Abstract
Although spliceosomal Sm proteins can assemble spontaneously onto UsnRNA in vitro, this process requires assisting factors in vivo. SMN, the protein involved in spinal muscular atrophy, is part of a complex that contains the Sm proteins and serves as a critical factor for this reaction. Here, we have reconstituted the SMN-dependent assembly of UsnRNPs in vitro. We demonstrate that the SMN complex is necessary and sufficient for the assembly reaction. The PRMT5 complex, previously implicated in methylation and storage of Sm proteins, interacts with the SMN complex and enhances its activity in an ATP-dependent manner. These data uncover the SMN–PRMT5 complex as a functional entity that promotes the assisted assembly of spliceosomal UsnRNPs, and potentially other, RNA–protein complexes.
Keywords: PRMT5/RNP assembly/SMN/spliceosomes
Introduction
Proximal spinal muscular atrophy (SMA) is a recessive neuromuscular disorder characterized by progressive loss of alpha motor neurons in the spinal cord (Schmalbruch and Haase, 2001; Sendtner, 2001). This monogenic disease is caused by mutations in the survival motor neuron (SMN) gene, which is present in two copies on chromosome 5 (termed SMN1 and SMN2). SMA-causing mutations are always found in SMN1 and prevent the expression of functional protein from this locus. Although SMA patients still express protein encoded by the SMN2 locus, the low level of functional SMN does not compensate for the mutations in SMN1. Thus, reduced expression, rather than the complete absence of SMN, leads to the disease phenotype (Lefebvre et al., 1995, 1997; Monani et al., 2000). In contrast to humans, most other eukaryotic organisms analyzed thus far harbor only one copy of the SMN gene that is essential for viability (Sendtner, 2001).
The ubiquitously expressed SMN protein localizes in the cytoplasm as well as in the nucleus, where it is highly concentrated in subnuclear domains termed Cajal bodies/Gems (Liu and Dreyfuss, 1996). SMN forms macromolecular complexes (‘SMN complexes’) in vivo, which contain Gemin2 (formerly SIP1), the putative ATPase/RNA helicase dp103 (also termed Gemin3), Gemin4 (also termed GIP), p175 (also termed Gemin5), hsc70, Gemin6 and unrip (Liu et al., 1997; Charroux et al., 1999, 2000; Grundhoff et al., 1999; Campbell et al., 2000; Meister et al., 2000, 2001b; Pellizzoni et al., 2002; for a comprehensive overview, see Meister et al., 2002). As these proteins associate tightly with SMN in the nucleus and the cytoplasm, they are believed to be integral components of all SMN complexes. In contrast, other components appear to associate transiently with SMN or may be part of a subpopulation of cellular SMN complexes. These include spliceosomal proteins and UsnRNAs (Fischer et al., 1997), a specific subset of hnRNP proteins, termed hnRNP-R and Q (Mourelatos et al., 2001; Rossoll et al., 2002), the small nucleolar (sno)RNP-associated proteins fibrillarin and GAR1 (Jones et al., 2001; Pellizzoni et al., 2001a) and the polymerase II-associated RNA helicase A (Pellizzoni et al., 2001b).
Studies on the function of SMN and its complexes revealed an unexpected link to the assembly of the small nuclear ribonucleoproteins (snRNPs) U1, U2, U4/U6 and U5, i.e. essential RNA–protein complexes of the spliceosome. In the assembly process of these particles, the m7G-capped snRNAs U1, U2, U4 and U5 are transiently exported from the nucleus. In the cytoplasm, the seven Sm proteins B/B′, D1, D2, D3, E, F and G bind in an apparently ordered process to the Sm site of these snRNAs to form a ring-shaped Sm core domain. The proper assembly of the Sm core domain is a pre-requisite for the subsequent m3G cap formation of the UsnRNA and import of the assembled particle into the nucleus. At an as yet to be defined step, additional factors are recruited to form the mature UsnRNP particles that function in splicing (for a recent review, see Will and Luhrmann, 2001). Among the spliceosomal UsnRNPs, only U6 does not bind to canonical Sm proteins. Instead, this particle contains the seven closely related like Sm (LSm) proteins 2–8, which form a heptameric ring on U6 snRNA that is structurally very similar to the Sm core domain. The biogenesis of the U6 snRNP is likely to occur in the nucleus (Hamm and Mattaj, 1989; Will and Luhrmann, 2001).
Although isolated Sm proteins bind spontaneously to UsnRNA in vitro (Raker et al., 1996, 1999), recent data revealed that assembly of the Sm core domain is an ATP-dependent process that requires assisting factors in vivo. In particular, studies in Xenopus laevis oocytes and in a cell-free assembly assay showed that SMN, as part of a complex containing all Sm proteins, is a critical factor in this reaction (Fischer et al., 1997; Buhler et al., 1999; Meister et al., 2001a). SMN binds directly to Sm proteins, an interaction that is strongly enhanced by modification of arginines in Sm proteins B/B′, D1 and D3 to symmetric dimethylarginines (sDMAs) (Buhler et al., 1999; Brahms et al., 2001; Friesen et al., 2001a). Interestingly, the blocking of this interaction by specific antibodies leads to an inhibition of UsnRNP assembly, and several disease-causing mutations in SMN prevent binding to Sm proteins (Buhler et al., 1999; Pellizzoni et al., 1999). It has therefore been suggested that the regulated association of Sm proteins with the SMN complex is a crucial step in the assembly of UsnRNPs. Whether the Sm proteins bound to the SMN complex are transferred onto the UsnRNA and hence serve as substrates for the assembly reaction, or whether they serve other functions in the context of this complex, is currently unclear.
Recent data suggest that not only the SMN complex, but also the methyltransferase complex that modifies Sm proteins, may be involved in the biogenesis of UsnRNPs. This complex, termed PRMT5 complex or methylosome, contains the type II (i.e. sDMA-generating) methyltransferase PRMT5, the novel WD-repeat protein WD45 and pICln, a putative regulator of UsnRNP assembly (Pu et al., 1999; Friesen et al., 2001b; Meister et al., 2001b). In X.laevis oocytes, most Sm proteins found in the cytoplasm are bound to the PRMT5 complex, and cannot be transferred directly onto the UsnRNA (Pu et al., 1999; Meister et al., 2001b). These observations suggested that the PRMT5 complex affects assembly at an early stage, possibly by regulating the transfer of Sm proteins onto UsnRNPs. However, a direct link between the PRMT5 complex and the formation of the Sm core domain remains to be demonstrated.
To gain insight into the mechanism by which SMN and PRMT5 complexes facilitate the biogenesis of UsnRNPs, we have reconstituted the SMN-dependent formation of the Sm core domain in vitro. We show that a purified SMN complex directly transfers Sm proteins onto the UsnRNA and hence is necessary and sufficient to promote the assembly of the Sm core domain. Furthermore, evidence is provided that the PRMT5 complex directly interacts with the SMN complex and stimulates its activity in an ATP-dependent manner. These data show that the SMN– PRMT5 complex functions as the assembly unit that promotes the active formation of spliceosomal UsnRNPs.
