Abstract
RNA silencing phenomena, known as post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference (RNAi) in animals, are mediated by double-stranded RNA (dsRNA) and mechanistically intersect at the ribonuclease Dicer. Here, we report cloning and expression of the 218 kDa human Dicer, and characterization of its ribonuclease activity and dsRNA-binding properties. The recombinant enzyme generated ∼21–23 nucleotide products from dsRNA. Processing of the microRNA let-7 precursor by Dicer produced an apparently mature let-7 RNA. Mg2+ was required for dsRNase activity, but not for dsRNA binding, thereby uncoupling these reaction steps. ATP was dispensable for dsRNase activity in vitro. The Dicer·dsRNA complex formed at high KCl concentrations was catalytically inactive, suggesting that ionic interactions are involved in dsRNA cleavage. The putative dsRNA-binding domain located at the C-terminus of Dicer was demonstrated to bind dsRNA in vitro. Human Dicer expressed in mammalian cells colocalized with calreticulin, a resident protein of the endoplasmic reticulum. Availability of the recombinant Dicer protein will help improve our understanding of RNA silencing and other Dicer-related processes.
Keywords: Dicer/dsRNA/ribonuclease III/RNA binding/RNA processing
Introduction
The emerging concept of the RNA silencing phenomena known as post-transcriptional gene silencing in plants (Baulcombe, 1996), quelling in fungi (Romano and Macino, 1992) and RNA interference (RNAi) in animals (Fire et al., 1998) indicates that they share a common mechanism (Hammond et al., 2001; Sharp, 2001; Tuschl, 2001; Hutvágner and Zamore, 2002). Double-stranded RNA (dsRNA) molecules, either produced endogenously or introduced exogenously, induce specific degradation of homologous messenger RNA (mRNA), thereby inhibiting the expression of the encoded proteins. The RNAi cascade is initiated by the processing of long dsRNA inducers into short ∼21–25 bp oligonucleotides, which have been named small interfering RNAs (siRNAs) (Zamore et al., 2000; Elbashir et al., 2001b). The siRNAs appear to serve as guide sequences that instruct the multicomponent nuclease RNA-induced silencing complex (RISC) to destroy specific mRNAs (Hammond et al., 2000). siRNA duplexes have been shown to specifically suppress gene expression in transfected mammalian cells (Elbashir et al., 2001a).
The structural features of the siRNAs—5′ phosphate and 3′ hydroxyl termini with two nucleotide (nt) 3′ overhangs (Elbashir et al., 2001b)—suggested that they were produced by a member of the RNase III family of double-strand-specific ribonucleases. RNase III was first characterized in Escherichia coli as an activity that degraded dsRNA into short oligonucleotide products (Robertson et al., 1968). The enzyme was subsequently found to participate in the processing of ribosomal RNA precursors and in the turnover of mRNA (Court, 1993). Eukaryotic RNase III homologs have been identified in fission (Xu et al., 1990; Iino et al., 1991) and budding (Abou Elela et al., 1996) yeast. These enzymes are very similar to their bacterial counterpart with respect to essential structural elements (Rotondo et al., 1995), mode of action (Rotondo and Frendewey, 1996) and role in ribosomal RNA synthesis (Abou Elela et al., 1996; Rotondo et al., 1997).
Genomic sequencing and similarity searches revealed open reading frames (ORFs) in nematodes (Wilson et al., 1994; Rotondo et al., 1995) and fission yeast (Rotondo and Frendewey, 1996) that predicted RNase III-like enzymes with non-canonical structures. These putative proteins are unusually large (>1000 amino acids) members of the RNase III family. They possess a C-terminal dsRNA-binding motif immediately adjacent to two tandem RNase III catalytic signatures (Rotondo and Frendewey, 1996). The unexpected feature of these proteins is a long N-terminal extension whose sequence predicts an RNA helicase activity. Similar protein sequences have been found in flowering plants (Jacobsen et al., 1999), insects (Bernstein et al., 2001) and mammals (Provost et al., 1999; Matsuda et al., 2000; Nicholson and Nicholson, 2002).
Based on the predicted properties of this new class of hybrid RNA helicase–RNase III, Bass (2000) proposed that this type of enzyme was likely to be the agent that catalyzed the triggering events in RNAi. This hypothesis was strongly supported by experimental evidence that implicated a similar enzyme, named Dicer, in the initiation step of RNAi in extracts from fruit fly cells (Bernstein et al., 2001). These results were confirmed by genetic and biochemical experiments in nematodes and human cells that established the requirement for Dicer in both RNAi and in the synthesis of the let-7 small temporal RNA (Grishok et al., 2001; Hutvágner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001).
In this study, we report the cloning, expression and characterization of human Dicer. Our results show that polyhistidine-tagged Dicer is a double-strand-specific RNase that converts dsRNA substrates into ∼21–23 nt fragments. The enzyme also processes let-7 precursor RNA to produce a product whose size and sequence are consistent with that expected for mature let-7 RNA. We describe the differential requirements of the Dicer protein for binding and processing dsRNA, and confirm the ability of its putative dsRNA-binding domain (dsRBD) to bind dsRNA.
Results
Human Dicer cDNA cloning
We reported previously the isolation of a partial human cDNA clone of 2063 bp (clone FL59) encoding a protein homologous to the hypothetical helicase K12H4.8 from Caenorhabditis elegans in a yeast two-hybrid screen for proteins that interact with 5-lipoxygenase (5LO) (Provost et al., 1999), which catalyzes the first two steps in leukotriene biosynthesis (Samuelsson, 1983). Clone FL59 was similar to the RNase III-like domain of K12H4.8, later identified as C.elegans Dicer, but lacked the putative N-terminal helicase domain. Extension of clone FL59 led to the isolation of clone H20277 (4140 bp) from a human fetal brain library, clone H21154 (1178 bp) from a human spleen library, and clone POH1 (6200 bp) from a human testis library.
Clone POH1 appeared to encode the full-length human Dicer by virtue of stop codons in all three frames upstream of the potential translation initiation site. The apparent 5′ untranslated region is composed of 271 bp, while the 190 bp 3′ untranslated region contains a potential polyadenylation signal (AATAAA) 15 bp upstream of the poly(A) tail. By taking into account the potential starting ATG positioned in the most favorable translation initiation context (Kozak, 1987), which is identical to that reported for mouse Dicer (Nicholson and Nicholson, 2002), the ORF encodes a protein of 1912 amino acids with a calculated molecular weight of 217 628 Da and a theoretical pI of 5.45 (see Supplementary figure S1 available at The EMBO Journal Online). This predicted amino acid sequence showed several discrepancies with that of the helicase-MOI (DDBJ/EMBL/GenBank accession No. AB028449), mostly near the N-terminus (22 substitutions and two insertions). However, it is identical to that reported by Zhang et al. (2002), with only one nucleotide substitution.
