Abstract
Yra1p and Sub2p are components of the TREX complex, which couples transcription elongation with nuclear export of mRNAs. Here, we report a genetic interaction between Yra1p and a conserved protein Sac3p, which previously was found to interact with Sub2p. In vivo, Sac3p forms a stable complex with Thp1p, which was reported to function in transcription elongation. In addition, Sac3p binds to the mRNA exporter Mex67p–Mtr2p and requires the nucleoporin Nup1p to dock at the nuclear side of the nuclear pore complex (NPC). Significantly, mutations in Sac3p or Thp1p lead to strong mRNA export defects. Taken together, our data suggest that the novel Sac3p–Thp1p complex functions by docking the mRNP to specific nucleoporins at the nuclear entrance of the NPC.
Keywords: Mex67p/mRNA export/nuclear pore complex/nucleoporin/Sac3p
Introduction
In eukaryotes, mRNAs need to be exported from the nucleus before they are translated in the cytoplasm. Transport of mRNAs occurs through nuclear pore complexes (NPCs), large macromolecular assemblies of ∼60 MDa in yeast, which consist of ∼30 different nucleoporins (Rout and Aitchison, 2001; Vasu and Forbes, 2001). The NPCs are made up of several distinct structural elements. These are the spoke complex, which spans the double nuclear membrane and forms the central gated channel, the cytoplasmic filaments, which are attached to the cytoplasmic ring, and the nuclear basket, which is attached to the nucleoplasmic ring of the NPC (Stoffler et al., 1999). While most nucleoporins occur on both sides, some are present on only one side of the NPC. Among the asymmetrically distributed nucleoporins are Nup1p, Nup60p and Nup2p, which are nuclear basket components (Rout et al., 2000; Solsbacher et al., 2000; Denning et al., 2001; Dilworth et al., 2001). These latter nucleoporins could provide docking or release sites for transport complexes to or from the NPCs, respectively.
Nucleocytoplasmic transport of proteins and nuclear export of rRNAs, tRNAs and SRP RNA is mediated predominantly by transport receptors of the importin-β family (Weis, 2002). These shuttling receptors bind to their cargos and transiently interact with phenylalanine– glycine (FG) repeats of nucleoporins. The binding of the β-family transport receptors to cargo is controlled by the small GTPase Ran (Weis, 2002). In contrast, the mechanism of nuclear export of bulk mRNA differs from the other nucleocytoplasmic transport routes as it appears not to be directly dependent on RanGTP (Clouse et al., 2001; Reed and Hurt, 2002). In addition, the general export receptor for mRNAs, Mex67p–Mtr2p in yeast and Tap–p15 in metazoans, does not belong to the importin-β family (Conti and Izaurralde, 2001; Reed and Hurt, 2002). However, like their importin β-family cousins, Mex67p and Tap can bind directly to FG repeat-containing nucleoporins (Conti and Izaurralde, 2001; Grant et al., 2002). Since the Mex67p–Mtr2p complex also binds to mRNA, this mRNA exporter can move the mRNP through the nuclear pore channel using the FG repeats of nucleoporins as transient docking sites (Sträßer et al., 2000).
Yra1p, another key player of the conserved mRNA export machinery, belongs to the conserved REF family. REF family members are nuclear at steady state and contain a central RNA recognition motif (RRM) and highly conserved N- and C-termini, which constitute binding sites for Mex67p/Tap as well as RNA (Sträßer and Hurt, 2000; Stutz et al., 2000; Rodrigues et al., 2001). Thus, Yra1p was proposed to recruit the mRNA exporter to the mRNA (Sträßer and Hurt, 2000; Zenklusen et al., 2001). This model was corroborated further by the characterization of Aly, the metazoan homolog of Yra1p. Aly was shown to be recruited to the mRNP during splicing and also binds directly to Tap (Zhou et al., 2000; Rodrigues et al., 2001). Furthermore, Aly is a component of the exon–exon junction complex (EJC) (Le Hir et al., 2000, 2001; Lejeune et al., 2002). This complex associates with the spliced mRNA ∼20 nucleotides upstream of the exon–exon junction in a splicing-dependent manner. Interestingly, Aly was identified initially as a coactivator of two transcription factors (LEF-1 and AML) (Bruhn et al., 1997). In addition, Yra1p is recruited to the mRNA during transcription (Lei et al., 2001). Moreover, Yra1p/Aly interacts directly with the conserved protein Sub2p in yeast (Sträßer and Hurt, 2001) and UAP56 in mammals (Luo et al., 2001). It was suggested that Sub2p/UAP56 plays a role in recruiting Yra1p/Aly to the mRNA during splicing and thereby performs its crucial role in mRNA export (Luo et al., 2001; Sträßer and Hurt, 2001). However, Sub2p is required for export of mRNAs derived from intronless genes as well as for export of spliced mRNAs (Jensen et al., 2001; Sträßer and Hurt, 2001).
