Abstract
The conserved, structure-specific flap endonuclease FEN1 cleaves 5′ DNA flaps that arise during replication or repair. To address in vivo mechanisms of flap cleavage, we developed a screen for human FEN1 mutants that are toxic when expressed in yeast. Two targets were revealed: the flexible loop domain and the catalytic site. Toxic mutants caused G2 arrest and cell death and were unable to repair methyl methanesulfonate lesions. All the mutant proteins retained flap binding. Unlike the catalytic site mutants, which lacked cleavage of any 5′ flaps, the loop mutants exhibited partial ability to cut 5′ flaps when an adjacent single nucleotide 3′ flap was present. We suggest that the flexible loop is important for efficient cleavage through positioning the 5′ flap and the catalytic site.
Keywords: FEN1/flap endonuclease/loop domain/repair/replication
Introduction
The flap endonuclease (FEN1) is central to several aspects of DNA replication and repair and belongs to a family of structure-specific nucleases evolutionarily conserved between Archaea and Eukarya (reviewed in Bambara et al., 1997; Lieber, 1997; Shen et al., 1998). Members of this family are highly homologous to the nucleotide excision repair XPG-like family of proteins and share structural and functional similarity to several 5′ nucleases of bacteriophages and to prokaryotic 5′ nucleases that are part of DNA polymerases (Shen et al., 1997). The FEN1 enzymes can recognize and cleave 5′ flap structures including RNA-containing flaps that are common intermediates in DNA replication (Kao et al., 2002). Flap removal by FEN1 is also essential in long-patch base excision repair (Klungland and Lindahl, 1997). Proliferating cell nuclear antigen (PCNA) stimulates FEN1 activity, enhancing its binding stability, resulting in increased cleavage efficiency in vitro (Li et al., 1995; Tom et al., 2000) as well as enhanced function in vivo (Gary et al., 1999).
The minimal requirement for cleavage in vitro is a bifurcated double-stranded molecule with a free 5′ end (pseudo Y) (Kaiser et al., 1999). Cleavage is more efficient with 5′ flap structures that have an upstream double-stranded region adjacent to the bifurcation. For all FEN1 and FEN1-like enzymes examined from Eubacteria, Archaea, yeast and mouse, the presence of a one-base overlap (resulting in a local double-flap structure) can stimulate activity (Harrington and Lieber, 1995; Kaiser et al., 1999; Xie et al., 2001; Kao et al., 2002). This stimulation has led Kaiser et al. (1999) to suggest that there is a region or pocket in the FEN1 protein that specifically recognizes a 3′ unpaired nucleotide at a junction in lagging strand replication to facilitate proper cutting and subsequent production of a nick that can be ligated by DNA ligase I.
Among several crystal structures of FEN1 proteins and its homologs that have been solved, all have a flexible loop containing a helical motif. The loop is thought to interact with the 5′ flap prior to cleavage. A flexible loop structure was identified in Methanococcus jannaschii and Pyrococcus furiosus (Hosfield et al., 1998; Hwang et al., 1998), as well as in T5 5′-exonuclease where the corresponding region adopts a helical arch similar to the loop of FEN1 proteins (Ceska et al., 1996). The loop consists of a number of positively charged and aromatic residues and may form a hole large enough to accommodate single-stranded DNA (Hwang et al., 1998). The flexible loop region of human FEN1 (hFEN1) probably spans 48 amino acids (from residue 87 to 134) based on sequence comparison with the L1 loop domain in M.jannaschii (Hwang et al., 1998). It has been proposed that the 5′ end of the DNA flap could thread through the hole of the loop, and that the junction between single- and double-stranded regions could be recognized by both the L1 loop and the catalytic active site metals in M.jannaschii FEN1, and the two domains might interact to accomplish the cleavage reaction (Hwang et al., 1998).
The importance of FEN1 in preserving genome integrity has been demonstrated by the dramatic consequences of defects in flap endonuclease activity in yeast and mammalian cells. Null mutants of the FEN1 gene RAD27 (also known as RTH or/YKL510) of the yeast Saccharomyces cerevisiae exhibit a strong mutator phenotype that is due primarily to small duplications involving 3–12 base repeat sequences, high recombination rates, growth inhibition at 37°C and extreme sensitivity to the DNA alkylating agent methyl methanesulfonate (MMS) (Tishkoff et al., 1997; and references therein). rad27 mutants also exhibit high rates of gross chromosome rearrangements (Chen and Kolodner, 1999). Similarly, human bladder carcinoma cells expressing nuclease-deficient FEN1 are sensitive to MMS and UV (Shibata and Nakamura, 2002). Based on work in yeast, FEN1 may also prevent trinucleotide repeat instability associated with several diseases such as mytonic dystrophy and fragile X syndrome (Schweitzer and Livingston, 1998; White et al., 1999).
The yeast S.cerevisiae has proven an ideal in vivo test tube for characterization of mammalian DNA metabolic genes (Resnick and Cox, 2000), particularly for the characterization of hFEN1. Within the FEN1 family, the structure-specific flap endonucleases are highly homologous. The S.cerevisiae Rad27 protein has 61 and 55% amino acid identity with human and Xenopus FEN1, respectively. The human and Xenopus FEN1 proteins complement the MMS sensitivity of a S.cerevisiae rad27 mutant (Bibikova et al., 1998; Frank et al., 1998). Moreover, the hFEN1 protein can (i) compensate for the rad27 defect in genetic stability; (ii) complement the synthetic lethality between rad27 and various repair and replication mutants (Greene et al., 1999); and (iii) complement the temperature-sensitive growth defect of a rad27 null mutant (Frank et al., 1998).
While there has been considerable analysis of the biochemical activities of mutant hFEN1 proteins, relatively little is known about in vivo consequences of mutations in this protein. Sequence alignment of 18 structure-specific nucleases within the FEN1 and the XPG family revealed seven highly conserved carboxyl residues essential for flap cleavage which are clustered in the two conserved N-terminal and intermediate (N and I) regions (Shen et al., 1996, 1997). In vivo structure–function analysis of hFEN1 has been confined to the catalytic site residues D181 and D160 (Frank et al., 1998; Greene et al., 1999). Interestingly, a D181A mutation in hFEN1 or the corresponding D179A change in S.cerevisiae Rad27, which eliminates nuclease activity while maintaining substrate binding capability, can result in toxicity (inhibition of survival and growth) even in the presence of the wild-type yeast gene (Greene et al., 1999).
Based on our observations with the D181A allele, we anticipated that other toxic mutants could provide insight into the relationship between in vivo function and the structure of hFEN1. We therefore developed a screen in the yeast S.cerevisiae for the isolation of hFEN1 mutants that are toxic when expressed from an inducible promoter. Remarkably, half of the toxic mutations were located in the predicted flexible loop domain, while all the others corresponded to the conserved catalytic residues. Through a combination of genetic approaches and biochemical analyses with various flap substrates, we established that the flexible loop is essential for flap cleavage by hFEN1.
Results
Development of a yeast reporter system and isolation of human FEN1 toxic mutants
Previously, we showed that the mutation D181A rendered hFEN1 toxic when the protein was expressed at high levels in a S.cerevisiae rad27Δ background (Greene et al., 1999). The toxicity was increased when the last 49 amino acids were replaced by the terminal 48 amino acids of Rad27, presumably due to increased binding to yeast PCNA and/or greater transport into the nucleus. We examined the consequences of the hFEN1 D181A allele and the corresponding chimeric (Ch) derivative when overexpressed from a GAL1-10-inducible promoter on a high copy plasmid (i.e. YEp195SpGALFEN1-n and YEp195SpGALFEN1-n,Ch) in the following strains: a rad27Δ mutant, a rad27 mutant defective in PCNA binding (rad27-p), a wild-type RAD27 strain and two strains (FS-1 and FS-3) where the RAD27 open reading frame (ORF) was replaced by hFEN1 and hFEN1-Ch ORF, respectively. The highest toxicity was observed for the D181A hFEN1 chimera mutant expressed in the rad27Δ background (order of toxicity in the chosen backgrounds: Δrad27>rad27-p>hFEN1 and hFEN1-Ch>wild-type RAD27). We therefore chose to mutagenize hFEN1-Ch and express this gene on the multicopy YEp195SpGALFEN1-Ch plasmid in the Δrad27 mutant strain.