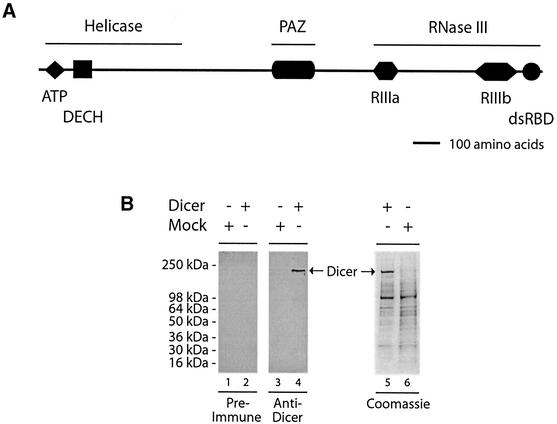
As illustrated in Figure 1A, analysis of the predicted amino acid sequence and database searches identified three major domains: an N-terminal helicase domain, a Piwi/Argonaute/Zwille (PAZ) domain, and a C-terminal RNase III domain, comprised of a dsRBD and tandem ribonuclease III motifs; 1303ERLEMLGDS1311 (RIIIa; residue divergent from the consensus sequence is underlined) and 1692QRLEFLGDA1700 (RIIIb). The ATP-binding site motif 54LNTGSGKT61 and a 165DECH168 box are also found within the helicase domain. Sequence alignment of Dicer proteins from Homo sapiens, Mus musculus, C.elegans, Drosophila melanogaster, Schizosaccharomyces pombe and Arabidopsis thaliana revealed a relatively high sequence similarity and conservation of these putative domains (see Supplementary figure S2), except for the S.pombe protein, which apparently lacks the PAZ domain.
Fig. 1. Domain structure and expression of human Dicer. (A) Schematic illustration of the domain structure of the human Dicer protein. (B) Polyhistidine-tagged human Dicer protein was expressed in a baculovirus-based system, partially purified by Ni2+-affinity chromatography, and analyzed by SDS–PAGE followed by western blotting with anti-Dicer antibody or pre-immune serum, or Coomassie Blue staining. A preparation from mock-infected Sf9 cells was used as a control.
Preparation of human Dicer protein
A baculovirus expressing a polyhistidine-tagged human Dicer protein was prepared, and the protein was expressed in Sf9 insect cells. This eukaryotic system uses many of the protein modification processes characteristic of higher eukaryotes (Murphy et al., 1997). Figure 1B shows immunoblot analysis of our Dicer preparation (partially purified by nickel affinity chromatography) with a rabbit anti-Dicer antibody. A single immunoreactive band of the expected molecular mass (∼218 kDa) is present in the Dicer (Figure 1B, lane 4), but not in the mock (Figure 1B, lane 3), preparation. No band was visible using pre-immune serum (Figure 1B, lanes 1 and 2). On Coomassie Blue-stained gels, the ∼218 kDa band (Figure 1B, lane 5), which is absent in the mock preparation (Figure 1B, lane 6), represented ∼25% of the total protein content. Amino acid sequencing, after in-gel digestion with trypsin (performed by the Protein Analysis Center, Karolinska Institute), identified this band as human Dicer.
dsRNA-binding properties
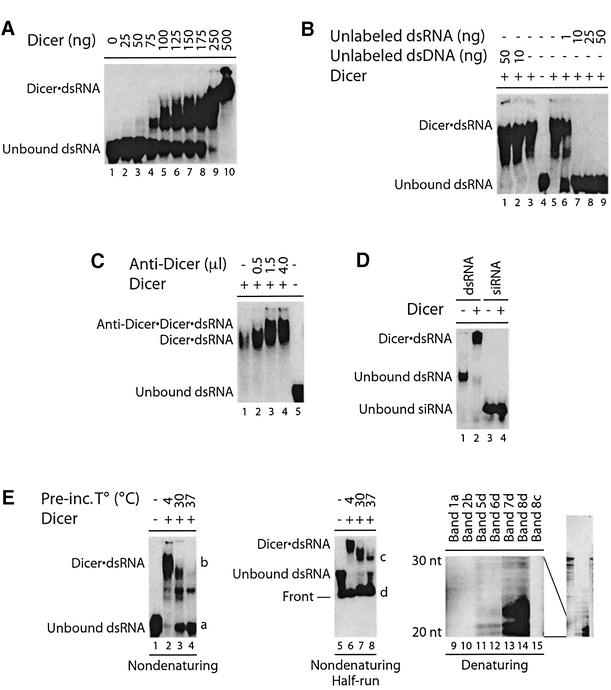
To determine whether the human Dicer protein was functional, we assayed its capacity to bind dsRNA by an electrophoretic mobility shift assay (EMSA). A 32P-labeled 5LO dsRNA (100 bp) was incubated with the human Dicer protein, and formation of the Dicer·dsRNA complex was analyzed by non-denaturing PAGE. Incubation of 32P-labeled dsRNA with increasing amounts of human Dicer induced formation of slower migrating bands indicative of Dicer·dsRNA complexes (Figure 2A). The pattern of complex formation suggests multiple binding sites for Dicer on the 100 bp dsRNA. Formation of Dicer·dsRNA complexes was prevented by co-incubation with unlabeled dsRNA (Figure 2B, lanes 6–9), but not with the synthetic dsDNA poly(dA·dT) (Figure 2B, lanes 1 and 2). The nature of the competing dsRNA had no influence, as dsRNA derived from either 5LO or coactosin-like protein (CLP) coding sequences could prevent binding to 5LO dsRNA (our unpublished data). Addition of an anti-Dicer antibody caused a further retardation of the electrophoretic mobility of the Dicer·dsRNA complex (Figure 2C, lanes 2–4), an effect reversed by adding the competing peptide antigen (our unpublished data).
Fig. 2. Dicer interacts with dsRNA in EMSA. (A) 32P-labeled dsRNA was incubated in the absence or presence of Dicer (no MgCl2 added), (B) without or with unlabeled dsRNA or dsDNA poly(dA·dT), or (C) without or with anti-Dicer antibody (0.5–4.0 µl; no MgCl2 added). (D) 32P- labeled CLP 23 nt siRNA or 5LO dsRNA (100 bp) was incubated in the absence or presence of Dicer. (E) Left panel, 32P-labeled dsRNA was incubated in the absence or presence of Dicer, at the indicated temperatures for 1 h prior to EMSA analysis. Middle panel, same as left panel, but run half way. Dicer·dsRNA complex formation was analyzed by non-denaturing PAGE and autoradiography. Right panel, RNA was extracted from the indicated bands and analyzed by denaturing PAGE and autoradiography. The 20–30 nt region is shown, with the full-length gel on the right.
We further substantiated the dsRNA binding of Dicer in polynucleotide binding assays. In these experiments, Dicer bound to poly(I)·poly(C) agarose beads in a dose-dependent manner; no binding to poly(C) agarose beads was observed (see Supplementary figure S3). Dicer also bound to poly(I)·poly(C) beads in the presence of 10% fetal bovine serum (FBS), supporting the specificity of Dicer binding. Used as a control, 5LO, which is not known to bind dsRNA, did not interact with poly(I)·poly(C) beads (our unpublished data). These data demonstrate that human Dicer preferentially binds dsRNA.
