Abstract
Structural maintenance of chromosomes (SMC) proteins play central roles in regulating higher order chromosome dynamics from bacteria to humans. As judged by electron microscopy, the SMC homodimer from Bacillus subtilis (BsSMC) is composed of two antiparallel, coiled-coil arms with a flexible hinge. Site-directed cross-linking experiments show here that dimerization of BsSMC is mediated by a hinge–hinge interaction between self-folded monomers. This architecture is conserved in the eukaryotic SMC2–SMC4 heterodimer. Analysis of different deletion mutants of BsSMC unexpectedly reveals that the major DNA-binding activity does not reside in the catalytic ATPase domains located at the ends of a dimer. Instead, point mutations in the hinge domain that disturb dimerization of BsSMC drastically reduce its ability to interact with DNA. Proper hinge function is essential for BsSMC to recognize distinct DNA topology, and mutant proteins with altered hinge angles cross-link double-stranded DNA in a nucleotide-dependent manner. We propose that the hinge domain of SMC proteins is not a simple dimerization site, but rather it acts as an essential determinant of dynamic SMC–DNA interactions.
Keywords: Bacillus subtilis/cohesin/coiled-coil/condensin/structural maintenance of chromosomes
Introduction
Structural maintenance of chromosomes (SMC) proteins are ubiquitous among three phyla of life and play fundamental roles in ensuring the faithful segregation of chromosomes during cell division (reviewed by Koshland and Strunnikov, 1996; Cobbe and Heck, 2000; Hirano, 2002). In eukaryotes, at least six members of the SMC protein family are found in individual organisms. SMC1 and SMC3 form a heterodimer that functions as the core of the cohesin complex involved in sister chromatid cohesion (Losada et al., 1998; Toth et al., 1999; Tomonaga et al., 2000), whereas SMC2 and SMC4 act as components of the condensin complex responsible for chromosome condensation (Hirano et al., 1997; Sutani et al., 1999; Freeman et al., 2000; Schmiesing et al., 2000). The third complex containing SMC5 and SMC6 is implicated in DNA repair and checkpoint responses, but its exact functions remain elusive (Fousteri and Lehmann, 2000; Taylor et al., 2001). Each of the three complexes has a unique set of non-SMC subunits, which confer additional structural and functional diversity on the eukaryotic SMC complexes. Most, if not all, of the bacterial and archaeal genomes contain a single smc gene, and its gene product functions as a homodimer. Disruption of the smc gene in Bacillus subtilis causes decondensation and mis-segregation of chromosomes, indicating that bacterial SMC proteins share related functions with their eukaryotic counterparts in vivo (Britton et al., 1998; Graumann et al., 1998; Moriya et al., 1998).
SMC proteins are large polypeptides (between 1000 and 1500 amino acids) with a unique structural organization. Two nucleotide-binding motifs, the Walker A and Walker B motifs, are located in the conserved N- and C-terminal domains, respectively. The central domain is composed of a moderately conserved ‘hinge’ sequence that is flanked by two long coiled-coil motifs. An electron microscopy (EM) study showed that the B.subtilis SMC (BsSMC) homodimer has a two-armed structure with a flexible hinge, and each arm is composed of an antiparallel coiled-coil (Melby et al., 1998). This antiparallel configuration allows association of the N- and C-terminal sequences to assemble an ATP-binding ‘catalytic’ domain at the distal end of each arm. This domain is structurally related to the nucleotide-binding domain (NBD) of the ATP-binding cassette (ABC) transporter family of proteins (Lowe et al., 2001). What is not fully established is how the two subunits are folded in the BsSMC dimer. The original model by Melby et al. (1998) proposed that dimerization is mediated by coiled-coil interactions between the two different subunits. However, point mutations in the hinge domain disturb dimerization, raising the alternative possibility that two self-folded monomers may dimerize by a hinge–hinge interaction (Hirano et al., 2001).
In recent years, substantial progress has been made in our understanding of the biochemical activities associated with SMC proteins. Perhaps the best characterized example is the eukaryotic condensin complex, which has been shown to possess an ability to introduce positive supercoils or positive knots into DNA in an ATP-dependent manner (Kimura and Hirano, 1997; Kimura et al., 1999, 2001; Hagstrom et al., 2002). The presence of the non-SMC subunits unique to condensin, as well as their cell cycle-dependent phosphorylation, are vital to these activities, suggesting that these are not universal activities shared by all SMC proteins (Sutani and Yanagida, 1997; Kimura et al., 1998; Kimura and Hirano, 2000). Consistent with this notion, the cohesin complex displays remarkably different DNA-binding properties from those of condensin (Losada and Hirano, 2001). The distinct biochemical functions of condensin and cohesin could be attributed, at least in part, to their different arm conformations and hinge angles (Anderson et al., 2002). On the other hand, the BsSMC homodimer binds preferentially to single-stranded DNA (ssDNA) and makes large protein–DNA aggregates in an ATP-dependent manner (Hirano and Hirano, 1998). It has been proposed that closing or opening of the central hinge allows association of two catalytic domains within a dimer or between different dimers, respectively, thereby modulating the ATPase cycle of BsSMC (Hirano et al., 2001).
Despite the increasing amount of information, an integrated molecular picture of how SMC proteins might work at a mechanistic level is still missing. In particular, very little is known about how the two-armed SMC proteins interact with DNA. In this study, we have used BsSMC as a model system to understand the common theme of SMC action. Site-directed, protein–protein cross-linking experiments show that a BsSMC dimer is formed by a hinge-mediated interaction of self-folded monomers. By characterizing a number of mutants with altered dimerization properties, we show that the two-armed conformation of BsSMC, and not its catalytic domains, is essential for proper binding to DNA. Our results emphasize the importance of the hinge domain, which not only acts as the dimerization site but also plays an active role in determining the mode of SMC–DNA interactions.