We used mutagenic PCR to introduce random point mutations into the entire FEN1-Ch ORF of YEp195Sp GALFEN1-Ch. The PCR products (∼1 kb) together with linearized YEp195SpGAL vector were co-transformed into the Δrad27 strain, and recombinants resulting from recombination in the overlapping regions of the PCR fragments and the plasmid (containing the URA3 gene) were recovered on Ura– medium (see Materials and methods in the Supplementary data, available at The EMBO Journal Online). Among ∼12 000 transformants tested, nearly 100 failed to grow on galactose medium (Figure 1).
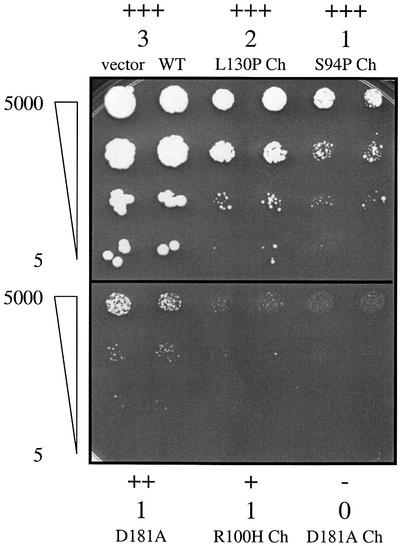
Fig. 1. Categories of toxic mutants based on differences in survival and growth. Survival and growth effects of toxic hFEN1 mutants highly expressed under the GAL1-10 promoter on plasmids pGALFEN1-Ch or pGALFEN1 in the Δrad27 strain (E275). Presented are 10-fold serial dilutions of cultures plated to Gal Ura– media after 3 days of incubation at 30°C. Survival categories: +++, 100% similar to wild-type; ++, 10–100%; +, 1–10%; –, <1%. Growth categories: 3, normal colonies, like wild-type; 2, small colonies; 1, very small colonies; 0, no detectable growth. Presented are the Δrad27 strain containing the vector YEp195SpGAL and the wild-type hFEN1 in the chimeric form (top left); mutant L130P and S94P in the chimeric form; mutant D181A in the non-chimeric form; and mutant R100H and D181A in the chimeric form. A difference in survival and growth (+/1 versus –/0) was clearly observed between R100H Ch and D181A Ch after an additional 3 days of growth. Survival and growth were the same (+++/3) for all clones on Ura– media (data not shown).
To evaluate toxicity better, we next crossed each of the clones containing a potential hFEN1 mutation on the plasmid with three isogenic strains containing either the Δrad27 allele, the PCNA binding-deficient allele rad27-p or the wild-type RAD27 gene. Single colony isolates were tested for toxicity after dilution pronging on galactose media lacking uracil. All Δrad27/RAD27 diploids grew normally on galactose. Some mutants, referred to as weak, grew normally when crossed with the rad27-p strain and showed partial growth inhibition when crossed with a Δrad27 strain. The strong mutants showed partial growth when crossed with the rad27-p strain and showed strong growth inhibition in a homozygous Δrad27 diploid strain (data not shown).
Fifty (39 strong and 11 weak) of the toxic mutant strains were chosen for sequence analysis. As shown in Supplementary table I, all had mutations in the hFEN1 sequence with repeated occurrence of some sites. Among 40 of the 50 clones, there was a mutation in one of the following amino acids: D34, D86, K93, R100, L111, L130, E158, E160, D179 or D181 (a previously identified toxic site; Greene et al., 1999) (see Figure 2A, and Supplementary table I). Twenty-one of these 40 were single mutants, and single mutations were found in each of the 10 residues listed above. These amino acids include all the reported catalytic residues, D34, D86, E160 and D181, which when mutated lack nuclease activity but retain substrate binding (Shen et al., 1997; Frank et al., 1998). Mutations in residues E158 and D179 that were described as leading to a deficiency both in nuclease activity and flap binding (Shen et al., 1997) were also isolated in our screen, although with different amino acid substitutions.
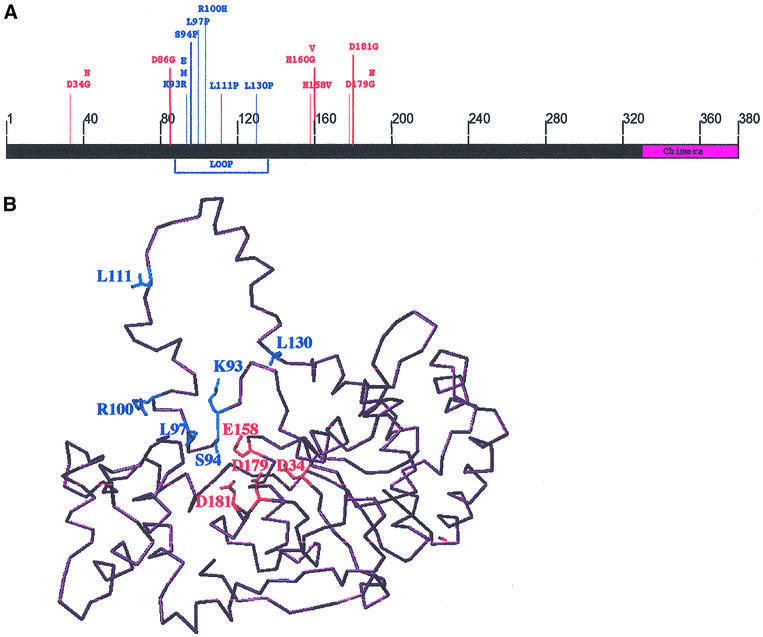
Fig. 2. Location of the human FEN1 toxic mutations. (A) The position of all toxic mutations in the hFEN1Ch protein. The residues affected are shown together with the substitution(s) identified in the screen for toxicity. Many of the toxic mutations are located in the loop domain (blue). (B) Three- dimensional model of hFEN1 protein based on X-ray structures of FEN1 proteins from M.jannaschii and P.furiosus (PDB1A76 and PDB1B43 are the respective accession numbers into the 3D databases), and the positions of toxic mutants. The model of hFEN1 was produced by an automatic program, after submitting the hFEN1 amino acid sequence and selecting the comparative three-dimensional sequences (Web location: www.expasy.ch/swissmod), the software used to label amino acids of interest in hFEN1 is Swiss-PdbViewer (v3.5b1; O version 6.2.1). The Swiss-PdbViewer file containing the model is provided in the Supplementary data. Amino acids involved in the catalytic site and the loop domain are represented in red and blue, respectively.
In addition to mutations affecting highly conserved residues, we repeatedly found mutations in amino acids K93, R100, L111 and L130 (8, 3, 5 and 4 times, respectively; see Supplementary table I) that are conserved in human, mouse and S.cerevisiae. These residues of hFEN1 lie in the region that aligns with the loop domain of M.jannaschii (Hwang et al., 1998). In a three-dimensional model of hFEN1 protein derived from X-ray structures of M.jannaschii (Hwang et al., 1998) and P.furiosus (Hosfield et al., 1998) FEN1 proteins, these residues are a part of the loop domain (Figure 2B). Moreover, hFEN1 protein has 34 and 38% amino acid identity with those of M.jannaschii and P.furiosus, respectively.
The remaining 10 clones contained more than one mutation and none of them corresponded to residues isolated as single amino acid changes. Altered S94, L97 and E106 residues were found in three toxic mutants. These amino acids also map to the loop domain. When these mutations were created as single changes in hFEN1, two of them (S94 and L97P) also had a toxic effect. The combined results suggest that the loop domain between amino acids 87 and 134 plays an important role in hFEN1 function.
