Abstract
Amyloid fibrils and prions are proteinaceous aggregates that are based on a unique form of polypeptide configuration, termed cross-β structure. Using a group of chemically distinct polyamino acids, we show here that the existence of such a structure does not require the presence of specific side chain interactions or sequence patterns. These observations firmly establish that amyloid formation and protein folding represent two fundamentally different ways of organizing polypeptides into ordered conformations. Protein folding depends critically on the presence of distinctive side chain sequences and produces a unique globular fold. By contrast, the properties of different polyamino acids suggest that amyloid formation arises primarily from main chain interactions that are, in some environments, overruled by specific side chain contacts. This side chain effect can be thought of as the inverse of the one that characterizes protein folding. Conditions including Alzheimer’s and Creutzfeldt–Jakob diseases represent, on this basis, pathological cases in which a natural polypeptide chain has aberrantly adopted the conformation that is primarily defined by main chain interactions and not the structure that is determined by specific side chain contacts that depend on the polypeptide sequence.
Keywords: aggregation/amyloid fibril/inverse side chain effect/polyamino acid/protein folding
Introduction
A group of highly debilitating conditions, including Alzheimer’s, Creutzfeldt–Jakob, Huntington’s and Parkinson’s diseases, are characterized by the deposition of proteinaceous aggregates, including amyloid fibrils and prions (Sunde and Blake, 1998; Dobson, 2001). These aggregates involve a specific type of polypeptide conformation, termed ‘cross-β structure’. This conformational state consists of intermolecular β-sheets in a distinctive orientation such that the strands from which these sheets are assembled extend transversely to the major axis of the fibril (Sunde and Blake, 1998). More than 20 natural polypeptides have been found to produce amyloid fibrils inside living organisms in an aberrant manner and thereby to give rise to disease (Sunde and Blake, 1998). In the case of prions, aggregates with cross-β structure can serve as templates that transmit specific steric information to another organism, thus propagating as stable molecular and clinical strains (Prusiner, 1998). It has remained unclear, however, to what extent our present understanding of protein structure might suffice to describe the formation of the particular polypeptide conformations that are specific for the formation of the amyloid core structure.
According to protein folding theory, the side chain sequence encodes the way in which a natural polypeptide chain folds up into a globular structure (Anfinsen, 1973; Dill et al., 1995; Dobson et al., 1998; Baker, 2000). On this basis, a given polypeptide chain is able to give rise to a single ordered conformation. In a similar manner, the existence of amyloid structures has been associated with some specific (albeit generally unrelated) sequence properties, such as the presence of side chain amide groups (Perutz et al., 1994; DePace et al., 1998), certain binary patterns (West et al., 1999; Rochet and Lansbury, 2000) or secondary structural propensities (Rochet and Lansbury, 2000). In contrast, we found that myoglobin, the textbook example of an α-helical and globular protein structure is able to adopt a conformation that is fundamentally different from the native state and shows the typical characteristics of pathological amyloid fibrils (Fändrich et al., 2001). Moreover, observations that numerous polypeptides can adopt amyloid structure under appropriate conditions in vitro, although these are not known to relate to any misfolding disease, have suggested that many, perhaps all, polypeptide chains are able to form amyloid fibrils (Chiti et al., 1999; Dobson, 1999, 2001; Fändrich et al., 2001). However, it follows from such proposals that specific sequences, despite their very significant influence on the rates and conditions under which such aggregates form (Dobson, 2001; Chiti et al., 2002), are not required. Using polyamino acids (PAAs), we set out to resolve these apparently paradoxical observations concerning the effect of the side chain sequence and to assess the relationship between amyloid formation and protein folding. PAAs are simple polypeptide model systems that lack a distinctive sequence and hence also the ability to fold. Their high molecular weight enables the study of the invariant polypeptide properties under conditions in which the effects of the chain ends are negligible.
Results and discussion
Polyamino acids can form fibrillar aggregates
PAAs have long been known to adopt different configurations depending on the physico-chemical environment in which they are found. One example is the structural variability of poly-l-lysine (PK) that is revealed clearly by using circular dichroism (CD) spectroscopy (Figure 1). PK is disordered in water at pH 7 but collapses readily into an α-helical structure if the pH is increased to 11.1. In contrast, heating at this high pH value irreversibly converts α-helical PK into a β-form (Greenfield et al., 1969). When we placed solutions containing β-PK at room temperature and monitored its conformation by CD, β-PK was not found to revert into α-PK within the time span investigated (up to 1 week). However, readjusting the pH from 11.1 to a near neutral value led to the immediate conversion of β-PK into a random coil state, as seen by CD. The characteristics of β-PK and the similarity of some of these procedures to protocols for the conversion of globular proteins, such as the α-helical myoglobin, into amyloid fibrils (Fändrich et al., 2001) prompted us to examine the ultrastructural organization of β-PK by electron microscopy (EM).
Fig. 1. Far-UV CD spectra of different conformations of poly-l-lysine. The far-UV CD spectra of 0.1 mg/ml poly-l-lysine in an α-helical (filled circles, freshly dissolved PK at pH 11.1), β-sheet (open circles, PK at pH 11.1 after heating for 15 min, 52°C) and random coil conformation (diamonds, freshly dissolved PK in pure water).