Results
Construction of BsSMC mutants
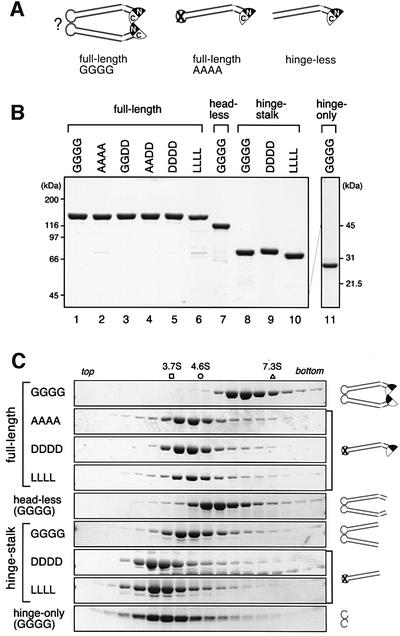
We had reported previously that mutations in the hinge domain synergistically perturb the global conformation of BsSMC and affect its ability to form a stable dimer (Hirano et al., 2001). In particular, when four conserved glycine residues (G657, G658, G662 and G663) were all mutated into alanines, the resulting mutant was found to form a single-armed monomer in solution, as judged by both electron microscopy and hydrodynamic analyses. For simplicity, wild-type BsSMC is referred to as GGGG (Figure 1A, left and B, lane 1) whereas the quadruple alanine mutant is called AAAA (Figure 1A, middle and B, lane 2). In the same study (Hirano et al., 2001), we also constructed a so-called hinge-less mutant, in which the N-terminal (1–476) and C-terminal (672–1186) fragments were co-expressed to make a single-armed BsSMC with no hinge sequence (Figure 1A, right). As shown below, we have now found that the hinge-less mutant displays an extremely poor DNA-binding activity, whereas the binding of AAAA to DNA is substantial. This observation was surprising because the only difference between the two mutant proteins was the presence or absence of the hinge domain. To understand better the DNA-binding properties of SMC proteins, we constructed a series of BsSMC derivatives that contain different point mutations in the hinge domain or deletions in the non-hinge regions. In the first set of mutants, some or all of the glycine residues conserved in the hinge domain were replaced by aspartic acids (D) or leucines (L) to test whether the acidic or hydrophobic residues might perturb hinge functions differently from alanines. The resulting mutants are referred to as GGDD, AADD, DDDD and LLLL (Figure 1B, lanes 3–6). As judged by sucrose gradient centrifugation (Figure 1C; data not shown), the four mutant proteins had a virtually identical sedimentation coefficient (4.2S), which was much smaller than that of GGGG (6.3S) but was indistinguishable from that of AAAA (4.1S). The results suggested that these hinge mutants are all single-armed monomers in solution.
Fig. 1. Design and characterization of BsSMC mutants. (A) Wild-type BsSMC (GGGG) and its mutant derivatives (AAAA and hinge-less) used in a previous study (Hirano et al., 2001). In this diagram, it is postulated that dimerization of BsSMC is mediated by a hinge–hinge interaction (shown by ?). (B) The wild-type and mutant BsSMC proteins were purified, fractionated by SDS–PAGE and stained with Coomassie Blue. (C) The purified BsSMC proteins were fractionated by centrifugation on 5–20% sucrose gradients. Fractions were resolved by SDS–PAGE and stained with Coomassie Blue. The positions of three protein standards [ovalbumin (3.7S), BSA (4.6S) and aldolase (7.3S)] are indicated. The predicted structure for each construct is shown on the right.
In the second set of mutants, the N- and C-terminal regions were deleted systematically. The head-less mutant (160–1037) lacked the catalytic domain (Figure 1B, lane 7), whereas the hinge–stalk mutant (262–861) further lacked the ‘neck’ region that connects the catalytic and coiled-coil stalk domains (Figure 1B, lane 8). The sedimentation coefficients of the head-less and hinge–stalk mutants were 5.5S and 4.4S, respectively (Figure 1C), values consistent with the prediction that they form dimers with elongated shapes. We also made variants of the hinge–stalk mutant, in which the conserved glycines in the hinge domain were replaced by aspartic acids (referred to as hinge–stalk DDDD; Figure 1B, lane 9) or leucines (hinge–stalk LLLL; Figure 1B, lane 10). The two mutants have a sedimentation coefficient of 3.0–3.1S (Figure 1C), again in agreement with the idea that the hinge mutations convert a dimer into monomers. Finally, the hinge domain with no coiled-coil stalk (476–672) was expressed (Figure 1B, lane 11). The sedimentation coefficient of this hinge-only mutant was 3.3S (Figure 1C), a value predicted from the formation of a globular dimer (note that elongated molecules display anomalously smaller sedimentation coefficients compared with globular molecules of the same molecular mass).
Architecture of BsSMC as revealed by site-directed, protein–protein cross-linking
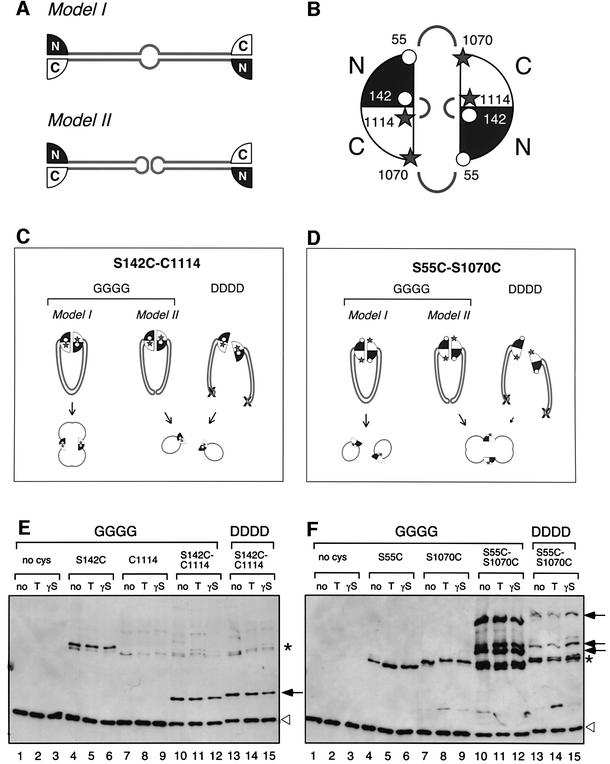
The original model by Melby et al. (1998) proposed that dimerization of BsSMC is achieved by coiled-coil interactions between the two different subunits (Figure 2A, model I). Our current and previous studies convincingly show that mutations introduced into the hinge domain of BsSMC (AAAA, DDDD and LLLL) disrupt its dimerization, resulting in the formation of self-folded monomers (Hirano et al., 2001). These observations raise the possibility that wild-type BsSMC may dimerize by a hinge–hinge interaction of two self-folded monomers (Figure 2A, model II). To distinguish between the two models, we performed site-directed, protein–protein cross-linking experiments using bis-maleimidohexane (BMH), a cross-linker specific for free sulfhydryl groups. Wild-type BsSMC has four cysteine residues (C119, C437, C826 and C1114) that are cross-linkable with this reagent. We first replaced three or all of the cysteines by serines to make mutants that contain either a single cysteine at 1114 (C1114) or no cysteine. We further mutated these polypeptides by replacing one or two serines by cysteines, and constructed mutants that have two cysteine residues in single polypeptides. The locations of these cross-linkable residues were selected on the basis of the crystal structure of an SMC catalytic domain from Thermotoga maritima (Lowe et al., 2001) (Figure 2B). The first mutant (S142C/C1114) was designed so that the N- and C-terminal sequences within a single catalytic domain can be cross-linked with each other. In the second mutant (S55C/S1070C), the two cysteines introduced into the N- and C-terminal sequences are too far apart to be cross-linked within a single catalytic domain. However, when the hinge is closed and two different catalytic domains interact with each other (Hopfner et al., 2000), the S55C residue in one domain would be close enough to be cross-linked to the S1070C residue in the other. If BsSMC dimerizes by a coiled-coil interaction between the two different subunits, cross-linking of the first mutant (S142C/C1114) would yield a dimeric circular polypeptide (Figure 2C, GGGG, model I) whereas the second mutant (S55C/1070C) would form circular monomers (Figure 2D, GGGG, model I). If dimerization of BsSMC is mediated by a hinge–hinge interaction, then the cross-linking patterns would be reversed (Figure 2C and D, GGGG, model II). To judge the electrophoretic mobilities of cross-linked products without ambiguity, we introduced the corresponding cysteine mutations into the single-armed DDDD mutant. In this case, cross-linking between S142C and C1114 would occur readily within a monomer (Figure 2C, DDDD), whereas cross-linking between S55C and S1070C would be greatly suppressed, because two catalytic domains are not in the same molecule and barely interact with each other (Figure 2D, DDDD).