Interestingly, Yra1p and Sub2p are members of the TREX (transcription and mRNA export) complex, which also contains the THO complex members Tho2p, Hpr1p, Mft1p and Thp2p (Sträßer et al., 2002). Deletion of any of the members of the THO complex leads to inhibition of transcription elongation and impairment of mRNA export (Aguilera, 2001; Sträßer et al., 2002). Conversely, it has been shown that mutation of components of the mRNA export machinery such as SUB2, YRA1, MEX67 and MTR2 leads to defective transcription (Jimeno et al., 2002). Since Hpr1p and Tho2p are recruited to activated genes during gene expression, it was suggested that the TREX complex couples transcription elongation with the export of the mRNA (Sträßer et al., 2002).
Here, we report the identification of two further genes isolated in a synthetic lethal screen with the yra1-ΔRRM allele, SAC3 and NUP60. Both components together with another nuclear basket nucleoporin Nup1p function in mRNA export at the nucleoplasmic side of the NPC. Importantly, in vivo, Sac3p forms a complex with Thp1p, which is also required for mRNA export. Moreover, Sac3p directly binds to the mRNA exporter Mex67p–Mtr2p. Thus, the Sac3p–Thp1p complex is involved in mRNA export with a distinct role at the nuclear site of the NPC.
Results
YRA1 interacts genetically with SAC3, NUP60 and NUP1
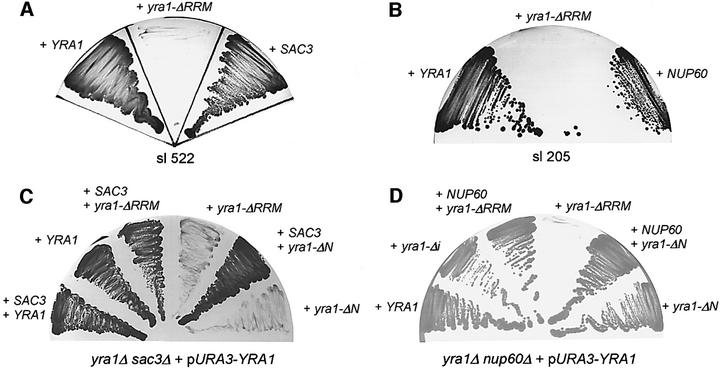
Previously, we performed a synthetic lethal (sl) screen with a yra1 mutant allele (yra1-ΔRRM) to find components of the mRNA export machinery in yeast. We identified Sub2p, an intranuclear RNA helicase that couples splicing with mRNA export (Luo et al., 2001; Sträßer and Hurt, 2001), and the TREX complex, which couples transcription elongation with export of mRNA (Sträßer et al., 2002). Here, we characterize two further sl mutants isolated in this screen, which are complemented by SAC3 (Figure 1A) and NUP60 (Figure 1B), respectively.
Fig. 1. Genetic interaction of YRA1 with SAC3 and NUP60. (A and B) Identification of SAC3 and NUP60 in the sl screen with yra1-ΔRRM. Strains sl 522 (A) and sl 205 (B) were transformed with plasmids harboring YRA1, yra1-ΔRRM, SAC3 or NUP60. They were grown for 5 days on 5-fluoro-orotic acid (FOA)-containing plates. Colony formation indicates complementation of the sl phenotype. (C and D) Genetic relationships between YRA1 and SAC3 or YRA1 and NUP60. The indicated double-disrupted strains harboring plasmid-borne YRA1 (pURA3-YRA1) were transformed with the indicated plasmid-borne gene constructs. Growth was analyzed on FOA-containing plates. No growth indicates synthetic lethality.
In order to confirm these genetic interactions, the yra1-ΔRRM mutant allele was combined directly with sac3Δ or nup60Δ. This revealed that yra1-ΔRRM and also yra1-ΔN are sl with sac3Δ (Figure 1C). Moreover, yra1-ΔRRM, but not yra1-ΔN, is sl with nup60Δ (Figure 1D). Interestingly, yra1-ΔRRM is also sl with nup1Δ and nup2Δ mutant alleles (Table I). Notably, Nup60p, Nup1p and Nup2p are asymmetrically located nucleoporins, all attached to the nuclear basket of the NPC (see Introduction). In contrast, yra1-ΔRRM is not sl with mutant alleles of NUP116, a nucleoporin crucially involved in mRNA export (Wente and Blobel, 1993; Sträßer et al., 2000) (Table I).
Table I. Genetic interactions between YRA1, SAC3, NUP1, NUP60 and THP1 alleles.
| Allele | sl | With |
|---|---|---|
| yra1-ΔRRM | + | sac3Δ, nup1Δ, nup60Δ, nup2Δ, thp1Δ |
| – | nup116Δ | |
| sac3Δ | + | yra1-ΔN, mex67-5, mtr2-9, nup116Δ, gle2Δ |
| – | sub2-85, mlp1Δ, mlp2Δ, nup1Δ, nup60Δ, nup2Δ, nup42Δ, thp1Δ | |
| nup1Δ | + | nup60Δ |
| – | sub2-85, thp1Δ | |
| nup60Δ | – | mex67-5, sub2-85, thp1Δ |
| thp1Δ | + | yra1-ΔN, mex67-5 |
To assess further the genetic network around SAC3, we tested for genetic interactions with known factors involved in mRNA export. We found that sac3Δ is sl with mutant alleles of MEX67, MTR2, NUP116 and GLE2, but not with mutant alleles of SUB2, MLP1, MLP2, NUP2, NUP60, NUP42 and NUP1 (see Table I). Previously, it was shown that sac3Δ is sl with cdc23-1, xpo1-1 and yrb2Δ (Jones et al., 2000). In contrast to sac3Δ, nup60Δ is not sl with mex67 or sub2 mutant alleles (see Table I).