Results
SMN-dependent assembly of UsnRNPs in HeLa cytosolic extracts
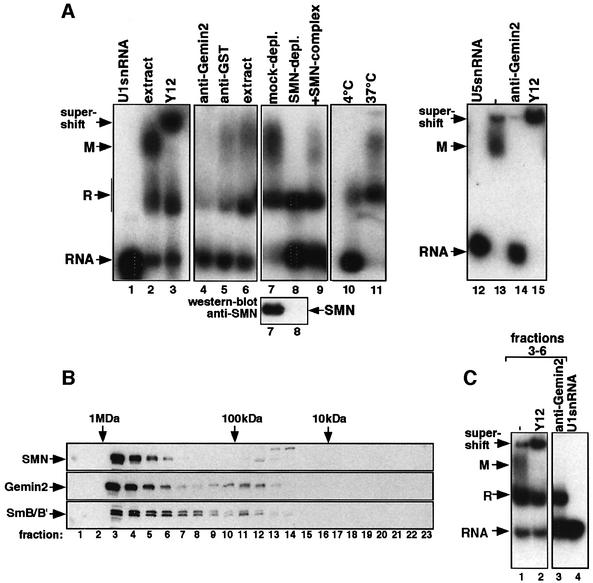
The SMN-dependent assembly of spliceosomal UsnRNPs has been reproduced recently in X.laevis egg extract (Meister et al., 2001a). As this extract is derived from a highly specialized cell type and allows only limited biochemical fractionation studies, we asked whether the assembly reaction could also be analyzed in a somatic cell extract. To test this, cytosolic extract was prepared from HeLa cells, incubated with 32P-labeled U1 snRNA and subsequently analyzed by native gel electrophoresis. U1 snRNA was incorporated efficiently into two major complexes designated R and M (Figure 1A, lane 2). Complex R contains the U1-specific A protein (U1-A) bound to U1 snRNA as this complex is also formed when isolated U1-A is incubated with U1 snRNA but not on a mutant lacking the binding site for this protein (data not shown). In contrast, complex M contained Sm proteins (and U1-A), as evident by the supershift of this complex by the anti-Sm antibody Y12 and the failure to form on a mutant U1 snRNA lacking the Sm-binding site (U1ΔSm) (Figure 1A, lane 3 and Supplementary figure 1 available at The EMBO Journal Online). Thus, the complexes formed in the HeLa cytosolic extract are similar in size and composition to those appearing in X.laevis egg extract. However, a complex termed R2 that is formed in the egg extract and contains only the Sm core domain could not be observed in the HeLa system (Meister et al., 2001a).
Fig. 1. In vitro reconstitution of UsnRNPs in HeLa cytosolic extract. (A) 32P-labeled U1 snRNA (lanes 2–11) or U5 snRNA (lanes 12–15) were incubated with HeLa cytosolic extract either at 4 (lane 10) or 37°C (all other lanes). The indicated antibodies were added either after (lanes 3 and 15) or prior to assembly (lanes 4, 5 and 14). Reactions shown in lanes 7–9 were carried out in extract that was either mock depleted, SMN immunodepleted or SMN immunodepleted and subsequently incubated with purified SMN complex. The western blot below indicates the level of SMN in both extracts. Lanes 1 and 12 show snRNAs U1 and U5 in the absence of extract. The assembly reactions were separated by native gel electrophoresis and complexes visualized by autoradiography. Arrows indicate the positions of complexes R and M as well as of the Y12 supershift. (B) Fractionation of HeLa cytosolic extract active in UsnRNP assembly on a Superose-6 gel filtration column. SMN, Gemin2 and B/B′ were detected in the individual fractions by western blotting. The molecular masses of marker proteins are indicated on the top of the western blots, and fraction numbers on the bottom. (C) Fractions 3–6 were pooled and incubated with 32P-labeled U1 snRNA (lane 1) in the presence of either Y12 (added after assembly, lane 2) or Gemin2 (added prior to assembly, lane 3). Lane 4 shows the free U1 snRNA. Samples were analyzed as in (A).
Several lines of evidence indicate that binding of Sm proteins to UsnRNAs in HeLa cytosol is dependent on the SMN complex. First, complex M was not formed in an extract that was incubated at 4°C (Figure 1A, lane 10). Previous studies in egg extracts had shown that the SMN-dependent assembly reaction was blocked at low temperature in X.laevis egg extracts (Meister et al., 2001a). Secondly, anti-Gemin2 antibodies, but not a non-related antibody (anti-GST antibody), completely inhibited binding of Sm proteins to the U1 snRNA (Figure 1A, lanes 4–6). Thirdly, and most importantly, cytosol immunodepleted of SMN failed to assemble Sm proteins onto the U1 snRNA, whereas re-addition of affinity-purified SMN complex reconstituted the assembly reaction (Figure 1A, lanes 7–9). Furthermore, the cytosolic extract used for these studies also promoted assembly of U2, U4 and U5 snRNPs in a similar manner (see U5 snRNP assembly in Figure 1A, lanes 12–15, and data not shown). Together, these data show that the SMN-dependent assembly of UsnRNPs can be reproduced in a cell-free assay based on extract from HeLa cells.
In vitro reconstitution of UsnRNP assembly from purified components
Previous studies performed in X.laevis egg extracts had shown that a macromolecular SMN complex mediates an essential step in the formation of the Sm core domain (Meister et al., 2001a). To identify the minimal set of factors that are sufficient for the assembly reaction, cytosolic extract from HeLa cells was initially fractionated by gel filtration. Figure 1B shows that the majority of SMN, Gemin2 and B/B′ co-migrate in a single peak corresponding to a mass of ∼0.9 MDa (lanes 3–6). Intriguingly, the SMN-containing fractions (but none of the other fractions; see Supplementary data) generated two complexes on U1 snRNA (Figure 1C, lane 1) whose identity as complexes R and M was corroborated by supershift analysis with anti-Sm antibody Y12 and inhibition by anti-Gemin2 antibody (Figure 1C, lanes 2 and 3). To exclude the possibility that the lack of assembly activity in the SMN-lacking fractions was due solely to the absense of significant amounts of Sm proteins, the Sm protein content in all fractions was normalized. As shown in Supplementary figure 2B, only the SMN-containing fractions promoted assembly under these conditions. Together, these results suggest that a high molecular weight fraction containing SMN is necessary and sufficient to facilitate the formation of the Sm core domain on U1 snRNA.