We assessed whether Dicer could bind siRNA. A 32P-labeled CLP siRNA duplex of 23 nt, with complementary strands of 21 nt each and 2 nt 3′ overhangs, was incubated with Dicer and complex formation was analyzed by EMSA. Whereas Dicer formed a complex with the 5LO dsRNA (100 bp) (Figure 2D, lane 2), used as a positive control, no Dicer·siRNA complex was observed (Figure 2D, lane 4). Similar results were obtained with a 5LO siRNA (our unpublished data).
Interestingly, pre-incubation of Dicer with the 5LO dsRNA (100 bp) and MgCl2 at 37°C for 1 h prior to analysis by EMSA eliminated the larger Dicer·dsRNA complexes (Figure 2E, lane 4 versus 2). In parallel, the amount of unbound dsRNA increased, together with a weaker band of intermediate mobility. To examine whether the disappearance of the Dicer·dsRNA complex was associated with the appearance of smaller RNA species, a similar gel was run half way (Figure 2E, middle panel) and RNA from the fastest migrating bands was extracted from the gel. Analysis by denaturing PAGE revealed that the loss of the larger Dicer·dsRNA complexes coincided with an increase in ∼20–24 nt RNA products (Figure 2E, right panel, lane 14 versus 12). No ∼20–24 nt RNA products were generated from the Dicer·dsRNA complex at 4°C (Figure 2E, right panel, lane 12). Like Dicer-bound (Figure 2E, right panel, lane 10) and unbound dsRNA (Figure 2E, right panel, lane 9), the band of intermediate mobility migrated as a 100 nt RNA species under denaturing conditions (Figure 2E, right panel, lane 15). This latter band may represent a different conformation of Dicer bound to dsRNA. We also observed an increase in unbound full-length dsRNA (Figure 2E, left panel, lanes 3 and 4) with temperature, indicating some dissociation of the Dicer·dsRNA complex.
dsRNase activity
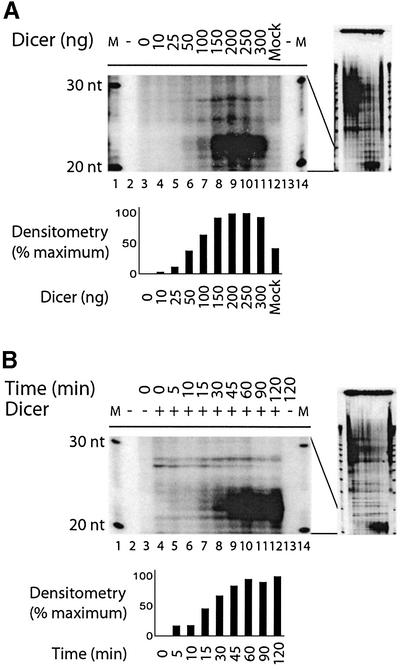
To investigate Dicer dsRNase activity, we incubated a 32P-labeled CLP dsRNA substrate (500 bp) with increasing amounts of human Dicer, and analyzed the reactions by denaturing PAGE and autoradiography. In these experiments, human Dicer (up to 150 ng) dose-dependently generated ∼21–23 nt RNA products from dsRNA (Figure 3A, lanes 3–11). Since Dicer is present in Drosophila, and we expressed human Dicer in insect cells, it was essential to confirm the absence of dsRNase activity in a preparation from mock-infected Sf9 cells (Figure 3A, lane 12). Product accumulated linearly with time for the first hour (Figure 3B, lanes 4–12), and the dsRNase activity of human Dicer increased with temperature (up to 37°C; see Supplementary figure S4), and was more efficient at low pH (see Supplementary figure S5). The proteinaceous nature of the dsRNase activity, as assayed by the formation of the ∼21–23 nt RNA products, was confirmed by its sensitivity to heat inactivation and proteinase K treatment (our unpublished data). Limited proteolysis, however, considerably enhanced dsRNase activity (our unpublished data).
Fig. 3. Dicer is catalytically active and generates ∼21–23 nt RNA products from dsRNA in vitro. 32P-labeled CLP dsRNA (500 bp) was incubated in the absence or presence of increasing amounts of Dicer (10–300 ng), in (A) a mock preparation (300 ng) in assay buffer (pH 7.5) at 30°C for 1 h, or (B) at 30°C for the indicated time. M, indicates a 10 nt RNA size marker. The samples were analyzed by denaturing PAGE and autoradiography. The 20–30 nt regions are shown, with the full-length gels on the right. The bar graphs show quantitation of product formation by densitometric analysis of the autoradiographs, after background subtraction.
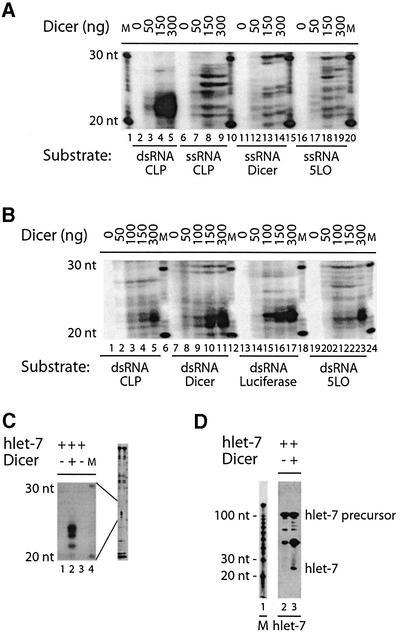
Next, we examined the substrate specificity for the dsRNase activity of human Dicer. Dicer dose-dependently generated ∼21–23 nt RNA products from a 500 bp dsRNA substrate, but not from three different single-stranded RNA (ssRNA) substrates (Figure 4A). Some ssRNA degradation was observed in the incubation mixtures, despite the presence of the RNase inhibitor Superase·In, but without accumulation of the characteristic ∼21–23 nt RNA products seen with dsRNA substrates. In addition, conversion of radioactive substrate was gradually competed by increasing amounts of unlabeled dsRNA (our unpublished data). Dicer was able to generate ∼21–23 nt RNA products from dsRNA substrates varying between 100 and 500 bp (see Supplementary figure S6). Moreover, Dicer was active on dsRNA from four different genes (Figure 4B). It is interesting to note a subtle difference in the apparent size of the RNA products of Dicer dsRNase activity, as cleavage of CLP, luciferase or 5LO dsRNA led to the formation of ∼22–23 nt RNA products, whereas ∼21–22 nt products were produced upon cleavage of Dicer dsRNA. There appears to be a slight difference in the efficiency of cleavage among the four different dsRNA substrates, but this is probably explained by differences in labeled uridine monophosphate content (Dicer, 58% AU; luciferase, 52% AU; CLP, 40% AU; 5LO, 43% AU).