In vivo toxicity and dominance of FEN1 loop mutations
To characterize the various single mutations further, the following site-specific changes were created in plasmids pGALFEN1-WT and pGALFEN1-ChWT: D34G (GAT→ GGT), D34N (GAT→AAT), D179G (GAC→GGC), D179N (GAC→AAC), E158V (GAG→GTG), K93R (AAG→AGG), R100H (CGC→CAC), L111P (CTG→ CCG), L130P (CTG→CCG), S94P (TCA→CCA), L97P (CTG→CCG), E106G (GAG→GGG) and D181A (the previously reported mutation in the catalytic site). The pGALFEN1-ChWT derivative mutant plasmids were transformed into Δrad27, rad27::FEN1-Ch (strain FS-3) and RAD27 strains. Since the hFEN1-n mutation is toxic in the rad51 background (Greene et al., 1999) suggesting DNA damage, we also introduced the same plasmids into a Δrad51 strain (DAG97). Similarly, we transformed Δrad27 and rad27::FEN1 (strain FS-1) strains with pGALFEN1-WT-derived mutant vectors. Strains FS-1 and FS-3 were used to evaluate dominance and toxicity of mutants. The plasmids YEp195SpGAL, pGALFEN1-WT and pGALFEN1-ChWT were used as controls.
Six transformants for each strain and plasmid combination were examined for survival and growth by dilution pronging on Gal Ura– medium. Overexpression of the hFEN1 mutants in the different backgrounds revealed a wide range of toxicities. Presented in Table I are the single mutants in increasing order of toxicity. Relative to catalytic site mutants, the loop domain mutants exhibited toxicities that ranged from comparable (L111P), to more (K93R and L97P) or less toxic (L130P, S94P and R100H). The lack of toxicity of E106G suggests that one of the other mutations, Q113R or T127P, which were identified in the same clone might be the source of toxicity (Supplementary table I).
Table I. Effects of toxic mutants on survival and growth when highly expressed on plasmids.
| Allele | Domaina | Δrad27 + pGALFEN1-Ch | Δrad27 + pGALFEN1 | rad27::hFEN1-Ch + pGALFEN1-Ch | rad27::hFEN1 + pGALFEN1 | Δrad51 + pGALFEN1-Ch | RAD27 + pGALFEN1-Ch |
|---|---|---|---|---|---|---|---|
| Vector | +++b | +++ | +++ | +++ | +++ | +++ | |
| 3c | 3 | 3 | 3 | 3 | 3 | ||
| Wild-type | +++ | +++ | +++ | +++ | +++ | +++ | |
| FEN1-Ch | 3 | 3 | 3 | 3 | 3 | 3 | |
| E106G | Loop | +++ | +++ | +++ | +++ | +++ | +++ |
| 3 | 3 | 3 | 3 | 3 | 3 | ||
| L130P | Loop | +++ | ++ | +++ | +++ | +++ | +++ |
| 2 | 1 | 2 | 2 | 1 | 3 | ||
| S94P | Loop | +++ | ++ | +++ | +++ | ++ | +++ |
| 1 | 1 | 2 | 1 | 1 | 3 | ||
| R100H | Loop | + | – | +++ | ++ | +++ | +++ |
| 1 | 0 | 1 | 1 | 1 | 3 | ||
| D34N | Catalytic | – | ++ | +++ | +++ | ++ | +++ |
| 0 | 2 | 2 | 2 | 1 | 3 | ||
| D181A | Catalytic | – | ++ | ++ | +++ | ++ | +++ |
| 0 | 1 | 1 | 2 | 1 | 3 | ||
| D34G | Catalytic | – | + | +++ | +++ | ++ | +++ |
| 0 | 1 | 1 | 1 | 1 | 3 | ||
| E158V | Catalytic | – | – | +++ | +++ | +++ | +++ |
| 0 | 0 | 1 | 2 | 2 | 3 | ||
| D179G | Catalytic | – | – | ++ | ++ | ++ | +++ |
| 0 | 0 | 1 | 1 | 1 | 3 | ||
| D179N | Catalytic | – | – | ++ | ++ | ++ | +++ |
| 0 | 0 | 1 | 1 | 1 | 3 | ||
| L111P | Loop | – | – | ++ | + | ++ | +++ |
| 0 | 0 | 1 | 1 | 1 | 3 | ||
| K93R | Loop | – | – | – | + | ++ | +++ |
| 0 | 0 | 0 | 1 | 1 | 3 | ||
| L97P | Loop | – | – | – | – | ++ | +++ |
| 0 | 0 | 0 | 0 | 1 | 3 |
Survival and growth effects of toxic mutants highly expressed (2% galactose) under the GAL1-10 promoter on plasmids pGALFEN1-Ch or pGALFEN1 in Δrad27, FS-3 (rad27::hFEN1-Ch), FS-1 (rad27::hFEN1), Δrad51 and RAD27 strains on Gal Ura– media.
aThe domain (catalytic or loop) of each mutation is shown.
bSurvival categories: +++, 100%, similar to wild-type; ++, ≥10% <100%; +, ≥1% <10%; –, <1%.
cGrowth categories: 3, normal colonies, like wild-type; 2, small colonies; 1, very small colonies; 0, no detectable growth. Survival and growth categories are illustrated in Figure 1. They were determined as the most frequent categories among six independent transformant clones in a dilution pronging experiment following 3 days incubation at 30°C. Variation within each group of six clones was never wider than one category and it was never observed for more than two of the six clones tested. After 6 days of incubation, survival did not change (not shown).
With the exception of E106G, all mutants were dominant over wild-type hFEN1-Ch and hFEN1 when overexpressed. While there was no apparent dominance over the wild-type RAD27 gene, there was partial dominance in a Δrad51 strain (Table I). This suggests that the toxic mutants can lead to DNA damage, some of which is subject to recombination repair.
Analysis of toxic mutants integrated into the genome
Since plasmid copy number and stability might be affected by the overexpressed hFEN1-Ch mutant proteins, we also characterized the effects of single copies of toxic alleles integrated in the genome. The hFEN1-Ch mutants under control of the GAL1-10 promoter were integrated such that they replaced the RAD27 ORF (Table II; Materials and methods). Effects on survival and growth were analyzed by dilution pronging of three isolates of each strain on 2% galactose medium. As described in Table II, the effects of overexpression of the integrated versus the multicopy toxic mutants were similar (Table II, third column).
Table II. Effects of integrated FEN1-Ch mutants on survival, growth and repair of MMS-induced change.
| Strain | Domaina | High expression | Low expression | Low expression | Very low expression | Very low expression | Basal expression | Basal expression | Basal expression |
|---|---|---|---|---|---|---|---|---|---|
| |
|
2% Gal |
0.01% Gal |
0.01% Gal 1 mM MMS |
0.004% Gal |
0.004% Gal 1 mM MMS |
0% Gal |
0% Gal 1 mM MMS |
0% Gal 2 mM MMS |
| RAD27 | +++b | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |
| 3c | 3 | 3 | 3 | 3 | 3 | 3 | 3 | ||
| Wild-type | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | |
| FEN1-Ch | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | |
| Δrad27 | +++ | +++ | ++ | +++ | ++ | +++ | ++ | – | |
| 3 | 3 | 2 | 3 | 2 | 3 | 2 | 0 | ||
| L130P | Loop | +++ | +++ | + | +++ | ++ | +++ | ++ | – |
| 2 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| S94P | Loop | +++ | +++ | + | +++ | ++ | +++ | ++ | – |
| 2 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| R100H | Loop | +++ | +++ | + | +++ | ++ | +++ | ++ | – |
| 1 | 2 | 1 | 2 | 2 | 3 | 2 | 0 | ||
| D34N | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| D181A | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 2 | 2 | 3 | 2 | 0 | ||
| D34G | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| E158V | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 2 | 2 | 3 | 2 | 0 | ||
| D179G | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| D179N | Catalytic | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 2 | 2 | 3 | 2 | 0 | ||
| L111P | Loop | + | ++ | + | +++ | ++ | +++ | ++ | – |
| 1 | 2 | 1 | 3 | 2 | 3 | 2 | 0 | ||
| K93R | Loop | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 2 | 1 | 3 | 2 | 0 | ||
| L97P | Loop | – | ++ | + | +++ | ++ | +++ | ++ | – |
| 0 | 2 | 1 | 2 | 1 | 3 | 1 | 0 |
Survival and growth effects of integrated toxic mutants expressed under the GAL1-10 promoter (rad27::GAL-FENCh-Mut) on COM media with different galactose concentrations (3% raffinose was used to examine the effect of low levels of induction) and 1 or 2 mM MMS. High expression media contained 2% galactose; low expression media contained 3% raffinose and 0.01% galactose; very low expression media contained 3% raffinose and 0.004% galactose. ‘Basal expression’ contained 3% raffinose and no galactose (replacing 3% raffinose with 2% glucose yielded no apparent difference in results). Data have been obtained from a dilution pronging experiment following 3 days of incubation at 30°C. Gal = galactose.
aThe domain (catalytic or loop) of each mutation is shown.
bSurvival and cgrowth categories are the same as in Table I.