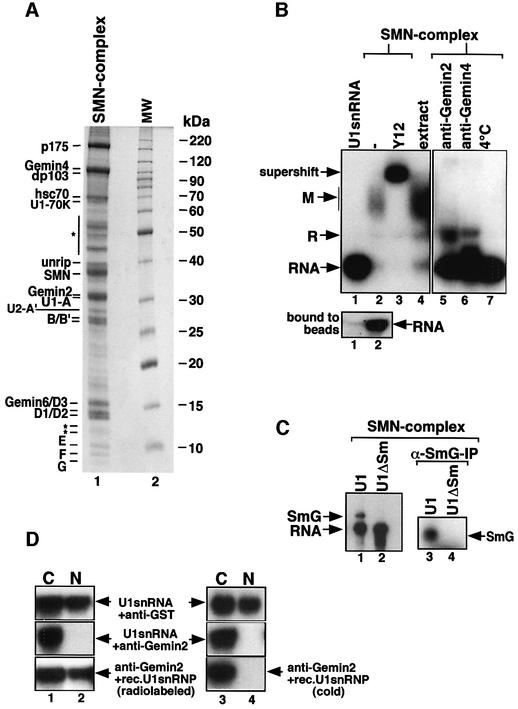
SMN was purified from these fractions by anti-SMN affinity chromatography and co-purifying proteins were identified by MALDI-TOF. Nineteen prominent proteins co-eluted specifically from the column, most of which had been described previously as components of SMN complexes (Figure 2A, lane 1). These include p175, dp103, Gemin4, hsc70, unrip, SMN, Gemin2 and all Sm proteins (i.e. B/B′, D1, D2, D3, E, F and G). Several additional proteins were identified, including the U1-specific proteins A and 70K, the U2-specific protein A′ and a novel component of 18 kDa (for the peptide sequence, see Supplementary data). This protein recently has also been identified in an independent study and termed Gemin6 (Pellizzoni et al., 2002).
Fig. 2. A purified SMN complex is sufficient to promote UsnRNP assembly. (A) HeLa nuclear extract was passed over an anti-SMN affinity column and proteins were analyzed by SDS–PAGE. Lane 1 shows the affinity-purified SMN complex, and lane 2 a protein size marker. Proteins of the SMN complex indicated on the left were identified by MALDI-TOF and/or western blotting. Bands indicated with an asterisk are either non-specific or have not yet been identified. (B) 32P-labeled U1 snRNA was incubated with Sepharose beads (lane 1) or anti-SMN affinity beads that contain the SMN complex shown in (A) (lanes 2–7). Assembly reactions in the presence of either anti-Gemin2 antibodies, anti-Gemin4 antibodies or at 4°C are shown in lanes 5, 6 and 7, respectively. The beads were pelleted and the supernatants analyzed by native gel electrophoresis as above. Y12 was added to the reaction after assembly had been completed (lane 3). The lower gel shows U1 snRNA binding to the Sepharose beads (lane 1) and to Sepharose beads containing the SMN complex (lane 2). (C) UV cross-linking of UsnRNA and the G-protein. 32P-labeled U1 snRNA (lanes 1 and 3) and U1ΔSm (lanes 2 and 4) were incubated with SMN complex for 1 h. The samples were then irradiated with UV light and either directly separated by SDS–PAGE (lanes 1 and 2) or immunoprecipitated with an anti-G antiserum (lanes 3 and 4). Bands were visualized by autoradiograpy. (D) Nuclear transport of U1 snRNP assembled by the SMN complex. 32P-labeled U1 snRNA (upper and middle panels, lanes 1–4) and U1 snRNP assembled by the SMN complex (lower panel, lanes 1 and 2) were injected into the cytoplasm of X.laevis oocytes. Oocytes were pre-injected with an anti-GST control antibody (upper panels, lanes 1–4) or with anti-Gemin2 antibody (middle and lower panels). As a further control, 32P-labeled U1 snRNA was co-injected together with cold U1 snRNP assembled by the SMN complex and Gemin2 antibodies (lower panel, lanes 3 and 4). After incubation for 12 h, RNA was extracted from the nuclear (N) or cytosolic (C) fractions, separated on a denaturing gel and visualized by autoradiography.
To test whether the identified set of proteins is sufficient to mediate the formation of the Sm core domain, SMN complex bound to the anti-SMN affinity matrix was incubated with 32P-labeled U1 snRNA. After incubation for 1 h, the matrix was pelleted and the supernatant analyzed by native gel electrophoresis. An RNP co-migrating with complex M was formed on U1 snRNA, which contained Sm proteins, as indicated by the supershift with anti-Sm antibody Y12 (Figure 2B, lanes 2 and 3). At this point, we asked whether complex M reflected the functional Sm core domain, or a dead-end product of assembly. Since previous reports stated that the G protein could be cross-linked to the Sm site of U1 snRNA in the context of a functional Sm core domain (Urlaub et al., 2001), we decided to employ this approach as a test criterion. Indeed, after irradiation of in vitro assembled U1 snRNP, a cross-link to the Sm site could be observed (Figure 2C, lanes 1 and 2). This cross-link corresponds to the G-protein bound to U1 snRNA, as it could be immunoprecipitated by an antibody specific for the G-protein (Figure 2C, lanes 3 and 4). As a second criterion for the functional composition of the Sm core domain, we analyzed by microinjection whether complex M formed in our purified system could be imported into the nucleus of X.laevis oocytes. This approach is based on the observation that only UsnRNPs containing properly assembled Sm core domains are imported into this compartment (Raker et al., 1996). Oocytes were pre-injected with large amounts of either a control antibody or an affinity-purified antiserum directed against Gemin2, to block the activity of the SMN complex in vivo. The same oocytes received a second injection of either 32P-labeled U1 snRNA (upper and middle panel) or U1 snRNP assembled by isolated SMN complex (lower panel), and transport was analyzed 12 h later. In agreement with previous studies (Fischer et al., 1997), transport of injected U1 snRNA occurred efficiently in control oocytes, whereas nuclear import was abolished in oocytes pre-injected with anti-Gemin2 antibodies (Figure 2D, upper and middle panels). However, transport inhibition by anti-Gemin2 antibody could be overcome by injecting assembled U1 snRNP generated by the SMN complex (lower panel, lanes 1 and 2). This was not due to the co-injection of isolated SMN complex together with the assembled particle, as U1 snRNA injected together with unlabeled in vitro reconstituted U1 snRNP was still exclusively nuclear (lower panel, lanes 3 and 4). Together, these data strongly suggest that the isolated SMN complex promoted the formation of the functional Sm core domain on U1 snRNA in vitro.
To exclude the possibility that the Sm proteins dissociated from the SMN complex and bound spontaneously to the UsnRNA, assembly was analyzed at low temperature and in the presence of antibodies directed against either Gemin2 or Gemin4. As shown in Figure 2B, in neither case was binding of Sm proteins to the U1 snRNA observed (lanes 5–7). In contrast, spontaneous assembly with purified Sm proteins occurred at 4°C and in the presence of these antibodies (see Supplementary data). Thus, Sm proteins must have been transferred onto the U1 snRNA in conjunction with the activity of the SMN complex. Based on these data, we conclude that the isolated SMN complex alone is sufficient to promote the assembly of the Sm core domain. We note, however, that assembly occurred in an ATP-independent manner and only in the presence of a large excess of purified SMN complex (data not shown). Moreover, only ∼30–50% of the U1 snRNA added to the anti-SMN affinity beads was incorporated into complex M, whereas the rest remained bound to the immobilized SMN complexes (Figure 2B, lower panel, lanes 1 and 2). This suggests that additional stimulatory factors are needed to ensure the efficient and ATP-dependent assembly of the Sm core domain (see below).