Sac3p, Nup60p and Nup1p are involved in mRNA export
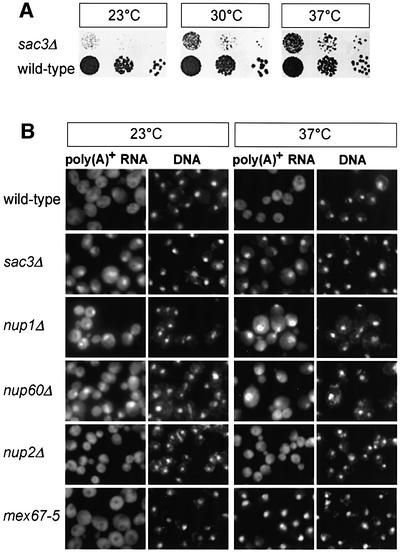
The genetic linkage of SAC3, NUP60 and NUP1 to YRA1 pointed to a role for these components in mRNA export. Therefore, we analyzed whether poly(A)+ RNA export is inhibited in cells disrupted for SAC3, NUP60 or NUP1. For SAC3, we tested two different sac3 knock-out strains, which vary slightly in their growth at different temperatures (see Supplementary table I, available at The EMBO Journal Online). As shown in Figure 2A, sac3Δ cells grow slowly at 23 and 30°C, but are less affected at 37°C. Importantly, sac3Δ strains strongly accumulate poly(A)+ RNA in the nucleus at all the tested temperatures (23 and 37°C; Figure 2B). This mRNA export defect is specific, since no nuclear accumulation of tRNA, SRP RNA or ribosomal 60S and 40S subunits was observed in sac3Δ cells, and nuclear import of nuclear localization sequence (NLS)–reporter constructs was also normal (Supplementary figure 1).
Fig. 2. mRNA export is defective in SAC3, NUP1 and NUP60 mutants. (A) SAC3 deletion causes slow cell growth. Wild-type and sac3Δ cells were spotted in 10–1 dilutions onto YPD plates and growth was analyzed after 3 days at the indicated temperatures. (B) Deletion of SAC3, NUP1 or NUP60 causes an mRNA export defect. Wild-type, sac3Δ, nup1Δ, nup60Δ, nup2Δ and mex67-5 cells were grown at the indicated temperatures and localization of poly(A)+ RNA was assessed by oligo(dT) in situ hybridization. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI).
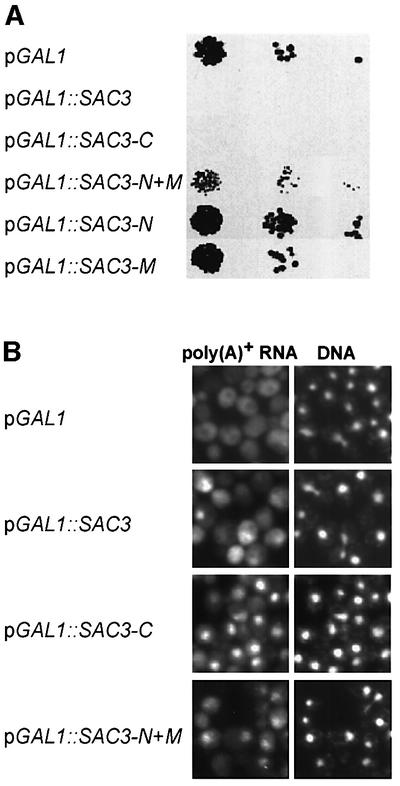
Since overexpression of SAC3 is known to be toxic for the cell (Jones et al., 2000), we studied whether it impairs mRNA export. After 3.5 h induction in galactose medium, GAL1::SAC3 cells showed a significant nuclear accumulation of poly(A)+ RNA (see below; Figure 4). Thus, both overexpression and deletion of SAC3 inhibit mRNA export, which suggests that Sac3p is a novel factor of the mRNA export machinery (see Discussion).
Fig. 4. Overexpression of Sac3p or its C-domain causes a dominant-negative phenotype and an mRNA export defect. (A) Overexpression of full-length Sac3p or the C-domain is toxic. Wild-type cells transformed with the indicated plasmid-borne constructs were spotted in 10–1 dilutions on a plate containing galactose (inductive for the GAL1 promoter). Growth was analyzed after 3 days at 30°C. (B) Overexpression of Sac3p, its C-domain or its N + M-domain causes an mRNA export defect. Poly(A)+ RNA localization was determined by in situ hybridization after 3.5 h induction of Sac3p expression in galactose-containing medium. Cells were also stained for DNA.