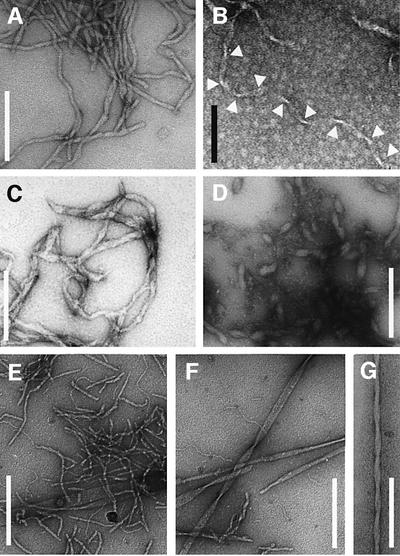
We found that heating at pH 11.1 readily induces the formation of large quantities of fibrillar aggregates (Figure 2A and B). These structures can be seen to be up to 11 nm wide and of a variable length (Figure 2A). Occasionally, we observe ribbon-like morphologies with a helical pitch of ∼100 nm (Figure 2B). We have found that other water-soluble PAAs can produce similar species by careful choice of conditions. For example, poly-l-glutamic acid (PE) was observed to form fibrils at pH values close to 4 (Figure 2C and D). Compared with the other PAAs, some PE aggregates appear less well organized, presumably because the high propensity to precipitate rapidly precludes the formation of highly ordered aggregates in saturated PE solutions. Increasing the pH value to near neutral values led to the dissociation of these PE aggregates. In contrast, the aggregates formed by the more soluble poly-l-threonine (PT) at pH 9 are better delineated and defined. Different morphologies can, however, be discerned (Figure 2E–G). We encountered thin filaments (diameter <5 nm), tapes (diameter 8–9 nm) with lengths of up to 200 nm, as well as highly ordered fibrils. The latter are generally longer than 1 µm (Figure 2F and G) and often occurred in the form of broad plates with diameters up to 70 nm or helical bundles with diameters of 20 nm and a pitch of ∼170 nm. Fibrillar structures (in particular the highly ordered PT fibrils) were found to form only after prolonged incubation (>1 day) and by rearrangement from an aggregated but less well organized (by EM) precursor that can already be detected (by IR spectroscopy) at much shorter incubation times (<15 min). In contrast to PK and PE, PT did not require heating to promote aggregation. The species observed correspond in their overall appearance to the morphologies of amyloid aggregates formed from peptides and proteins, including those associated with specific diseases (Sunde and Blake, 1998).
Fig. 2. Morphology of PAA fibrils defined by EM. PK: (A) 2.5 mg/ml, H2O, pH 11.2, 65°C, 4 days or (B) 1 mg/ml, H2O, pH 10.1, 65°C, 1 week. Arrowheads emphasize the constrictions of a ribbon-like morphology. PE: (C) 1 mg/ml, D2O, pD 4.08, 65°C, 2 days or (D) H2O, pH 4.1, 65°C, 6 days. PT: 10 mg/ml (E and F) or 1 mg/ml (G) PT in 50 mM sodium borate pH 9.0 was exposed for 4 days to 65°C. Samples in (E) and (F) subsequently were kept at room temperature for 6 weeks. Scale bars: 200 nm (white), 100 nm (black).
We have, in addition, examined several hydrophobic PAAs including the polymers of alanine, valine, lecuine, isoleucine, phenylalanine and tyrosine. We found that the commercially available polymers of very high molecular weight were extremely prone to undergo precipitation reactions, thus preventing the study of aggregate formation. Some dissolved in concentrated trifluoroacetic acid (TFA) or dichloroacetic acid, but they precipitated effectively instantaneously upon addition of small quantities of aqueous solutions, acetonitrile or methanol. However, early studies using organic solvents have already demonstrated that several of these PAAs can readily aggregate into β-structures under specific conditions or as small peptides (Arnott et al., 1967; Glenner et al., 1972; Komoto et al., 1974; Lotz, 1974). Interestingly, we found that the dimensions reported for these aggregated structures correspond closely to those of disease-related amyloid fibrils or the aggregates of water-soluble PAAs (see below).
Fibrillar PAA aggregates possess the characteristics of amyloid structures
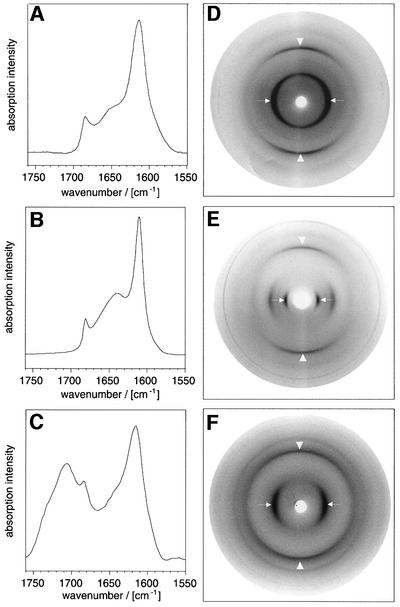
In order to assess whether or not these fibrils show the characteristic polypeptide chain arrangement of aggregated β-sheet structures, we examined them by Fourier transform infrared (FTIR) spectroscopy. The spectra obtained showed that the three PAAs produce very similar amide I′ regions (Figure 3A–C) that contained absorption maxima at 1611 cm–1 for PK, 1616 cm–1 for PE and 1614 cm–1 for PT, demonstrating the presence of aggregated β-sheets. Using X-ray diffraction techniques, we then tested whether these β-sheets possess the orientation properties characteristic of amyloid fibrils. These methods provide the most specific and direct test for the presence of a cross-β structure; if fibrils containing such structure are aligned and oriented with their main axes perpendicular to the X-ray beam, a highly characteristic diffraction pattern can be observed. At least two reflections must be present in this pattern to be defined as cross-β structure: a sharp meridional one at almost precisely 4.75 Å corresponding to the separation of two hydrogen-bonded chains (the ‘main chain reflection’) and an equatorial one that is more variable and that is often seen to be close to 10 Å (Sunde et al., 1997). This so-called ‘side chain reflection’ depends on the packing distance between two juxtaposed β-sheets. The typical anisotropy of the diffraction patterns of amyloid fibrils implies a β-sheet orientation in which the hydrogen bonds and the plane of the sheet are parallel to the main fibril axis, whereas backbone and side chains extend transversely to this axis.
