-independent import pathway. To achieve this, it may be necessary to identify the exact sequence of importin α that is required for targeting to the NPC.
Materials and methods
Cell culture
HeLa cells or Madin–Darby bovine kidney (MDBK) cells were incubated in DMEM (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and penicillin/streptomycin (100 IU/ml and 100 µg/ml, respectively) at 37°C in a 5% CO2 atmosphere.
Constructs for transfection and transfection experiments
Full-length mouse NPI2 cDNA (1–534 amino acids) was constructed in pEGFP-C2 (Clontech). The mutants of mNPI2 were amplified from pEGFP-C2/mNPI2 (1–534) by PCR using the following synthetic oligonucleotides: forward primers, 5′-TTCGAATTCGAGCTGATTAA TGAAGAAGCTGCC-3′, 5′-TTCGAATTCTTCCGGAAACTCCTGT CCAAAGAGCCG-3′ and 5′-TTCGAATTCTATGTCTTGAATTGCT CCATCC-3′ for mutants lacking 1–50, 1–100 and 1–200 amino acids, respectively; and the reverse primer 5′-CCGTCGACTTTATAG CTGGAAGCCCTCCATGGG-3′. The PCR products were digested with EcoRI and SalI, and ligated into the EcoRI and SalI sites of pEGFP-C2. The expression vectors of the mNPI2 (101–466), (101–413) and (101–353) mutants were constructed from pEGFP-C2/mNPI2 (101–534) by cutting out the fragments of EcoNI–BamHI, KpnI–KpnI and HindIII–SacII, respectively, in which the HindIII site in pEGFP-C2 had been cut and blunted beforehand, followed by blunting and ligation. The expression vector of the mNPI2 (1–413) mutant was constructed from pEGFP-C2/mNPI2 (1–534) by cutting out the KpnI–KpnI fragment, followed by ligation. HeLa cells were plated onto 18 × 18 mm micro cover glass (Matsunami) in 6-well dishes 1–2 days prior to use. The vectors were transfected using the Effecten Transfection Reagent (Qiagen) under the conditions recommended by the supplier, for 12 h.
Expression and purification of recombinant proteins
The GFP chimeras of each recombinant importin α were generated as follows. Full-length mouse Rch1, mouse NPI2, mouse Qip1 and IBB domain (1–35 amino acids)-deleted human Rch1 cDNA were constructed into pGEX 6P-2-GFP vector, which is an expression vector of GST-fused GFP. The expression vectors were transformed into Escherichia coli strain BL21(DE3), and the resulting cells grown in Luria–Bertani medium containing 50 µg/ml ampicillin. Expression and lysis of the bacteria and purification of the fusion proteins were performed as described previously (Imamoto et al., 1995b). The GST portion of the chimeras was cleaved with PreScission protease (Amersham Biosciences) for 5 h at 4°C in buffer (50 mM phosphate pH 7.2, 50 mM NaCl, 2 mM DTT, containing 1 µg/ml each aprotinin, leupeptin and pepstatin). The GFP-fused proteins were further purified on a MonoQ column (Pharmacia), and then dialyzed against 20 mM HEPES pH 7.3, 110 mM CH3COOK, 2 mM DTT, containing 1 µg/ml each aprotinin, leupeptin and pepstatin. GST–NLS–GFP recombinant protein (Yokoya et al., 1999), a MBP–IBB recombinant protein (Nagoshi and Yoneda, 2001), Q69LRan recombinant protein (Tachibana et al., 2000), GST–NPI1 (full-length and ΔIBB) recombinant protein (Sekimoto et al., 1997), GFP–importin β (full-length) and the importin β mutant (1–449 amino acids) (Kose et al., 1997) were constructed and prepared as described previously.
Constructs for yeast experiments
Full-length importin α and ΔIBB importin α (amino acid residues 89–542) were subcloned in-frame into CEN URA3 plasmids derived from pRS316 (Sikorski and Hieter, 1989) that express GFP or myc (3×) at the C-terminus. To create a mutant ΔIBB importin α that has a significantly decreased interaction with NLS cargo, we mutated two residues within the NLS binding pocket of ΔIBB importin α (E402 to R and D203 to K) (Conti et al., 1998; Gruss et al., 2001) to generate ΔIBB ED importin α–GFP.
Cell-free import assay
Digitonin-permeabilized HeLa or MDBK cells were prepared as described previously (Adam et al., 1990). In order to decrease the remaining factors in the cytoplasm after permeabilization, cells were incubated for 10 min at 4°C and then washed three times with transport buffer [TB; 20 mM HEPES pH 7.3, 110 mM CH3COOK, 5 mM CH3COONa, 2 mM (CH3COO)2Mg, 0.5 mM EGTA, 2 mM DTT, 1 µg/ml each leupeptin, pepstatin and aprotinin]. The cells were incubated with TB containing the test proteins and 2% BSA, in 10 µl, under each condition indicated in the respective figure legends. After the import reaction, the cells were fixed with 3.7% formaldehyde in TB. Ehrlich ascites tumor cell cytosol was prepared as described previously (Imamoto et al., 1995a). ATP regeneration system (1 mM ATP, 20 U/ml creatine phosphokinase, 5 mM creatine phosphate) was adjusted with TB.
