Abstract
The multiple functions of the p97/Cdc48p ATPase can be explained largely by adaptors that link its activity to different cellular pathways, but how these adaptors recognize different substrates is unclear. Here we present evidence that the mammalian adaptors, p47 and Ufd1–Npl4, both bind ubiquitin conjugates directly and so link p97 to ubiquitylated substrates. In the case of Ufd1–Npl4, which is involved in endoplasmic reticulum (ER)-associated degradation and nuclear envelope reassembly, binding to ubiquitin is mediated through a putative zinc finger in Npl4. This novel domain (NZF) is conserved in metazoa and is both present and functional in other proteins. In the case of p47, which is involved in the reassembly of the ER, the nuclear envelope and the Golgi apparatus, binding is mediated by a UBA domain. Unlike Ufd1–Npl4, it binds ubiquitin only when complexed with p97, and binds mono- rather than polyubiquitin conjugates. The UBA domain is required for the function of p47 in mitotic Golgi reassembly. Together, these data suggest that ubiquitin recognition is a common feature of p97-mediated reactions.
Keywords: CDC48/NZF/TAB2/UBA/VPS36
Introduction
The abundant AAA protein, p97/Cdc48p, is an essential ATPase present in the cytoplasm, on membranes and in the nucleus of eukaryotes from yeast to man (Patel and Latterich, 1998). Since its discovery as a cell cycle arrest mutant in yeast (Moir et al., 1982), a large number of reports have shown its requirement or involvement in a wide variety of cellular processes, including membrane fusion events (Acharya et al., 1995; Latterich et al., 1995; Rabouille et al., 1995; Roy et al., 2000; Hetzer et al., 2001), apoptotic regulation (Madeo et al., 1999; Shirogane et al., 1999), ubiquitin–proteasome-dependent degradation of cytosolic proteins (Ghislain et al., 1996; Dai and Li, 2001), endoplasmic reticulum (ER)-associated degradation (ERAD) (Bays et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002; Rabinovich et al., 2002) and regulated ubiquitin-dependent processing (RUP) (Hitchcock et al., 2001; Rape et al., 2001). Though many of these cellular processes appear to be unrelated, common aspects are emerging that point to a general mechanism of p97/Cdc48p function.
One widely accepted aspect of p97 function is its ability to exert mechanical force on substrate proteins. This is true for other AAA-ATPases which can disassemble macromolecular complexes and unfold proteins (Ogura and Wilkinson, 2001), and is particularly true for the p97 homolog, NSF, an AAA-ATPase that unravels stable SNARE complexes during the membrane fusion cycle (Söllner et al., 1993). The unfolding activity of p97 was first shown for its archaebacterial homolog, VAT (Golbik et al., 1999), and then for p97/Cdc48p in the ERAD and RUP pathways, where it mediates the retro-translocation or mobilization of substrate proteins by either pulling them through the translocation pore into the cytoplasm or separating them from integral membrane proteins (reviewed in Bays and Hampton, 2002; Tsai et al., 2002).
Another aspect is the involvement of cofactors. Although the relevance of p97/Cdc48p binding to partners such as the yeast and mammalian Ufd3p/PLAP (Ghislain et al., 1996; Seigneurin-Berny et al., 2001) and Ufd2 (Koegl et al., 1999; Meyer et al., 2000) has remained elusive, at least two binding partners have been shown to be crucial for p97-mediated reactions. The first, p47, is required for p97-mediated membrane fusion events during post-mitotic reassembly of Golgi stacks (Kondo et al., 1997). It is thought to link p97 to the t-SNARE, syntaxin-5, thereby catalyzing changes in SNARE complexes needed for p97-mediated Golgi membrane fusion (Rabouille et al., 1998). The second binding partner is a heterodimeric complex of Ufd1 and Npl4 (UN), first isolated from rat liver cytosol using affinity methods (Meyer et al., 2000). UN and p47 bind p97 in a mutually exclusive manner. This observation raised the idea that they represent adaptors that direct the unfolding activity of p97 to substrates in different reactions, p47 functioning in membrane fusion and UN in ubiquitin-dependent processes (Meyer et al., 2000). This role for UN was first suggested by the fact that mutations in UFD1 prevented degradation of a ubiquitin fusion protein in yeast (Johnson et al., 1995) and recently has been corroborated by experiments showing that UN is required for the p97-mediated reactions in two ubiquitin-dependent processes, ERAD (Bays et al., 2001; Ye et al., 2001; Braun et al., 2002; Jarosch et al., 2002) and RUP (Hitchcock et al., 2001; Rape et al., 2001).
It was, therefore, a surprise to find that UN could also be involved in membrane fusion events. p97 is needed for the assembly of the nuclear envelope in Xenopus egg extracts in a two-step process (Hetzer et al., 2001). The first step requires p97–UN and leads to the formation of a closed nuclear envelope. The second step involves the expansion of this nuclear envelope and requires p97–p47. Since both steps involve topologically identical fusion events, one could argue that both adaptors carry out essentially the same reaction, the need for different adaptors reflecting subtle differences in detail such as the need to close fenestrae in the first step but not the second (Burke, 2001).
In this study, we have, therefore, examined the relationship between p97 adaptors and ubiquitin conjugates. Since UN has been implicated in ubiquitin-related processes, we carried out experiments showing that this adaptor does indeed bind to ubiquitin conjugates. Similar experiments were then carried out using p47, with the surprising result that this adaptor also bound ubiquitin conjugates, though the binding domain and substrate specificity were different. We also show that this binding domain is needed for p97–p47-mediated reassembly of Golgi cisternae.