Fig. 3. Internal conformations of fibrillar PAAs. FTIR amide I′ region of PT (A), PK (B) and PE (C) in D2O. Maxima: 1614 and 1685/cm (A); 1611 and 1680/cm (B); and 1616, 1683 and 1706/cm (C). X-ray diffraction pictures of films cast from PT (D), PK (E) and PE (F). To obtain the β-form of PE, the PAA was dissolved at 25 mg/ml in D2O, pD 4.1 and heated for 4 h to 95°C. Arrowheads are parallel to the plane of the film and show the main chain spacing; the arrows point to the side chain spacing.
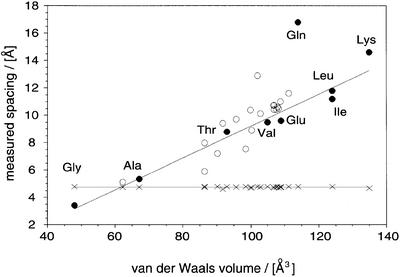
Alignment of the PAA fibrils was achieved by drying aliquots of the sample solution onto a glass slide (Perutz et al., 1994). When films cast from such materials were exposed to X-rays in such a way that the plane of the film was mounted parallel to the beam, diffraction patterns of the typical cross-β type could be observed (Figure 3D–F). The orientation of the main chain and side chain spacings, together with their breadth ratio, demonstrates an organization of β-crystallites in a fibrillar arrangement characteristic of amyloid structure. On the meridian, two reflections were conserved in all patterns (Table I). These were close to 4.7 Å (the main chain spacing) and 3.7 Å. Both diffraction spots are encountered commonly in the patterns of amyloid fibrils (Sunde et al., 1997), and the clear conservation of the main chain spacing reflects the invariant backbone geometry of β-sheets. In contrast, the value of the side chain spacing varied considerably between different PAAs and ranged from 8.77 Å (PT) and 9.6 Å (PE) to 14.6 Å (PK) (see Table I). Consistent with its association with intersheet distances, we find that this spacing correlates closely with the van der Waals volumes of the amino acid residue concerned (Figure 4). In addition, Figure 4 shows that our measurements on fibrillar PAAs are in excellent agreement with the dimensions reported in some very early studies for the β-structure of PAA aggregates (Arnott et al., 1967; Glenner et al., 1972; Komoto et al., 1974; Lotz, 1974). Moreover, if we compare the side chain spacings of amyloid fibrils from peptides having a heterogeneous sequence with their average residual volume, we observe, despite a greater scatter, the same type of correlation as for PAAs (Figure 4).
Table I. Observed Bragg spacings of β-PAAs (in Å).
| PE | PK | PT | PE + PK |
|---|---|---|---|
| 4.73 m | 4.67 m | 4.77 m | 4.67 |
| 3.87 m | 3.79 m | 3.62 m | 3.74 |
| 9.6 e | 14.6 e | 8.77 e | 13.7 |
| 7.5 e | 6.9 | ||
| 5.0 e |
m, meridional reflection; e, equatorial reflection.
Fig. 4. Comparison of the internal structure of PAA fibrils and other amyloid structures. Correlation of the main chain spacing (crosses) and the side chain spacing (circles) with the residual volume (Creighton, 1993). Closed symbols, PAAs (Arnott et al., 1967; Komoto et al., 1974; Lotz, 1974; Perutz et al., 1994); open symbols, heterogeneous sequences (Glenner et al., 1974; Inouye et al., 1993, 2000; Nguyen et al., 1995; Weaver, et al., 1996; Symmons et al., 1997; Kirschner et al., 1998; Malinchik et al., 1998; Groß et al., 1999; Luckey et al., 2000; Serpell et al., 2000). A linear fit to the PAA side chain spacings (excluding glutamine) is shown.
These findings provide strong support for previous ideas in which the fundamental polypeptide arrangement of amyloid fibrils has been represented as a cross-β structure (Sunde and Blake, 1998). Moreover, the quantitative manner of the correlation described here is particularly interesting as it may provide a means to assess the nature of the sequence regions within natural proteins from which the core structure of amyloid fibrils is formed. Of the various PAAs, only the proposed overall structure of poly-l-glutamine aggregates deviates significantly from this relationship. The published intersheet distance of poly-l-glutamine aggregates (16.8 Å) clearly exceeds the value expected from the dimensions of glutamine side chains (see Figure 4). The reasons for these deviations remain to be established, but intercalation of other species, hydration effects or specific alterations in the overall aggregate structure are possible explanations (Perutz et al., 2002). The fibrillar PAA aggregates that we have examined in this study, however, are based on the same fundamental type of structure as those amyloid fibrils that are constructed from heterogeneous sequences and that are associated with clinical conditions.
Amyloid formation and protein folding are distinct processes
Amyloid formation has often been described as an aberrant pathway within protein folding reactions that diverges from the routes leading to the native state after some initial folding events have produced a partially folded intermediate state. Although this description might well reflect, for some polypeptide systems, the order of events that can be observed experimentally within the particular physico-chemical environment chosen for analysis, we believe that amyloid formation differs from protein folding in a more radical way than has been generally recognized. Protein folding depends critically on a distinctive sequence and produces a globular structure that is specific for the respective polypeptide sequence (Anfinsen, 1973; Dill et al., 1995; Dobson et al., 1998; Baker, 2000). In contrast, PAAs do not possess any specific sequence and are, for this reason, intrinsically unable to fold to specific globular structures (Dill et al., 1995). Our observation that different PAAs are able to form amyloid structures nevertheless demonstrates that this configuration is not the product of a protein folding reaction and represents, instead, a generic type of chain arrangement that is defined by interactions that are common to different sequences, in particular those involving the main chain (Fändrich et al., 2001). Although the nature of, and interactions between, the side chains of the polypeptide involved will influence the stability of the amyloid state relative to others including the folded state, and indeed can influence the details of the fibrillar asembly, they do not dictate the fundamental organisation of the cross-β structure. Amyloid formation does not therefore relate to protein folding, and represents, instead, an entirely different and unique way of organizing the polypeptide chain into an ordered conformation.