Since hFEN1 can complement the RAD27 repair function in yeast (Greene et al., 1999), we examined the ability of mutants to complement or interfere with repair of MMS-induced damage. As shown in Table II, the toxic hFEN1 alleles that had replaced the RAD27 locus resulted in MMS sensitivities comparable with those of a Δrad27 mutant under conditions of low (glucose) or somewhat higher basal expression (raffinose). When expression was increased by a low level (0.01%) of galactose, the toxic hFEN1 alleles sensitized the Δrad27 cells to MMS. We conclude that the toxic hFEN1 alleles, even those causing modest toxicity, are not only defective in DNA repair, but they actually increase the sensitivity of a rad27 mutant.
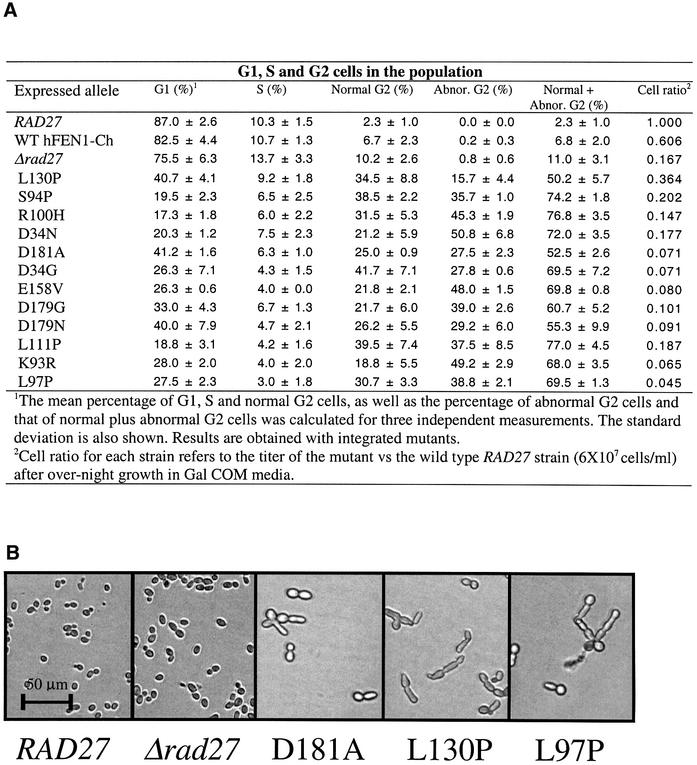
Since the toxic mutants affect growth, we analyzed their impact on cell progression. Arrests at specific points in the cell cycle are indicative of checkpoint responses to lesions. Strains with integrated toxic alleles were incubated in 2% galactose overnight, and the frequencies of G1 cells (unbudded), S-phase cells (small buds) and G2 or G2/M cells (large buds) were determined microscopically (Weinert and Hartwell, 1988). Consistent with the toxic phenotype, there was little growth of the mutants during this period (last column in Figure 3A). As shown in Figure 3A, only a small fraction (up to ∼10%) of the wild-type, the Δrad27 mutant and the wild-type hFEN1-Ch strains formed large buds, typical of G2/M cells. This increased from 18.8 to 41.7% for the toxic mutants. More over, a high percentage of cells had abnormal morphologies, indicative of altered cell division (Figure 3A and B). The reduction in S-phase cells indicates that if DNA lesions arise during replication, they are subject to G2/M checkpoint controls.
Fig. 3. Cell cycle distribution and cell morphology of toxic mutants. Cells (∼105) of strains with integrated FEN1 alleles, wild-type RAD27 and Δrad27 strains were grown overnight in a microtiter plate (200 µl) in medium (Gal COM) providing high levels of induction of the wild-type and toxic hFEN1 proteins. (A) Percentage of toxic mutant cells in G1, S and G2 based on microscopic analysis (Weinert and Hartwell, 1988). Also shown is the ‘cell ratio’, which corresponds to the relative cell titer (mutant to wild-type). (B) Examples of cell morphology after overnight growth in liquid Gal COM media of the wild-type RAD27 strain, and the mutant strains Δrad27 D181A (catalytic), weak toxic L130P (loop) and strong toxic L97P (loop).
Flap cleavage by wild-type human FEN1 and the role of single and double flaps
All the hFEN1 mutant proteins used in this study had His6 tags on their C-termini to facilitate purification. To determine whether this modification affects enzyme activity, wild-type hFEN1 was cloned without the tag and purified according to the methods of O’Jónsson et al. (1998). No differences in activity (DNA binding and catalysis) compared with the His6 tag counterpart were observed (data not shown).
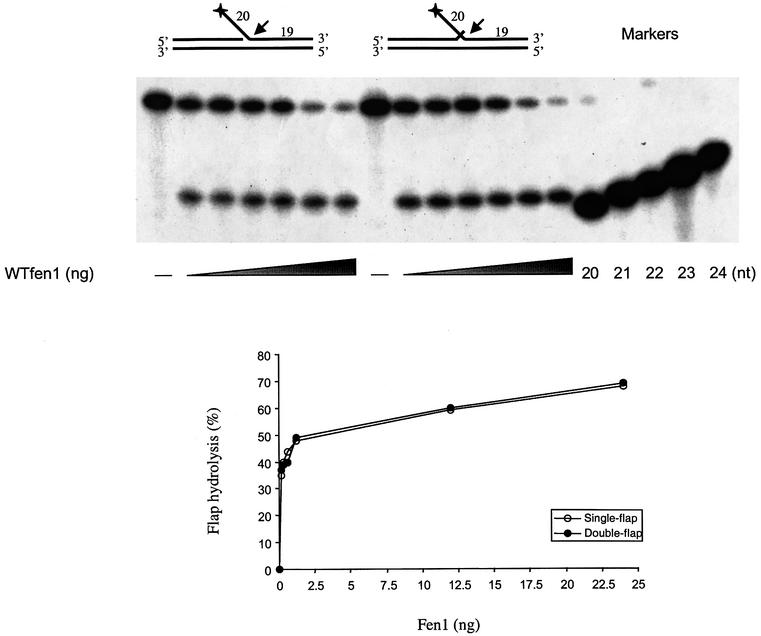
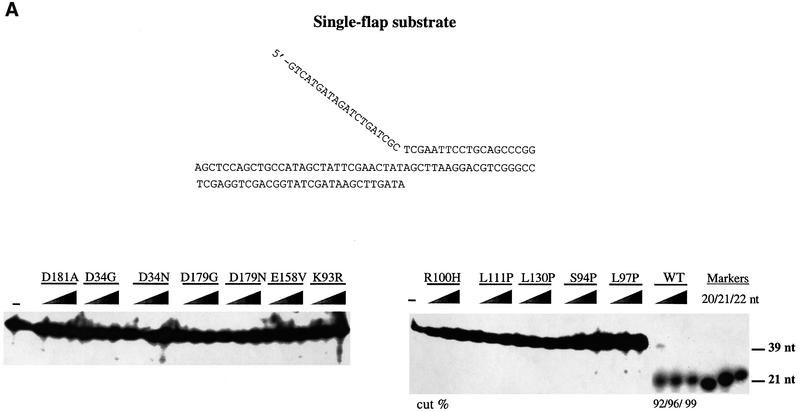
Flap cleavage at a 5′ flap is stimulated by an overlapping 3′ flap for the mouse (Harrington and Lieber, 1995), yeast (Kao et al., 2002), bacterial and archaeal nucleases (Kaiser et al., 1999). We therefore examined the ability of the wild-type tagged hFEN1 protein to cleave a 5′ flap in the presence or absence of a one-base overlapping 3′ flap. Cutting was determined at low salt concentrations, where nuclease activity can be observed in the absence of its accessory protein PCNA (O’Jónsson et al., 1998) and was detected by the release of the radioactively labeled flap as described in Figure 4 (see Materials and methods). Hydrolyzed products were quantified by phosphoimaging. In contrast to FEN1 protein from other organisms, hFEN1 did not exhibit substrate preference for single- versus double-flap structures. The amount of cutting was comparable over the range of nuclease concentrations tested (0.15–25 ng; 3.6–600 fmol), and the rate of cleavage was similar with single- and double-flap substrates. In both cases, the 5′ flap was cut at a single position (21 nucleotide product) between the first two base pairs in the downstream duplex, as reported previously (O’Jónsson et al., 1998). Previous reports indicating two sites of cleavage, one base into the duplex as we observed, plus another site at the flap junction (Xie et al., 2001; Kao et al., 2002), could be due to differences in substrate sequence at the junction.