Results
Mammalian Ufd1–Npl4 mediates p97 binding to polyubiquitin conjugates
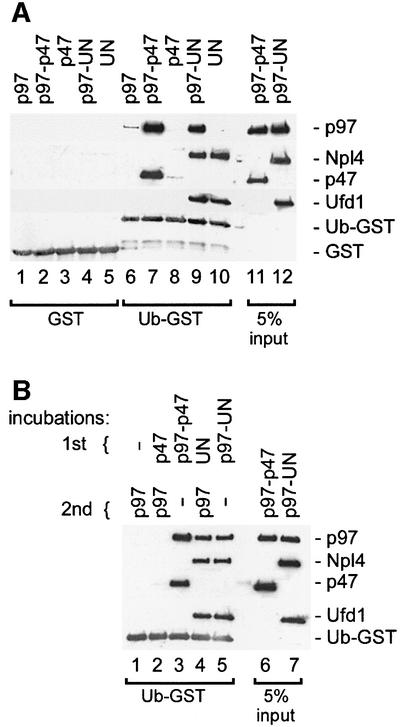
To study the binding of p97 adaptor complexes with ubiquitin conjugates, we first tested their ability to bind naturally occurring polyubiquitylated proteins (Figure 1). Recombinant p97, p47 or UN were biotinylated and immobilized on streptavidin–beads. p97 beads were used either directly or after incubation with either p47 or UN. The pre-assembled UN dimer was used in this and most of the following experiments since Ufd1 and Npl4 bind p97 cooperatively (Meyer et al., 2000) and only the heterodimer is functional (Hetzer et al., 2001). The coated beads were incubated with a detergent extract from cultured HeLa cells, and bound polyubiquitylated proteins were detected by western blot analysis using anti-ubiquitin antibodies (Figure 1, upper panel). The filters were also stained for total protein to show the amount of bait protein used (Figure 1, lower panel). Control and p97-coated beads bound very little, if any, polyubiquitylated proteins. In contrast, UN, either alone or in a complex with p97, was most effective, with ∼10% of the input polyubiquitylated proteins bound. On the other hand, p47, alone or in a complex with p97, bound much less polyubiquitylated proteins than UN, but slightly more than background amounts. This experiment clearly shows that UN binds to polyubiquitylated proteins, though there remains the possibility that this interaction is indirect. It should also be noted that the beads coated with p47 or UN extracted p97 from the lysate, as expected (Figure 1, lower panel, lanes 5 and 6; and immunodetection, data not shown).
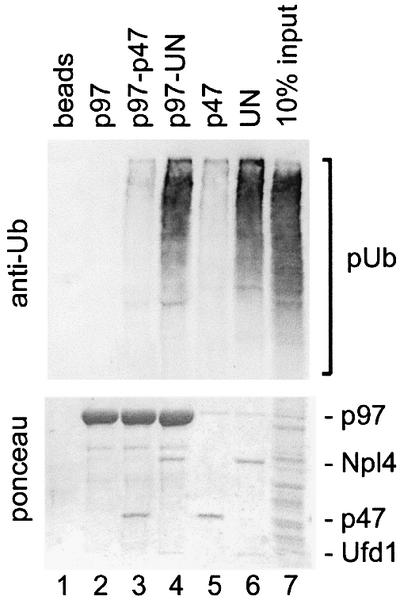
Fig. 1. The p97 adaptor, Ufd1–Npl4, mediates binding to polyubiquitylated proteins. Recombinant mammalian p97 (lanes 2–4), p47 (lane 5) and Ufd1–Npl4 (UN, lane 6) were biotinylated and immobilized on streptavidin–beads. The p97–beads were either used without further treatment (lane 2) or first saturated with recombinant p47 or UN (lanes 3 and 4, respectively). The immobilized protein complexes were then incubated with a Triton X-100 extract from HeLa cells and bound proteins analyzed by western blotting using a ubiquitin-specific antibody (upper panel). The filter was also stained with Ponceau S to visualize the immobilized proteins (lower panel). Note that only UN efficiently binds polyubiquitylated proteins (pUb) either alone or in combination with p97.
UN and p47 bind directly to a mono-ubiquitylated substrate
To test whether the adaptors could bind ubiquitin directly, we carried out experiments using a model mono-ubiquitylated substrate, ubiquitin fused to the N-terminus of GST (Ub–GST). Ub–GST was expressed in bacteria and therefore enabled us to study the interaction of the adaptors with ubiquitin using pure proteins.
GST or Ub–GST was immobilized on glutathione beads, incubated with different combinations of pure, recombinant p97, p47 and UN, and analyzed for bound proteins. As shown in Figure 2A, Ub–GST bound the p97–p47 complex very efficiently (lane 7), typically removing ∼10% of the complex from the incubation. This contrasts with the very weak binding of polyubiquitylated proteins to p97–p47 (Figure 1), suggesting that it has a much higher specificity for mono- than for polyubiquitylated substrates. Furthermore, neither p97 nor p47 alone bound significantly to Ub–GST (Figure 2A, lanes 6 and 8). Since mapping experiments below locate the ubiquitin-binding site to p47, this shows that binding of p47 to Ub–GST is regulated by p97. The p97–UN complex could also bind Ub–GST (lane 9) although slightly less efficiently than p97–p47. Unlike p47, however, UN could bind Ub–GST independently of p97 (lane 10).
Fig. 2. The binding of p47 and UN to mono-ubiquitin is direct but differentially regulated. (A) The indicated combinations of p97, p47 and UN were incubated with either GST (lanes 1–5) or a ubiquitin–GST fusion protein (Ub–GST, lanes 6–10) immobilized on glutathione– beads. Bound material was analyzed by western blotting using specific antibodies against the proteins indicated on the right; 5% of the input was loaded as a reference (lanes 11 and 12). Note that p97 binds Ub–GST efficiently only as a complex with either p47 (lane 7) or UN (lane 9), and that UN (lane 10), but not p47 (lane 8), can bind Ub–GST independently of p97. (B) Binding and analysis were carried out as in (A) but using sequential incubations. Ub–GST was first incubated with p47 (lane 2) or UN (lane 4), and with p97–47 or p97–UN as a reference (lanes 3 and 5). The beads were then washed and incubated with p97 as indicated. Note that UN (lane 4), but not p47 (lane 2), binds Ub–GST, stays bound during the washes and can then recruit p97 in the second incubation (lane 4).