Fig. 2. The folding of BsSMC as revealed by site-directed, protein–protein cross-linking. (A) Two models for the folding of BsSMC. Dimerization may be mediated by coiled-coil interactions between two different subunits (model I). Alternatively, the two subunits may be self-folded to form two separate coiled-coil arms, which in turn dimerize by a hinge-mediated interaction (model II). (B) The positions of cysteine residues used in the cross-linking experiments. Two catalytic domains, each of which is composed of N- and C-terminal sequences, are shown. The open circles indicate the positions of the cysteine residues introduced into the N-terminal sequence (S55C and S142C). The filled stars indicate the positions of the naturally occurring cysteine (C1114) and the artificially introduced cysteine (C1070S) in the C-terminal sequence. Cross-linking is expected to occur between the two residues connected by the arches. (C and D) Predicted results from the site-directed cross-linking of the two-armed (GGGG) or single-armed (DDDD) protein. (E) Purified proteins were treated with BMH in the presence or absence of the indicated nucleotides (no, no nucleotide; T, 1 mM ATP; γS, 1 mM ATPγS), fractionated by 2.5–7.5% SDS–PAGE and analyzed by immunoblotting with an anti-BsSMC antibody. The no-cysteine mutant (lanes 1–3), single cysteine mutants (S142C, lanes 4–6; C1114, lanes 7–9) and the two-cysteine mutant (S142C-C1114, lanes 10–15) were used. The two-armed (GGGG; lanes 1–12) and single-armed (DDDD; lanes 13–15) versions were tested. Specific cross-linking products involving two cysteines at different positions (S142C and C1114) are shown by arrows. Background products involving cysteines at the same position (e.g. S142C of one polypeptide and S142C of the other) are shown by asterisks. These bands probably correspond to linear dimers, which are not depicted in (C) or (D). Non-cross-linked BsSMC polypeptides are shown by open triangles. (F) The same experiment was performed with a different set of mutants: the no-cysteine mutant (lanes 1–3); single-cysteine mutants (S55C, lanes 4–6; S1070C, lanes 7–9); and the two-cysteine mutant (S55C-S1070C, lanes 10–15). The two-armed (GGGG; lanes 1–12) and single-armed (DDDD; lanes 13–15) versions were tested.
The mutant proteins were purified and cross-linked with BMH in the presence or absence of ATP or ATPγS. After quenching the cross-linking reaction, protein samples were analyzed by SDS–PAGE followed by immunoblotting. When two cysteines were present at positions 142 and 1114 in the two-armed GGGG form, a specific band appeared just above the linear monomer (Figure 2E, lanes 10–12). A similar product was obtained with the single-armed DDDD form (Figure 2E, lanes 13–15), suggesting that this band corresponds to a circular monomer. In contrast, when cysteines were present at positions 55 and 1070, a subset of high molecular weight bands appeared (Figure 2F, lanes 10–12). These bands are likely to be the products in which two different subunits are cross-linked together. Consistent with this interpretation, a similar set of products was observed in the single-armed form, but the cross-linking reaction was far less efficient (Figure 2F, lanes 13–15). The specificity of the cross-linking reactions was confirmed by using mutants with none or a single cysteine residue (Figure 2E and F, lanes 1–9). The presence or absence of the nucleotides had little, if any, effect on the cross-linking properties in any cases. Thus, the results from the two complementary sets of cross-linkable mutants are consistent with model II (Figure 2A), in which two self-folded monomers dimerize by a hinge-mediated interaction.
DNA-binding properties of BsSMC mutants
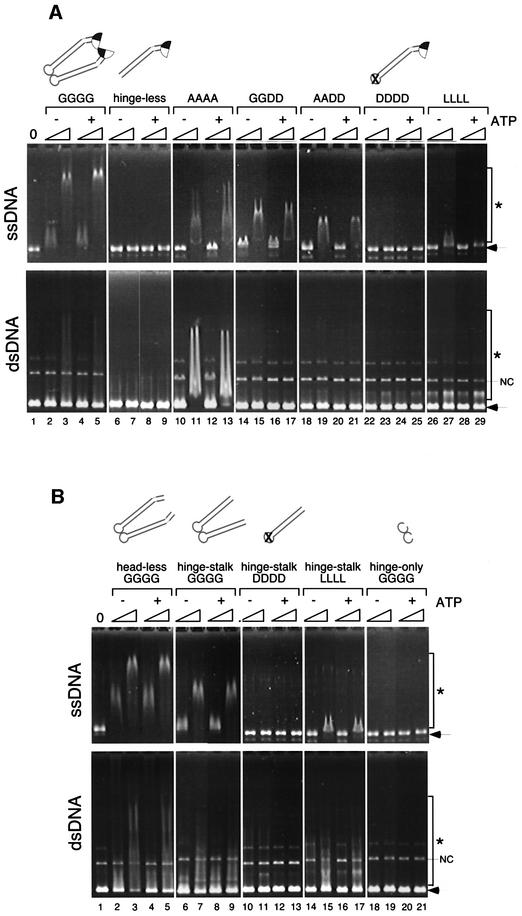
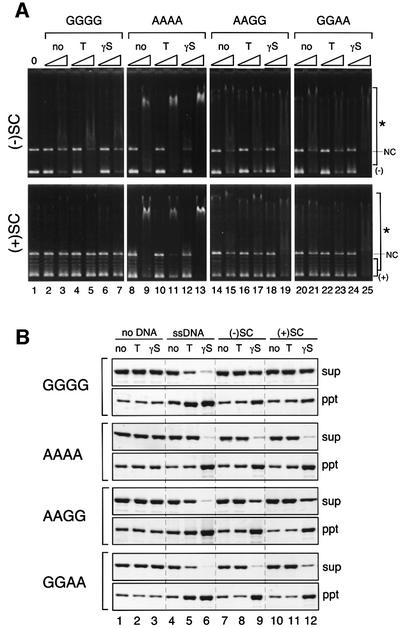
Having established the folding pattern of the BsSMC dimer, we used a gel shift assay to examine the role of the hinge domain in the DNA-binding activity of BsSMC (Figure 3A). The wild-type BsSMC protein (GGGG) displayed a strong affinity for ssDNA (Figure 3A, upper panel, lanes 2–5) and a weaker affinity for negatively supercoiled double-stranded DNA (dsDNA; Figure 3A, lower panel, lanes 2–5). The binding to both substrates was independent of ATP, as described previously (Hirano and Hirano, 1998). The hinge-less mutant failed to shift both forms of DNA under the same condition (Figure 3A, lanes 6–9). Interestingly, we found that point mutations in the hinge domain greatly affected the ability of BsSMC to bind to ssDNA. The affinity of the hinge mutants for ssDNA was reduced progressively in the order AAAA, GGDD, AADD and DDDD (Figure 3A, upper panel, lanes 10–25). Binding to dsDNA was even better in AAAA than in GGGG, but was extremely low or negligible in the other mutants (Figure 3A, lower panel, lanes 10–25). One possible interpretation of these data is that a specific DNA-binding site resides in the hinge domain, and the introduction of acidic residues into this domain disturbs its DNA-binding activity due to electrostatic hindrance. This apparently is not the case, however, because the LLLL mutant displayed a very weak binding activity comparable with that of DDDD (Figure 3A, lanes 26–29).