Fig. 4. Cleavage efficiency and specificity of wild-type human FEN1 toward single- and double-flap DNA substrates. An enzyme titration assay of wild-type hFEN1 (0, 0.15, 0.3, 0.6, 1.2, 12 and 24 ng) was performed with 50 fmol of 5′-radiolabeled substrates in a single- and double-flap cutting assay. Products were separated on 20% urea polyacrylamide gels and visualized by autoradiography (top panel). Quantification was obtained with a PhosphorImager (bottom graph). The sequences of the 20, 21, 22, 23 and 24 oligonucleotide markers correspond to the potential cleavage products of the 5′ flap region.
Toxic human FEN1 mutants retain DNA binding
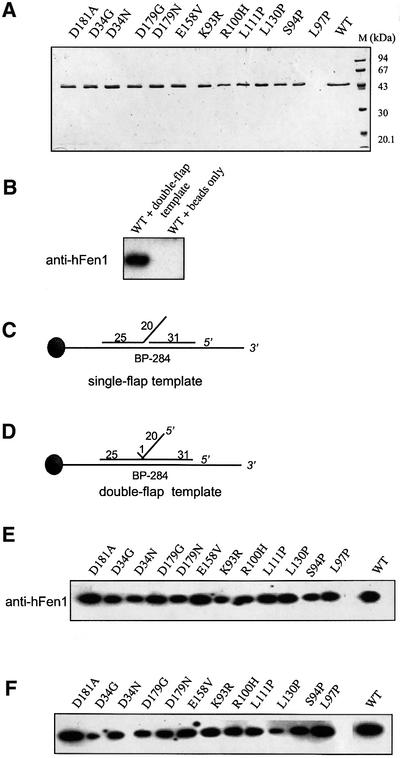
Twelve hFEN1 mutant proteins were expressed in Escherichia coli and purified extensively (Figure 5A). Wild-type and hFEN1 mutants were incubated with single- (Figure 5C) or double-flap (Figure 5D) DNA–beads and tested as outlined in Materials and methods. Bound hFEN1 mutant proteins were detected by immunoblotting using antibody to hFen1. There was no binding to beads without attached DNA for wild-type or mutant proteins (Figure 5B; data not shown). All 12 hFEN1 mutants exhibited DNA binding to both substrates (Figure 5E and F), at levels similar to that of the wild-type hFEN1 even in the presence of excess unlabeled double-stranded DNA (Supplementary figure 1).
Fig. 5. Effects of human FEN1 catalytic site and loop domain mutations on DNA binding. (A) SDS–PAGE analysis of purified wild-type and mutant hFEN1. A 1 µg aliquot of each hFEN1 preparation was separated on a 12% SDS–polyacrylamide gel and stained with Coomassie Blue R-250. (B) Control binding to the beads, where the ability of FEN1 to bind to ‘double-flap template’ or ‘beads only’ is assessed with anti-hFEN1 antibody. Presented are schematic views of the single- (C) and double-flap (D) structures used for the hFEN1 DNA-binding assays. DNA-binding activity of hFEN1 was tested by using DNA substrates attached to magnetic beads via a biotin– streptavidin linkage. The single- and double-flap templates were incubated in a stepwise fashion and processed for immunoblot analysis using anti-hFEN1 antibody. The autoradiograms correspond to proteins bound to the single-flap (E) and double-flap (F) DNA–beads, respectively.
We also found that all mutant and wild-type hFEN1 proteins were able to bind PCNA (data not shown). The ability to bind both PCNA and flap substrates indicates that the mutant proteins are not misfolded.
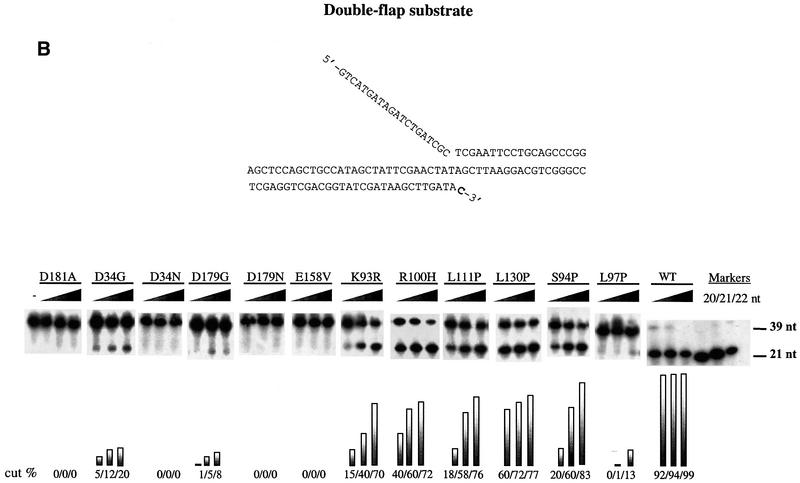
Loop and catalytic site mutants lack single-flap cleavage but differ in cutting at a double flap
The ability of the toxic mutants to cleave single- and double-flap structures was compared with that of wild-type hFEN1. There was almost no cleavage of the single-flap substrate by any of the mutant proteins under conditions where the wild-type protein hydrolyzed 92% (0.5 ng = 0.012 pmol), 96% (5 ng = 0.12 pmol) and 99% (50 ng = 1.2 pmol) of the molecules (Figure 6A). Unlike the situation with the single flap, there was double-flap cleavage for eight of the mutants, as shown in Figure 6B. Mutants could be grouped according to cutting efficiency. There was no activity with catalytic site mutant proteins D34N, D179N, E158V and D181A. All the loop mutants retained at least some cutting activity towards the double flap, from low (L97P, together with catalytic site mutants D34G and D179G) to medium (K93R S94P, R100H, L111P), to medium–high (L130P), when compared with wild-type. The position of cutting was the same as that for wild-type protein (Supplementary figure 2A). Clearly there is reduced activity since, under conditions that allow only 40% cleavage by the wild-type enzyme, there was very little cutting by any of the mutant proteins (Supplementary figure 2B).
Fig. 6. Cleavage efficiency of hFEN1 mutants towards a single- and a double-flap substrate. Activities of buffer alone (negative control, ‘–’), wild-type hFEN1 (positive control, WT) and the 12 mutants were measured in assays designed to examine cleavage of single and double flaps. Proteins were titrated to 50 fmol of substrate and the reaction products were separated on 15% urea polyacrylamide gels and visualized by autoradiography. Quantification was carried out using a PhosphorImager. (A) Single-flap endonucleolytic activity. (B) Double-flap endonucleolytic activity. The 3′ overlapping base is in bold. The amounts of hFEN1 protein used were 0.5 ng (12 fmol), 5 ng (120 fmol) and 50 ng (1.2 pmol).
Thus, the loop appears to have an important role in cleavage of both single and double flaps, with single flaps being much more sensitive to changes in the loop. The toxicity is inversely related to the ability to cut double flaps, based on the results in Tables I and II, where mutants are ranked from top to bottom in increasing order of toxicity and the cleavage activities of the mutants (Figure 6B).
Discussion
FEN1 plays an important role in DNA replication and repair through its cleavage of 5′ flap structures (Bambara et al., 1997; Lieber, 1997). However, the molecular mechanisms by which FEN1 recognizes, binds and cleaves these substrates have not been clarified. Based on our previous analysis of the human and yeast FEN1 genes, we anticipated that a screen of toxic mutants would provide structure–function insights into the role of FEN1 in DNA metabolism. Through a combination of genetics and biochemistry, we have established that the flexible loop, in addition to the catalytic site, is essential for flap cleavage. Moreover, the inverse relationship between extent of toxicity and flap cutting ability suggested that a double flap is the DNA structure that is acted upon by FEN1 in vivo.