To confirm that UN could recruit p97 to Ub–GST, we carried out a two-step reaction in which Ub–GST was first incubated with or without UN and then with p97. As shown in Figure 2B, p97 was recruited only if UN was present in the first incubation (compare lanes 1 and 4). Similar incubations with p47 gave no binding (compare lanes 1 and 2), in agreement with earlier results (Figure 2A).
Binding to ubiquitin conjugates was also found to be ATP independent since the absence or presence of ATP, ATPγS or ADP did not influence complex formation between Ub–GST and either p97–p47 or p97–UN (data not shown).
Ubiquitin binding of p97–p47 is mediated by the p47-UBA domain
p47 contains a peptide sequence of 45 amino acids at the N-terminus (Figure 3A) with homology to the UBA motif (Hofmann and Bucher, 1996) that was shown in the Rad23 protein and others to bind ubiquitin (Bertolaet et al., 2001; Wilkinson et al., 2001). To test the role of this peptide sequence in ubiquitin binding, we generated a p47 mutant that lacked this domain [p47(ΔUBA)]. As shown in Figure 3B (upper panels), this mutant failed to bind ubiquitin. This was not due to decreased binding of p47(ΔUBA) to p97, since the mutant could bind equally well to immobilized p97 as the wild-type p47 (Figure 3B, lower panels). The UBA domain is also sufficient for ubiquitin binding and able to compete with p97–p47(wt). This was shown using a bacterially expressed peptide representing the UBA domain of p47 (Figure 3C). When increasing amounts of this peptide were titrated into incubations containing p97–p47(wt) and immobilized Ub–GST, the p97–p47 complex was displaced and the peptide became bound (lanes 2–4). A control peptide, p47-UBA(F41A), that contained a mutation equivalent to a ubiquitin-binding mutant of the UBA2 domain in Rad23 (Bertolaet et al., 2001), bound less efficiently and competed with p97–p47 to a lesser extent (Figure 3C, lanes 5–7). Conversely, the peptide representing the UBA2 domain of hhRad23 had the same effect as the p47 UBA(wt) peptide (compare lanes 2–4 with lanes 8–10).
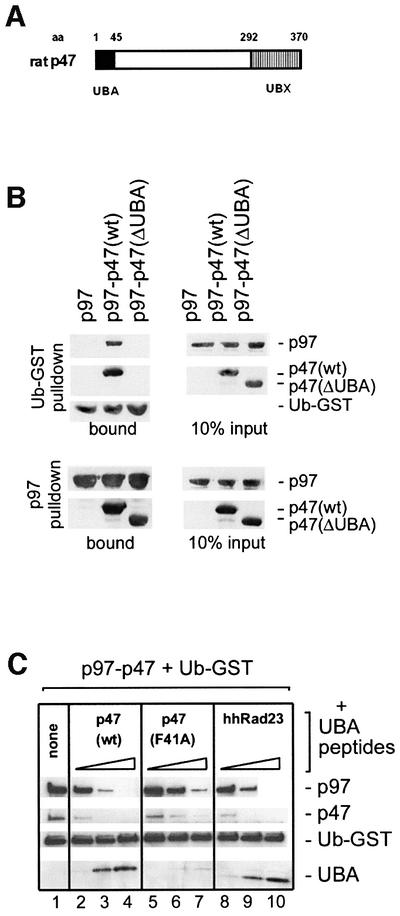
Fig. 3. The UBA domain of p47 is both necessary and sufficient for mono-ubiquitin binding. (A) Schematic representation of the p47 domain structure with amino acid (aa) numbers indicated. p47 contains a UBX domain at the C-terminus, that has a ubiquitin fold which contributes to p97 binding (Yuan et al., 2001). The N-terminal 45 amino acids constitute the UBA domain. (B) Upper panels: p97 was incubated with immobilized Ub–GST either alone or in the presence of p47(wt) or p47 lacking the UBA domain [p47(ΔUBA)]. Bound proteins were analyzed by western blotting using specific antibodies to the proteins indicated on the right. Note that the UBA domain is needed for binding to Ub–GST. Lower panels: biotinylated p97 was incubated with p47 or p47(ΔUBA) and complexes retrieved using streptavidin–beads. Note that the UBA domain is not required for binding to p97. The panel on the right shows 10% of input material used. (C) p97–p47 was bound to immobilized Ub–GST in the absence (lane 1) or presence of increasing amounts (5, 25 and 125 mol excess over p47) of peptides representing different UBA domains: p47(wt) (lanes 2–4), p47(F41A) mutant (lanes 5–7) and the human homolog of Rad23 (hhRad23, lanes 8–10). Bound material was analyzed by western blotting. Note that the p47(wt) and the hhRad23 peptides bind Ub–GST directly and compete with the p97–p47 complex, whereas the p47(F41A) mutant does so much less efficiently.
Together, these data show that the UBA domain of p47 is both necessary and sufficient for binding to mono-ubiquitin–GST and that it does not contribute to the interaction between p97 and p47.