Fig. 3. DNA-binding activities of BsSMC mutants. (A) A fixed amount of ssDNA (top; 15.6 µM nucleotides) or negatively supercoiled dsDNA (bottom; 15.6 µM nucleotides) was incubated with two different concentrations of proteins (210 and 420 nM arms) in a buffer containing 5 mM KCl in the presence or absence of 1 mM ATP. No protein was added in lane 1. The reaction mixtures were fractionated on a 0.7% agarose gel and visualized by ethidium bromide stain. Protein–DNA complexes are indicated by asterisks, and free DNAs are indicated by arrows. NC indicates a nicked-circular population present in the dsDNA substrate. (B) The same assay was performed as above using a series of deletion mutants.
To determine how the dimeric form of BsSMC interacts with DNA, we tested the DNA-binding properties of the deletion mutants described in Figure 1. Unexpectedly, we found that the head-less and hinge–stalk mutants have DNA-binding activities comparable with that of wild-type BsSMC (Figure 3B, lanes 2–9). When dimerization of the hinge–stalk mutant was perturbed by combining with the DDDD mutations, however, the DNA-binding activity was completely abolished (Figure 3B, lanes 10–13). A combination with LLLL also substantially reduced the binding activity (Figure 3B, lanes 14–17). Finally, the dimeric hinge-only mutant failed to bind to DNA (Figure 3B, lanes 18–21). These data show that the catalytic domains may not play a major role in DNA binding of BsSMC. Instead, hinge-mediated dimerization of coiled-coil stalks appears to be essential. We suggest that, even though all of the hinge mutants are single-armed monomers in solution (Figure 1C), the hinge of AAAA (and to a lesser extent that of GGDD and AADD) is partially active and is able to support dimerization only when DNA is present.
ATPase activities of hinge mutants
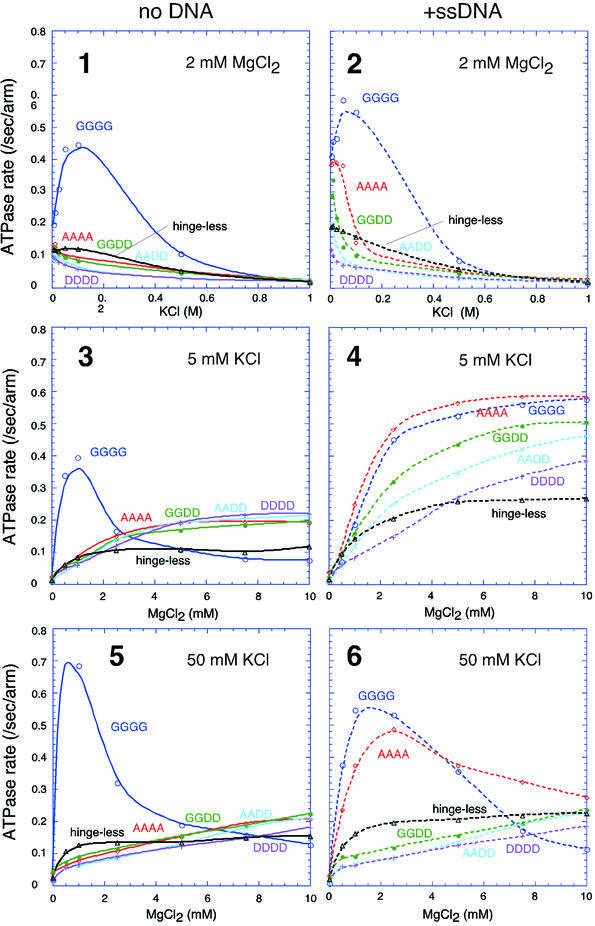
The ATPase activity of BsSMC has a very unique and complex character. It has DNA-independent and DNA-stimulated modes that respond differently to KCl or MgCl2 titration (Figure 4, panels 1–6, GGGG; Hirano and Hirano, 1998). Although the AAAA mutant largely loses its DNA-independent ATPase activity (Figure 4, panels 1, 3 and 5, AAAA), it retains a normal level of ssDNA-stimulated ATPase under a low salt condition (Figure 4, panel 4, AAAA; Hirano et al., 2001). This is consistent with the finding that AAAA can bind to ssDNA. Thus, stimulation of ATP hydrolysis by ssDNA offers a highly sensitive assay for BsSMC–DNA interactions. To understand better the role of the hinge domain in the action of BsSMC, we performed KCl and MgCl2 titration experiments with the new series of hinge mutants. In the absence of DNA, the titration curves of the hinge mutants were almost indistinguishable from one another, although they were strikingly different from those of wild-type BsSMC (Figure 4, panels 1, 3 and 5). In the presence of ssDNA, however, the different hinge mutants displayed different titration curves. In a KCl titration experiment, the rate of ATP hydrolysis decreased in the order AAAA, GGDD, AADD, DDDD (Figure 4, panel 2). This is in excellent agreement with their decreasing affinities for ssDNA as judged by the gel shift assay (Figure 3A). Similarly, MgCl2 titration curves of the hinge mutants at 5 mM KCl corresponded well to their DNA-binding activities (Figure 4, panel 4). At 50 mM KCl, virtually no ssDNA-dependent stimulation was observed in GGDD, AADD and DDDD, although AAAA retained a substantial activity that was comparable with that of GGGG (Figure 4, panel 6). The behavior of the hinge-less mutant was similar, if not identical, to that of DDDD (Figure 4, panels 1–6, hinge-less). These results provide additional evidence that the hinge of the AAAA mutant is partially active in the presence of DNA. In contrast, replacement of the conserved glycine residues by aspartic acids disrupts the dimerization interface more completely, thereby impairing the DNA-binding and DNA-stimulated ATPase activities of BsSMC.
Fig. 4. ATPase activities of wild-type protein (GGGG), hinge mutants (AAAA, GGDD, AADD and DDDD) and the hinge-less mutant under different conditions. KCl titration at 2 mM MgCl2 (panels 1 and 2), MgCl2 titration at 5 mM KCl (panels 3 and 4) and MgCl2 titration at 50 mM KCl (panels 5 and 6) in the absence (panels 1, 3 and 5) or presence (panels 2, 4 and 6) of ssDNA (φX174 virion DNA; 31.2 µM nucleotides) are shown. A fixed protein concentration of 300 nM arms (equivalent to 150 nM dimers in the case of wild-type BsSMC) was used for all the proteins. The rate of ATP hydrolysis is expressed as the number of ATP molecules hydrolyzed per second per arm.