Sm protein association with the SMN complex precedes incorporation into UsnRNPs in vivo
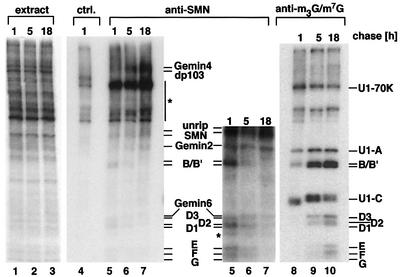
The in vitro studies described above strongly implied that direct transfer of Sm proteins from the SMN complex onto the UsnRNA is an essential step in the assisted assembly of the Sm core domain. To test whether a similar mechanism accounts for the assembly process in vivo, we performed pulse–chase experiments in living HeLa cells. Cells were pulsed for 1.5 h with [35S]methionine and [35S]cysteine to allow in vivo labeling of nascent proteins. After a chase in non-radioactive medium for 1, 5 or 18 h, extracts were prepared from the cells, and subjected to immunoprecipitation with anti-SMN antibody 7B10 and anti-m7G/m3G cap antibody H-20, respectively. 7B10 precipitates SMN complexes and hence monitors the binding of newly synthesized (i.e. 35S-labeled) Sm proteins to SMN (Meister et al., 2000). H-20, in contrast, co-precipitates only Sm proteins that are bound to UsnRNAs and hence monitors the assembly of spliceosomal UsnRNPs (Bringmann and Luhrmann, 1986). As shown in Figure 3, labeled Sm proteins could be detected readily in the SMN complex after a chase for 1 h, but the signal gradually disappeared within a time frame of 18 h (compare lanes 5–7; for a better view, see a longer exposure of the lower part of the gel). The opposite was detected when the H-20 antibody had been used for immunoprecipitation, as evident by marginal amounts of labeled Sm proteins after a 1 h chase and a gradual increase over time (lanes 8–10). These experiments, in conjunction with the results obtained with the isolated SMN complex in vitro, strongly suggest that the Sm proteins associate prior to their incorporation into UsnRNPs with the SMN complex.
Fig. 3. Transfer of Sm proteins from the SMN complex onto UsnRNPs in vivo. HeLa cells were pulsed with [35S]methionine/[35S]cysteine for 1.5 h. After a chase with non-labeled amino acids, extracts were prepared at the indicated times and immunoprecipitated with anti-SMN antibody 7B10 (lanes 5–7), anti-m3G/m7G cap antibody H-20 (lanes 8–10) and a non-related control antibody (lane 4). Proteins were separated by SDS–PAGE and visualized by fluorography. The lower part of the anti-SMN immunoprecipitation is shown as a longer exposure. One-fiftieth of the amount of proteins used for the immunoprecipitations is shown in lanes 1–3. Bands indicated by an asterisk were immunoprecipitated non-specifically.
A partial SMN complex lacking B/B′ and D3 fails to promote assembly of U1 snRNPs
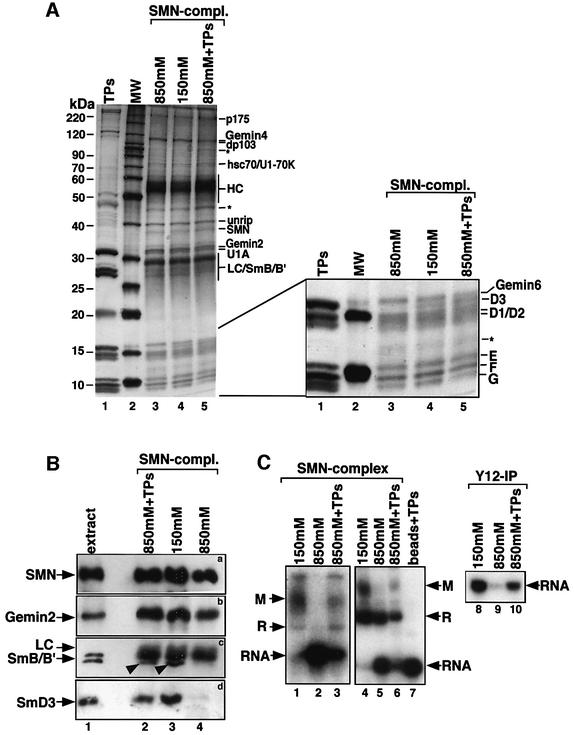
In an approach to determine the stringency of protein– protein interactions within the SMN complex, we have subjected this complex to increasing concentrations of sodium chloride and analyzed the resulting protein composition. Intriguingly, the first proteins that dissociated from the SMN complex were the Sm proteins B/B′ and D3 (Figure 4A and B, lanes 3 and 4). This observation was particularly interesting in the light of previous reports on the spontaneous assembly of Sm proteins in vitro (Raker et al., 1996). Under the conditions of these assays, the Sm proteins were shown to pre-form a so-called subcore particle that is composed of UsnRNA bound by Sm proteins D1, D2, E, F and G. Our ability to prepare SMN complexes devoid of B/B′ and D3 gave us a tool at hand to test whether the subcore particle is also formed in the SMN-dependent assembly pathway. If so, we should be able to observe an UsnRNA-containing RNP using the ΔB/B′–D3 complex. To analyze this, we have employed native gel electrophoresis and anti-Sm immunoprecipitation. Interestingly, as opposed to the complete SMN complex, the ΔB/B′–D3 complex failed to transfer Sm proteins onto the UsnRNA whereas formation of complex R was only slightly affected (Figure 4C, compare lanes 1, 4 and 8 with lanes 2, 5 and 9). We next asked whether the back-addition of B/B′ and D3 to the ΔB/B′–D3 complex could restore its activity. As a source for these proteins, we used Sm proteins derived from isolated spliceosomal UsnRNPs (termed TPs). This preparation was added to the ΔB/B′–D3 complex immobilized on anti-SMN affinity beads and, after an incubation for 30 min, the beads were washed to remove non-bound proteins. The protein composition of the resulting complex was then compared by Coomassie Blue staining and immunoblot analysis with the original SMN complex and the ΔB/B′–D3 complex. These analyses revealed specific re-loading of the ΔB/B′–D3 complex with B/B′ and D3, whereas the stoichiometry of the other Sm proteins remained unchanged (Figure 4A, lane 5, and B, lane 2). Importantly, the complementation of the ΔB/B′–D3 complex substantially restored its activity (Figure 4C, lanes 3, 6 and 10), indicating that B/B′ and D3 were the critical proteins required for successful transfer. Taken together, these data suggest that all Sm proteins need to be present on the SMN complex to allow the formation of the Sm core domain. Furthermore, in the assay system used here, a stable subcore particle cannot be formed in the absence of B/B′ and D3.