Next, we tested whether Nup1p and Nup60p play a role in mRNA export. Previously, the nup1Δ deletion strain was shown to be defective in mRNA export (Bogerd et al., 1994; Schlaich and Hurt, 1995), which was confirmed in this study (Figure 2B). Moreover, the nup60 null mutant is impaired in poly(A)+ RNA export (Figure 2B). Notably, the sl mutants mapping in NUP60 (see above) exhibit nuclear accumulation of poly(A)+ RNA, which was abolished by transformation with the intact NUP60 gene (data not shown). The export defect observed in the nup1Δ and nup60Δ strains is specific for mRNA, since no significant nuclear accumulation of tRNA or ribosomal 60S and 40S subunits was observed in these cells (Supplementary figure 2). In contrast to NUP1 and NUP60, the NUP2 disruption strain did not exhibit nuclear accumulation of poly(A)+ RNA (Figure 2B). The mex67-5 temperature-sensitive mutant served as positive control, which strongly accumulates poly(A)+ RNA in the nucleus upon shift to 37°C (Figure 2B). We conclude that Nup1p and Nup60p are required for nuclear mRNA export, which could explain their genetic relationship to the mRNA export factor Yra1p.
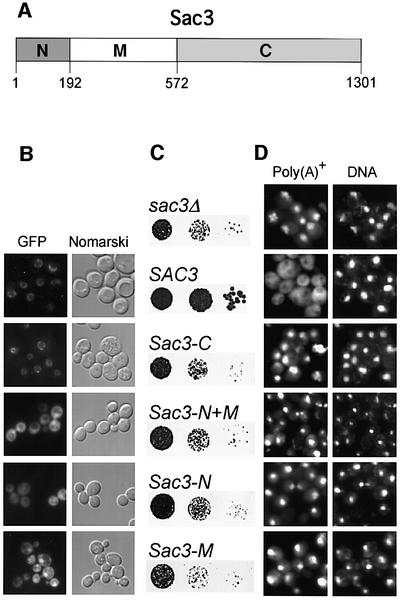
Sac3p has a nuclear pore targeting activity in its C-terminal domain
Interestingly, Sac3p was shown to associate with nuclear pores and nucleoporins such as Nsp1p (Jones et al., 2000). We therefore wondered whether association of Sac3p with the nuclear pore is crucial for mRNA export. To find out which part of Sac3p binds to nuclear pores, we expressed different Sac3p domains in yeast. Notably, an ∼400 amino acid sequence in the middle part of Sac3p (amino acids 193–572) is strongly conserved, whereas the flanking N- (amino acids 1–192) and C-terminal domains (amino acids 573–1301) are variable (Figure 3A; see also Jones et al., 2000). Accordingly, Sac3p was subdivided into an N-, M- and C-domain. The different green fluorescent protein (GFP)-tagged Sac3p domains were expressed in sac3Δ cells and their localization was determined by fluorescence microscopy (Figure 3B). Only the Sac3p C-domain localizes to the nuclear pores, whereas Sac3p-N, Sac3p-M and Sac3p-N + M are localized to the nucleoplasm and cytoplasm (Figure 3B). Importantly, all tested Sac3p domains were unable to complement either the slow growth phenotype of the sac3Δ strain (Figure 3C) or the mRNA export defect (Figure 3D). Thus, an intact Sac3p protein is required for efficient mRNA export in yeast.
Fig. 3. The C-domain of Sac3p contains a nuclear pore-targeting sequence. (A) Schematic of the Sac3p protein indicating the N-, M- and C-domain and their amino acid boundaries. (B) The C-domain of Sac3p mediates nuclear pore targeting. Fluorescence microscopy of full-length Sac3p and its different domains, all tagged with GFP and expressed in sac3Δ cells. (C) Individual Sac3p domains cannot rescue the growth defect of sac3Δ cells. Strain sac3Δ was transformed with the indicated plasmid constructs and cells were spotted in 10–1 dilutions on selective plates and incubated for 3 days at 30°C. (D) Individual Sac3p domains cannot rescue the mRNA export defect of sac3Δ cells. Localization of poly(A)+ RNA was analyzed by oligo(dT) in situ hybridization. DNA was stained with DAPI.
To assess which domains of Sac3p, when overexpressed, are responsible for the dominant-negative phenotype and mRNA export defect, we overproduced the various domains in yeast cells. This analysis showed that overexpression of the Sac3p C-domain is toxic to the cells (Figure 4A) and causes a strong mRNA export defect (Figure 4B). Overexpression of the N + M-domain causes a milder growth impairment and a less strong mRNA export inhibition (Figure 4A and B).
The C-domain of Sac3p can be replaced by Nup60p
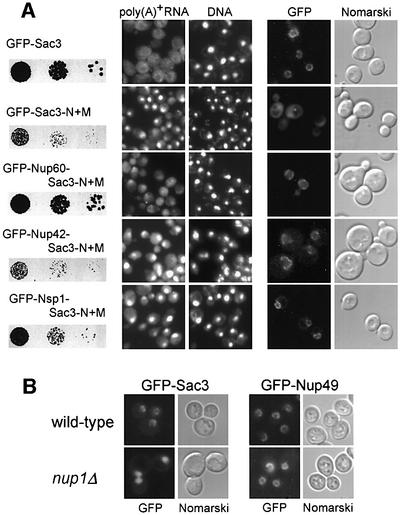
To find out whether the crucial function of the C-domain is to target the N + M-domain of Sac3p to the nuclear pores, we fused Sac3p-N + M to three different nucleoporins: (i) Nup60p, which is located asymmetrically at the nuclear basket (see Introduction); (ii) Nup42p, which is located asymmetrically at the cytoplasmic face of the NPC (Strahm et al., 1999); and (iii) Nsp1p (only the C-domain), which is found in the central channel of NPC (Fahrenkrog et al., 1998). The derived fusion proteins were also tagged with GFP to follow their in vivo location. All three nucleoporins were able to target Sac3p-N + M to the nuclear pores (Figure 5A, right panel). However, only Nup60p, but not Nup42p or Nsp1p, was able to complement the growth defect (Figure 5A, left panel) and mRNA export inhibition caused by deletion of the C-domain of Sac3p (Figure 5A, middle panel). We conclude that Sac3p has to be targeted to the nucleoplasmic side of the NPC to perform its function in mRNA export.