The p47 UBA domain is required for reassembly of Golgi cisternae
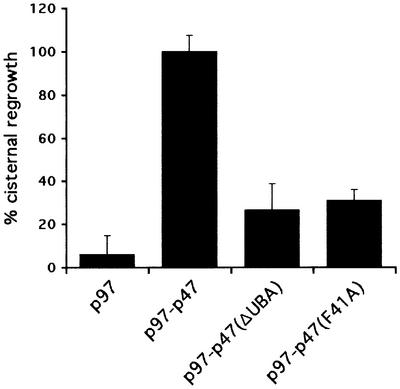
The identification of p47 as a ubiquitin-binding protein was surprising, since the only known function of p47 is in membrane fusion reactions and this has never been linked to the ubiquitin system. We therefore tested whether its ability to bind ubiquitin is required for its activity in Golgi membrane fusion. Mitotic Golgi fragments (MGFs) were generated by treatment of rat liver Golgi membranes with mitotic HeLa cytosol. After re-isolation, the membranes were treated with N-ethylmaleimide (NEM) to inactivate the two ATPases (NSF and p97) that independently can catalyze cisternal reassembly (Rabouille et al., 1995). These MGFs were then incubated with buffer or p97, alone or in combination with p47(wt), p47(ΔUBA) or p47(F41A), for 60 min at 37°C. The samples were processed for electron microscopy and the extent of cisternal regrowth determined using stereological procedures. As shown in the representative experiment in Figure 4, only a combination of p97 and p47(wt) could reassemble cisternae efficiently. Deletion of the UBA domain [p47(ΔUBA)] reduced reassembly by 75–80% compared with p47(wt). Even a single point mutation in the full-length protein [p47(F41A)] that reduces the affinity of the UBA domain for ubiquitin (Figure 3C) had almost the same effect. These data strongly suggest that ubiquitin recognition by the UBA domain is crucial for the function of p47 in mediating Golgi membrane fusion.
Fig. 4. The p47 UBA domain is required for p97–p47-mediated reassembly of cisternal membranes from MGFs. Rat liver Golgi membranes were fragmented by treatment with mitotic HeLa cell lysate, and MGFs were isolated. After incubation with purified p97 alone or in combination with p47(wt), p47(ΔUBA) or p47(F41A), the samples were processed for electron microscopy and the percentage of Golgi membranes in cisternae determined. Results are presented as the mean percentage of cisternal regrowth (± SEM), where 0% represents incubations with buffer alone (38 ± 3% of membrane in cisternae) and 100% represents incubations with p97–p47(wt) (66 ± 2% of membrane in cisternae). Note that both deletion and mutation of the UBA domain reduced the reassembly activity of p97–p47 by 75–80%.
A zinc finger motif in Npl4 represents a novel ubiquitin-binding domain
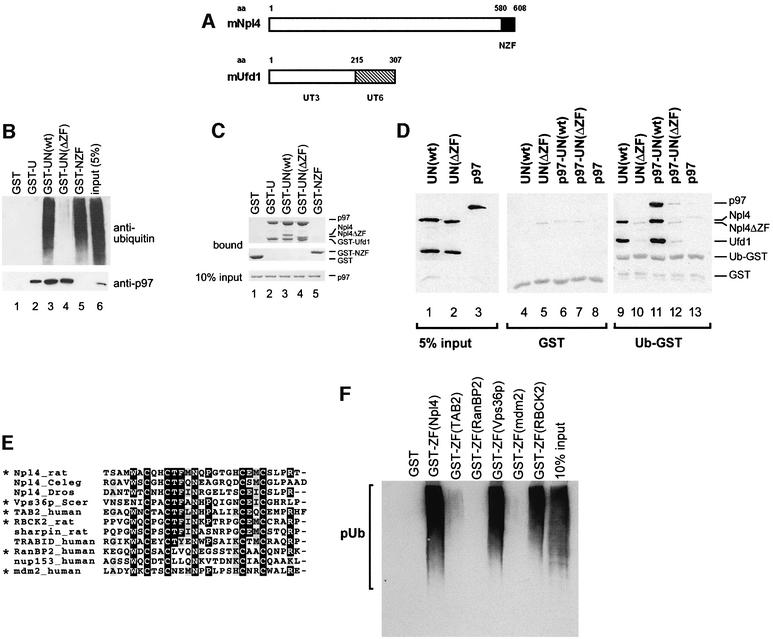
We conducted a series of binding experiments with fragments of Ufd1 and Npl4 to identify domains that were crucial for ubiquitin binding. We narrowed down the region to the C-terminus of Npl4 which contained a putative zinc finger domain (NZF, Figure 5A), and an experiment characterizing this region is shown in Figure 5B. GST–Ufd1 was immobilized on beads and either used alone or saturated with Npl4(wt) or Npl4 with a deletion of the zinc finger [Npl4(ΔZF)]. The zinc finger alone, fused to GST (GST–NZF), was also immobilized. The beads were then incubated with HeLa cell extract and analyzed for binding of polyubiquitylated proteins. As shown in Figure 5B, deletion of the zinc finger abolished the ability of UN to bind ubiquitylated proteins (compare lanes 3 and 4) which were also bound to the same extent by the zinc finger alone (lane 5). The zinc finger did not mediate the binding of UN to p97, as shown by two experiments. First, western blot analysis of the precipitates in Figure 5B showed that Npl4 stimulated the binding of p97 to Ufd1 (compare lanes 2 and 3) (Meyer et al., 2000) even when the zinc finger was lacking (lane 4). The zinc finger alone did not bind any detectable p97 (lane 5). Secondly, we performed the same binding experiment using pure p97 instead of lysate and obtained essentially the same result (Figure 5C). Npl4 with or without the zinc finger stimulated the binding of p97 to Ufd1 to the same extent (compare lane 2 with lanes 3 and 4), whereas the zinc finger domain alone bound little if any p97 (lane 5).