Synthetic peptide and preparation of peptide conjugate
BSA (Sigma) was chemically conjugated to synthetic peptides containing wild-type (CYGGPKKKRKVEDP) or reverse (CYGGPDEVKRKKKP) sequence of SV40 large T-antigen NLS as described previously (T-BSA or reverse T-BSA) (Imamoto et al., 1995a).
Time-lapse photography
Cultured cells were grown on a glass bottom dish (MatTek Corporation; Part No. P35G-1.50-10-C). Before the cells were permeabilized with digitonin, Hoechst 33342 (1 µg/ml) was added to the dishes, followed by incubation for 20 min at 37°C in a 5% CO2 atmosphere. The cells were permeabilized as described above and then fixed to the stage of an inverted confocal microscope LSM510 (Carl Zeiss, Inc.). The fluorescent scans were started in the time-lapse mode and the focal plane was adjusted to the nuclei. The import reaction was then initiated by the addition of the import mixtures containing GFP fusion proteins. The import reaction was performed at 30°C in TB.
Microscopy
Cultured cells were fixed with 3.7% formaldehyde in PBS for 15 min at room temperature. After permeabilization with 0.5% Triton X-100 in PBS for 5 min at room temperature, the cells were blocked with 3% skim milk in PBS and incubated with each antibody described in the respective figure legends. The samples were examined using a Zeiss Axiophot fluorescent microscope (Carl Zeiss).
Indirect immunofluorescence microscopy on yeast cells was performed essentially as described previously (Spector et al., 1998), with the following conditions. The myc antibody was used at 1:100 dilution (Santa Cruz Biotechnology) and incubated with cells overnight at 4°C. The Texas Red-labeled anti-mouse secondary antibody (Jackson ImmunoResearch; 1:1000 dilution) was incubated with cells for 2 h at room temperature. DNA was stained with DAPI (1 µg/ml). Samples were viewed through a Texas Red-optimized filter (Chroma Technology) using an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera.
Direct fluorescence microscopy was used to localize GFP fusion proteins in live yeast cells. Wild-type cells (ACY192) were transformed with constructs encoding the fusion proteins expressed from the endogenous importin α promoter. Cells were grown to early log phase in synthetic media at 30°C. Cells were stained with DAPI to visualize the DNA and confirm the location of the nucleus. The localization of the fusion proteins was monitored by directly viewing the GFP signal in living cells through a GFP-optimized filter as described for indirect immunofluorescence microscopy.
Acknowledgments
Acknowledgements
We thank Dr N.Imamoto and Dr S.Kose for valuable discussions, and members of our laboratory for useful discussions. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas (B) (No. 11237202), Grant-in-Aid for Scientific Research (B) (No. 12480215), Grant-in-Aid for COE Research (No. 12CE2007) from the Japanese Ministry of Education, Science, Sports and Culture, the Mitsubishi Foundation and the Human Frontiers Science Program.
References
- Adam S.A., Marr,R.S. and Gerace,L. (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol., 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R. and Ponstingl,H. (1991) Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature, 354, 80–82. [DOI] [PubMed] [Google Scholar]
- Booth J.W., Belanger,K.D., Sannella,M.I. and Davis,L.I. (1999) The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem., 274, 32360–32367. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy,M., Leighton,L., Blobel,G. and Kuriyan,J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Cortes P., Ye,Z.S. and Baltimore,D. (1994) RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl Acad. Sci. USA, 91, 7633–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo C.A., Kirch,S.A., Gyuris,J., Brent,R. and Oettinger,M.A. (1994) Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl Acad. Sci. USA, 91, 6156–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1991) Nuclear targeting sequences—a consensus? Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Fontes M.R., Teh,T. and Kobe,B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol., 297, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Funayama N., Fagotto,F., McCrea,P. and Gumbiner,B.M. (1995) Embryonic axis induction by the armadillo repeat domain of β-catenin: evidence for intracellular signaling. J. Cell Biol., 128, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Mattaj,I.W. (1996) Nucleocytoplasmic transport. Science, 271, 1513–1518. [DOI] [PubMed] [Google Scholar]
- Görlich D., Henklein,P., Laskey,R.A. and Hartmann,E. (1996) A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J., 15, 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J. et al. (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell, 104, 83–93. [DOI] [PubMed] [Google Scholar]
- Herold A., Truant,R., Wiegand,H. and Cullen,B.R. (1998) Determination of the functional domain organization of the importin α nuclear import factor. J. Cell Biol., 143, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. and Mattaj,I.W. (2000) An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B” spliceosome proteins. J. Cell Biol., 148, 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M., Meyer,H.H., Walther,T.C., Bilbao-Cortes,D., Warren,G. and Mattaj,I.W. (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol., 3, 1086–1091. [DOI] [PubMed] [Google Scholar]
- Hood J.K., Casolari,J.M. and Silver,P.A. (2000) Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J. Cell Sci., 113, 1471–1480. [DOI] [PubMed] [Google Scholar]