Fig. 5. Nucleoporin Nup60p attached to Sac3p-N + M rescues the mRNA export defect. (A) Complementation of the growth defect of sac3Δ cells by Sac3p-N + M fused to Nup60p. GFP-tagged Sac3p-N + M was fused to Nup60p, Nup42p or the C-terminal domain of Nsp1p. sac3Δ cells expressing the indicated plasmid-borne constructs were grown in 10–1 dilutions on selective plates for 4 days at 30°C (left panel), tested for mRNA export defects by in situ hybridization (middle panel) or analyzed by fluorescence microscopy for subcellular protein location (right panel). (B) Nuclear pore location of Sac3p requires Nup1p. GFP–Sac3p or GFP–Nup49 (control) were expressed in wild-type or nup1Δ cells and inspected with the fluorescence microscope.
To determine whether Nup1p or Nup60p are involved in docking Sac3p to the nuclear pores in vivo, we analyzed the location of Sac3p–GFP in nup1Δ or nup60Δ cells. Strikingly, GFP–Sac3p is no longer associated with the nuclear pore in nup1Δ cells (Figure 5B), whereas its nuclear envelope association is not significantly altered in nup60Δ cells (data not shown). In contrast, a bona fide nucleoporin, Nup49p, is still associated with the nuclear envelope in nup1Δ cells (Figure 5B). We conclude that the nuclear basket protein Nup1p plays a crucial role in localizing Sac3p to the NPC.
Sac3p, Nup1p and Nup60p interact with the Mex67p–Mtr2p complex in vitro
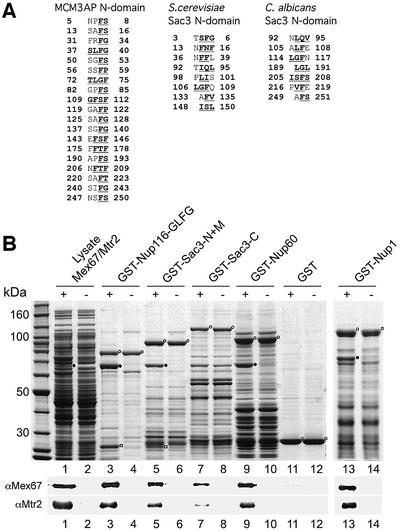
Our data suggested that Sac3p targeting to the nuclear face of the NPC is crucial for mRNA export. However, it remained open how Sac3p participates in this process. Interestingly, mammalian homologs of Sac3p [MCM3AP and germinal center-associated nuclear protein (GANP); Kuwahara et al., 2001; Takei et al., 2001] exhibit several degenerate FG, FS, FP and FSF peptide motifs in their N-terminal domains, which typically are found in FG repeat nucleoporins (Figure 6A). The N-domains of Saccharomyces cerevisiae and Candida albicans Sac3p might also contain a few of these types of degenerate FG, FS, (F,I,L)XF motifs. However, these repeats are very degenerate and thus it is not clear whether they serve as FG motifs in vivo (Figure 6A). This prompted us to test in vitro whether Sac3p binds to the conserved mRNA exporter Mex67p–Mtr2p, which is known to associate with FG repeat nucleoporins (Sträßer et al., 2000). These studies revealed that Mex67p–Mtr2p binds efficiently to GST–Sac3p-N + M, but only marginally to GST–Sac3p-C (Figure 6B). GST–Nup116 GLFG repeats served as positive, and GST alone as negative control (Figure 6B). Thus, Sac3p can associate directly with the Mex67p–Mtr2p mRNA exporter.
Fig. 6. In vitro binding of Sac3p, Nup60p and Nup1p to Mex67p–Mtr2p. (A) The Sac3p N-domains of several Sac3p homologs contain degenerate FG repeats. Peptide sequences from the Sac3p N-domains of human, S.cerevisiae and C.albicans, which exhibit degenerate FG motifs (bold and underlined) are depicted. (B) Recombinant GST fusion proteins (indicated by open circles) of Nup116-GLFG (positive control), the Sac3p N + M- and C-domains, Nup60p and Nup1p, or GST alone were immobilized on glutathione beads and incubated with an E.coli lysate (+) containing recombinant Mex67p (closed circle) and Mtr2p (open square). As negative control, the same GST fusion proteins were incubated with an E.coli lysate lacking recombinantly expressed proteins (–). Proteins bound to the beads were eluted with SDS buffer and analyzed by SDS–PAGE and Coomassie staining (upper panel). The presence of Mex67p and Mtr2p was confirmed by western blot using antibodies specific for Mex67p and Mtr2p, respectively (lower panel).