Fig. 5. The zinc finger of mammalian Npl4 (NZF) represents a novel, conserved ubiquitin-binding domain. (A) Domain structure of rat Ufd1 and Npl4: Npl4 contains a single putative zinc finger motif in the C-terminal 29 amino acids (NZF). Ufd1 contains the N-terminal UT3 domain that putatively folds as a double-ψβ-barrel and a UT6 domain that binds both p97 and Npl4. (B) GST–Ufd1 (GST-U) was immobilized on glutathione–beads and saturated with either wild-type Npl4 [N(wt)] or Npl4 with a deletion of the zinc finger [N(ΔZF)]. A fusion protein comprising the Npl4 zinc finger alone (GST–NZF) was also immobilized on glutathione–beads. The beads were then incubated with a detergent lysate from HeLa cells. GST served as a control. Bound proteins were analyzed by western blotting for the presence of either polyubiquitin (upper panel) or p97 (lower panel). Note that the NZF was both necessary and sufficient to mediate binding to polyubiquitylated proteins (pUb). (C) Beads prepared as in (B) were incubated with pure p97 and bound fractions were analyzed by SDS–PAGE followed by Coomassie Blue staining. Ten percent of the input p97 used is shown in the lower panel. Note that the NZF domain does not participate in p97 binding to UN. (D) GST or Ub–GST was immobilized on glutathione–beads and incubated with different combinations of Ufd1–Npl4 [UN(wt)], Ufd1–Npl4ΔZF [UN(ΔZF)] and p97 as indicated. Complexes were isolated and analyzed by western blotting using antibodies specific to the proteins indicated on the right. Note that deletion of the zinc finger abolished binding to Ub–GST. (E) CLUSTAL_W alignment of related zinc finger peptide sequences of different proteins indicated on the left. The rat sequence is identical to that for mouse and humans and is probably representative of all mammals. (F) Peptides indicated with an asterisk in (E) were expressed as fusions with GST and incubated with a HeLa cell lysate. Their ability to bind polyubiquitylated proteins was tested by western blot analysis. See Materials and methods for details.
To eliminate the possibility that ubiquitin binding by the NZF domain was mediated indirectly via an unknown factor in the lysate (Figure 5B), we performed experiments using pure Ub–GST as a model substrate (Figure 5D). Once again, the absence of the zinc finger prevented binding of UN to Ub–GST (lane 10), which in turn prevented recruitment of p97 to this mono-ubiquitylated substrate (lane 12). Together, these data strongly suggest that the zinc finger domain of Npl4 is both necessary and sufficient for ubiquitin binding.
Homology searches for similar zinc fingers led us to speculate that it might have the same function as putative zinc finger motifs in the nuclear pore proteins, RanBP2 and nup153, which bind RanGDP via this domain (Nakielny et al., 1999; Yaseen and Blobel, 1999). Database searches identified more proteins with similar motifs, and some of these sequences are aligned in Figure 5E. To test the functional relevance of this homology, we expressed the indicated peptides of Npl4, TAK1-binding proteins (TAB2), RanBP2, Vps36p, mdm2 and RBCK2 as GST fusion proteins and performed binding experiments to polyubiquitylated proteins (Figure 5F). Intriguingly, two of the peptides with highest homology to Npl4 (Vps36p and RBCK2) bound polyubiquitin as well as Npl4, whereas more distantly related peptides (RanBP2 and mdm2) did not bind. TAB2 bound weakly, though consistently. Based on these findings, we conclude that the NZF motif is not functionally related to the RanBP2 zinc finger, but represents a novel ubiquitin-binding domain conserved in other proteins.
The p47 UBA domain, but not the Npl4 NZF domain, can inhibit polyubiquitylation of Ub–GST
The previous experiments suggested that p97–p47 binds to mono-ubiquitylated proteins. Mono-ubiquitylation has been shown to regulate the function of some substrates rather than trigger degradation (Hicke, 2001). It has also been shown that the UBA domain-containing protein Rad23 can inhibit further ubiquitylation, thereby possibly preventing degradation of its substrate by the proteasome (Ortolan et al., 2000). We wanted to test this ability for the p47-UBA and Npl4-NZF domains and took advantage of the fact that Ub–GST is modified in vivo and in vitro by multi-ubiquitin chains that are attached to specific lysine residues within the ubiquitin moiety of the fusion protein (Johnson et al., 1995; Koegl et al., 1999). Polyubiquityl ation also occurs when Ub–GST is expressed in a reticulocyte lysate (Figure 6, lane 1). We confirmed that the bands of higher molecular weight were indeed ubiquitylated forms of Ub–GST by addition of methylated ubiquitin (me-Ub) that cannot in turn be ubiquitylated, and so reduces polyubiquitylation (lane 2). Addition of the UBA peptide significantly inhibited the polyubiquitylation of Ub–GST (lane 3), whereas the mutant UBA(F41A), that has a lower affinity for Ub–GST (see Figure 3B), did not (lane 4). The NZF domain, either alone or fused to GST, had no effect (lanes 5 and 6), even when the concentration was raised to three times that of the UBA peptide (data not shown). These data suggest that the two domains have different modes of interaction with this ubiquitin substrate such that the UBA domain blocks further ubiquitylation, whereas the NZF still allows it.
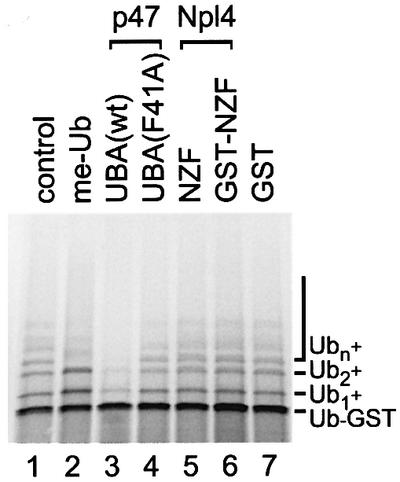
Fig. 6. The UBA domain of p47, but not the zinc finger domain of Npl4, inhibits polyubiquitylation of Ub–GST in vitro. Ub–GST was expressed in a rabbit reticulocyte lysate in the absence or presence of methylated ubiquitin (me-Ub, 20 µM) or 35 µM of the indicated peptides: p47-UBA(wt), p47-UBA(F41A), Npl4-NZF peptide, a GST–NZF fusion or GST. Samples were fractionated using SDS–PAGE and visualized using a phosphoimager. The radiolabeled Ub–GST fusion protein was polyubiquitylated under control conditions (Ub1–n) by factors present in the lysate. Note that p47-UBA(wt) significantly inhibited polyubiquitylation.