Polypeptides are organic polymers that are constructed from units containing the same basic structure, that of an α-amino acid. Natural polypeptides, however, are highly selected sequences and contrast to many of the synthetic polymers in the remarkable structural and chemical diversity that is manifested in their side chains and which has conferred on them the ability to fold into proteins, i.e. to adopt highly specific and globular states. Nevertheless, we believe that natural polypeptides can adopt, in addition to their specific globular protein conformation, a generic structure that reflects their character as organic polymers. This structure is the one that is seen in amyloid fibrils. Three observations provide strong support to this concept. First, amyloid structures are not inherently the product of specific side chain interactions. The PAAs used in this study vary in the structure and properties of their side chains, including such features as their secondary structure propensities, hydrophobicity and electrostatic properties. Most notably, they all lack asparagine and glutamine residues, demonstrating that, despite their frequent occurrence in clinical polypeptide aggregates (Perutz et al., 1994; DePace et al., 1998), these specific residues and their associated properties are not necessary for amyloid formation. Secondly, mutations that alter the main chain properties, such as proline insertions, are found generally to perturb amyloid formation (Moriarty and Raleigh, 1999; Wigley et al., 2002). Thirdly, some synthetic polymers (polyamides or nylons) which lack side chains are known to adopt a chain arrangement similar to the aggregated polypeptide states discussed here (Bunn and Garner, 1947). Although polyamides have different energetic properties compared with polypeptides, both organic polymer chains possess a similar molecular structure.
The kinetic partitioning between aggregates and alternative conformations
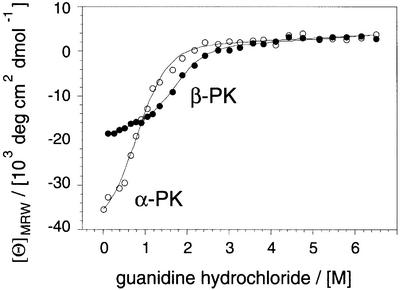
Despite the generic nature of the amyloid structure, different polypeptide chains can differ considerably in their aggregation behaviour within the same physico-chemical environment. Moreover, PAAs require, as do other polypeptides, distinctive conditions in order to form amyloid aggregates. We have therefore utilized our set of PAAs to explore the effects of several physico-chemical parameters on the aggregation reaction and to assess, more generally, the relationship between the common and the specific properties of different polypeptide chains. For example, the aggregation reactions of α-helical PK, PE and poly-l-alanine (Blondelle et al., 1997) are accelerated by heating. This α to β transition is macroscopically irreversible, i.e. if these PAA solutions are placed at room temperature the aggregated chains will not revert spontaneously to an α-helical species. In fact, β-PK is, at room temperature, even more resistant to chaotropic denaturation using guanidine hydrochloride (transition midpoint 1.76 M) than the α-helical form (0.77 M), as shown in Figure 5. Yet, the α-helical configuration forms most readily under these conditions, suggesting that the formation of this structure is due to a kinetic rather than a thermodynamic preference. In contrast to these clear effects on the polypeptide conformation, we could not obtain any evidence that heating promotes the formation of any covalent modifications. All three PAAs were incubated in their respective buffers for up to 4 weeks at 65°C and examined by amino acid analysis. On the basis of their UV profiles in reversed phase chromatography, no samples used in this study contained significant quantities of covalently modified residues, even after this prolonged heating.
Fig. 5. Stability of α-PK and β-PK against denaturation. Guanidine hydrochloride denaturation of α-PK (222 nm, open symbols) and β-PK (217 nm, closed symbols) monitored by CD and fitted to a two-state transition (Santoro and Bolen, 1988).
From these data, we conclude that the partitioning of these PAAs between the α-helical and the amyloid form is critically determined by the stability of the α-helical form. In an environment that supports the conformational stability of this state, α-helices can prevent the polypeptide chain from converting into amyloid structures. These findings on PAAs resemble closely the case of proteins such as myoglobin that form amyloid fibrils at elevated temperatures where the globular structure is disrupted (Fändrich et al., 2001). Taken together, we believe that the presence of an α-helical or globular conformations represents one way, amongst others (Richardson and Richardson, 2002), to suppress amyloid formation by making the polypeptide backbone inaccessible for intermolecular interactions. Further support for this view comes from the observations that PT, poly-l-asparagine and poly-l-glutamine, residues with lower α-helical propensities such that PAAs do not adopt stable α-helical structures in water, form amyloid structures at room temperature (Perutz et al., 2002). Under conditions in which α-helices or globular proteins are unstable, the main chain of a polypeptide will become disordered, solvent exposed and prone to aggregate.
These schemes imply that α-helical or globular polypeptide conformations are able to form faster than amyloid structures. By analogy to globular proteins, in which fast folding correlates with a high proportion of short-range interactions (Baker, 2000), we believe that a kinetic preference for α-helical or globular species is plausible on the basis of the higher proportion of short-range interactions stabilizing these conformations compared with the intermolecular β-sheet structure of aggregates. These ideas are supported further by findings that the formation of the cellular α-helical isoform of the prion protein is under kinetic control (Baskakov et al., 2000) as well as observations that high α-helical propensities can suppress amyloid formation (Villegas et al., 2000). The very fast precipitation reactions of hydrophobic PAAs discussed above and the common insolubility of polypeptides at their isoelectric points (Zurdo et al., 2001) represent further examples of reaction pathways that compete with, and are able to block, amyloid formation.
An inverse side chain effect governs amyloid formation by polyamino acids
The simple presence of a disordered polypeptide chain does not, however, generally suffice for aggregation. Neither PK nor PE aggregates in pure water at a pH near to neutral; under these conditions, unfavourable coulombic interactions between the side chains will make both α-helical and aggregated β-structures unfavourable (Figure 6A and B). In contrast, it has been shown previously that specific counterions that compensate for these electrostatic effects promote the aggregation of these PAAs. X-ray analysis of these aggregates reveals intersheet spacings larger than those observed in the present analysis. For example, the intersheet distance of PE increases from 9.6 to 12.6 Å in the presence of Ca2+ (Keith et al., 1969) and, in the case of PK solutions containing HPO42– ions, from 14.6 to 17.0 Å (Padden et al., 1969). The larger values of side chain spacings suggest an intercalation of these counterions between otherwise densly packed β-sheets. Hence, the side chain interactions across these sheets must be diminished. Uncompensated electrostatic groups therefore represent one example of a specific side chain contact that can depend on the physico-chemical environment and that can repress amyloid formation by destabilizing the amyloid core structure.