Features of toxic mutants
The toxic mutant screen that was developed utilized a Δrad27 background and expression of hFEN1 which had the terminal region replaced by the corresponding yeast region. This replacement allows for more efficient binding by the yeast PCNA (Gary et al., 1999). As proposed for the rad27-n mutant (Gary et al., 1999), the interaction of a nuclease-deficient mutant with PCNA in vivo may ‘freeze’ the mutant protein at a flap leading somehow to toxicity, possibly by preventing the action of an alternative nuclease.
We identified 17 (Figure 2A), and characterized 12 toxic mutants that resulted in various levels of growth inhibition and reduced survival. The toxicity appeared to result from interference with replication similar to the rad27-n mutation of RAD27 that exhibits a stronger mutator phenotype than a complete deletion of RAD27 when overexpressed in wild-type RAD27 (Greene et al., 1999). The mutants had a strong impact on cell cycle progression, consistent with a defect in replication. The toxic mutants also appeared to impact on repair, since low expression from integrated toxic alleles in a Δrad27 background resulted in MMS sensitivity comparable with, or greater than, that of a Δrad27 mutant (Table II). There may even be a synergistic effect of MMS and toxicity on yeast growth and survival; however, this requires further analysis. Even the weakest mutants (L130P, S94P and R100H) had no apparent positive effect on DNA repair when expressed at low levels. Similar results were also obtained for mutants expressed on plasmids (data not shown).
While there was no apparent dominance in a RAD27 wild-type background, the toxic mutants exhibited partial dominance when expressed in a RAD27 Δrad51 mutant, which is deficient in the recombinational repair of double-strand breaks (DSBs), indicating that toxicity may involve the production of DSBs. The enrichment of G2 cells (Figure 3A) is consistent with DSBs being produced in the mutants. The toxic mutants also exhibited dominance over the mutant rad27-p, which is defective in PCNA binding and which has little phenotypic effect on its own (Gary et al., 1999; data not shown), and over wild-type hFEN1 and hFEN1-Ch (Table I).
Relationship between toxic mutations, flaps and human FEN1 structure
The screen for hFEN1 toxic mutants identified the catalytic sites plus several amino acids within the loop domain. The fact that there were several examples where more than one mutation was recovered at the same site suggested that the screen was nearly saturated for all sites that could result in toxicity. It is possible, however, that there are additional sites in the loop that are important in function, and these might be revealed through other screens.
Catalytic site
In our screen for toxic mutants, we identified all the highly conserved catalytic residues of the N and I hFEN1 conserved regions except D233 (Shen et al., 1997; Frank et al., 1998): D34, D86, E158, E160, D179 and D181. In hFEN1, these seven conserved amino acids were assigned to the active site pocket, surrounding two tightly bound Mg2+ ions (Shen et al., 1997; Hosfield et al., 1998; Hwang et al., 1998). No other residues in these domains were isolated in our screen, which suggests that toxicity is due to a lack of nucleolytic function under conditions where flap binding is retained. A simple loss of nuclease function (such as found for a RAD27 deletion mutant) would not be expected to result in toxicity.
Loop domain
The flexible loop has been proposed to be required for specific recognition of a Y junction between the flap and double-stranded DNA and/or cleavage (Hwang et al., 1998), based on the consequences of 22 and 32 amino acid deletions in the loop domain. However, because of the sizes of the deletions, the specific role of the loop could not be addressed. In addition to the mutations of conserved amino acids in the catalytic domain, we found a comparable number of single amino acid changes within residues across the loop region that could alter endonuclease activity to varying extents (K93, S94, L97, R100, L111 and L130, Figure 2). The Q113 and T127 amino acids may also be important since toxic mutants containing these mutations also had an E106G mutation, which on its own retains wild-type activity. Since the toxic Rad27 and FEN1 mutant proteins retained the ability to bind flap DNA (Figure 5E and F), as well as PCNA (Gary et al., 1999; and data not shown), the proteins in vitro are likely to be folded correctly. The toxic mutant proteins were unable to cut single flaps and had reduced activity towards double flaps. In fact, there was an approximate inverse relationship between toxicity and double-flap cleavage for the loop mutants. This leads us to conclude that the loop domain has an essential role in hFEN1 cleavage activity, since not only large loop deletions (Hwang et al., 1998) but even single amino acid substitutions in the loop domain can significantly impair hFEN1 cleavage. The sites of loop mutations are conserved in human, mouse and S.cerevisiae. We note that there is not a complete inverse correlation between toxicity and double-flap cleavage since some of the catalytic site mutants, which exhibited no cleavage, were not the most toxic. There may be additional factors that can determine the specific level of toxicity. Possibly, changes in the flexibility of hFEN1 due to the changes in the loop might amplify the toxicity.
Based on the size of the flexible loop and on the basic charge distribution on the inside of the loop, it has been proposed that this domain could interact with DNA flaps in two ways, either by threading of the single-stranded DNA flap through the loop or by FEN1 adopting a ‘thumb’ structure that can partially encompass the flap and slide towards the 5′ junction (Bornarth et al., 1999). Unlike the structure of the N-terminal fragment of E.coli DNA topoisomerase I that folds to generate a hole large enough to encircle a double-stranded DNA (Lima et al., 1994), the flexible loop structures in FEN1 and T5 exonuclease could only form a hole large enough to accommodate a single-stranded DNA (Ceska et al., 1996; Hosfield et al., 1998; Hwang et al., 1998). The inability of FEN1 to process partially double-stranded flaps (Murante et al., 1995) or flaps adopting secondary structure (Henricksen et al., 2000) is consistent with limited space in the loop. In either case, the loop may also serve to orient the flap properly with the catalytic domain. The 4% of α-helical conversion which has been detected in the hFEN1 protein after substrate binding has been attributed to the tightening of the flexible loop around the DNA substrate (Kim et al., 2001). The flexible loop region of hFEN1 extends from amino acid 87 to 134, based on sequence comparison with the M.jannaschii FEN1 (Hwang et al., 1998) and on the model presented in Figure 2B. From crystal structure models of FEN1 homologs, the 48 amino acids of the flexible loop represent the only stretch of disordered structure long enough to accommodate the turns associated with α-helical conversion that are observed when hFEN1 binds to a flap substrate. It remains to be investigated whether the single amino acid changes we identified affect the α-helical conversion.
Flap structure
A stimulatory effect of a small 3′ flap in the cleavage of a 5′ flap has been reported for mouse FEN1 (Harrington and Lieber, 1995), yeast Rad27 (Xie et al., 2001; Kao et al., 2002), and P.furiosus and A.fulgidus FEN1 (Kaiser et al., 1999). It is the presence, rather than the identity of the 3′ flap that activates the cleavage. The fact that the 3′ nucleotide of the upstream primer is unpaired during the cleavage reaction suggests that there is a region or pocket in the enzyme that specifically recognizes the 3′ protrusion. Such a pocket may help to position the catalytic site of FEN1 proteins properly relative to the flap (Kaiser et al., 1999). We found that the 3′ flap did not stimulate 5′ flap cutting by human wild-type FEN1. The position of flap cleavage was one base from the Y junction in the duplex DNA region regardless of the flap structure. All the toxic mutant proteins could bind single- and double-flap DNA (Figure 5), and lacked nuclease activity toward single-flap structures (Figure 6A). Surprisingly, however, the loop mutants exhibited different levels of double-flap cutting, although the position of cutting was the same as for wild-type protein (Figure 6B and Supplementary figure 2A). This finding suggested that the double-flap structure is an important intermediate in DNA metabolism that is required for the proper function of hFEN1.