Discussion
Many lines of evidence link ubiquitin with p97/Cdc48p-mediated processes, though the exact role of ubiquitin in linking substrate recognition to the unfolding activity of p97/Cdc48p is still controversial. It is also not clear whether ubiquitin is required for all p97-mediated processes. In this report, we have focused on two p97 adaptors, UN and p47, and provide direct, biochemical evidence that both mediate the binding of p97 to ubiquitin conjugates, albeit in different ways.
p47 binds to ubiquitin via the UBA domain at its N-terminus. The UBA motif was discovered originally as a motif present in a variety of proteins (Hofmann and Bucher, 1996). It was found to bind ubiquitin in Rad23 and other proteins (Bertolaet et al., 2001; Wilkinson et al., 2001), although it is also involved in other protein–protein interactions (Withers-Ward et al., 1997; Suyama et al., 2000). We show that the UBA domain of p47 is both necessary and sufficient to bind ubiquitin. The full-length p47, however, can bind ubiquitin only when present in a complex with p97, which suggests that binding to p97 activates the ubiquitin-binding site in p47. The p97–p47 complex has high specificity for the model mono-ubiquitylated substrate, Ub–GST, and binds polyubiquitin from cell lysates very inefficiently when compared with p97–UN. We cannot exclude the possibility that the small amount of polyubiquitin bound to p47 in Figure 1 represents a specific subclass of conjugates since the specificity of UBA-containing proteins for polyubiquitin complexes of different size and linkages is still under debate (Buchberger, 2002).
The UBA domain of p47 is also essential for its function in reassembling Golgi cisternae from MGFs. Both deletion of the UBA domain and a point mutation that decreases its affinity for ubiquitin reduced its activity by 75–80%, arguing that ubiquitin recognition by p97–p47 plays a crucial role in Golgi reassembly.
The ubiquitin-binding site in mammalian UN, on the other hand, comprises a novel zinc finger motif at the C-terminus of Npl4 that we term the NZF domain. The NZF is a conserved motif present in Npl4 proteins of higher eukaryotes and also found in other proteins. We show here that homologous peptide sequences of Vps36p, RBCK2 and, to some extent, TAB2 can bind ubiquitin. We speculate that the NZF motif plays an important role in the function of these proteins. Vps36 was identified in a screen for vacuolar protein sorting mutants and is likely to be involved in the sorting of proteins into multivesicular bodies, a process that has been shown to be ubiquitin dependent (Katzmann et al., 2001). RBCK2 is also known to be a ubiquitin conjugase-7-interacting protein, and TAB2 is involved in the activation of IκB kinase (Wang et al., 2001), a process dependent on the assembly of ubiquitin chains with Lys63 linkages. It will be interesting to determine whether the polyubiquitin conjugates bound by TAB2 (Figure 5D) comprise chains with Lys63 linkages.
Ubiquitin binding by mammalian UN is regulated differently to that of p47 in that it can bind mono- and polyubiquitylated proteins and can do so independently of p97, which can then be recruited to the conjugate. To date, we have been able to detect little if any direct binding of ubiquitin conjugates to p97 alone, in contrast to previous reports (Dai and Li, 2001; Rape et al., 2001). This may reflect the different substrates and different systems that were used. p97 binding to ubiquitin conjugates in cell extracts could always be mediated by adaptors in the extract, so model substrates have to be used to demonstrate direct binding. The problem then is the physiological relevance of the model substrates that are used. Further work is clearly needed before any general conclusions can be drawn. For the moment, what we can say is that the two adaptors we have studied dramatically enhance the ability of p97 to bind to ubiquitin conjugates.
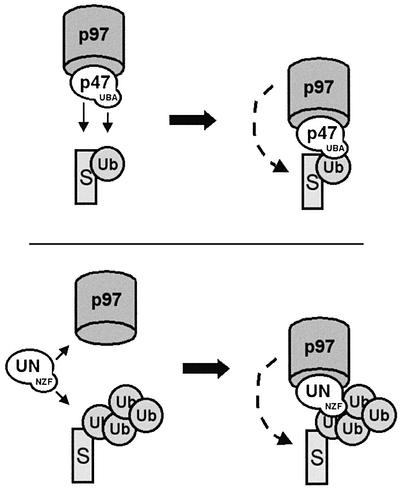
Our findings have important implications for the role of these p97 adaptors. One possible view would be that both help to recruit p97 to ubiquitylated substrates, however in differently regulated manner (Figure 7). In the case of p97–p47, our results are consistent with a role for mono-ubiquitylation which has been found to be important for the regulation of protein function rather than degradation (Hicke, 2001). We show that p97–p47 binds Ub–GST more efficiently than polyubiquitylated proteins when compared with UN. Furthermore, the p47-UBA domain can inhibit further ubiquitylation of Ub–GST. The same mechanism has been shown for the UBA domain protein Rad23 (Ortolan et al., 2000), a protein involved in DNA repair. It is important, however, to remember that Ub–GST represents a particular type of degradation substrate (Johnson et al., 1995), and until more work is done it will not be possible to generalize these results to physiological mono-ubiquitylated substrates.
Fig. 7. Hypothetical model for the role played by p97 adaptors in ubiquitin-mediated processes. p47 bound to p97 would bind to substrates both directly and indirectly via mono-ubiquitin (using the UBA domain). Binding would prevent further ubiquitylation. UN would bind mono- or polyubiquitylated substrates via the zinc finger on Npl4 and permit further ubiquitylation. UN would either bind as a complex with p97 or would recruit p97 to the substrate (S).
Ubiquitin binding by p47 was somewhat surprising since p47 has also been shown to bind recombinant syntaxin 5 (Rabouille et al., 1998), which should lack bound ubiquitin. This suggests that p47 has two specificities, for syntaxin 5 and ubiquitin. An attractive, but highly speculative, model would be that during the course of the p97–p47-mediated membrane fusion, SNAREs are ubiquitylated and then serve as substrates for p97–p47. This would implicate ubiquitin as an unfolding signal. Interestingly, a similar mechanism has been proposed for Tsg101, a protein that recruits retroviral gag proteins into membrane buds in a ubiquitin-dependent manner. Like p47, Tsg101 can bind both ubiquitin and its substrate, the gag protein, independently. It binds the conjugate, however, with a much higher affinity (Garrus et al., 2001).