Fig. 4. Dicer cleaves dsRNA and miRNA let-7 precursor into ∼21–24 nt RNA products. (A) 32P-labeled CLP dsRNA (500 bp), or CLP ssRNA, Dicer ssRNA or 5LO ssRNA (500 nt) was incubated in the absence or presence of Dicer. (B) 32P-labeled CLP dsRNA, Dicer dsRNA, luciferase dsRNA or 5LO dsRNA (500 bp) was incubated in the absence or presence of Dicer. (C) 32P-labeled miRNA let-7 precursor was incubated in the absence or presence of Dicer. The samples were analyzed by denaturing PAGE and autoradiography. (D) miRNA let-7 precursor was incubated in the absence or presence of Dicer and analyzed by northern hybridization with a probe recognizing the mature let-7 RNA. The hybridized probe was visualized by autoradiography. M, indicates a 10 nt RNA size marker.
In addition to dsRNA, we also tested the precursor for the human let–7 microRNA (miRNA; Pasquinelli et al., 2000) as a substrate. This RNA, which is predicted to form a stem–loop structure (Pasquinelli et al., 2000), was efficiently converted by Dicer into ∼22–24 nt RNA products (Figure 4C, lane 2). A ∼24 nt Dicer-dependent RNA product was detected by northern hybridization using a probe complementary to the mature let-7 (Figure 4D, lane 3). A probe that recognizes the sequence complementary to the mature let-7 in the folded precursor failed to hybridize to a ∼24 nt RNA product (our unpublished data).
Requirements for dsRNase activity
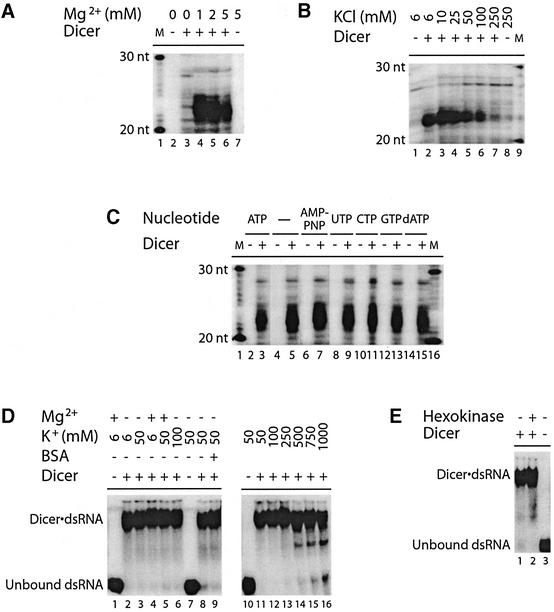
We verified the divalent cation requirement of the Dicer dsRNase activity, which was initially tested by pre-incubation in the absence or presence of MgCl2 (1–5 mM) for 5 min. As shown in Figure 5A, Dicer required MgCl2 for the production of ∼21–23 nt RNA products. Mg2+ could not be substituted by any other divalent cations, including the chloride salts of Ca2+, Mn2+, Co2+, Ni2+, Cu2+ and Zn2+ (see Supplementary figure S7). We also observed that Dicer dsRNase activity was sensitive to increased ionic strength. A gradual decrease in dsRNase activity was observed with increasing KCl concentration from 6 to 250 mM, with almost complete inhibition at 250 mM (Figure 5B).
Fig. 5. Dicer dsRNase activity requires magnesium ion, but not ATP, and is sensitive to increased ionic strength. (A–C) dsRNase assays. 32P-labeled CLP dsRNA (500 bp) was incubated in the absence or presence of Dicer, (A) without or with MgCl2 (1–5 mM), (B) with increasing concentrations of KCl (6–250 mM), or (C) without or with the indicated nucleotides (1 mM). M, indicates a 10 nt RNA size marker. The samples were analyzed by denaturing PAGE and autoradiography. (D and E) EMSAs. 32P-labeled 5LO dsRNA (100 bp) was incubated in the absence or presence of Dicer, (D) without or with MgCl2 (5 mM), KCl (6–1000 mM) and/or BSA (1 µg), or (E) that was pre-treated with glucose (2 mM) and/or hexokinase (0.4 U) at 30°C for 20 min prior to incubation in the absence of ATP (no KCl added). Dicer·dsRNA complex formation was analyzed by non-denaturing PAGE and autoradiography.
Considering the presence of the consensus ATP-binding site in Dicer, we asked whether ATP was required for dsRNase activity. Dicer was able to generate ∼21–23 nt RNA products from dsRNA in the absence of added ATP (Figure 5C, lane 5), and addition of ATP did not increase product formation (Figure 5C, lane 3). Moreover, replacement of ATP by equimolar amounts of AMP-PNP, UTP, CTP, GTP or dATP in the incubation mixtures had no effect (Figure 5C, lanes 7–15), thus arguing against an obligatory role of ATP in Dicer dsRNase activity in vitro.
Requirements for dsRNA binding
We asked whether the loss of activity observed in the absence of Mg2+ or presence of high KCl concentrations was related to decreased dsRNA binding. 32P-labeled dsRNA was incubated in the presence of Dicer, without or with MgCl2 or various concentrations of KCl, and Dicer·dsRNA complex formation was analyzed by EMSA. Dicer·dsRNA complex formation did not require MgCl2, nor was it influenced by an excess of bovine serum albumin (BSA) protein (Figure 5D, left panel). The Dicer·dsRNA complex was relatively insensitive to KCl concentrations, as only a slight dissociation was observed between 500 mM and 1 M (Figure 5D, right panel), indicative of a resistance to increased ionic strength. In addition, no effect on complex formation was observed upon removal of ATP by pre-treatment with hexokinase and glucose (Figure 5E, lane 2), as compared with treatment with glucose alone (Figure 5E, lane 1). These data indicate that Mg2+ and ATP are dispensable for dsRNA binding by Dicer.
Dicer dsRBD binds dsRNA
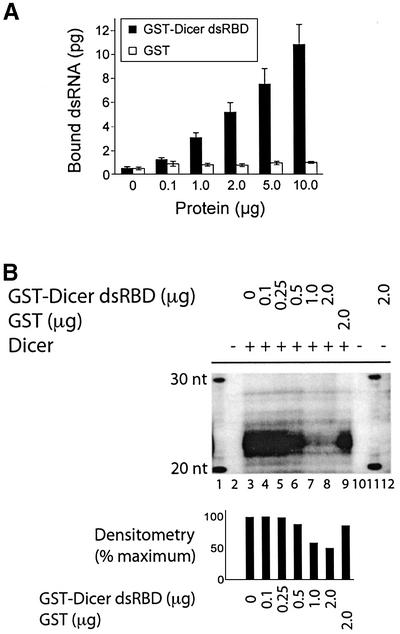
To examine whether the putative C-terminal dsRBD (amino acids 1772–1912) functions as presumed, Sepha rose 4B beads carrying the glutathione S-transferase (GST)–Dicer dsRBD protein were prepared and incubated with 32P-labeled CLP dsRNA (500 bp) in GST pull-down assays. The GST–Dicer dsRBD protein dose-dependently bound 32P-labeled dsRNA in vitro, while no significant binding of dsRNA to GST beads was observed (Figure 6A). Binding of the 32P-labeled dsRNA by the GST–Dicer dsRBD protein was gradually prevented by addition of increasing amounts of soluble, unlabeled dsRNA to the incubation mixtures (our unpublished data).