Finally, we wanted to test whether Nup60p and Nup1p, which also contain degenerate FG repeat sequences (Davis and Fink, 1990; Denning et al., 2001), can associate with Mex67p–Mtr2p and could thus provide additional docking sites for the mRNA exporter at the nuclear basket. When recombinant GST–Nup60p or GST–Nup1p were tested for binding to Mex67p–Mtr2p in vitro, both directly bound to the Mex67p–Mtr2p heterodimer (Figure 6B).
In vivo Sac3p is associated with Sub2p and Nup1p and forms a complex with Thp1p
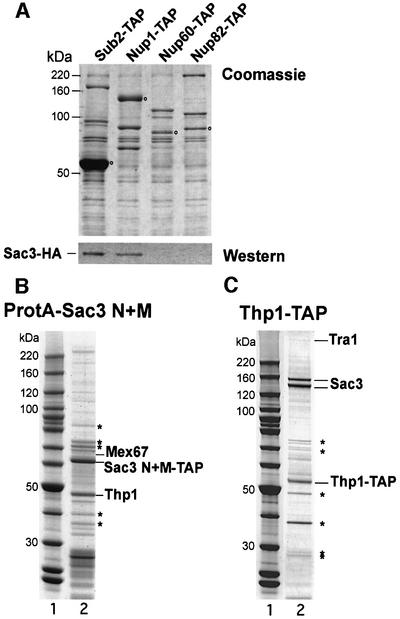
Previously, we reported that Sac3p co-purifies, although substoichiometrically, with Sub2p purified by tandem affinity purification (TAP) (Sträßer et al., 2002). In this same preparation, Sub2p purified the components of the TREX complex, i.e. Tho2p, Hpr1p, Mft1p, Thp2p, Tex1p and Yra1p. We wanted to confirm this finding and in addition test whether Sac3p can be found in association with other proteins relevant to this study. We therefore purified TAP-tagged Sub2p, Nup1p and Nup60p from yeast cells that express hemagglutinin (HA)-tagged Sac3p. Interestingly, in addition to the Sub2p-TAP, we find Sac3p in the Nup1p-TAP preparation (Figure 7A). In contrast, we could not detect Sac3p-HA in the Nup60p-TAP or Nup82p-TAP (further negative control) preparations (Figure 7A). These findings correlate well with the observed in vivo location of GFP–Sac3p in nup1Δ and nup60Δ cells (see above).
Fig. 7. Sac3p interacts in vivo with Sub2p, Nup1p, Mex67p, and Thp1p. (A) Sub2p, Nup1p, Nup60p and Nup82p, all tagged with TAP, were purified from a strain expressing HA-tagged Sac3p (Sac3p-HA). TEV eluates were analyzed by SDS–PAGE and visualized by Coomassie staining (20 µl of eluates; top panel) or analyzed by western blotting using anti-HA antibodies (5 µl of Sub2p eluate and 20 µl of Nup1p, Nup60p and Nup82p eluates; lower panel). The open circles indicate the TAP-tagged bait proteins. (B) Affinity purification of protein A-tagged Sac3p-N + M reveals Thp1p as the major co-purifying protein. The TEV eluate was analyzed by SDS–PAGE and Coomassie staining. The indicated bands were identified by mass spectrometry. A protein molecular weight standard is also shown. Other bands identified by mass spectrometry are most likely contaminants (indicated by asterisks from top to bottom: heat shock Ssa1p, Hsp70p, Hsp70p, Rpl4p, Rpl5p ribosomal protein). (C) Affinity purification of TAP-tagged Thp1p reveals association with Sac3p. TAP-purified Thp1p was analyzed by SDS–PAGE and Coomassie staining, and the indicated bands, including a weak band at ∼400 kDa (Tra1p), were identified by MS. Other bands identified by MS most probably are contaminants [indicated by asterisks from top to bottom: major coat L-A protein, Pab1p, eEF-1, E.coli matrix protein (contaminant from calmodulin beads), Rps1p ribosomal protein]. A protein molecular weight standard is also shown.
To identify proteins that stably associate with Sac3p in vivo, we also TAP tagged Sac3p. However, full-length Sac3p-TAP could be purified only very inefficiently (data not shown). To circumvent this problem, we tagged the different domains of Sac3p. The protein A-tagged Sac3p N + M-domain was significantly enriched during affinity purification (Figure 7B). In contrast, protein A-tagged Sac3p-C, perhaps due to its toxic character (see above), was recovered inefficiently (data not shown). The bands co-purifying with protein A-Sac3p-N + M were visualized by SDS–PAGE and Coomassie staining and identified by mass spectrometry (MS). A prominent co-purifying band at ∼47 kDa corresponds to Thp1p (Figure 7B), a protein that previously was shown to be involved in transcription elongation and transcription-dependent recombination (Gallardo and Aguilera, 2001). Moreover, we identified Mex67p by MS in the purified Sac3p-N + M preparation (Figure 7B), consistent with the in vitro binding studies (see above). To determine whether Thp1p is associated specifically with Sac3p, we purified TAP-tagged Thp1p (Figure 7C). Thp1p was enriched efficiently after the second purification step together with several other proteins. The most prominent co-purifying bands are a doublet at ∼160 kDa, which was identified to be Sac3p (the lower band is most probably a C-terminal degradation product of Sac3p). Another conspicuous, although weak band at ∼400 kDa is Tra1p, a component of the SAGA transcriptional activator–histone acetyltransferase complex and the NuA4 histone acetyltransferase complex (Grant et al., 1998; see Discussion).