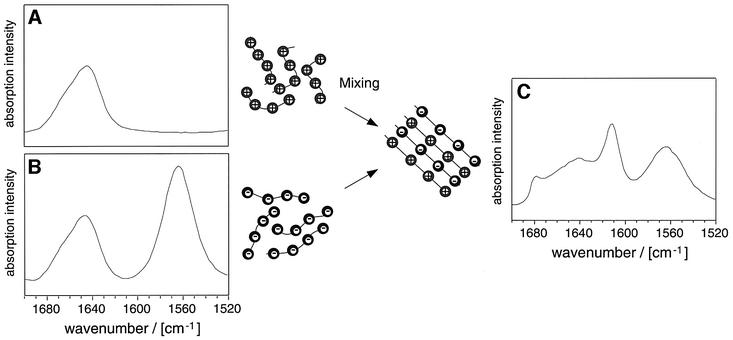
Fig. 6. Formation of mixed aggregates from PE and PK. PK (A) and PE (B) do not aggregate in D2O, pD 7.5. The FTIR spectra show a random coil structure. On mixing the two solutions, the PE and PK aggregates are formed (C). FTIR spectra were recorded in D2O, pD 7.5. The total free amino acid concentrations were all 0.1 M. Amide I′ maxima: 1646 cm–1 (PE and PK in isolation); 1612, 1679 and 1642 cm–1 (mixed aggregates). The carboxylate group gives rise to a peak at 1565 cm–1.
Based on these observations, we speculated that complementary charged side chains might mutually suppress the unfavourable electrostatic properties of the isolated chains by forming mixed aggregates. Indeed, when a 1:1 mixture of PK and PE was exposed to conditions that do not promote the aggregation of either PAA in isolation, namely in aqueous solution at near neutral pH and at room temperature, large quantities of aggregates were found to form (Figure 6C). Although well-defined fibrils were not detected in such experiments by EM, presumably as a result of the insolubility of these species and their strong propensity to precipitate, X-ray diffraction analysis reveals an intersheet distance of 13.7 Å (Table I). This value differs from the side chain spacings of PK (14.6 Å) and PE (9.6 Å) aggregates, and is also larger than their average (12.1 Å), suggesting that lysine residues are present in all these densely packed sheets and that the two PAAs form mixed aggregates. These observations not only support the idea of a generic polypeptide structure, but they are also in accord with our proposals concerning the influence of charged side chains on aggregation reactions.
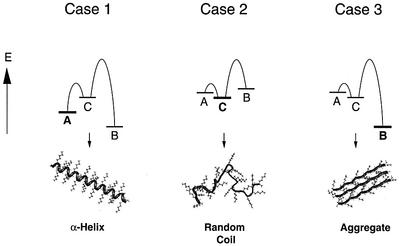
Figure 7 illustrates schematically the possible thermodynamic relationship between the three distinct conformational states of PK (here abbreviated as A, B and C). State C corresponds to a random coil structure, and states A and B represent two ordered conformations. We assume that A (here the α-helical conformation) contains many more local contacts than B (corresponding to amyloid structures). By analogy with a computational analysis of the kinetic partitioning between different folded conformations in a cubic-lattice system (Abkevich et al., 1998), the energy barriers of the formation of A from the unstructured state C will be lower than for the reaction from C to B. The relative energies of the three states and their interconnecting transition states will be determined, for a given polypeptide chain such as PK, by the physico-chemical environment. Although temperature will also affect the height of the energy barrier of the different transition states, aggregation will depend mainly on the extent to which the conditions make potentially competing states of lower contact order unfavourable (Figure 7, case 3).
Fig. 7. The partitioning between folding and amyloid formation. Polypeptides that can adopt ordered states with low contact order (A or α-PK), an ordered state with high contact order (B or β-PK) and a random coil state (C) show different relative energies under different physico-chemical conditions (cases 1–3). For example, the PK side chains have properties that destabilize both states A and B at neutral pH (case 2). Upon increasing the pH, unfavourable electrostatic effects are reduced. Since A is kinetically favourable over B (case 1), the latter state can only be observed under conditions that destabilize A, such as mild heating (case 3).
It follows from these ideas that the side chains influence, in these cases, amyloid formation in a manner that contrasts radically with their effect on the process of protein folding. Protein folding reactions depend on a productive side chain effect, i.e. the polypeptide sequence determines the structure of the reaction product (Anfinsen, 1973; Dill et al., 1995; Dobson et al., 1998; Baker, 2000). A similar assumption, namely that polypeptides cannot convert into amyloid structures unless specific sequences encode this state, has been implied by many previous hypotheses concerning amyloid formation. We believe, however, that our present data demonstrate, at least for the PAAs used in this study, an effect of the side chains that is effectively the inverse of such proposals, namely that the common ability of polypeptide chains to aggregate is, in some circumstances, overruled by specific side chain interactions. Such a suppressive effect is exerted either by destabilizing aggregated structures or by promoting alternative states that are kinetically favourable compared with amyloid formation. Stable α-helical structures or co-operative globular protein conformations represent examples of such alternative states. Destabilization of the interactions that disfavour amyloid structure, for example by changing the experimental conditions, can result in a higher population of disordered chains that are competent to convert into fibrillar states. Once formed, these states resist ready dissociation, largely as a result of substantive transition state barriers or desolvation. The various PAAs differ therefore in their apparent ‘amyloidogenicity’, and the general observation that any polypeptide chain requires particular physico-chemical conditions to promote fibril formation also reflects the fact that not all solution conditions inhibit amyloid formation to the same extent. It is of note that polyglycine and polyamides might therefore be expected to form this structure very readily. Indeed, these polymers have been found to form such fibrillar aggregates (Bunn and Garner, 1947; Lotz, 1974). However, the absence of any specific side chain leads to a highly solvated polypeptide backbone that has strong effects on the energetics of the unfolded state (Avbelj and Baldwin, 2002). Hence, the properties of glycine residues that determine how readily such a structure is adopted may not readily compare with those of chiral residues.