Fig. 6. The putative dsRBD of Dicer interacts with dsRNA and interferes with Dicer dsRNase activity. (A) GST-binding assay. The GST–Dicer dsRBD fusion protein, or GST alone, coupled to GSH–Sepharose 4B beads, was incubated with a 32P-labeled CLP dsRNA (500 bp) (n = 5–8 experiments). The bound 32P-labeled dsRNA was quantified by Cerenkov counting. Results are expressed as mean ± SEM. (B) dsRNase assay. 32P-labeled CLP dsRNA (500 bp) was incubated in the absence or presence of Dicer, without or with GST–Dicer dsRBD or GST. M, indicates a 10 nt RNA size marker. The samples were analyzed by denaturing PAGE and autoradiography. The bar graph shows quantitation of product formation by densitometric analysis of the autoradiographs, after background subtraction.
Given its ability to bind dsRNA, we tested whether the dsRBD could interfere with the cleavage of 32P-labeled CLP dsRNA by Dicer. In these experiments, radiolabeled dsRNA substrate was incubated in the presence of Dicer with GST–Dicer dsRBD or GST alone in dsRNase assays. As shown in Figure 6B, GST–Dicer dsRBD (Figure 6B, lanes 4–8), but not GST alone (Figure 6B, lane 9), prevented dsRNA cleavage by Dicer. GST–Dicer dsRBD (Figure 6B, lane 12) and GST (our unpublished data) themselves had no catalytic activity towards the dsRNA substrate.
Dicer localizes to the endoplasmic reticulum
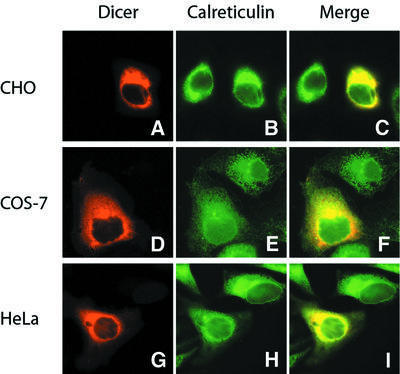
We expressed human Dicer in mammalian cells to study its intracellular localization. Immunofluorescence microscopy was performed in CHO, Cos-7 or HeLa cells transiently transfected with constructs encoding Myc-tagged Dicer. Expression of the Myc-Dicer protein was confirmed by western blotting (our unpublished data). In cells harvested 18–24 h after transfection, human Dicer colocalized with calreticulin, a resident protein of the endoplasmic reticulum (ER), in the three cell lines (Figure 7C, F and I). No Dicer staining of the nucleus could be observed (Figure 7A, D and G). Similar results were obtained at 96 h post-transfection or when expressing a FLAG-tagged Dicer protein (our unpublished data).
Fig. 7. Human Dicer colocalizes with the endoplasmic reticulum marker calreticulin in transiently transfected mammalian cells. (A–I) Immunofluorescence microscopy. (A–C) CHO, (D–F) Cos-7, or (G–I) HeLa cells grown on coverslips were transfected with plasmid constructs encoding a Myc-tagged human Dicer. (A, D and G) Myc-Dicer was stained with anti-Myc antibody (TRITC channel, red). (B, E and H) Calreticulin was labeled with anti-calreticulin antibody (FITC channel, green). (C, F and I) Merged images demonstrates colocalization of human Dicer with calreticulin.
Discussion
Powerful genetic approaches and biochemical studies of crude extracts have provided key insights into the RNA silencing phenomena. Fewer reports examined the function of purified components of these processes. In this study, we have cloned, expressed and characterized the 218 kDa recombinant human Dicer protein as a double-strand-specific RNase that processed dsRNA into ∼21–23 nt RNA products, recapitulating dsRNA processing observed in Drosophila and C.elegans cell lysates or immunoprecipitates (Zamore et al., 2000; Bernstein et al., 2001; Elbashir et al., 2001b; Ketting et al., 2001). A recombinant Dicer enzyme having similar endonucleolytic activity has been independently prepared by Zhang et al. (2002).
We observed cleavage of the human let-7 miRNA precursor by Dicer in vitro. The size of the fragments generated is consistent with previous reports that human let-7 exists in 21 and 22 nt forms that differ at their 5′ ends (Pasquinelli et al., 2000), and that the 21 nt let-7 RNA migrates as a 23 nt species (Hutvágner et al., 2001). By northern hybridization analysis, we identified the ∼24 nt RNA product as the mature let-7 RNA. The absence of an RNA product smaller than ∼50 nt corresponding to the strand complementary to the mature let-7 RNA suggests that Dicer does not cleave the let-7 precursor into an siRNA-like duplex in vitro. This result raises the interesting possibility that Dicer processes the let-7 RNA precursor by a different mode of action than it uses on generic dsRNA substrates; Dicer produces a single-stranded product from the let-7 RNA precursor, but double-stranded siRNAs from long, simple dsRNA substrates. This apparent difference in Dicer function, which is reminiscent of the variable actions of bacterial and yeast RNase III enzymes on their natural substrates versus synthetic dsRNAs (Li et al., 1993; Abou Elela et al., 1996; Chanfreau et al., 1997; Zhou et al., 1999), requires confirmation by further detailed investigation. The recent reports on the accumulation of let-7 precursor in animals lacking Dicer (Grishok et al., 2001; Ketting et al., 2001) or upon silencing of Dicer in cultured human cells (Hutvágner et al., 2001) suggest that the precursors of miRNAs, a growing family of small RNAs (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001) typified by let-7 and lin-4, likely represent the endogenous substrates of Dicer.
The noticeable difference in Dicer binding to dsRNA and siRNA duplexes that we observed is in agreement with the current notion that siRNAs, produced upon cleavage of dsRNA substrates by Dicer, are released and transferred to another protein complex, such as the RISC, which, in addition to directing the silencing process, may also protect siRNAs from cellular nucleases. Our observations are not compatible with Dicer serving as a route of entry for siRNAs in the RNAi pathway. Given our current state of knowledge, we anticipate that synthetic siRNAs, used as a tool for gene silencing, may enter the RNAi pathway downstream of Dicer.