Thp1p localizes to nuclear pores and is needed for efficient mRNA export
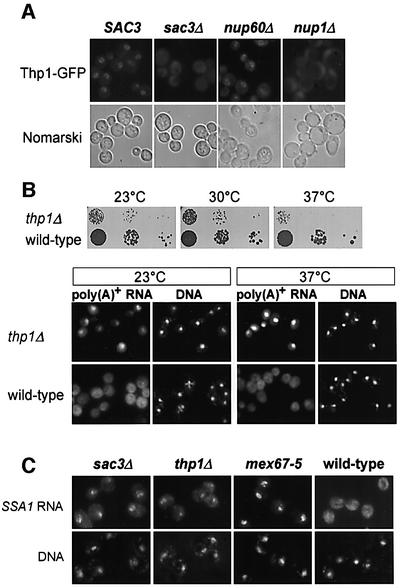
To find out whether Thp1p is associated with nuclear pores in living cells, we GFP tagged Thp1p by chromosomal integration at the C-terminus. When these cells were inspected with the fluorescence microscope, Thp1p, like Sac3p, exhibits a nuclear envelope location (Figure 8A). Interestingly, Thp1p–GFP is no longer associated with the nuclear envelope in sac3Δ cells (Figure 8A). This shows that Sac3p is needed for association of Thp1p with the nuclear pores. In addition, we examined the localization of Thp1p–GFP in nup1Δ and nup60Δ cells. Similarly to Sac3p, Thp1p significantly detaches from the nuclear envelope in nup1Δ cells, but not in nup60Δ cells (Figure 8A). Thus, Nup1p provides a binding site for the Sac3p–Thp1p complex at the nuclear pores.
Fig. 8. Thp1p is located at the nuclear envelope and required for mRNA export. (A) GFP-tagged Thp1p exhibits a nuclear envelope staining, which is lost in sac3Δ and nup1Δ cells. Cells were analyzed by fluorescence microscopy and by Nomarski optics. (B) Deletion of THP1 causes growth inhibition and inhibition of mRNA export. Serial dilutions of thp1Δ and wild-type cells were spotted onto YPD plates and grown for 4 days at the indicated temperatures (upper panel). Poly(A)+ RNA accumulates in the nucleus of thp1Δ cells at 23 and 37°C (lower panel). (C) SSA1 mRNA accumulates in the nucleus of sac3Δ and thp1Δ cells. Cells were shifted to 42°C for 30 min to induce the expression of the SSA genes, and localization of the SSA1 transcript was assessed in comparison to mex67-5 cells by in situ hybridization with a fluorescently labeled probe specific for SSA1 mRNA.
Finally, nuclear export of mRNA was analyzed in cells lacking Thp1p. The thp1-null mutant is viable but exhibits a slow growth at 23 and 30°C and is temperature-sensitive for growth at 37°C (Figure 8B). Similarly to sac3Δ cells, thp1Δ cells exhibit a strong nuclear accumulation of poly(A)+ RNA at the tested temperatures (Figure 8B; 23 and 37°C). To confirm that the nuclear signal for poly(A)+ RNA corresponds to mRNA, we performed in situ hybridization with a fluorescently labeled probe specific for SSA1 mRNA. SSA1 encodes the heat shock protein Hsp70. Transcription of this gene is strongly induced upon shift of the cells to 42°C. Whereas in wild-type cells the SSA1 mRNA is exported efficiently to the cytoplasm, it accumulates in the nucleus of sac3Δ as well as thp1Δ cells (Figure 8C). Since Thp1p is conserved in evolution, we conclude that we identified another factor of the conserved mRNA export machinery.
Discussion
The conserved mRNA export machinery consists of an mRNA exporter (Mex67p–Mtr2p in yeast; Tap–p15 in metazoans) and several components that act ‘upstream’ to couple the intranuclear steps in mRNA formation with nuclear export (Reed and Hurt, 2002). However, it is not known how the mRNA export machinery docks to the nuclear face of the NPC before entrance into the pore channel. Here, we identified several new factors, which participate in this process. Two of these factors, Nup60p and Nup1p, are nucleoporins of the nuclear basket. The other two components, Sac3p and Thp1p, may associate with the export machinery in the nucleoplasm before they mediate docking to the nuclear face of the NPC.
Initially, Sac3p was identified as a suppressor of an actin mutation, act1-1 (Novick et al., 1989), and subsequently was shown to be involved in cell cycle progression (Bauer and Kölling, 1996). Later on, Sac3p was found to be localized at the nuclear pores with a role in nuclear protein export of a nuclear export sequence (NES)-containing reporter construct (Jones et al., 2000). Our data suggest that Sac3p performs a crucial and specific role in nuclear export of mRNAs. Evidence for such a function is that (i) Sac3p is genetically linked to key components of the conserved mRNA export machinery; (ii) deletion or overexpression of Sac3p leads to strong mRNA export defects; and (iii) Sac3p interacts directly with the mRNA exporter Mex67p–Mtr2p. Thus, Sac3p is a novel component of the mRNA export machinery in yeast. Whether higher eukaryotic Sac3p homologs also function in mRNA export remains to be determined.