Fig. 4. An SMN complex that lacks B/B′ and D3 fails to assemble the Sm core domain. (A) Cytosolic extract active in UsnRNP assembly was incubated with an anti-SMN affinity matrix and washed with either 850 or 150 mM NaCl. Proteins bound to the matrix were eluted from the column, separated by SDS–PAGE and visualized by Coomassie Blue staining (lanes 3 and 4, respectively). In lane 5, the SMN complex was first treated with 850 mM NaCl and subsequently incubated at 150 mM NaCl with Sm proteins isolated from UsnRNPs (TPs). Unbound proteins were removed by repeated washes at 150 mM NaCl. TPs used for this procedure and a protein size marker are seen in lanes 1 and 2, respectively. A magnification of the lower part of the gel is shown on the right. The positions of co-eluting 7B10 heavy chain (HC) and light chain (LC) are indicated. (B) Western blot analysis of SMN complexes shown in (A). Monoclonal antibody 7B10 was used to detect SMN (a), and affinity-purified rabbit antisera were used to detect Gemin2 (b), B/B′ (c, indicated by arrowheads) and D3 (d). The major band seen in (c) above B/B′ corresponds to the light chain that was co-eluted from the column. (C) In vitro assembly of U1 snRNP with SMN complex isolated at 150 mM NaCl (lanes 1, 4 and 8), at 850 mM NaCl (lanes 2, 5 and 9) and after re-loading with Sm proteins (lanes 3, 6 and 10). Lane 7 shows a control assembly reaction on beads that had been incubated with TPs and was washed subsequently with PBS. Assembly of the Sm core domain was analyzed by native gel electrophoresis as above (lanes 1–7) and by immunoprecipitation using Y12 monoclonal antibody (lanes 8–10).
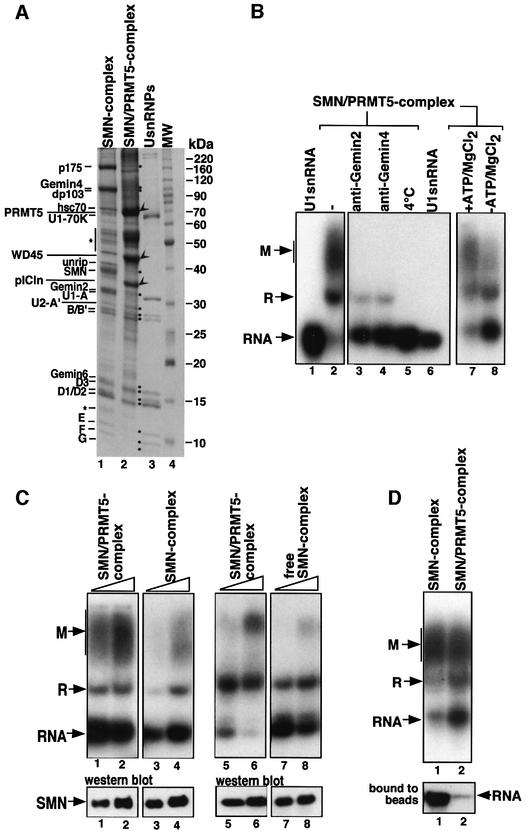
The PRMT5 complex interacts with the SMN complex and stimulates assembly of the Sm core domain
Although assembly of the Sm core domain can be mediated in vitro by the SMN complex alone, this process is moderately efficient and occurs without the addition of ATP to the in vitro assembly reaction (see above). We therefore reasoned that the activity of the SMN complex might be stimulated by additional factors. A candidate for this potential function is the PRMT5 complex, which previously has been implicated in the modification and storage of Sm proteins, i.e. in processes that are believed to occur prior to the assembly reaction (Pu et al., 1999; Friesen et al., 2001b; Meister et al., 2001b). The PRMT5 complex was affinity purified using an antiserum directed against pICln to address its potential function in the assembly reaction (Figure 5A, lane 2). As reported earlier, this complex contains three prominent proteins, namely pICln, WD45 and PRMT5, and, in addition, the Sm proteins. Apart from these proteins, a confined set of less prominent components specifically co-eluted from the column. Interestingly, MALDI-TOF and western blot analysis identified these proteins as the components of the SMN complex (Figure 5A, lanes 1 and 2).
Fig. 5. Identification and characterization of an SMN–PRMT5 complex. (A) Affinity purification of the SMN complex (lane 1) and the SMN–PRMT5 complex (lane 2) on anti-SMN and anti-pICln affinity columns, respectively. Proteins that were identified by MALDI-TOF are indicated on the left. SMN complex components and the Sm proteins are marked by dots, and the PRMT5 complex proteins pICln, WD45 and PRMT5 by arrowheads. A UsnRNP protein marker and a molecular size marker were loaded in lanes 3 and 4. (B) The SMN–PRMT5 complex facilitates the ATP-dependent assembly of U1 snRNP. 32P-labeled U1 snRNA was incubated with affinity-purified SMN–PRMT5 complex (bound to the anti-pICln affinity beads, lanes 2–5 and 7–8). Anti-Gemin2 and anti-Gemin4 antibodies were added prior to the assembly reaction shown in lanes 3 and 4. In lane 8, assembly was carried out in the absence of ATP, whereas all other reactions contained 5 mM ATP. In lane 5, the SMN–PRMT5 complex was incubated with U1 snRNA at 4°C instead of 37°C; in lane 6, U1 snRNA was incubated with control beads. The beads were removed from all reactions and the supernatants analyzed by native gel electrophoresis as described in Figure 1A. (C) Increasing amounts of affinity beads containing either the SMN or the SMN–PRMT5 complex were incubated with 32P-labeled U1 snRNA. After 1 h at 37°C, the beads were pelleted and the supernatant analyzed by native gel electrophoresis (lanes 1–4, upper panel). The amount of SMN present in the assembly reactions was determined by western blotting and is shown in the lower panel. Comparison of the assembly efficiency between the SMN–PRMT5 complex (lanes 5 and 6) and the SMN complex that was dissociated from the SMN–PRMT5 complex (lanes 7 and 8). The lower western blot shows the amount of SMN in the assembly reactions. (D) Enhanced dissociation of U1 snRNA from the SMN–PRMT5 complex. In vitro assembly of U1 snRNP with either SMN complex (lane 1) or SMN–PRMT5 complex (lane 2). The RNA that remained bound to the beads in both assembly reactions is shown in the lower panel.
The observation that the PRMT5 complex co-purifies, and hence physically interacts with the SMN complex led us to next test whether this complex was active in UsnRNP assembly. Indeed, affinity-purified SMN–PRMT5 complex promoted the assembly of the Sm core domain whereas the PRMT5 complex alone had no activity (Figure 5B, lane 2 and data not shown). Importantly, this reaction exhibited key features of the assembly mediated by the SMN complex, as evident by the inhibition of complex M formation in the presence of antibodies against either Gemin2 or Gemin4 and at 4°C (lanes 3–5). However, in contrast to the SMN complex alone, assembly mediated by the SMN–PRMT5 complex was strongly reduced when ATP and MgCl2 were omitted from the reaction (Figure 5B, compare lanes 7 and 8) (assembly in the presence of only MgCl2 was comparable with the reaction without ATP and MgCl2; data not shown). Furthermore, the SMN–PRMT5 complex was more active in promoting the assembly reaction than the SMN complex alone (Figure 5C, compare lanes 1 and 2 with lanes 3 and 4). Dissociation of both complexes, in contrast, resulted in the reduction of assembly efficiency (Figure 5C, compare lanes 5 and 6 with lanes 7 and 8). Lastly, in comparison with the SMN complex, much less U1 snRNA was stably bound on the SMN–PRMT5 complex, suggesting that the release of assembled particles from the latter was enhanced (Figure 5C, lower panel, lanes 1 and 2). Thus, the SMN–PRMT5 complex promotes assembly of UsnRNPs in a manner that is indistinguishable from assembly in vivo or in cellular (egg or HeLa) extracts. We conclude that the SMN complex and the PRMT5 complex join to form a functional entity, which is sufficient to promote the assisted assembly of spliceosomal UsnRNPs.