A similar model can be proposed for UN (Figure 7), with the added flexibility that it can bind to both mono- and polyubiquitylated substrates. Thus it could bind to mono-ubiquitylated substrates during nuclear reassembly, catalyzing fusion reactions that differ subtly from those catalyzed by p97–p47 (Burke, 2001). Alternatively, it could bind to polyubiquitylated substrates such as those that need to be removed from membranes during ERAD or RUP. However, this simple model can only hold true for higher eukaryotes, since yeast Npl4 does not have the NZF motif. Such differences between yeast and higher eukaryotes are not uncommon. We have shown that yeast Vps36p contains the NZF domain but it is not present in the human homolog CGI-145/EAP45. Furthermore, two orthologs in the endocytic pathway, yeast Ede1p and human Eps15, have non-homologous ubiquitin-binding domains (Shih et al., 2002), indicating that recombination of these domains between different proteins could have occurred during evolution.
In conclusion, our data clearly show that both p47 and UN mediate complex formation of p97 with ubiquitin conjugates that are likely to be crucial intermediates in p97–UN- and p97–p47-mediated reactions. They point to ubiquitylation and ubiquitin recognition as a common theme in p97-mediated processes. Present efforts are now focused on identifying the ubiquitylated substrates.
Materials and methods
Expression and purification of proteins
All proteins used were expressed in Escherichia coli and purified to homogeneity: p97 (Meyer et al., 2000), GST–Ufd1 and Ufd1–Npl4 (Hetzer et al., 2001) were generated as described. Npl4(ΔZF) was expressed with a deletion of amino acids 580–608 and purified as for wild-type Npl4. p47(wt) and mutants were all expressed as His-tagged proteins using pTrcHis-p47(wt) (Kondo et al., 1997) or derived constructs: p47(ΔUBA) contains amino acids 46–370, p47-UBA contains amino acids 1–45, and p47-UBA(F41A) represents p47-UBA with an alanine at position 41. p47(wt) and mutants were purified from bacterial lysates using Ni–agarose followed by gel filtration. DNA encoding amino acids 319–363 of hhRAD23 was cloned into pTrcHisA (Invitrogen) to generate UBA(hhRAD23). Ub–GST was expressed using the pET-Ub-V-GST construct and codes for mouse ubiquitin(G76V) followed by a lacI spacer sequence described in Johnson et al. (1995), fused to the N-terminus of GST. DNAs encoding the following peptide sequences were cloned into pGEX-4T (Amersham Pharmacia) to express them as GST fusions: rat Npl4, amino acids 580–608; human TAB2, amino acids 664–693; human RanBP2, amino acids 1479–1507; Saccharomyces cerevisiae Vps36p, amino acids 177–205; human mdm2 protein, amino acids 299–327; and rat RBCK2, amino acids 181–209. The NZF peptide was generated by thrombin cleavage of GST–NZF and subsequent purification.
Binding assays
All binding experiments were carried out at 4°C in 0.2 ml of binding buffer (150 mM KCl, 50 mM HEPES pH 7.3, 2.5 mM MgCl2, 5% glycerol, 2 mM β-mercaptoethanol, 0.1% Triton X-100, 1 mg/ml bovine serum albumin). If not otherwise stated, 2 µg of GST or Ub–GST were immobilized on 5 µl of glutathione–Sepharose (Amersham Pharmacia), washed and incubated with 5 µg of p97 and/or 2 µg of p47, UN or mutants thereof for 1 h. If streptavidin beads were used, proteins were biotinylated and immobilized as described (Meyer et al., 2000). When nucleotide was used, it was added at 2 mM to the binding and washing buffers. The matrix was then washed and bound protein was subjected to western blot analysis. Specific antibodies were used as described (Meyer et al., 2000). For studies of binding to polyubiquitylated proteins, HeLa cells were grown exponentially, treated with 50 µM MG132 (Calbiochem) for 30 min and suspended in lysis buffer (150 mM KCl, 50 mM HEPES pH 7.3, 2.5 mM MgCl2, 5% glycerol, 0.5% Triton X-100, 2.5 mM NEM) for 30 min. Lysates were cleared for 15 min at 10 000 g and NEM was quenched with 5 mM dithiothreitol (DTT). A 100 µg aliquot of lysate protein typically was incubated with 2 µg of immobilized protein. Ubiquitin conjugates were detected by western blot using an anti-ubiquitin antibody (Sigma U5379).
Ubiquitylation assay
Ub–GST was expressed and radiolabeled with [35S]methionine for 45 min in rabbit reticulocyte lysate using the Promega TNT Kit according to the manufacturer’s instructions. pET-Ub-V-GST (see above) was used as a DNA template. Peptide inhibitors were present as indicated from the start of the reaction. Unmodified and ubiquitylated Ub–GST was analyzed by SDS–PAGE and detected with a phosphoimager (Molecular Dynamics). Methylated ubiquitin was purchased from Calbiochem.
Golgi reassembly assay
The Golgi reassembly assay was performed as described previously (Rabouille et al., 1995). Endogenous NSF and p97 were inactivated by treatment with 2.5 mM NEM for 15 min, followed by quenching with 5 mM DTT, and the mitotic membranes then reisolated. After incubation for 60 min at 37°C with pre-formed complexes of rat liver p97 (100 ng/µl) and recombinant wild-type p47 or mutants (25 ng/µl), samples were fixed, processed for electron microscopy and cisternal regrowth quantitated as described in Rabouille et al. (1995).
Acknowledgments
Acknowledgements
We thank Lelia Delamarre, Cynthia He, Al Price and James Shorter for critical reading. This work was supported by grants from the Human Frontiers Science Program, the NIH and the Ludwig Institute for Cancer Research.