Hinge mutations alter the mode of DNA recognition by BsSMC
The finding that AAAA has an altered, but not completely defective, hinge function prompted us to test whether such mutations might affect the mode of DNA recognition by BsSMC. A gel shift assay using different forms of dsDNA revealed that the wild-type BsSMC protein binds to negatively supercoiled DNA but not to positively supercoiled DNA (Figure 5A, lanes 2–7). The addition of ATP or ATPγS (a slowly hydrolyzable ATP analog) had little effect on the dsDNA-binding properties of wild-type BsSMC. In contrast, the hinge mutant AAAA bound equally well to negative and positive supercoils, and its binding to both substrates was slightly enhanced in the presence of ATPγS (Figure 5A, lanes 8–13). Interestingly, two intermediate mutants, AAGG and GGAA (Hirano et al., 2001), displayed an affinity for positive supercoils only in the presence of ATPγS (Figure 5A, lanes 14–25).
Fig. 5. Altered DNA-binding properties of hinge mutants. (A) Gel shift assay. A fixed amount of negatively supercoiled DNA (top; 15.6 µM nucleotides) or positively supercoiled DNA (bottom; 15.6 µM nucleotides) was incubated with two different concentrations of proteins (210 and 420 nM arms) in a buffer containing 5 mM KCl in the presence or absence of the indicated nucleotides (no, no nucleotide; T, 1 mM ATP; γS, 1 mM ATPγS). The reaction mixtures were fractionated on a 0.7% agarose gel and visualized by ethidium bromide stain. Protein–DNA complexes are indicated by asterisks. The positions of nicked circular DNA (NC), negative (–) and positive (+) supercoiled DNA are also shown. (B) Spin-down assay. A fixed amount of BsSMC proteins was mixed with either no DNA, ssDNA, negatively supercoiled DNA [(–)SC] or positively supercoiled DNA [(+)SC], in the presence or absence of the indicated nucleotides (no, no nucleotide; T, 1 mM ATP; γS, 1 mM ATPγS). After incubation, the mixtures were spun at 16 000 g for 15 min. The supernatants (sup) and pellets (ppt) were separated, fractionated by 7.5% SDS–PAGE and stained with silver.
To test further the differential responses to nucleotides between wild-type BsSMC and the hinge mutants, we performed a spin-down assay. In this assay, BsSMC was first incubated with different forms of DNA in the presence or absence of nucleotides, and then the mixture was spun by low-speed centrifugation (Hirano and Hirano, 1998). When incubated with ssDNA, wild-type BsSMC formed precipitable complexes in the presence of ATP or ATPγS (Figure 5B, GGGG, lanes 4–6). AAAA supported a similar reaction in the presence of ATPγS (Figure 5B, AAAA, lanes 4–6). Remarkable differences were found between the two proteins when dsDNAs were used as binding substrates. Although wild-type BsSMC formed very few precipitable complexes with dsDNA (Figure 5B, GGGG, lanes 7–12), AAAA readily assembled aggregates with both negative and positive supercoils in the presence of ATPγS (Figure 5B, AAAA, lanes 9 and 12). The AAGG and GGAA mutants exhibited intermediate phenotypes (Figure 5B, AAGG and GGAA), as expected. These results show that the hinge mutations synergistically alter the DNA-binding property of BsSMC. We suggest that two factors, an open hinge structure and binding of ATPγS to the catalytic domains, stabilize a protein–protein interaction that promotes cross-linking of dsDNA (see Discussion).
Folding of eukaryotic SMC2–SMC4 heterodimers
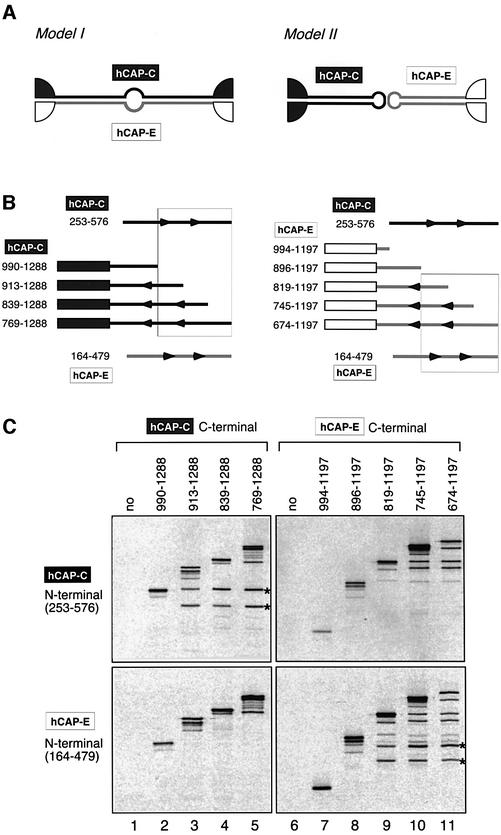
In eukaryotic organisms, CAP-C/SMC4 and CAP-E/SMC2 form an SMC heterodimer that acts as the core of the condensin complex (Hirano et al., 1997; Schmiesing et al., 2000). We wished to test whether the conclusion regarding the architecture of BsSMC is also applicable to that of the eukaryotic dimer. Taking advantage of the heterodimeric nature of the eukaryotic SMC proteins, we set up a coiled-coil interaction assay. If dimerization is mediated by coiled-coil interactions between the two different subunits, the N-terminal half of the CAP-C would associate with the C-terminal half of CAP-E to form an intermolecular antiparallel coiled-coil (Figure 6A, model I). Likewise, the C-terminal half of CAP-C would interact with the N-terminal half of CAP-E. Alternatively, both CAP-C and CAP-E may self-fold by an intramolecular coiled-coil interaction (Figure 6A, model II). In this case, the N-terminal half of CAP-C (or CAP-E) would associate with its own C-terminal half. To distinguish between the two models, C-terminal fragments of human CAP-C (hCAP-C) of different lengths were co-translated in vitro with an N-terminal fragment of either hCAP-C or hCAP-E (Figure 6B, left). A specific antibody was then used to immunoprecipitate the C-terminal fragments of hCAP-C from the reaction mixtures. We found that the N-terminal fragment of hCAP-C, but not of hCAP-E, co-precipitated with a subset of the C-terminal fragments of hCAP-C (Figure 6C, left panels). We also set up a reciprocal experiment (Figure 6B, right) and obtained consistent results: a subset of the C-terminal fragments of hCAP-E associated with the N-terminal fragment of hCAP-E, but not with that of hCAP-C (Figure 6C, right panels). These results show that, like BsSMC, the eukaryotic SMC2–SMC4 heterodimer is composed of two antiparallel coiled-coils that are folded intramolecularly. No hinge sequences were included in the current assays, suggesting that the coiled-coil sequences contain information that determines their binding specificity.