Discussion
Using proteins prepared from cell extracts, the assembly of spliceosomal Sm proteins onto UsnRNA in vitro previously has been shown to occur spontaneously (Raker et al., 1996, 1999). Nonetheless, the ability of antibodies against SMN or its interacting protein Gemin2 to inhibit this process in X.laevis oocytes provided initial evidence for a more complex scenario in vivo (Fischer et al., 1997; Buhler et al., 1999). Consistently, it was shown that SMN and Gemin2 are part of a large complex, and depletion of this complex from X.laevis egg extract resulted in a complete block of UsnRNP assembly. We recently have isolated this complex from HeLa cell extracts and shown that it contains a set of specific proteins and all canonical Sm proteins (Meister et al., 2001a). In an effort to analyze the proposed function of this complex in UsnRNP assembly, we have chosen a biochemical approach for the reconstitution of UsnRNPs from purified components. We demonstrate that the SMN complex alone is necessary and sufficient to promote the formation of the Sm core domain. Further evidence is provided for an interaction between the SMN complex and the PRMT5 complex, whose methyltransferase activity is known to increase the affinity of Sm proteins for SMN (Brahms et al., 2001; Friesen et al., 2001a). We show that both complexes form a functional entity that efficiently promotes the assembly of the Sm core domain. The SMN–PRMT5 complex can therefore be envisaged as an assembly and modification center that brings together key activities required for the formation of UsnRNPs.
Several lines of evidence support the view that the biochemical reconstitution system described here recapitulates major aspects of the UsnRNP assembly pathway in vivo. First, assembly in our in vitro assay occurs only on a U1 snRNA containing the Sm-binding site, i.e. the RNA element that previously has been shown to be essential for the formation of the Sm core domain in vivo. Secondly, antibodies against the integral SMN complex components Gemin2 and Gemin4 inhibit binding of Sm proteins to UsnRNA. Consistently, these antibodies previously have been shown to block UsnRNP assembly in X.laevis oocytes and egg extracts (Fischer et al., 1997; Meister et al., 2001a). Thirdly, reduced Sm core formation at low temperatures or in the absence of ATP indicates that assembly in vitro, like the situation in cell extracts, is an active process. Finally, the efficient nuclear import of in vitro assembled particles injected into X.laevis oocytes indicates that the SMN– PRMT5 complex generates functional Sm core domains. As the aforementioned features are hallmarks of the UsnRNP assembly reaction in vivo, we are confident that our system faithfully mimics this process in vitro.
Much of our previous knowledge on the assembly of Sm proteins is based on in vitro assays using isolated proteins. These studies have revealed a propensity of Sm proteins to form three hetero-oligomeric complexes that associate in an ordered manner (Raker et al., 1996, 1999). When assayed in a reconstitutive system, an association of E–F–G and D1–D2 complexes precedes the binding of Sm proteins to UsnRNAs, thus forming the so-called subcore particle. The subsequent addition of a B/B′–D3 complex to this subcore particle then yields the Sm core domain. Remarkably, we have not succeeded in detecting subcore particle formation on an SMN complex depleted of B/B′ and D3. While this observation argues against the occurrence of a subcore particle in the assembly pathway mediated by the SMN–pICln complex, we cannot conclusively rule out the possibility that such an intermediate is formed when all Sm proteins are present.
The joining of SMN and PRMT5 complexes to a fully functional assembly unit raises further questions of how both complexes interplay to facilitate UsnRNP formation. Although each complex alone is able to bind Sm proteins, it is only the SMN complex that can transfer the Sm substrates onto UsnRNA. However, the enhanced assembly activity observed in the SMN–PRMT5 complex indicates that the PRMT5 complex can stimulate this process. Since the content of Sm proteins is not significantly altered in the SMN–PRMT5 complex, as compared with isolated SMN complex (Figure 5), this stimulation is more likely to be due to the biochemical properties of the PRMT5 complex rather than the mere addition of substrate. This idea is supported by the consumption of ATP that occurs in the SMN–PRMT5 complex (Figure 5), suggesting that conformational rearrangements are facilitated in the presence of the PRMT5 complex. The putative ATP-dependent RNA helicase dp103, a component of the SMN complex, may be a candidate to be activated in response to an interaction with the PRMT5 complex.
The PRMT5 complex has, in addition to its stimulatory activity in the SMN–PRMT5 complex, a strong potential to interfere with the spontaneous assembly of Sm proteins in vitro (Pu et al., 1999; Meister et al., 2001b). If this proved to be valid in vivo, then free PRMT5 complex would indeed force assembly to be an assisted, and thus controlled, rather than a spontaneous event. Another important property of the PRMT5 complex can be delineated from its protein composition. PRMT5, an integral protein of the PRMT5 complex, catalyzes the formation of sDMA residues in Sm proteins, and this modification has been shown to greatly increase the affinity of Sm proteins for binding to SMN (Brahms et al., 2001; Friesen et al., 2001a; Meister et al., 2001b). Thus, the PRMT5 complex may not only prevent spontaneous assembly, but also activate Sm proteins for assisted assembly through modification. According to this view, we may speculate that all Sm proteins undergo initial binding to the PRMT5 complex before they are subject to assembly. In fact, these data also suggest that Sm proteins are, in the context of the SMN–PRMT5 complex, transferred from the PRMT5 complex to the SMN complex. This view is consistent with a recent report by Friesen et al. (2001b), who demonstrated in vitro the release of Sm protein D3 from a 20S complex containing pICln and its transfer onto the SMN complex.
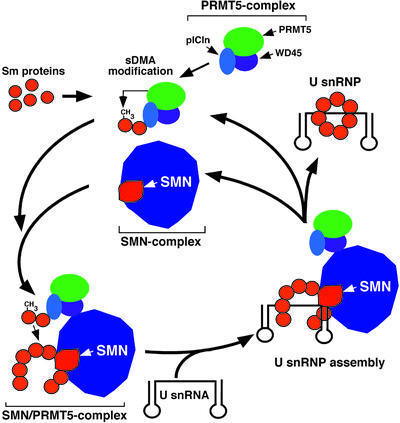
Based on the present body of data, we envisage a model for the assembly pathway of UsnRNPs mediated by the SMN–PRMT5 complex (Figure 6). In this model, Sm proteins are sequestered by the PRMT5 complex and thereby directed to assisted assembly. The PRMT5 complex subsequently catalyzes the sDMA modification of the Sm proteins B/B′, D1 and D3 through the action of PRMT5, and docks onto the SMN complex, giving rise to the SMN–PRMT5 complex. Here, the Sm proteins are transferred to the SMN complex, possibly in a reaction that requires ATP. By an as yet unknown mechanism, the SMN complex (as part of the SMN–PRMT5 complex) will pass on the complete set of seven Sm proteins to the UsnRNA. Ultimately, the resulting UsnRNP is released and targeted to the nucleus, whereas the SMN–PRMT5 complex may dissociate before its components unite again to engage in a new round of assembly.