Dicer is similar to common RNases III in that it can bind and cleave dsRNA irrespective of its sequence. A recent crystallographic study by Blaszczyk et al. (2001) shed new light on the mechanism of dsRNA cleavage by RNases III. In their model for Aquifex aeolicus RNase III, long dsRNA duplexes were cut at four places by a dimeric class 1 RNase III, such as E.coli RNase III. The RNA product had a total length of 13 nt, created by two overlapping strands of 11 nt each, with 2 nt 3′ overhangs (Blaszczyk et al., 2001). As a signature of class 3 RNase III activity, Dicer consistently cleaved dsRNA into ∼21–23 nt products which, incidentally, corresponds to two turns of the dsRNA helix. As suggested by Blaszczyk et al. (2001), this characteristic may emanate from inactivation of one of the two RNase III domains. It may be speculated that Dicer, by acting as a dimer, may have alternate disabled active centers leading to production of dsRNA fragments with two strands of ∼22 nt (Blaszczyk et al., 2001). In the model for A.aeolicus RNase III, it was also suggested that Mg2+ may stabilize dimerization. This is in agreement with previous reports for class 1 RNases III, which require divalent cations (preferably Mg2+) for cleavage, but not for binding, of dsRNA substrate (Li et al., 1993; Conrad et al., 1998). We obtained similar findings with Dicer. However, the lack of any complementation of Dicer dsRNase activity by Mn2+ diverge with E.coli RNase III (Li et al., 1993) and S.pombe Pac1 (Rotondo and Frendewey, 1996). In contrast to Dicer dsRNase activity, dsRNA binding was resistant to KCl concentrations as high as 1 M. Under such conditions, Dicer binds dsRNA, but the Dicer·dsRNA complex is catalytically inactive, suggesting a role for electrostatic interactions in dsRNase activity. The S.pombe Pac1 RNase III shows a similar salt sensitivity (Rotondo and Frendewey, 1996).
Recently, the ATP requirements in the RNAi pathway were investigated by Nykänen et al. (2001). They reported that dsRNA processing into siRNA is ATP-dependent in Drosophila embryo lysates. However, in the present study, the dsRNase activity of recombinant Dicer exhibited no ATP requirement in vitro, similar to common RNases III. Similar findings have been reported independently by Zhang et al. (2002). Our observations suggest that direct dsRNA cleavage by Dicer may not involve ATP, but do not preclude the implication of a distinct ATP-dependent catalytic activity in the RNAi pathway.
The Dicer C-terminal dsRBD was shown to bind dsRNA. This function may explain the ability of this fragment to interfere with cleavage of dsRNA into ∼21–23 nt RNA products, likely through dsRNA substrate sequestration. Whether this domain is essential for dsRNase activity remains uncertain, since E.coli RNase III lacking the dsRBD could process dsRNA (Sun et al., 2001).
Information regarding the intracellular localization of Dicer and other proteins involved in RNAi is lacking. In the present study, we found that human Dicer colocalized with calreticulin at the ER. Neither in transiently transfected mammalian cells (this study) nor in S.pombe (our unpublished data) was Dicer immunostaining of the nucleus observed. This observation is corroborated by Billy et al. (2001) in embryonal carcinoma and HeLa cells. In their study, a cyan fluorescent protein–Dicer fusion protein showed a cytoplasmic localization in transfected HeLa cells (Billy et al., 2001). Given the 5LO-binding properties of Dicer (our unpublished data), it is of interest that 5LO is sparsely, but consistently, observed in the ER and nuclear membrane invaginations (Woods et al., 1993, 1995). Thus, our results position Dicer at a site that could be a strategic advantage for an enzyme that produces miRNAs, which may participate in translational control, and siRNAs, which promote the destruction of targeted mRNAs. In addition, such a positioning may confer an additional level of specificity to RNA silencing processes in facilitating the discrimination of silencing-prone dsRNA from dsRNA structures essential for proper translation and other RNA-dependent processes. In support of the notion that RNAi may occur at the ER–cytosol interface are the findings that (i) nuclear pre-mRNAs are resistant to RNAi (Fire et al., 1998), and (ii) the RISC complex has been found to be associated with ribosomes (Hammond et al., 2000). Determination of the localization of other components of the RNA silencing pathways may help to better understand these processes and their possible ramifications in the control of gene expression, including miRNA processing (Lagos-Quintana, 2001; Lau et al., 2001; Lee and Ambros, 2001).
In a separate study, we have observed that human Dicer partially rescued the defect in chromosome segregation observed in Dicer knockout S.pombe cells (P.Provost, R.Silverstein, D.Dishart, J.Walfridsson, I.Djupedal, B.Kniola, A.Wright, B.Samuelsson, O.Rådmark and K.Ekwall, submitted), suggesting an evolutionary conserved role of Dicer in chromosome function. It will be interesting to study the involvement of Dicer in chromosome function in mammalian cells and, conversely, whether Dicer can initiate efficient RNA silencing in fission yeast.
This study contributes to a biochemical and molecular comprehension of the mechanisms underlying RNA silencing and other Dicer-related phenomena. Moreover, the availability of recombinant Dicer protein may facilitate the development of strategies for interfering with these processes.
Materials and methods
Cloning, expression and purification of human Dicer
The human Dicer full-length cDNA was isolated from a testis cDNA library and both strands were sequenced as described previously (Provost et al., 1999). The human Dicer cDNA was amplified by PCR and cloned into the SalI/HindIII sites of pFastBac-HTa vector (Invitrogen), which introduced a His6 tag at the N-terminus. Baculovirus was prepared and the human Dicer protein was expressed in Sf9 insect cells according to the manufacturer’s instructions.
Briefly, Sf9 cells (2 × 106/ml) were infected with recombinant or empty (mock) baculovirus, grown for 3–4 days, harvested, and resuspended in ice-cold lysis buffer [50 mM Tris–HCl, 100 mM KCl, 5 mM β-mercaptoethanol, 1% Nonidet P-40 (NP-40), 1 mM phenylmethanesulfonyl fluoride, complete protease inhibitor cocktail (Roche), pH 8.5]. After 15 min at 4°C, the cell lysate was sonicated on ice, and centrifuged at 10 000 g for 10 min at 4°C. The supernatant was incubated with Ni-NTA (nitrilo triacetic acid) resin (Qiagen) on a rotator for 1 h at 4°C. The resin was then successively washed with 10 volumes of buffer A [20 mM Tris–HCl, 1 M KCl, 20 mM imidazole, 10% (v/v) glycerol, 5 mM 2-mercaptoethanol, pH 8.5], 2 volumes of buffer B [20 mM Tris–HCl, 1 M KCl, 10% (v/v) glycerol, 5 mM 2-mercaptoethanol, pH 8.5], and 2 volumes of buffer A. The bound Dicer protein was eluted with buffer C [20 mM Tris–HCl, 100 mM KCl, 200 mM imidazole, 10% (v/v) glycerol, 5 mM 2-mercaptoethanol, pH 8.5]. Active fractions were collected and pooled, and the protein concentration was determined by the method of Bradford (Bradford, 1976) using the Bio-Rad dye reagent, with BSA as a standard.