- Hübner S., Smith,H.M., Hu,W., Chan,C.K., Rihs,H.P., Paschal,B.M., Raikhel,N.V. and Jans,D.A. (1999) Plant importin α binds nuclear localization sequences with high affinity and can mediate nuclear import independent of importin β. J. Biol. Chem., 274, 22610–22617. [DOI] [PubMed] [Google Scholar]
- Imamoto N., Tachibana,T., Matsubae,M. and Yoneda,Y. (1995a) A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J. Biol. Chem., 270, 8559–8565. [DOI] [PubMed] [Google Scholar]
- Imamoto N., Shimamoto,T., Takao,T., Tachibana,T., Kose,S., Matsubae,M., Sekimoto,T., Shimonishi,Y. and Yoneda,Y. (1995b) In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J., 14, 3617–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R.H. and Gerace,L. (2000) Phosphorylation of the nuclear transport machinery down-regulates nuclear protein import in vitro. J. Biol. Chem., 275, 17848–17856. [DOI] [PubMed] [Google Scholar]
- Kobe B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol., 6, 388–397. [DOI] [PubMed] [Google Scholar]
- Köhler M., Ansieau,S., Prehn,S., Leutz,A., Haller,H. and Hartmann,E. (1997) Cloning of two novel human importin-α subunits and analysis of the expression pattern of the importin-α protein family. FEBS Lett., 417, 104–108. [DOI] [PubMed] [Google Scholar]
- Kose S., Imamoto,N., Tachibana,T., Shimamoto,T. and Yoneda,Y. (1997) Ran-unassisted nuclear migration of a 97 kD component of nuclear pore-targeting complex. J. Cell Biol., 139, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Görlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Eickbush,T.H. and Goldfarb,D.S. (1997) Evolutionary specialization of the nuclear targeting apparatus. Proc. Natl Acad. Sci. USA, 94, 13738–13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra K. and Parnaik,V.K. (1995) Essential role of protein phosphorylation in nuclear transport. Exp. Cell Res., 216, 124–134. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y., Imamoto,N., Sekimoto,T., Tachibana,T., Seki,T., Tada,S., Enomoto,T. and Yoneda,Y. (1997) Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem., 272, 26375–26381. [DOI] [PubMed] [Google Scholar]
- Moore M.S. and Blobel,G. (1993) The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature, 365, 661–663. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel,G. and Radu,A. (1997) RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc. Natl Acad. Sci. USA, 94, 9699–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler S.G., Tritschler,D., Haffar,O.K., Blake,J., Bruce,A.G. and Cleaveland,J.S. (1997) Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. J. Biol. Chem., 272, 4310–4315. [DOI] [PubMed] [Google Scholar]
- Nagoshi E. and Yoneda,Y. (2001) Dimerization of sterol regulatory element-binding protein 2 via the helix–loop–helix–leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol. Cell. Biol., 21, 2779–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut M.E. and Macara,I.G. (2000) Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol., 149, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I., Weis,K., Klebe,C., Mattaj,I.W. and Dingwall,C. (1996) RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J. Cell Biol., 133, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M. and Gerace,L. (1995) Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol., 129, 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieve M.G., Guttridge,K.L., Munguia,J.E. and Waterman,M.L. (1996) The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J. Biol. Chem., 271, 7654–7658. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Imamoto,N., Nakajima,K., Hirano,T. and Yoneda,Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J., 16, 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer,P., Vogel,F. and Schlenstedt,G. (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol., 20, 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Goldman,R.D. and Leinwand,L.A. (eds) (1998) Cells: A Laboratory Manual. Vol. 3. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 106.1–106.7.
- Tabb M.M., Tongaonkar,P., Vu,L. and Nomura,M. (2000) Evidence for separable functions of Srp1p, the yeast homolog of importin α (karyopherin α): role for Srp1p and Sts1p in protein degradation. Mol. Cell. Biol., 20, 6062–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana T., Hieda,M., Miyamoto,Y., Kose,S., Imamoto,N. and Yoneda,Y. (2000) Recycling of importin α from the nucleus is suppressed by loss of RCC1 function in living mammalian cells. Cell Struct. Funct., 25, 115–123. [DOI] [PubMed] [Google Scholar]
- Tsuji L., Takumi,T., Imamoto,N. and Yoneda,Y. (1997) Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-targeting complex and their tissue-specific expression. FEBS Lett., 416, 30–34. [DOI] [PubMed] [Google Scholar]
- Weis K., Ryder,U. and Lamond,A.I. (1996) The conserved N-terminal domain of hSRP1α is essential for nuclear protein import. EMBO J., 15, 1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Yano R., Oakes,M., Yamaghishi,M., Dodd,J.A. and Nomura,M. (1992) Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 5640–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Oakes,M.L., Tabb,M.M. and Nomura,M. (1994) Yeast Srp1p has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc. Natl Acad. Sci. USA, 91, 6880–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoya F., Imamoto,N., Tachibana,T. and Yoneda,Y. (1999) β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell, 10, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]