We propose that the 3′-terminal nucleotide of the upstream strand in the double-flap substrate is sequestered by the hydrophobic pocket of hFEN1 (Kaiser et al., 1999) to prevent replication extension and reduce strand displacement by DNA polymerases during DNA replication or repair. The interaction with the 3′ nucleotide could also help to position the flap precisely relative to the catalytic site via the loop region. This would ensure efficient cutting at a precise site in the Y structure so as to create a ligatable nick for DNA ligase I with the 3′ flap (Maga et al., 2001). Moreover, this is consistent with our observation that, under the conditions used, wild-type and mutant FEN1 proteins cleave one nucleotide downstream in the duplex (Figures 4 and 6B; Supplementary figure 2A), and this double-flap cutting activity was inversely proportional to its toxicity in vivo (compare the results in Figure 6B with those in Tables I and II). This model suggests that the formation of a catalytically competent substrate–enzyme complex requires structural rearrangement of hFEN1, and this is mediated by the flexible loop. The approximate inverse relationship between toxicity and double-flap cleavage by loop mutants might reflect differences in flexibility of hFEN1. Further characterization using additional mutants along with crystal structure analysis of FEN1 bound to substrate will help to elaborate the role of FEN1 in DNA metabolic processes.
Implications of toxic human FEN1 mutants
The characterization of toxic hFEN1 mutants in yeast has enabled us to identify several sites in the flexible loop required for flap processing. Altering the loop structure may lead to toxicity of hFEN1 comparable with or even stronger than the toxicity of catalytic mutants, where the protein remains bound to a flap. There are several ways that the toxic mutants may be useful in addressing issues of damage-induced or replication-related genome instability. For example, the toxic mutants could be employed to identify factors that can impact on hFEN1 function. Based on the results of expressed toxic mutants in yeast, it will be interesting to investigate their consequences in mammalian cells. Since altered hFEN1 can affect genome stability in yeast, the mutants might be expected to increase chromosome alterations and mutations in human cells. While FEN1 polymorphisms have not been identified, such mutants could be associated with genetic diseases that are revealed through epidemiology studies.
Materials and methods
Strains and plasmids
The S.cerevisiae strains used in this study are isogenic to a wild-type strain E133 (MATα, ade5-1, lys2-12A, trp1-289, his7-2, leu2-3,112, ura3-52), a derivative of CG379 containing the A12 microsatellite in LYS2 described in Greene et al. (1999). Strains E275 and DAG158 contain the Δrad27-null mutation and the rad27-p mutation F346A/F347A, which prevents binding to PCNA, respectively (Gary et al., 1999). All genetic and molecular procedures were performed as previously described (Greene et al., 1999; Storici et al., 2001).
The vector YEp195SpGAL used in several constructions is a 2 µm derivative, high-copy, expression plasmid, containing the URA3 selectable marker (Greene et al., 1999). FEN1-n and FEN1-n,Ch represent the human FEN1 and its chimeric derivative carrying the D181A nuclease mutation (Greene et al., 1999). Plasmids YEp195SpGALFEN1, YEp195SpGALFEN1-Ch, YEp195SpGALFEN1-n and YEp195Sp GALFEN1-n,Ch contain the sequence of the human FEN1 ORF, the hFEN1-Ch, the hFEN1-n and the hFEN1-n,Ch sequence, respectively (Greene et al., 1999). Plasmids YEp195SpGALFEN1 and YEp195Sp GALFEN1-Ch were modified by replacing the XbaI–BstXI hFEN1 fragment with the same XbaI–BstXI DNA fragment from pET23dhFen1-C-His (A.Dutta, personal communication) in order to eliminate the Y83H polymorphism present in the hFEN1 sequence in the original vectors. The new constructs were termed: pGALFEN1-WT and pGALFEN1-ChWT.
Toxic mutations identified with the yeast screen were reproduced on pET23dhFen1-C-His, pGALFEN1-WT and pGALFEN1-ChWT by in vitro mutagenesis (Quick Change Mutagenesis Kit, Stratagene, La Jolla, CA). All constructs were confirmed by sequence analysis. Replacement of the RAD27 ORF with that of hFEN1 and hFEN1-Ch, expressed under the endogenous RAD27 promoter (strains FS-1 and FS-3), was achieved using the recently developed delitto perfetto approach (Storici et al., 2001). Similarly, all mutants expressed under the GAL1-10 promoter on pGALFEN1-Ch were integrated into the yeast genome in place of RAD27 ORF. Correct integration was determined by colony PCR followed by sequencing of the PCR products, as described in Storici et al. (2001). From previous studies, it has been shown that the level of gene expression from the GAL1-10 promoter can be regulated by altering the concentration of galactose (Kokoska et al., 2000; A.Inga and M.A.Resnick, personal communication). Nucleotide sequences of all the oligonucleotides used in this study are available upon request.
Plasmid pGALHOT (Bennett et al., 1993) was used to switch the mating type of strains E133, E275 and DAG158.
Random generation of hFEN1 toxic mutants
Mutations were introduced into the FEN1-Ch sequence by PCR mutagenesis using YEp195SpGALFEN1-Ch as template and using primers complementary to sequences ∼100 nucleotides upstream from the ATG and 60 nucleotides downstream from the stop codon of FEN1-Ch ORF, respectively (see the Supplementary data for details).
Nucleic acids and proteins
Single- and double-flap duplex molecules were prepared from oligonucleotides purchased from Microsynth GmbH, Switzerland. All oligonucleotides used for the biochemical experiments are described in Supplementary table II. Prior to annealing, oligonucleotide Ft02_01 was labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. Free ATP was removed on a MicroSpin™ G-25 column. To generate the substrates, the appropriate primers were mixed at a 1:1 molar ratio in 20 mM Tris–HCl pH 7.4, 150 mM NaCl, heated to 75°C, and slowly cooled to room temperature. The single-flap duplex molecule contains the upstream primer Ft02_02 and the radiolabeled downstream Ft02_01 primer annealed to the complementary Ft02_04 oligonucleotide. The double-flap duplex molecule contains the upstream primer Ft02_06 and the radiolabeled downstream primer Ft02_01 annealed to the complementary Ft02_04 oligonucleotide. The pGEX-3X plasmid expressing human wild-type GST–PCNA fusion protein was provided by R.Freire (personal communication).
Expression and purification of hFEN1-C-His and hFEN1-C-His mutants
The plasmid pET23d/hFen1-C-His was used as a template for site-directed mutagenesis to generate hFEN1 mutants. Wild-type and mutant hFEN1C-His were expressed in E.coli BL21(DE3)pLysS. For purification, 250 ml cultures were grown to an A600nm of ∼0.5 in LB with 100 µg/ml ampicillin at 37°C. Expression was induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mM followed by growing for 4 h. The bacterial pellets were thawed for 7 min at 37°C and resuspended in 3.5 ml of buffer A (50 mM NaPO4 pH 8.0, 300 mM NaCl, 20 mM imidazole-HCl, 1 mM phenylmethylsulfonide fluoride). Cells were disrupted by a brief sonication and insoluble material was removed by centrifugation for 15 min at 15 000 g. The supernatant was loaded onto Ni-NTA spin columns (Qiagen) previously equilibrated with buffer A. The proteins were eluted with 300 µl of buffer B (50 mM HEPES–NaOH pH 7.5, 300 mM NaCl, 250 mM imidazole-HCl). The purity of the proteins was evaluated on 12% SDS– polyacrylamide gels. Protein concentrations were determined by the method of Bradford (1976) using the Bio-Rad reagent, and bovine serum albumin (BSA) as a standard. Endonuclease contaminations were tested by incubating 0.5 µg of each mutant with 0.5 µg of supercoiled pBlueScript DNA in buffer C [50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol (DTT)] for 30 min at 37°C followed by separation on a 1% (w/v) agarose gel. No degradation was detected.
DNA binding assays
A biotinylated 284 nucleotide fragment homologous to M13mp18 DNA from 6140 to 6423 bp was PCR amplified with upstream biotinylated primer BP-6140 (30 nucleotides) and downstream primer BP-6423 (25 nucleotides). The biotinylated 284 nucleotide fragment was immobilized on streptavidin–magnetic beads (Dynabeads) in buffer D (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 2 M NaCl) for 2 h 30 min at room temperature. Unbound products were washed off the beads twice with buffer E (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 M NaCl), and the duplex DNA was denaturated with 0.1 M NaOH. Non-biotinylated nucleotides were removed with buffer E. The beads-bound biotinylated 284 nucleotide fragment was annealed with D-45 and F-31 or F-32 and F-31 primers (see Supplementary table II) to create the single- and double-flap templates, respectively. DNA–beads-bound templates were washed with buffer E and resuspended in buffer F (30 mM HEPES–NaOH pH 7.5, 8 mM magnesium acetate, 0.2 mg/ml BSA, 1 mM DTT). Binding assays were carried out in a final volume of 16 µl. A 50 fmol concentration of DNA templates was coated with 1.5 pmol of replication protein A (RPA) for 2 min on ice and washed with buffer G [30 mM HEPES–NaOH pH 7.5, 5% (v/v) glycerol, 0.1 mg/ml BSA, 1 mM DTT, 0.01% (v/v) NP-40, 0.1 mM EDTA]. Reactions were performed in buffer F with 1.2 pmol of wild-type and mutant hFen1 for 1 min at 30°C. Then, magnetic beads were washed off with buffer G and immediately resuspended into SDS sample buffer. DNA-binding proteins were resolved on a 12% SDS– polyacrylamide gel and subsequently transferred to a PVDF membrane and probed with anti-hFen1 rabbit purified antibody (produced at the animal facility in the Institute of Animal Breeding of the University of Zürich) followed by horseradish peroxidase (HRP)-conjugated horse anti-rabbit IgG and enhanced chemiluminescence (ECL).