Fig. 6. The folding of the SMC2–SMC4 heterodimer as revealed by a coiled-coil interaction assay. (A) Two models for the folding of hCAP-C/SMC4 and hCAP-E/SMC2. Dimerization may be mediated by coiled-coil interactions between hCAP-C and hCAP-E (model I). Alternatively, the hCAP-C and hCAP-E subunits may be self-folded to form two separate coiled-coil arms, which in turn dimerize by a hinge-mediated interaction (model II). (B) Left: a deletion series of fragments containing the C-terminal domain of hCAP-C was co-translated in vitro with an N-terminal coiled-coil fragment of hCAP-C (253–576) or an N-terminal coiled-coil fragment of hCAP-E (164–479). Right: alternatively, a deletion series of fragments containing the C-terminal domain of hCAP-E was co-translated in vitro with one of the two N-terminal coiled-coil fragments. (C) The translation reactions were immunoprecipitated with antibodies that specifically recognize the C-terminal domain of hCAP-C (lanes 1–5) or hCAP-E (lanes 6–11). After washing, the immunoprecipitates were fractionated by SDS–PAGE and analyzed by autoradiography. Co-precipitated N-terminal fragments are indicated by the asterisks. In vitro translation of the N-terminal domain of hCAP-C or hCAP-E produced two bands, the smaller one of which is likely to be a translation product starting from an internal methionine. The combinations of fragments that interact with each other are boxed in the diagrams shown in (B).
Discussion
Architecture of SMC and related proteins
The current study is aimed at understanding the structure– function relationship of SMC proteins. First of all, we wanted to determine unambiguously how SMC subunits fold to make a dimer. On the basis of their primary sequence, Saitoh et al. (1994) predicted that SMC proteins have the potential to make antiparallel coiled-coils by either intermolecular or intramolecular interactions. A subsequent EM study by Melby et al. (1998) demonstrated that BsSMC has a two-armed structure in which two antiparallel coiled-coils are connected by a flexible hinge. Although the original images were interpreted so that dimerization is mediated by an intermolecular coiled-coiled interaction, our recent mutational analysis of the hinge domain argued in favor of an alternative model in which self-folded subunits dimerize by a hinge–hinge interaction (Hirano et al., 2001). Site-directed, protein– protein cross-linking experiments presented in the current study provide compelling evidence that the second model is correct. We have also extended this conclusion to the eukaryotic heterodimer of CAP-C/SMC4 and CAP-E/SMC2 using a coiled-coil interaction assay. Our results are in good agreement with those of Haering et al. (2002), who have very recently reported the architecture of the yeast cohesin complex (containing SMC1 and SMC3) based on a pull-down assay using epitope-tagged recombinant fragments. Thus, hinge-mediated dimerization of self-folded coiled-coils is applicable to all SMC proteins, from bacterial SMC homodimers to eukaryotic SMC1–SMC3 and SMC2–SMC4 heterodimers.
Rad50 is an SMC-like protein that is implicated in double-strand DNA break repair (reviewed by Connelly and Leach, 2002; D’Amours and Jackson, 2002). A study using scanning force EM reported that, like SMC, the Rad50 polypeptide folds intramolecularly to form an antiparallel coiled-coil (de Jager et al., 2001). Very recently, the hinge domain of Rad50 has been shown to form a zinc-binding hook that can flexibly support either dimerization or tetramerization of the Mre11–Rad50 complex and thereby bridge two broken DNA ends (Hopfner et al., 2002). The hinge domain of SMC has no zinc-binding motif, and is much bigger (∼150 amino acids) than that of Rad50 (∼30 amino acids). Thus, the hinge domains of SMC and Rad50 appear to be different both structurally and functionally.
The two-armed structure of SMC is essential for its DNA binding
A previous study using small fragments of yeast SMC proteins fused with GST reported that the major DNA-binding site of SMC proteins may reside in the C-terminal globular domain (Akhmedov et al., 1998). A subsequent and similar approach showed that some internal coiled-coil fragments could also bind to DNA (Akhmedov et al., 1999). Given our current knowledge of the SMC architecture, however, it seems that the constructs used in those early studies were too artificial and inappropriate to test the DNA-binding properties of SMC proteins. The current study therefore represents the first systematic analysis of DNA-binding properties of SMC proteins using physiologically relevant constructs. Our results show that neither the catalytic domain nor the hinge domain alone is sufficient to bind to DNA, as judged by gel shift assays. Instead, two coiled-coil stalks constitute the major DNA-binding surface only when they are linked together by a flexible hinge. Deletion of the hinge domain, which creates single-armed monomers, results in complete loss of the ability of BsSMC to interact with DNA. Another remarkable observation is that point mutations in the hinge domain differentially and synergistically affect the DNA-binding properties of BsSMC. The crystal structure of an SMC hinge domain from the bacterium T.maritima (Haering et al., 2002) has demonstrated recently that the conserved glycine residues mutated in our previous and current studies are clustered at the dimerization interface. Replacement of these glycines by alanines perturbs the interface, thereby altering the global conformation of BsSMC and its ability to interact with DNA. Replacement by aspartic acids has a more drastic effect as it results in the formation of single-armed monomers whose functional properties are comparable with those of the hinge-less mutant. Taking all these results together, we suggest that there may be no local high affinity site for DNA in BsSMC molecules. It is most likely that the two arms connected by a flexible hinge act like a ‘hook’ to trap DNA strands. The catalytic domains at the ends of a dimer would certainly play an important regulatory role in the subsequent manipulation of DNA, but may not be essential for the direct interaction with DNA. This DNA-binding property of SMC is clearly different from that of Rad50, which requires ATP-dependent dimerization of the two catalytic domains (Hopfner et al. 2000).
Does the hinge angle modulate the DNA-binding mode of SMC proteins?
Our results suggest that the hinge of BsSMC is not a simple dimerization site but has an important regulatory function in its DNA-binding activity. Comparison between the wild-type protein (GGGG) and the partially active hinge mutants (GGAA, AAGG and AAAA) is particularly informative. Previous hydrodynamic analysis showed that these mutations synergistically decrease the sedimentation coefficient of BsSMC, being indicative of arm opening (Hirano et al., 2001). Having established the folding problem in the current study, we can now speculate that these mutations make the hinge angle wider, promoting increasingly open conformations (and eventually loosening the hinge-mediated dimerization). Conceivably, a closed conformation of the wild-type hinge or its structural flexibility contributes to the specific recognition of negative supercoils of DNA, although the underlying mechanism remains elusive. Such flexibility would allow BsSMC to take a variety of conformations and support dynamic and plastic interactions with DNA (Figure 7, left). On the other hand, the constitutively open conformation of AAAA leads to deregulated DNA binding, and places the two catalytic domains far apart from each other. This fixed conformation would then allow the catalytic domains of one dimer to interact stably with those of another dimer in the presence of ATPγS, resulting in robust cross-linking of dsDNA (Figure 7, right). Thus, we propose that the hinge angle per se (or its flexibility) may have the potential to modulate the DNA-binding mode of BsSMC.
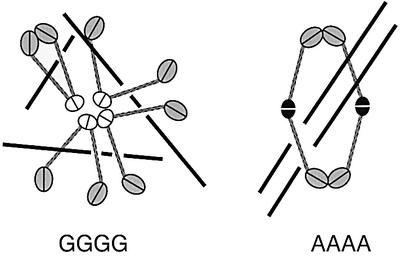
Fig. 7. A hypothetical model for BsSMC–DNA interactions. Wild-type BsSMC (GGGG), which has a flexible hinge (shown by the white ovals), displays a variety of conformations and supports dynamic interactions with DNA. The AAAA mutant, which has a fixed and open hinge (shown by the black ovals), supports a robust dimer–dimer interaction when the catalytic domains of two different dimers associate with each other in the presence of ATPγS.