Implications for aggregation reactions involved in conformational diseases
The formation of amyloid structures is the underlying molecular event of a range of human diseases (Sunde and Blake, 1998; Dobson 2001). Our proposals concerning the question of how amyloid formation can be explained from the chemical and structural properties of the polypeptide chain therefore address the common molecular basis for these different pathological conditions. According to these ideas, there are two fundamental modes in which polypeptide chains can be organized into a stable conformation. One of them, the amyloid structure, reflects the common polymer properties of polypeptide chains, whereas the other state (the globular protein conformation) is specific for each polypeptide chain. The latter form, however, is the one that generally has been utilized by nature for functional roles in living organisms. Amyloidoses therefore represent pathological conditions that can arise if a natural polypeptide chain adopts this generic polymer structure rather than the specific (protein) conformation that is encoded in its sequence.
Many apparently unrelated observations concerning the formation of specific pathological fibrils can readily be seen to represent specific cases of a more general structural principle. For example, amyloid formation has been associated with unstable globular protein conformations (Kelly et al., 1996; Dobson, 2001; Fändrich et al., 2001), hydrophobicity (Rochet and Lansbury, 2000; Chiti et al., 2002), enrichment in glutamine or asparagine residues (Perutz et al., 1994; DePace et al., 1998) or the absence of the specific ionic interactions characteristic of edge strands (Richardson and Richardson, 2002). It is particularly interesting that an increasing number of different PAAs has been described to give rise to disease or pathological fibrils. Poly-l-glutamine was the first PAA to be identified in this context. It was found that genetic mutations within the open reading frame of cellular proteins can lead to aberrant poly-l-glutamine stretches, intranuclear fibrous aggregates and diseases such as Huntington’s (Perutz et al., 1994). The formation of these aggregates has been explained previously on the basis of the specific polarity of glutamine side chains (Perutz et al., 1994). However, poly-l-alanine extensions have also been observed to cause disease (oculopharyngeal muscle dystrophy) and to be associated with pathological fibrils (Calado et al., 2000). Moreover, there is the recent report of a disorder termed ‘Huntington disease-like 2’ that depends on abnormal poly-l-leucine repeats (Holmes et al., 2001).
Whereas the wild-type sequences of natural polypeptide chains are likely to be selected in order to suppress the formation of generic polypeptide states by means of the inverse sequence effect described here, specific cases have been documented in which amyloid structures play a functional role in vivo or increase the evolutionary fitness of the affected organism. Examples include the yeast prions that represent the inheritance mechanism of a specific phenotype [PSI+] (True and Lindquist, 2000) or the ‘curli fibrils’ of Escherichia coli (Chapman et al., 2002). Downstream events can therefore determine the specific effect that a particular polypeptide aggregate will produce within its cellular environment. We believe, however, that the concepts discussed here concerning the molecular basis of amyloid formation can provide the key to understand the underlying structural origins of these diseases and, indeed, of other phenotypes that arise from aggregated polypeptides.
Materials and methods
Polyamino acids and amino acid hydrolysis
PE (74.4 kDa), PT (7.6 kDa) and PK (77.3 kDa) were purchased from Sigma (Gillingham, UK). PT and PK were dialysed and lyophilized before use, whereas PE was used without further purification. To test for covalent modifications, PAA aliquots (20 µg) were hydrolysed in 6 M HCl at 150°C, dried and re-dissolved in 2:1 methanol:diisopropylethylamine (DIEA). After a further drying step, the material was derivatized in 15 µl of a 7:2:2 mixture of methanol/DIEA/(5% phenylisothiocyanate in heptane). Reversed phase analyses were carried out at 254 nm using a Vydac 218TP54 column and acetonitrile gradients (0.1% TFA). These results were confirmed independently by an amino acid analysis carried out in the laboratory of L.Packman (Department of Biochemistry, University of Cambridge).
Electron microscopy
For negative staining, specimens were prepared on carbon-coated mica sheets and stained with 2% uranyl acetate solution using the floating carbon method. A freshly cleaved mica sheet was coated with carbon. Aliquots of the sample solution were diluted (typically 10-fold in buffer or pure water), inserted between the carbon layer and the mica and left for 3 min to enable binding to the carbon film. The mica was then transferred to the top of an uranyl acetate droplet (50 µl) so that the carbon film would fully dissociate from the mica and float on the uranyl acetate. Using a 400 square mesh grid, the carbon film was removed immediately from the staining solution and air dried. The samples were analysed with a JEM 1010 transmission electron microscope at 80 kV excitation voltage.
Circular dichroism and Fourier transform infrared spectroscopy
The CD measurements were carried out in a Jasco J-720 spectropolarimeter (Great Dunmow, UK). Solutions of 0.1 mg/ml were exposed to the respective guanidine hydrochloride concentration for 10 h before the analysis. The guanidine concentration in each sample was determined by refractometry. FTIR samples were prepared in D2O, and the pD value was adjusted using DCl or NaOD (Sigma). Quoted pD values were not corrected for isotope effects. Sample aliquots were placed between two CaF2 windows separated by a 50 µm spacer. Spectra were recorded at room temperature in a Bio-Rad FTS 175C FT-IR spectrometer equipped with an MCT detector cooled with liquid nitrogen. The sample compartment was purged thoroughly with dry nitrogen, and all spectra shown represent the average of 256 acquisitions.