Endonuclease assays
Endonuclease assays were performed in a final volume of 12.5 µl containing 40 mM Tris–HCl pH 7.5, 10 mM MgCl2, 5 mM DTT, 200 µg/ml BSA and 50 fmol of DNA substrates (single- and double-flap substrates). After the addition of wild-type or hFen1 mutants, the reactions were incubated at 30°C for 15 min and stopped with 2.5× stop buffer [95% formamide, 20 mM EDTA, 0.05% (w/v) each of bromophenol blue and xylene cyanol]. The products were separated on 15 or 20% denaturating gels, visualized by autoradiography and quantified on a PhosphorImager using the Image-Quant software (Molecular Dynamics).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Bennett Van Houten, Farid Kadyrov and Thomas Darden for critically reading the manuscript, Laura Weston for technical help, and Yong Hwang Jin, Alberto Inga, Kirill Lobachev and Kartikeyan Gopalakrishnan for advice and helpful discussions. This work was supported by the Swiss National Science Foundation (grant 31.61361.00) and by the Canton of Zürich to G.H. and U.H.
References
- Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- Bennett C.B., Lewis,A.L., Baldwin,K.K. and Resnick,M.A. (1993) Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl Acad. Sci. USA, 90, 5613–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Wu,B., Chi,E., Kim,K.H., Trautman,J.K. and Carroll,D. (1998) Characterization of FEN-1 from Xenopus laevis. cDNA cloning and role in DNA metabolism. J. Biol. Chem., 273, 34222–34229. [DOI] [PubMed] [Google Scholar]
- Bornarth C.J., Ranalli,T.A., Henricksen,L.A., Wahl,A.F. and Bambara,R.A. (1999) Effect of flap modifications on human FEN1 cleavage. Biochemistry, 38, 13347–13354. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Ceska T.A., Sayers,J.R., Stier,G. and Suck,D. (1996) A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature, 382, 90–93. [DOI] [PubMed] [Google Scholar]
- Chen C. and Kolodner,R.D. (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet., 23, 81–85. [DOI] [PubMed] [Google Scholar]
- Frank G., Qiu,J., Somsouk,M., Weng,Y., Somsouk,L., Nolan,J.P. and Shen,B. (1998) Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J. Biol. Chem., 273, 33064–33072. [DOI] [PubMed] [Google Scholar]
- Gary R., Park,M.S., Nolan,J.P., Cornelius,H.L., Kozyreva,O.G., Tran,H.T., Lobachev,K.S., Resnick,M.A. and Gordenin,D.A. (1999) A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol., 19, 5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene A.L., Snipe,J.R., Gordenin,D.A. and Resnick,M.A. (1999) Functional analysis of human FEN1 in Saccharomyces cerevisiae and its role in genome stability. Hum. Mol. Genet., 8, 2263–2273. [DOI] [PubMed] [Google Scholar]
- Harrington J.J. and Lieber,M.R. (1995) DNA structural elements required for FEN-1 binding. J. Biol. Chem., 270, 4503–4508. [DOI] [PubMed] [Google Scholar]
- Henricksen L.A., Tom,S., Liu,Y. and Bambara,R.A. (2000) Inhibition of flap endonuclease 1 by flap secondary structure and relevance to repeat sequence expansion. J. Biol. Chem., 275, 16420–16427. [DOI] [PubMed] [Google Scholar]
- Hosfield D.J., Frank,G., Weng,Y., Tainer,J.A. and Shen,B. (1998) Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J. Biol. Chem., 273, 27154–27161. [DOI] [PubMed] [Google Scholar]
- Hwang K.Y., Baek,K., Kim,H.Y. and Cho,Y. (1998) The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.Nat. Struct. Biol., 5, 707–713. [DOI] [PubMed] [Google Scholar]
- Kaiser M.W., Lyamicheva,N., Ma,W., Miller,C., Neri,B., Fors,L. and Lyamichev,V.I. (1999) A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem., 274, 21387–21394. [DOI] [PubMed] [Google Scholar]
- Kao H.I., Henricksen,L.A., Liu,Y. and Bambara,R.A. (2002) Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem., 277, 14379–14389. [DOI] [PubMed] [Google Scholar]
- Kim C.Y., Park,M.S. and Dyer,R.B. (2001) Human flap endonuclease-1: conformational change upon binding to the flap DNA substrate and location of the Mg2+ binding site. Biochemistry, 40, 3208–3214. [DOI] [PubMed] [Google Scholar]
- Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska R.J., Stefanovic,L., DeMai,J. and Petes,T.D. (2000) Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol. Cell. Biol., 20, 7490–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li,J., Harrington,J., Lieber,M.R. and Burgers,P.M. (1995) Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem., 270, 22109–22112. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- Lima C.D., Wang,J.C. and Mondragon,A. (1994) Three-dimensional structure of the 67K N-terminal fragment of E.coli DNA topoisomerase I. Nature, 367, 138–146. [DOI] [PubMed] [Google Scholar]
- Maga G., Villani,G., Tillement,V., Stucki,M., Locatelli,G.A., Frouin,I., Spadari,S. and Hübscher,U. (2001) Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase δ by the concerted action of replication protein A, proliferating cell nuclear antigen and flap endonuclease-1. Proc. Natl Acad. Sci. USA, 98, 14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murante R.S., Rust,L. and Bambara,R.A. (1995) Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem., 270, 30377–30383. [DOI] [PubMed] [Google Scholar]
- O’Jónsson Z.O., Hindges,R. and Hübscher,U. (1998) Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J., 17, 2412–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M.A. and Cox,B.S. (2000) Yeast as an honorary mammal. Mutat. Res., 451, 1–11. [DOI] [PubMed] [Google Scholar]
- Schweitzer J.K. and Livingston,D.M. (1998) Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet., 7, 69–74. [DOI] [PubMed] [Google Scholar]
- Shen B., Nolan,J.P., Sklar,L.A. and Park,M.S. (1996) Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J. Biol. Chem., 271, 9173–9176. [DOI] [PubMed] [Google Scholar]
- Shen B., Nolan,J.P., Sklar,L.A. and Park,M.S. (1997) Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res., 25, 3332–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Qiu,J., Hosfield,D. and Tainer,J.A. (1998) Flap endonuclease homologs in archaebacteria exist as independent proteins. Trends Biochem. Sci., 23, 171–173. [DOI] [PubMed] [Google Scholar]
- Shibata Y. and Nakamura,T. (2002) Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem., 277, 746–754. [DOI] [PubMed] [Google Scholar]
- Storici F., Lewis,L.K. and Resnick,M.A. (2001) In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol., 19, 773–776. [DOI] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S.cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- Tom S., Henricksen,L.A. and Bambara,R.A. (2000) Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem., 275, 10498–10505. [DOI] [PubMed] [Google Scholar]
- Weinert T.A. and Hartwell,L.H. (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science, 241, 317–322. [DOI] [PubMed] [Google Scholar]
- White P.J., Borts,R.H. and Hirst,M.C. (1999) Stability of the human fragile X (CGG)(n) triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol. Cell. Biol., 19, 5675–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Liu,Y., Argueso,J.L., Henricksen,L.A., Kao,H.I., Bambara,R.A. and Alani,E. (2001) Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability and flap cleavage. Mol. Cell. Biol., 21, 4889–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]