Fig. 6. A model for the SMN–PRMT5 complex-mediated assembly pathway of spliceosomal UsnRNPs. For details see text.
It is worth noting that, like U1, the incubation of U2, U4 or U5 snRNAs in cellular extracts results in Sm core formation that is susceptible to anti-Gemin2 antibodies (Figure 1 and data not shown). We may thus presume that assembly of these spliceosomal UsnRNPs follows a common, SMN-dependent, principle. Whether the formation of an LSm core structure on U6 snRNA also requires SMN is currently unclear. However, evidence of an interaction between SMN and LSm4 (Friesen and Dreyfuss, 2000; Brahms et al., 2001), together with the similar architectures of canonical (Sm-based) and LSm core domains (Toro et al., 2001), indeed supports the idea that formation of the LSm core likewise requires the SMN–pICln complex. Intriguingly, the U7 snRNP, whose function relates to histone mRNA processing, was found recently to contain five canonical Sm proteins (B/B′, D3, E, F and G) and additional proteins, two of which are proposed to replace SmD1 and SmD2 (Pillai et al., 2001). These proteins may thus form a heptameric ring similar to the spliceosomal Sm or LSm structures known so far. It will be interesting, therefore, to see whether the SMN–PRMT5 complex indeed assembles RNPs of various functions.
The functional characterization of the SMN–PRMT5 complex may also provide insight into the molecular mechanisms leading to SMA. The principle cause of this disease is the reduction of functional SMN protein in all cells of the body. Based on our finding that the majority of endogenous SMN is part of large complexes (most likely the SMN complex and the SMN–PRMT5 complex), we may rationalize that the cellular abundance of these complexes is critical for UsnRNP formation in SMA patients. In keeping with this notion, we have observed that extracts prepared from cells with reduced levels of SMN exhibit impaired UsnRNP assembly activity (our unpublished observation). It will therefore be interesting to analyze whether defects in the biogenesis of UsnRNPs indeed cause the pathophysiological events that lead to the SMA phenotype.
Materials and methods
Preparation of cellular extracts and gel filtration
HeLa cells were washed with phosphate-buffered saline (PBS) pH 7.4, resuspended in two pellet volumes of PBS and homogenized by douncing. The cytoplasm was subsequently separated from the nuclei by centrifugation at 1000 g for 10 min at 4°C. The supernatant (cytosolic extract) was fractionated by a Sprint FPLC (PE Biosystems) using a Superose-6 gel filtration column (Amersham-Biosciences). Extracts used for western blotting and for immunoprecipitations were obtained from freshly grown cells upon lysis in RIPA buffer [150 mM KCl, 25 mM Tris–HCl pH 7.5, 2 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.1% NP-40].
Antibodies, immunoprecipitations and western blotting
Polyclonal antibodies were generated by repeated injections of recombinant Sm complexes B–D3 and E–F–G into rabbits. Antibodies were affinity purified on CNBr-activated Sepharose columns (Amersham-Biosciences) coupled with the respective antigens. For immunoprecipitation experiments, ∼100 µg of the indicated antibody used in this study, anti-SMN (7B10), anti-m3/m7G cap (H-20) or anti-Sm proteins (Y12), were coupled onto protein G–Sepharose beads (Amersham-Biosciences) and incubated for 2 h. The beads were washed with PBS pH 7.4, and bound proteins analyzed by SDS–PAGE. For the analysis of RNA, beads were treated with phenol and the bound RNA was precipitated from the aqueous phase. The RNA was analyzed by denaturing RNA gel electrophoresis followed by autoradiography. Western blotting was performed as described in Meister et al. (2001a). Native HeLa TPs were obtained from affinity-purified UsnRNPs using a technique described by Raker et al. (1996).
Purification of SMN and SMN–PRMT5 complexes
For SMN complex isolation, cytosolic HeLa extract obtained from 5 × 109 cells was centrifuged at 25 000 g for 30 min and subsequently passed over an anti-SMN affinity column (Meister et al., 2001a). The column was washed with PBS and the bound proteins were eluted with 100 mM glycine pH 2.3, precipitated with trichloroacetic acid and separated by SDS–PAGE. Bands were visualized by Coomassie Blue staining. The SMN–PRMT5 complex was isolated by anti-pICln affinity chromatography using the same HeLa extract and the same elution conditions. Protein identification by mass spectrometry was carried out as described (Meister et al., 2000). The experiments shown in Figure 5C (lanes 7 and 8) were performed with SMN complex that had been dissociated from the affinity-purified SMN–PRMT5 complex. For this, SMN–PRMT5 complex bound to affinity beads was incubated for 30 min in PBS. The beads subsequently were pelleted and the supernatant, which contains released SMN complex, was used for the assembly reactions. PRMT5 complex lacking the SMN complex was obtained by washing the anti-pICln affinity column with 450 mM NaCl. Under these conditions, the SMN complex is quantitatively stripped from the column and pure PRMT5 complex is retained.
In vitro transcription of UsnRNAs and reconstitution of UsnRNPs
32P-labeled and unlabeled U1 snRNAs were generated by transcription in vitro and gel purified as described (Meister et al., 2000). In a standard reconstitution assay, 25 fmol of [32P]UsnRNA was incubated with 2 µl of HeLa extract (containing ∼25 mg/ml protein) in the presence of 2.5 pmol of unlabeled tRNA. After incubation for 35 min, the samples were mixed with 1 vol. of sample buffer (16% glycerol, 10 mg/ml heparin) and analyzed by native RNA gel electrophoresis as described (Meister et al., 2001a). For assembly experiments with isolated complexes, affinity beads containing either the SMN complex or the SMN–PRMT5 complex were incubated in PBS with 75 fmol of [32P]snRNA, 7.5 pmol of unlabeled tRNA, 5 mM ATP and 10 mM MgCl2. In the experiment shown in Figure 5B, ATP was omitted from the reaction. The beads subsequently were pelleted and the supernatant analyzed by native gel electrophoresis. For antibody inhibition studies, the assembly reactions were pre-incubated with 10 µg of the indicated antibody for 15 min. To prepare the ΔB/B′–D3 complex, immobilized SMN complex was treated with buffer containing 850 mM NaCl, 50 mM Tris–HCl pH 7.5 and 5 mM MgCl2. For re-loading with Sm proteins, the ΔB/B′–D3 complex was incubated with PBS containing purified Sm proteins (TPs) for 30 min. Unbound components were removed by several washes with PBS. In the experiment shown in Supplementary figure 2B, gel filtration fractions were normalized for their Sm protein content by adding TPs (20 ng/µl) before assembly reactions were carried out as above.