Does this idea have any implications for the action of the eukaryotic SMC proteins? A recent EM study has revealed that condensin and cohesin display different arm conformations with characteristic hinge angles (Anderson et al., 2002). The hinge of condensin is largely closed whereas the hinge of cohesin is wide open with an average angle of ∼90°. The current mutational analysis of BsSMC supports the hypothesis that the different hinge angles of condensin and cohesin may be one of the key factors that determines their drastically different DNA-binding properties (Losada and Hirano, 2001). If this is indeed the case, the AAAA mutant would be expected to display ‘cohesin-like’ rather than ‘condensin-like’ functions. The ATPγS-dependent dsDNA cross-linking activity supported by AAAA could reflect one such activity. Clearly, future work is required to understand exactly how two-armed structures with specific hinge angles might be able to recognize different forms of DNA.
Toward a unified understanding of SMC action
Although the current study provides important insights into the role of the hinge domain in SMC functions, it remains to be established how this class of two-armed ATPases works at a mechanistic level. A variety of different approaches are now beginning to catch a glimpse of their modes of action. By electron spectroscopic imaging, Bazett-Jones et al. (2002) have demonstrated that a single condensin complex is able to trap two positive supercoils of DNA in an ATP hydrolysis-dependent manner. Yoshimura et al. (2002) have used atomic force microscopy to show that condensin has an ability to form DNA loops. On the other hand, Haering et al. (2002) have proposed, on the basis of biochemical data, that cohesin might hold two sister chromatids together by ‘embracing’ duplicated DNA strands within its arms. Whatever the mode of SMC–DNA interactions might be, it is most likely that the two catalytic ATPase domains modulate the opening and closing of the two coiled-coil arms (Hirano et al., 2001). In the case of condensin, there is evidence that the non-SMC subunits that bind to the catalytic domains directly modulate the ATP binding and hydrolysis cycle of the SMC heterodimer (Kimura and Hirano, 2000; Anderson et al., 2002; Yoshimura et al., 2002). Until very recently, BsSMC has been thought to function as a simple homodimer in B.subtilis. A recent genetic study, however, suggests that two non-SMC components (termed ScpA and ScpB) may work together with the SMC protein in vivo (Mascarenhas et al., 2002). It is of great interest to determine whether these proteins interact with the BsSMC dimer in vitro and modulate its ATPase and ATP-dependent activities. Such efforts will further clarify the relationship between the eukaryotic and prokaryotic SMC protein machines, and will enhance our understanding of the action of this class of chromosomal ATPases that undoubtedly holds the secrets of higher order chromosome dynamics.
Materials and methods
Construction of BsSMC mutants
A plasmid (pSO133) that expresses wild-type BsSMC with a His6 tag at its C-terminal end was described previously (Hirano et al., 2001). Point mutations were introduced into this construct by a using QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing.
Deletion mutants were constructed as follows. For the head-less mutant, a DNA sequence encoding amino acids 160–1037 was amplified by PCR and inserted into the BamHI site of pET23b(+) (Novagen). For the hinge–stalk and hinge-only mutants, DNA sequences encoding amino acids 262–861 and 476–672, respectively, were amplified and inserted into the NdeI–BamHI site of the same vector. All deletion constructs have a His6 tag at their C-termini.
Purification of BsSMC mutants
BsSMC mutant proteins were expressed in the Escherichia coli strain BL21(DE3)pLysS and purified as described previously (Hirano et al., 2001), with the following modifications. Typically, 10 ml of lysate were obtained from a 250 ml culture. After a spin, the supernatant was loaded onto a 2 ml Ni-NTA metal affinity column (Qiagen; column size, 0.8 × 4 cm). The column was washed, and bound proteins were eluted with 8 ml of buffer containing 50 mM Na-phosphate pH 7.5, 300 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol and 500 mM imidazole. The peak fractions were pooled (∼4 ml) and dialyzed against buffer M [20 mM K-HEPES pH 7.7, 1 mM EDTA, 10% glycerol and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] containing 50 mM KCl and 5 mM 2-mercaptoethanol. The dialysate was applied to a 1 ml HiTrapQ column (Amersham Pharmacia Biotech), and fractionated with a 5 ml KCl gradient (100–600 mM) in buffer M containing 5 mM 2-mercaptoethanol. The peak fractions were pooled (∼0.5 ml), dialyzed against buffer M containing 50 mM KCl and 1 mM 2-mercaptoethanol, aliquoted and stored at –70°C. The typical yield of BsSMC from a 250 ml culture was ∼0.4 mg. Purification of the hinge-only mutant was performed in the same way except that HiTrapQ was replaced with a 1.5 ml SP-Sepharose FF column (Amersham Pharmacia Biotech; 1 × 1.9 cm). The concentrations of purified BsSMC were determined by SDS–PAGE followed by Coomassie Blue stain using bovine serum albumin (BSA) as a standard. To compare the activities of the two-armed and single-armed proteins directly, all protein concentrations are expressed in moles of BsSMC arms rather than moles of dimers or monomers.
Site-directed, protein–protein cross-linking
Protein–protein cross-linking was performed as described previously (Hirano et al., 2001) except that BMH (Pierce) was used at a final concentration of 0.08 mM.
In vitro transcription/translation of hCAP-C and hCAP-E fragments
DNA fragments encoding different regions of hCAP-C or hCAP-E were amplified by PCR and inserted into pRSETA (Invitrogen). In vitro transcription/translation reactions were performed using TNT Quick Coupled Transcription/Translation Systems (Promega) according to the manufacturer’s instructions. On the assumption that a coiled-coil fragment can fold properly only when a folding partner is present, we expressed two coiled-coil fragments simultaneously by adding two different plasmid constructs into a single reaction tube. Reaction mixtures (50 µl) contained two plasmid DNAs (10–20 µg/ml each) and [35S]methionine (1000 Ci/mmol) at a final concentration of 0.4 mCi/ml. After incubation at 30°C for 60 min, 250 µl of dilution buffer (20 mM HEPES pH 7.7, 100 mM KCl, 2.5 mM MgCl2, 0.1% Tween-20 and 0.5 mg/ml BSA) was added and the mixtures were spun at 10∼000 g at 4°C for 15 min. The supernatants were used for immunoprecipitation with anti-hCAP-C or anti-hCAP-E. Affinity-purified rabbit antibodies that recognize the C-terminal peptide sequences of hCAP-C and hCAP-E were described previously (Kimura et al., 2001). The precipitated polypeptides were resolved by SDS–PAGE and analyzed by autoradiography.