Sequence analysis
Sequence assembly, analysis and alignment were performed using the Wisconsin Package Version 9.1 [Genetics Computer Group (GCG), Madison, WI], or via the European Bioinformatics Institute (EBI) server (Hinxton Hall, UK, http://www.ebi.ac.uk) or the Swiss Institute of Bio informatics (SIB) server (Geneva, Switzerland, http://www.expasy.ch).
Immunoblot analysis
Protein suspensions were analyzed by SDS–PAGE and immunoblotting as described previously (Provost et al., 1999, 2001a). A polyclonal anti-human Dicer serum was raised in rabbits against the synthetic peptide 1382SNTDKWEKDEMTKDC1396 of human Dicer (Innovagen, Lund, Sweden). Immunoreactive proteins were visualized using alkaline phosphatase conjugates and substrates (nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate).
dsRNase assays
High specific radioactivity dsRNA substrates were prepared for dsRNase assays as described previously (Rotondo and Frendewey, 2001). Briefly, CLP (nucleotides 150–649; accession No. L54057), Dicer (nucleotides 272–771; accession No. AJ132261), 5LO (nucleotides 35–534; accession No. J03571) or luciferase (nucleotides 3788–4304; accession No. AF053461) sequences were amplified by PCR to introduce opposing T7 RNA polymerase promoters at both ends. For ssRNA substrates, a T7 promoter was introduced at one end of the cDNA fragments. For the human miRNA let-7 precursor, its sequence (Pasquinelli et al., 2000) was inserted downstream of a T7 RNA polymerase promoter between EcoRI sites into vector pCR2.1 (Invitrogen), and the template was released by EcoRI digestion. The DNA templates were gel-purified; the dsRNA, ssRNA and human let-7 precursor were synthesized by in vitro transcription and uniformly labeled with [α-32P]UTP. A 10 nt RNA size marker (Decade Markers, Ambion) was prepared with [γ-32P]ATP.
The dsRNase assay was performed in 20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP, 5% Superase·In (Ambion), with 10–500 ng human Dicer protein (12–600 fmol, assuming 25% purity) and ∼1–5 fmol (106 c.p.m.) 32P-labeled CLP dsRNA (500 bp) at the indicated temperature in a volume of 10 µl, unless otherwise indicated. Reactions were stopped on ice and an equal volume of gel loading buffer (95% formamide, 18 mM EDTA, 0.025% SDS, 0.1% xylene cyanol and 0.1% bromphenol blue) was added. After heating at 85°C for 5 min, the samples were analyzed by electrophoresis on a 10% polyacrylamide–7M urea gel run in TBE buffer (Rotondo and Frendewey, 2001). The gel was fixed, dried and the products detected by autoradiography.
Northern hybridization analysis
The Dicer cleavage products of the human miRNA let-7 precursor were analyzed by northern hybridization. Briefly, the dsRNase reactions were separated by electrophoresis on a 10% denaturing polyacrylamide gel and electroblotted onto Hybond-XL membrane (Amersham Pharmacia Biotech). The deoxyoligonucleotide probes recognizing the mature let-7 RNA (5′-ACTATACAACCTACTACCTCA-3′) or its complementary strand (5′-AAGACAGTAGATTGTATAGT-3′) were radioactively labeled with polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer). The membrane was hybridized with the probes at 42°C in 60 mM NaH2PO4, 900 mM NaCl, 6 mM EDTA, 0.1% SDS and 0.1 mg/ml E.coli tRNA (Roche), and washed. The hybridized probes were visualized by autoradiography.
Electrophoretic mobility shift assays
Proteins were incubated in 20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 50 mM KCl, 1 mM DTT, 1 mM ATP and 5% Superase·In for 10 min at room temperature, in a total volume of 10 µl, unless indicated otherwise. In some experiments, human Dicer was pre-treated with glucose (2 mM) without or with hexokinase (0.4–0.8 U) at 30°C for 20 min. The reaction mixtures were placed on ice, and mixed with ∼1 fmol (75–80 000 c.p.m.) 32P-labeled 5LO dsRNA (100 bp) (nucleotides 35–134; accession No. J03571) or CLP 23 nt siRNAs (nucleotides 244–266; accession No. L54057) (Xeragon) that were 5′ end-labeled with polynucleotide kinase and [γ-32P]ATP. After 30 min on ice, 0.1 volumes of glycerol (60%)/bromophenol blue (0.2%) was added, and the reactions were immediately loaded onto pre-run non-denaturing 5% polyacrylamide TBE gels (29:1; acrylamide:bisacrylamide). Electrophoresis was performed at 100 V for 90–125 min. Gels were sealed in cellophane sheets, and formation of the protein–RNA complex was detected by autoradiography.
In some experiments, RNA was extracted from the gel in extraction buffer (0.5 M ammonium acetate, 1 mM EDTA and 0.2% SDS) at 37°C for 12–16 h, precipitated and resuspended in water. After addition of an equal volume of gel loading buffer, the samples were heated at 85°C for 5 min and analyzed by electrophoresis on a 10% denaturing polyacrylamide gel. The gel was fixed, dried and the products detected by autoradiography.
Expression of GST–Dicer dsRBD and GST-binding assays
The Dicer cDNA encoding the dsRBD (amino acids 1772–1912) was amplified by PCR and cloned in-frame into the BamHI/XhoI sites of pGEX-5X-1 (Amersham Pharmacia Biotech) to generate pGEX-5X-1-Dicer dsRBD. This construct and the empty pGEX-5X-1 vector were used to transform E.coli BL21 for expression and purification of the GST–Dicer dsRBD fusion protein and GST, according to the manufacturer’s instructions. For dsRNA-binding studies, 0.1–10 µg of the GST–Dicer dsRBD fusion protein, or GST alone, coupled to GSH–Sepharose 4B beads (10 µl), was incubated with 32P-labeled CLP dsRNA (500 bp) (∼1.5–5 fmol, 106 c.p.m.) in 20 mM Tris–HCl pH 7.5, 5 mM MgCl2, 100 mM KCl, 1 mM DTT, 1 mM ATP, 0.5 mg/ml BSA and 0.01% NP-40 supplemented with 1% Superase·In. After incubation at 4°C for 30 min, the beads were washed twice with 75 volumes of incubation buffer, and the Dicer dsRBD-bound dsRNA was eluted with 10 mM glutathione for 30 min. The beads were sedimented by centrifugation, and the presence of 32P-labeled dsRNA in the eluate was quantified by Cerenkov counting.