Microinjection in Xenopus laevis oocytes
Injections into X.laevis oocytes were carried out as described (Fischer et al., 1997). In brief, ovaries were incubated at 20°C for 3 h in OR2 buffer containing 0.2% collagenase type II (Sigma) to obtain defolliculated oocytes. For injection, 30–50 nl of 32P-labeled U1 snRNA or U1 snRNP assembled in vitro was injected into the cytoplasm of oocytes. A 50 nl aliquot of anti-Gemin2 antiserum (1 mg/ml) was pre-injected into the cytoplasm and incubated for 1 h before they received a second injection of 32P-labeled RNA/RNP. RNAs were isolated from nuclear and cytosolic fractions, separated on denaturing urea gels and visualized by autoradiography.
UV cross-linking
For UV cross-linking experiments, isolated SMN complex was incubated with either 32P-labeled U1 snRNA or U1ΔSm under the reconstitution conditions described above. The assembly reactions were placed as drops on parafilm (Pechiney) and irradiated for 20 min with 840 mJ of UV light (254 nm). To determine whether the cross-link corresponded to G-protein bound to U1 snRNA, the reaction was first denatured by incubation with 2% SDS for 10 min at 70°C. After cooling to room temperature, Triton X-100 was added to a final concentration of 5%. Immunoprecipitation was carried out with an antiserum specific for the G-protein followed by separation by 12% SDS–PAGE and detection by autoradiography (Urlaub et al., 2001).
In vivo labeling experiments
For pulse–chase experiments, HeLa cells were incubated in starvation medium (medium 11970 without amino acids; Gibco-BRL) containing 10% fetal calf serum, 2 mM l-alanyl-l-glutamine and 25 mM HEPES-KOH pH 7.4 for 45 min. The cells were labeled in starvation medium containing 1 mCi of [35S]methionine/[35S]cysteine (Promix, Amersham-Biosciences) for 1.5 h. Cells subsequently were incubated with complete medium (Gibco-BRL) for 1, 5 and 18 h. Extracts were prepared as described above and used for immunoprecipitations with the indicated antibodies. Proteins were visualized by SDS–PAGE followed by fluorography.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to B.Laggerbauer and R.Lührmann for critical reading of the manuscript and continuous discussions, E.Keidel, G.Sowa and C.Thomas for technical support, and R.Lührmann, C.Kambach and I.Mattaj for providing reagents. This work was funded by the MPIB and by DFG grants Fi 573/2-2 and SFB 596 TP B7.
References
- Brahms H., Meheus,L., de Brabandere,V., Fischer,U. and Lührmann,R. (2001) Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4 and their interaction with the SMN protein. RNA, 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P. and Luhrmann,R. (1986) Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J., 5, 3509–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler D., Raker,V., Luhrmann,R. and Fischer,U. (1999) Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet., 8, 2351–2357. [DOI] [PubMed] [Google Scholar]
- Campbell L., Hunter,K.M., Mohaghegh,P., Tinsley,J.M., Brasch,M.A. and Davies,K.E. (2000) Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum. Mol. Genet., 9, 1093–1100. [DOI] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni,L., Perkinson,R.A., Shevchenko,A., Mann,M. and Dreyfuss,G. (1999) Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol., 147, 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni,L., Perkinson,R.A., Yong,J., Shevchenko,A., Mann,M. and Dreyfuss,G. (2000) Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol., 148, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Liu,Q. and Dreyfuss,G. (1997) The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Friesen W.J. and Dreyfuss,G. (2000) Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem., 275, 26370–26375. [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Massenet,S., Paushkin,S., Wyce,A. and Dreyfuss,G. (2001a) SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell, 7, 1111–1117. [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin,S., Wyce,A., Massenet,S., Pesiridis,G.S., Van Duyne,G., Rappsilber,J., Mann,M. and Dreyfuss,G. (2001b) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol., 21, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A.T., Kremmer,E., Türeci,Ö., Glieden,A., Gindorf,C., Atz,J., Müller-Lantzsch,N., Schubach,W.H. and Grässer,F.A. (1999) Characterisation of DP103, a novel DEAD box protein that binds to the Epstein–Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem., 274, 19136–19144. [DOI] [PubMed] [Google Scholar]
- Hamm J. and Mattaj,I.W. (1989) An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J., 8, 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.W., Gorzynski,K., Hales,C.M., Fischer,U., Terns,R.M. and Terns,M.P. (2001) Direct interaction of the spinal muscular atrophy disease protein SMN with the core snoRNP protein fibrillarin. J. Biol. Chem., 276, 38645–38651. [DOI] [PubMed] [Google Scholar]
- Lefebvre S. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burlet,P., Liu,Q., Bertrandy,S., Clermont,O., Munnich,A., Dreyfuss,G. and Melki,J. (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet., 16, 265–269. [DOI] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss,G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J., 15, 3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer,U., Wang,F. and Dreyfuss,G. (1997) The spinal muscular atrophy disease gene product, SMN and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Meister G., Buhler,D., Laggerbauer,B., Zobawa,M., Lottspeich,F. and Fischer,U. (2000) Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet., 9, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Meister G., Buhler,D., Pillai,R., Lottspeich,F. and Fischer,U. (2001a) A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol., 3, 945–949. [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert,C., Buehler,D., Brahms,H., Kambach,C. and Fischer,U. (2001b) Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol., 11, 1990–1994. [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert,C. and Fischer,U. (2002) SMN-mediated assembly of spliceosomal U snRNPs: a complex story. Trends Cell Biol., 12, 472–478. [DOI] [PubMed] [Google Scholar]
- Monani U.R. et al. (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(–/–) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet., 9, 333–339. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Abel,L., Yong,J., Kataoka,N. and Dreyfuss,G. (2001) SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J., 20, 5443–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux,B. and Dreyfuss,G. (1999) SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA, 96, 11167–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon,J., Charroux,B. and Dreyfuss,G. (2001a) The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol., 11, 1079–1088. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux,B., Rappsilber,J., Mann,M. and Dreyfuss,G. (2001b) A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol., 152, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon,J., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem., 277, 7540–7545. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Will,C.L., Luhrmann,R., Schumperli,D. and Muller,B. (2001) Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J., 20, 5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W.T., Krapivinsky,G.B., Krapivinsky,L. and Clapham,D.E. (1999) pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol. Cell. Biol., 19, 4113–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Plessel,G. and Luhrmann,R. (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth,K., Kastner,B. and Luhrmann,R. (1999) Spliceo somal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W., Kroning,A.K., Ohndorf,U.M., Steegborn,C., Jablonka,S. and Sendtner,M. (2002) Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum. Mol. Genet., 11, 93–105. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H. and Haase,G. (2001) Spinal muscular atrophy: present state. Brain Pathol., 11, 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendtner M. (2001) Molecular mechanisms in spinal muscular atrophy: models and perspectives. Curr. Opin. Neurol., 14, 629–634. [DOI] [PubMed] [Google Scholar]
- Toro I., Thore,S., Mayer,C., Basquin,J., Seraphin,B. and Suck,D. (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J., 20, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H., Raker,V.A., Kostka,S. and Luhrmann,R. (2001) Sm protein–Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J., 20, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]