X-ray diffraction
Sample solutions were dried onto glass slides. The resulting films were lifted off and mounted with their planes parallel to the beam. X-ray images were obtained by using an 18 cm Mar imaging plate detector (MarResearch, Norderstedt, Germany) mounted on a Rigaku RU200 rotating anode source of Cu Kα (1.5418 Å). The sample to detector distance was 100 mm and the image files were displayed using IPDISP software from the CCP4 package (CCP4, 1994).
Acknowledgments
Acknowledgements
We thank Minakshi Gosh for valuable assistance in collecting the X-ray data, the laboratory of Len Packman for amino acid analysis, and Thomas Appel and Stephan Diekmann for critically reading the manuscript. M.F. acknowledges a scholarship from the Rhodes Trust. The research of C.M.D. is funded in part by a Programme Grant from the Wellcome Trust. The Oxford Centre for Molecular Sciences is supported by BBSRC, EPSRC and MRC.
References
- Abkevich V.I., Gutin,A.M. and Shakhnovich,E.I. (1998) Theory of kinetic partitioning in protein folding with possible applications to prions. Proteins Struct. Funct. Genet., 31, 335–344. [PubMed] [Google Scholar]
- Anfinsen C.B. (1973) Principles that govern the folding of protein chains. Science, 181, 223–230. [DOI] [PubMed] [Google Scholar]
- Arnott S., Dover,S.D. and Elliott,A. (1967) Structure of β-poly-l-alanine: refined atomic co-ordinates for an anti-parallel β-pleated sheet. J. Mol. Biol., 30, 201–208. [DOI] [PubMed] [Google Scholar]
- Avbelj F. and Baldwin,R.L. (2002) Role of backbone solvation in determining thermodynamic β propensities of the amino acids. Proc. Natl Acad. Sci. USA, 99, 1309–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. (2000) A surprising simplicity to protein folding. Nature, 405, 39–42. [DOI] [PubMed] [Google Scholar]
- Baskakov I.V., Legname,G., Prusiner,S.B. and Cohen,F.E. (2000) Folding of prion protein to its native α-helical conformation is under kinetic control. J. Biol. Chem., 276, 19687–19690. [DOI] [PubMed] [Google Scholar]
- Blondelle S.E., Forood,B., Houghten,R.A. and Pérez-Payá,E. (1997) Polyalanine-based peptides as models for the self-associated β-pleated-sheet complexes. Biochemistry, 36, 8393–8400. [DOI] [PubMed] [Google Scholar]
- Bunn C.W. and Garner,E.V. (1947) The crystal structure of two polyamides (‘nylons’). Proc. R. Soc. Lond. Ser. A, 189, 39–68. [Google Scholar]
- Calado A., Tome,F.M.S., Brais,B., Rouleau,G.A., Kühn,U., Wahle,E. and Carmo-Fonseca,M. (2000) Nuclear inclusions in oculopharyngeal muscular dystrophy of poly(A)binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet., 9, 2321–2328. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chapman M.R., Robinson,L.S., Pinkner,J.S., Roth,R., Heuser,J., Hammar,M., Normark,S. and Hultgren,S.J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science, 295, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Webster,P., Taddei,N., Clark,A., Stefani,M., Ramponi,G. and Dobson,C.M. (1999) Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl Acad. Sci. USA, 96, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Taddei,N., Baroni,F., Capanni,C., Stefani,M., Ramponi,G. and Dobson,C.M. (2002) Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol., 9, 137–143 [DOI] [PubMed] [Google Scholar]
- Creighton T.E. (1993) Proteins: Structure and Molecular Properties, 2nd edn. W.H. Freeman and Company, New York, NY.
- DePace A.H., Santoso,A., Hillner,P. and Weissman,J.C. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Dill K.A., Bromberg,S., Yue,K., Fiebig,K.M., Yee,D.P., Thomas,P.D. and Chan,H.S. (1995) Principles of protein folding—a perspective from simple exact models. Protein Sci., 4, 561–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M. (1999) Protein misfolding, evolution and disease. Trends Biochem. Sci., 24, 329–332. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. (2001) The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. Ser. B., 356, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M., Sali,A. and Karplus,M. (1998) Protein folding: a perspective from theory and experiment. Angew. Chem. (Int. Ed.), 37, 868–893. [DOI] [PubMed] [Google Scholar]
- Fändrich M., Fletcher,M.A. and Dobson,C.M. (2001) Amyloid fibrils from muscle myoglobin. Nature, 410, 165–166. [DOI] [PubMed] [Google Scholar]
- Glenner G.G., Eanes,E.D. and Page,D.L. (1972) The relation of the properties of Congo red-stained amyloid fibrils to the β-conformation. J. Histochem. Cytochem., 20, 821–826. [DOI] [PubMed] [Google Scholar]
- Glenner G.G., Eanes,E.D., Bladen,H.A., Linke,R.P. and Termine,J.D. (1974) β-Pleated sheet fibrils: a comparison of native amyloid with synthetic protein fibrils. J. Histochem. Cytochem., 22, 1141–1158. [DOI] [PubMed] [Google Scholar]
- Greenfield N. and Fasman,G.D. (1969) Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry, 8, 4108–4116. [DOI] [PubMed] [Google Scholar]
- Groß M., Wilkins,D.K., Pitkeathly,M.C., Chung,E.W., Higham,C., Clark,A. and Dobson,C.M. (1999) Formation of amyloid fibrils by peptides derived from the bacterial cold shock protein CspB. Protein Sci., 8, 1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S.E. et al. (2001) A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat. Genet., 29, 377–378. [DOI] [PubMed] [Google Scholar]