Cell culture, transfections and immunofluorescence microscopy
The human Dicer ORF was amplified by PCR and cloned into a modified mammalian expression vector pcDNA3.1 (Invitrogen) containing an N-terminal Myc or FLAG epitope. CHO, Cos-7 and HeLa cells were grown on sterile glass coverslips, as described previously (Provost et al., 2001b), and transfected using Lipofectamine Plus (Invitrogen) in serum-free DMEM for 3 h. For double immunofluorescence staining, transfected cells were washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 20 min and washed again in PBS. After quenching in 50 mM NH4Cl for 15 min, the cells were permeabilized with 0.1% Triton X-100 for 5 min and incubated in blocking buffer (0.1% BSA in PBS) for 1 h. The epitope-tagged human Dicer was labeled with anti-Myc mouse monoclonal antibody (clone 9E10, 2 µg/ml; Santa Cruz Biotechnology) or anti-FLAG M2 mouse monoclonal antibody (2 µg/ml; Sigma) for 30 min. The ER was stained with anti-calreticulin rabbit polyclonal antibody (1/400 dilution; Affinity BioReagents). After extensive washing in PBS, the cells were incubated with tetramethylrhodamine isothiocyanate (TRITC)- or fluorescein isothiocyanate (FITC)- conjugated goat anti-mouse or goat anti-rabbit IgG antibodies (Sigma). After extensive washing in PBS, the coverslips were mounted on slides. The cells were viewed on an Axiophot (Zeiss) microscope, and the images were analyzed with the program QED CCD Camera StandAlone (QED Imaging).
Accession number
The sequence of human Dicer cDNA have been submitted to the DDBJ/EMBL/Genbank databases under accession number AJ132261.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We acknowledge Witold Filipowicz for sharing data prior to publication. We thank Isabelle Plante, Agneta Nordberg, Rostislav Chernomorsky and Jinsop Om for excellent technical assistance. P.P. was supported by Fellowships from the Canadian Institutes of Health Research and the Karolinska Institute, and by a grant from the Centre de Recherche du CHUL. This work was supported by grants from the Swedish Medical Research Council (03X-217), the European Union (QLG1-CT-2001-01521) and the Verum foundation.
References
- Abou Elela S., Igel,H. and Ares,M.,Jr (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Baulcombe D.C. (1996) RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol., 32, 79–88. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Billy E., Brondani,V., Zhang,H., Muller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk J., Tropea,J.E., Bubunenko,M., Routzahn,K.M., Waugh,D.S., Court,D.L. and Ji,X. (2001) Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure (Camb.), 9, 1225–1236. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Abou Elela,S., Ares,M.,Jr and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C., Rauhut,R. and Klug,G. (1998) Different cleavage specificities of RNases III from Rhodobacter capsulatus and Escherichia coli. Nucleic Acids Res., 26, 4446–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court D. (1993) RNA processing and degradation by RNase III. In Belasco,J.G. and Brawerman,G. (eds), Control of Messenger RNA Stability. Academic Press, San Diego, CA, pp. 71–116.
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Grishok A. et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- Hutvágner G. and Zamore,P.D. (2002) RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev., 12, 225–232. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Iino Y., Sugimoto,A. and Yamamoto,M. (1991) S. pombe pac1+, whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J., 10, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S.E., Running,M.P. and Meyerowitz,E.M. (1999) Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development, 126, 5231–5243. [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer,S.E., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S.W. and Bass,B.L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science, 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- Li H.L., Chelladurai,B.S., Zhang,K. and Nicholson,A.W. (1993) Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res., 21, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S., Ichigotani,Y., Okuda,T., Irimura,T., Nakatsugawa,S. and Hamaguchi,M. (2000) Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta, 1490, 163–169. [DOI] [PubMed] [Google Scholar]
- Murphy C.I., Piwnica-Worms,H., Grunwald,S. and Romanow,W.G. (1997) Expression of proteins in insect cells using baculovirus vectors. In Ausubel,F. et al. (eds), Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York, NY, pp. 16.9.1–16.9.10.
- Nicholson R.H. and Nicholson,A.W. (2002) Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm. Genome, 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Nykänen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E. et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Provost P., Doucet,J., Hammarberg,T., Gerisch,G., Samuelsson,B. and Rådmark,O. (2001a) 5-lipoxygenase interacts with coactosin-like protein. J. Biol. Chem., 276, 16520–16527. [DOI] [PubMed] [Google Scholar]
- Provost P., Doucet,J., Stock,A., Gerisch,G., Samuelsson,B. and Rådmark,O. (2001b) Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem. J., 359, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P., Samuelsson,B. and Rådmark,O. (1999) Interaction of 5-lipoxygenase with cellular proteins. Proc. Natl Acad. Sci. USA, 96, 1881–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.D., Webster,R.E. and Zinder,N.D. (1968) Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem., 243, 82–91. [PubMed] [Google Scholar]
- Romano N. and Macino,G. (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol., 6, 3343–3353. [DOI] [PubMed] [Google Scholar]
- Rotondo G. and Frendewey,D. (1996) Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res., 24, 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo G. and Frendewey,D. (2001) Pac1 ribonuclease of Schizosaccharomyces pombe. Methods Enzymol., 342, 168–193. [DOI] [PubMed] [Google Scholar]
- Rotondo G., Gillespie,M. and Frendewey,D. (1995) Rescue of the fission yeast snRNA synthesis mutant snm1 by overexpression of the double-strand-specific Pac1 ribonuclease. Mol. Gen. Genet., 247, 698–708. [DOI] [PubMed] [Google Scholar]
- Rotondo G., Huang,J.Y. and Frendewey,D. (1997) Substrate structure requirements of the Pac1 ribonuclease from Schizosaccharomyces pombe. RNA, 3, 1182–1193. [PMC free article] [PubMed] [Google Scholar]
- Samuelsson B. (1983) Leukotrienes: mediators of immediate hypersensitivity reactions and inflammation. Science, 220, 568–575. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference–2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Sun W., Jun,E. and Nicholson,A.W. (2001) Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry, 40, 14976–14984. [DOI] [PubMed] [Google Scholar]
- Tuschl T. (2001) RNA interference and small interfering RNAs. Chembiochem., 2, 239–245. [DOI] [PubMed] [Google Scholar]
- Wilson R. et al. (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- Woods J.W., Evans,J.F., Ethier,D., Scott,S., Vickers,P.J., Hearn,L., Heibein,J.A., Charleson,S. and Singer,I.I. (1993) 5-lipoxygenase and 5-lipoxygenase-activating protein are localized in the nuclear envelope of activated human leukocytes. J. Exp. Med., 178, 1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods J.W., Coffey,M.J., Brock,T.G., Singer,I.I. and Peters-Golden,M. (1995) 5-lipoxygenase is located in the euchromatin of the nucleus in resting human alveolar macrophages and translocates to the nuclear envelope upon cell activation. J. Clin. Invest., 95, 2035–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.P., Riggs,M., Rodgers,L. and Wigler,M. (1990) A gene from S.pombe with homology to E.coli RNase III blocks conjugation and sporulation when overexpressed in wild type cells. Nucleic Acids Res., 18, 5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb,F.A., Brondani,V., Billy,E. and Filipowicz,W. (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J., 21, 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Frendewey,D. and Lobo Ruppert,S.M. (1999) Pac1p, an RNase III homolog, is required for formation of the 3′ end of U2 snRNA in Schizosaccharomyces pombe. RNA, 5, 1083–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]