References
- Acharya U., Jacobs,R., Peters,J.-U., Watson,N., Farquhar,M.G. and Malhotra,V. (1995) The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell, 82, 895–904. [DOI] [PubMed] [Google Scholar]
- Bays N.W. and Hampton,R.Y. (2002) Cdc48–ufd1–npl4: stuck in the middle with ub. Curr. Biol., 12, R366–R371. [DOI] [PubMed] [Google Scholar]
- Bays N.W., Wilhovsky,S.K., Goradia,A., Hodgkiss-Harlow,K. and Hampton,R.Y. (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell, 12, 4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolaet B.L., Clarke,D.J., Wolff,M., Watson,M.H., Henze,M., Divita,G. and Reed,S.I. (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol., 8, 417–422. [DOI] [PubMed] [Google Scholar]
- Braun S., Matuschewski,K., Rape,M., Thoms,S. and Jentsch,S. (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J., 21, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A. (2002) From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol., 12, 216–221. [DOI] [PubMed] [Google Scholar]
- Burke B. (2001) The nuclear envelope: filling in gaps. Nat. Cell Biol., 3, 273–274. [DOI] [PubMed] [Google Scholar]
- Dai R.M. and Li,C.C. (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin–proteasome degradation. Nat. Cell Biol., 3, 740–744. [DOI] [PubMed] [Google Scholar]
- Garrus J.E. et al. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell, 107, 55–65. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Dohmen,R., Levy,F. and Varshavsky,A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J., 15, 4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Golbik R., Lupas,A., Koretke,K., Baumeister,W. and Peters,J. (1999) The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol. Chem., 380, 1049–1062. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Meyer,H.H., Walther,T.C., Bilbao-Cortes,D., Warren,G. and Mattaj,I.W. (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol., 3, 1086–1091. [DOI] [PubMed] [Google Scholar]
- Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol., 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Hitchcock A.L., Krebber,H., Frietze,S., Lin,A., Latterich,M. and Silver,P.A. (2001) The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell, 12, 3226–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. and Bucher,P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Jarosch E., Taxis,C., Volkwein,C., Bordallo,J., Finley,D., Wolf,D.H. and Sommer,T. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol., 4, 134–139. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Ma,P.C., Ota,I.M. and Varshavsky,A. (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem., 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst,M. and Emr,S.D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell, 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe,T., Schlenker,S., Ulrich,H., Mayer,T. and Jentsch,S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Kondo H., Rabouille,C., Newman,R., Levine,T.P., Pappin,D., Freemont,P. and Warren,G. (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature, 388, 75–78. [DOI] [PubMed] [Google Scholar]
- Latterich M., Fröhlich,K.-U. and Schekman,R. (1995) Membrane fusion and the cell cycle: cdc48p participates in the fusion of ER membranes. Cell, 82, 885–893. [DOI] [PubMed] [Google Scholar]
- Madeo F., Frohlich,E., Ligr,M., Grey,M., Sigrist,S.J., Wolf,D.H. and Frohlich,K.U. (1999) Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol., 145, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.H., Shorter,J.G., Seemann,J., Pappin,D. and Warren,G. (2000) A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J., 19, 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D., Stewart,S., Osmond,B. and Botstein,D. (1982) Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties and pseudoreversion studies. Genetics, 100, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T. and Wilkinson,A.J. (2001) AAA+ superfamily ATPases: common structure—diverse function. Genes Cells, 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L., Schauber,C. and Madura,K. (2000) The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- Patel S. and Latterich,M. (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol., 8, 65–71. [PubMed] [Google Scholar]
- Rabinovich E., Kerem,A., Frohlich,K.U., Diamant,N. and Bar-Nun,S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol., 22, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Levine,T.P., Peters,J.M. and Warren,G. (1995) An NSF-like ATPase, p97 and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell, 82, 905–914. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Kondo,H., Newman,R., Hui,N., Freemont,P. and Warren,G. (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell, 92, 603–610. [DOI] [PubMed] [Google Scholar]
- Rape M., Hoppe,T., Gorr,I., Kalocay,M., Richly,H. and Jentsch,S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell, 107, 667–677. [DOI] [PubMed] [Google Scholar]
- Roy L. et al. (2000) Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol. Biol. Cell, 11, 2529–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Verdel,A., Curtet,S., Lemercier,C., Garin,J., Rousseaux,S. and Khochbin,S. (2001) Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol., 21, 8035–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.C., Katzmann,D.J., Schnell,J.D., Sutanto,M., Emr,S.D. and Hicke,L. (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol., 4, 389–393. [DOI] [PubMed] [Google Scholar]
- Shirogane T., Fukada,T., Muller,J.M., Shima,D.T., Hibi,M. and Hirano,T. (1999) Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity, 11, 709–719. [DOI] [PubMed] [Google Scholar]
- Söllner T., Bennett,M.K., Whiteheart,S.W., Scheller,R.H. and Rothman,J.E. (1993) A protein assembly–disassembly pathway in-vitro that may correspond to sequential steps of synaptic vesicle docking, activation and fusion. Cell, 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Suyama M., Doerks,T., Braun,I.C., Sattler,M., Izaurralde,E. and Bork,P. (2000) Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO rep., 1, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Ye,Y. and Rapoport,T.A. (2002) Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol., 3, 246–255. [DOI] [PubMed] [Google Scholar]
- Wang C., Deng,L., Hong,M., Akkaraju,G.R., Inoue,J. and Chen,Z.J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature, 412, 346–351. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Seeger,M., Hartmann-Petersen,R., Stone,M., Wallace,M., Semple,C. and Gordon,C. (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol., 3, 939–943. [DOI] [PubMed] [Google Scholar]
- Withers-Ward E.S. et al. (1997) Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J. Virol., 71, 9732–9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen N.R. and Blobel,G. (1999) Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc. Natl Acad. Sci. USA, 96, 5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer,H.H. and Rapoport,T.A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature, 414, 652–656. [DOI] [PubMed] [Google Scholar]
- Yuan X., Shaw,A., Zhang,X., Kondo,H., Lally,J., Freemont,P.S. and Matthews,S. (2001) Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J. Mol. Biol., 311, 255–263. [DOI] [PubMed] [Google Scholar]