- Inouye H., Fraser,P.E. and Kirschner,D.A. (1993) Structure of β-crystallite assemblies formed by Alzheimer β-amyloid protein analogues: analysis by X-ray diffraction. Biophys. J., 64, 502–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye H., Bond,J., Baldwin,M.A., Ball,H.L., Prusiner,S.A. and Kirschner,D.A. (2000) Structural changes in a hydrophobic domain of the prion protein induced by hydration and by Ala→Val and Pro→Leu substitutions. J. Mol. Biol., 300, 1283–1296. [DOI] [PubMed] [Google Scholar]
- Keith H.D., Padden,F.J.,Jr and Giannoni,G. (1969) Crystal structures of β-poly-l-glutamic acid and its alkaline earth salts. J. Mol. Biol., 43, 423–438. [DOI] [PubMed] [Google Scholar]
- Kelly J.W. (1996) Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol., 6, 11–17. [DOI] [PubMed] [Google Scholar]
- Kirschner D.A., Elliott-Bryant,R., Szumowski,K.E., Gonnerman,W.A., Kindy,M.S., Sipe,J.D. and Cathcart,E.S. (1998) In vitro amyloid fibril formation by synthetic peptides corresponding to the amino terminus of apoSAA isoforms from amyloid-susceptible and amyloid-resistant mice. J. Struct. Biol., 124, 88–98. [DOI] [PubMed] [Google Scholar]
- Komoto T., Kim,K.Y., Oya,M. and Kawai,T. (1974) Crystallization of polypeptides in the course of polymerisation, 5. Effect of the steric hindrance on the crystal growth by the side chains. Makromol. Chem., 175, 283–299. [Google Scholar]
- Lotz B. (1974) Crystal structure of polyglycine I. J. Mol. Biol., 87, 169–180. [DOI] [PubMed] [Google Scholar]
- Luckey M., Hernandez,J.F., Arlaud,G., Forsyth,V.T., Ruigrok,R.W.H. and Mitraki,A. (2000) A peptide from the adenovirus fibre shaft forms amyloid-type fibrils. FEBS Lett., 468, 23–27. [DOI] [PubMed] [Google Scholar]
- Malinchik S.B., Inouye,H., Szumowski,K.E. and Kirschner,D.A. (1998) Structural analysis of Alzheimer’s β(1–40) amyloid: protofilament assembly of tubular fibrils. Biophys. J., 74, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty D.F. and Raleigh,D.P. (1999) Effects of sequential proline substitutions on amyloid formation by human amylin20–29. Biochemistry, 38, 1811–1818. [DOI] [PubMed] [Google Scholar]
- Nguyen J.T., Inouye,H., Baldwin,M.A., Fletterick,R.J., Cohen,F.E., Prusiner,S.B. and Kirschner,D.A. (1995) X-ray diffraction of scrapie prion rods and PrP peptides. J. Mol. Biol., 252, 412–422. [DOI] [PubMed] [Google Scholar]
- Padden F.J. Jr, Keith,H.D. and Giannoni,G. (1969) Single crystals of poly-l-lysine. Biopolymers, 7, 793–804. [Google Scholar]
- Perutz M.F., Johnson,T., Suzuki,M. and Finch,J.T. (1994) Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA, 91, 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz M.F., Pope,B.J., Owen,D., Wanker,E.E. and Scherzinger,E. (2002) Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid β-peptide of amyloid plaques. Proc. Natl Acad. Sci. USA, 99, 5596–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.S. and Richardson,D.C. (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA, 99, 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochet J.-C. and Lansbury,P.T.,Jr (2000) Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol., 10, 60–68. [DOI] [PubMed] [Google Scholar]
- Santoro M.M. and Bolen,D.W. (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry, 27, 8063–8068. [DOI] [PubMed] [Google Scholar]
- Serpell L.C., Blake,C.C.F. and Fraser,P.E. (2000) Molecular structure of a fibrillar Alzheimer’s Aβ fragment. Biochemistry, 39, 13269–13275. [DOI] [PubMed] [Google Scholar]
- Sunde M. and Blake,C.C.F. (1998) From the globular to the fibrous state: protein structure and structural conversion in amyloid formation. Q. Rev. Biophys., 31, 1–39. [DOI] [PubMed] [Google Scholar]
- Sunde M., Serpell,L.C., Bartlam,M., Fraser,P.E., Pepys,M.B. and Blake,C.C.F. (1997) Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol., 273, 729–739. [DOI] [PubMed] [Google Scholar]
- Symmons M.F., Buchanan,S.G.S.C., Clarke,D.T., Jones,G. and Gay,N.J. (1997) X-ray diffraction and far-UV CD studies of filaments formed by a leucine-rich repeat peptide: structural similarity to the amyloid fibrils of prions and Alzheimer’s disease β-protein. FEBS Lett., 412, 397–403. [DOI] [PubMed] [Google Scholar]
- True H.L. and Lindquist,S.L. (2000) A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature, 407, 477–483. [DOI] [PubMed] [Google Scholar]
- Villegas V., Zurdo,J., Filimonov,V.V., Avilés,F.C., Dobson,C.M. and Serrano,L. (2000) Protein engineering as a strategy to avoid formation of amyloid fibrils. Protein Sci., 9, 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L., Stagsted,J., Behnke,O., Matthews,B.W. and Olsson,L. (1996) β-Sheet models for the ordered filamentous structure formed by a peptide that enhances the action of insulin. J. Struct. Biol., 117, 165–172. [DOI] [PubMed] [Google Scholar]
- West M.W., Wang,W., Patterson,J., Mancias,J.D., Beasley,J.R. and Hecht,M.H. (1999) De novo amyloid proteins from designed combinatorial libraries. Proc. Natl Acad. Sci. USA, 96, 11211–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley W.C., Corboy,M.J., Cutler,T.D., Thibodeau,P.H., Oldan,J., Lee,M.G., Rizo,J., Hunt,J.F. and Thomas,P.J. (2002) A protein sequence that can encode native structure by disfavouring alternate conformations. Nat. Struct. Biol., 9, 381–388. [DOI] [PubMed] [Google Scholar]
- Zurdo J., Guijarro,J.I., Jimenez,J., Saibil,H.R. and Dobson,C.M. (2001) Dependence on solution conditions of aggregation and amyloid formation by an SH3 domain. J. Mol. Biol., 311, 325–340. [DOI] [PubMed] [Google Scholar]