Other assays
Gel shift and ATPase assays were performed as described previously (Hirano and Hirano, 1998; Hirano et al., 2001). Positively supercoiled DNA was prepared by using a recombinant form of the archaeal histone HMfB as described previously (Starich et al., 1996; LaMarr et al., 1997). Sucrose gradient centrifugation was carried out as described previously (Hirano et al., 2001) with minor modifications. A 5 ml sucrose gradient (5–20%) was made in buffer containing 20 mM Tris–HCl pH 7.5, 5 mM KCl and 2 mM MgCl2, and spun at 189 000 g for 18 h in an SW50.1 rotor (Beckman).
Acknowledgments
Acknowledgements
We are grateful to Kyoko Yokomori for hCAP-C and hCAP-E cDNAs, and Kathleen Sandman for an rHMfB expression plasmid. We thank members of the Hirano lab for critically reading the manuscript. This work was supported by grants from the National Institutes of Health (to T.H.).
References
- Akhmedov A.T., Frei,C., Tsai-Pflugfelder,M., Kemper,B., Gasser,S.M. and Jessberger,R. (1998) Structural maintenance of chromosomes protein C-terminal domains bind preferentially to DNA with secondary structure. J. Biol. Chem., 273, 24088–24094. [DOI] [PubMed] [Google Scholar]
- Akhmedov A.T., Gross,B. and Jessberger,R. (1999) Mammalian SMC3 C-terminal and coiled-coil protein domains specifically bind palindromic DNA, do not block DNA ends and prevent DNA bending. J. Biol. Chem., 274, 38216–38224. [DOI] [PubMed] [Google Scholar]
- Anderson D.E., Losada,A., Erickson,H.P. and Hirano,T. (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol., 156, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Kimura,K. and Hirano,T. (2002) Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell, 9, 1183–1190. [DOI] [PubMed] [Google Scholar]
- Britton R.A., Lin,D.C.-H. and Grossman,A.D. (1998) Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev., 12, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbe N. and Heck,M.M. (2000) SMCs in the world of chromosome biology: from prokaryotes to higher eukaryotes. J. Struct. Biol., 129, 123–143. [DOI] [PubMed] [Google Scholar]
- Connelly J.C. and Leach,D.R. (2002) Tethering on the brink: the evolutionarily conserved Mre11/Rad50 (MR) complex. Trends Biochem. Sci., 27, 410–418. [DOI] [PubMed] [Google Scholar]
- D’Amours D. and Jackson,S.P. (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol., 3, 317–327. [DOI] [PubMed] [Google Scholar]
- de Jager M., van Noort,J., van Gent,D.C., Dekker,C., Kanaar,R. and Wyman,C. (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell, 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Fousteri M.I. and Lehmann,A.R. (2000) A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J., 19, 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide,L. and Strunnikov,A.V. (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol., 149, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L., Losick,R. and Strunnikov,A.V. (1998) Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol., 180, 5749–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H., Lowe,J., Hochwagen,A. and Nasmyth,K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell, 9, 773–788. [DOI] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes,V.F., Cozzarelli,N.R. and Meyer,B.J. (2002) C.elegans condensin promotes mitotic chromosome architecture, centromere organization and sister chromatid segregation during mitosis and meiosis. Genes Dev., 16, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M. and Hirano,T. (1998) ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J., 17, 7139–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Anderson,D.E., Erickson,H.P. and Hirano,T. (2001) Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J., 20, 3238–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. (2002) The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion and repair. Genes Dev., 16, 399–414. [DOI] [PubMed] [Google Scholar]
- Hirano T., Kobayashi,R. and Hirano,M. (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell, 89, 511–521. [DOI] [PubMed] [Google Scholar]
- Hopfner K.-P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Hopfner K.-P. et al. (2002) The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature, 418, 562–566. [DOI] [PubMed] [Google Scholar]
- Kimura K. and Hirano,T. (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell, 90, 625–634. [DOI] [PubMed] [Google Scholar]
- Kimura K. and Hirano,T. (2000) Dual roles of the 11S regulatory subcomplex in condensin functions. Proc. Natl Acad. Sci. USA, 97, 11972–11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano,M., Kobayashi,R. and Hirano,T. (1998) Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science, 282, 487–490. [DOI] [PubMed] [Google Scholar]
- Kimura K., Rybenkov,V.V., Crisona,N.J., Hirano,T. and Cozzarelli,N.R. (1999) 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell, 98, 239–248. [DOI] [PubMed] [Google Scholar]
- Kimura K., Cuvier,O. and Hirano,T. (2001) Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem., 276, 5417–5420. [DOI] [PubMed] [Google Scholar]
- Koshland D. and Strunnikov,A. (1996) Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol., 12, 305–333. [DOI] [PubMed] [Google Scholar]
- LaMarr W.A., Sandman,K.M., Reeve,J.N. and Dedon,P.C. (1997) Large scale preparation of positively supercoiled DNA using the archaeal histone HMf. Nucleic Acids Res., 25, 1660–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. and Hirano,T. (2001) Intermolecular DNA interactions stimulated by the cohesin complex in vitro: implications for sister chromatid cohesion. Curr. Biol., 11, 268–272. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano,M. and Hirano,T. (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev., 12, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Cordell,S.C. and van den Ent,F. (2001) Crystal structure of the SMC head domain: an ABC ATPase with 900 residues antiparallel coiled coil inserted. J. Mol. Biol., 306, 25–35. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J., Soppa,J., Strunnikov,A.V. and Graumann,P.L. (2002) Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J., 21, 3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T.E.G., Ciampaglio,C.N., Briscoe,G. and Erickson,H.P. (1998) The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol., 142, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S., Tsujikawa,E., Hassan,A.K., Asai,K., Kodama,T. and Ogasawara,N. (1998) A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition and condensation. Mol. Microbiol., 29, 179–187. [DOI] [PubMed] [Google Scholar]
- Saitoh N., Goldberg,I., Wood,E.R. and Earnshaw,W.C. (1994) ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J. Cell Biol., 127, 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing J.A., Gregson,H.C., Zhou,S. and Yokomori,K. (2000) A human condensin complex containing hCAP-C–hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol. Cell. Biol., 20, 6996–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich M.R., Sandman,K., Reeve,J.N. and Summers,M.F. (1996) NMR structure of HMfB from the hyperthermophile Methanothermus fervidus confirms that this archaeal protein is a histone. J. Mol. Biol., 255, 187–203. [DOI] [PubMed] [Google Scholar]
- Sutani T. and Yanagida,M. (1997) DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature, 388, 798–801. [DOI] [PubMed] [Google Scholar]
- Sutani T., Yuasa,T., Tomonaga,T., Dohmae,N., Takio,K. and Yanagida,M. (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes Dev., 13, 2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.M., Moghraby,J.S., Lees,J.H., Smit,B., Moens,P.B. and Lehmann,A.R. (2001) Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol. Biol. Cell, 12, 1583–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T. et al. (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev., 14, 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Ciosk,R., Uhlmann,F., Galova,M., Schleiffer,A. and Nasmyth,K. (1999) Yeast cohesion complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev., 13, 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S.H., Hizume,K., Murakami,A., Sutani,T., Takeyasu,K. and Yanagida,M. (2002) Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr. Biol., 12, 508–513. [DOI] [PubMed] [Google Scholar]