How Sac3p functions exactly in mRNA export is not known. Notably, the murine Sac3p homolog contains degenerate FG repeats in its N-domain (see Figure 6A), which could allow for direct binding to the mRNA exporter (Mex67p–Mtr2p or Tap–p15). Even though the presence of FG repeats in the N-terminal domains of the S.cerevisiae and C.albicans Sac3p protein is not as evident, S.cerevisiae Sac3p does indeed bind to the Mex67p–Mtr2p heterodimer. It was shown recently that Tap, via its M- and C-domains, can bind to the phenylalanine-rich cores of FG nucleoporins via hydrophobic surface patches analogous to those of NTF2 and importin–β (Conti and Izaurralde, 2001; Grant et al., 2002). Through its ability to interact with the nucleoporins Nup1p and Nup60p, Sac3p could mediate recruitment of the mRNA export machinery to the nuclear side of the NPC. However, Sac3p appears not to be a bona fide nucleoporin and its association with the nuclear pores may be transient. Notably, a fraction of Sac3p was found to co-purify with Sub2p, an intranuclear coupling protein, which is linked to Yra1p and Mex67p. In mammalian cells, the Sac3p ortholog GANP was reported to be an intranuclear protein (Kuwahara et al., 2001). Moreover, another human Sac3p splice variant (MCM3AP) contains a HAT motif that acetylates MCM3, a subunit of the MCM complex (Takei et al., 2001). MCM proteins not only function in DNA replication (Tye, 1999), but were also shown to bind to the C-terminal domain of RNA polymerase II (Pol II) and function in transcription (Yankulov et al., 1999). In contrast, yeast Sac3p does not have a HAT motif in its C-domain but instead acquired an NPC-targeting sequence (Takei et al., 2001). This could explain why the Sac3p–Thp1p complex is associated with Tra1p, a factor present in different complexes with histone acetyltransferase activity (Grant et al., 1998). Taken together, the data suggest that Sac3p functionally overlaps with Yra1p to couple intranuclear mRNA biogenesis with export through the nuclear pores. For this function, Sac3p cooperates with Thp1p that previously was shown to function in Pol II-dependent transcription elongation and transcription-dependent DNA recombination (Gallardo and Aguilera, 2001). Interestingly, mutations in components of the TREX complex, as well as in the Mex67p–Mtr2p mRNA exporter also confer a transcription elongation defect and transcription-dependent hyper-recombination (Jimeno et al., 2002). Thus, the entire gene expression pathway, from transcription to mRNA export, is tightly coupled and intertwined. It is noteworthy that Thp1p exhibits homology to Rpn3p (a component of the 19S regulatory particle of the proteasome that also functions in Pol II transcription elongation; Gonzalez et al., 2002) (Supplementary figure 3). This homology is restricted to a sequence that is also found in other proteins and known as the PINT domain.
The association of Sac3p with the nuclear pores is mediated by its C-terminal domain. Interestingly, only a nucleoporin that is located at the nuclear basket (i.e. Nup60p) could functionally replace the Sac3p C-domain. This suggests that an asymmetric location of Sac3p at the nuclear side of the NPC is essential for the mRNA export function of the Sac3p–Thp1p complex. However, the mRNA export defect is not fully abolished by substituting the Sac3p C-domain with Nup60p. This could imply that the C-domain has an additional function apart from localizing Sac3p to the nuclear pores. Another explanation could be that Sac3p requires a more dynamic interaction with nucleoporins of the nuclear basket (see above). In vivo, the NPC-targeting activity of the C-domain is mediated predominantly by Nup1p, since Sac3p associates with Nup1p in vivo and Sac3p mislocalizes to the nucleus in nup1Δ cells. Importantly, Nup1p, Nup2p and Nup60p are nucleoporins that are located asymmetrically at the nuclear face of the NPC (Rout et al., 2000; Solsbacher et al., 2000; Denning et al., 2001; Dilworth et al., 2001). Previously, Nup60p in conjunction with Nup2p was proposed to serve as a docking site for import receptors Kap95, Kap60 and Kap123 (Solsbacher et al., 2000; Denning et al., 2001; Dilworth et al., 2001). Our data show that Nup1p and Nup60p are also important for efficient mRNA export. Since the Mex67p–Mtr2p heterodimer interacts directly with Nup1p and Nup60p, it is possible that these two nucleoporins, in conjunction with Sac3p, help to recruit the mRNA exporter to the nucleoplasmic face of the NPC. Nup1p and Nup60p perform a redundant function, since disruption of one of these genes already causes an mRNA export defect, and deletion of both of these proteins is lethal. Interestingly, metazoan Nup153p is a nuclear basket protein that is strongly implicated in mRNA export (Ullman et al., 1999). Thus, Nup153p could be a docking site at the nuclear basket for the conserved mRNA export machinery in metazoans.
Our work has revealed novel interactions between the Sac3p–Thp1p complex, two nuclear basket nucleoporins, Nup1p and Nup60p, and the conserved mRNA exporter Mex67p–Mtr2p. Importantly, Sac3p binds via its C-domain to the nuclear pores, and via its N-domain to Mex67p–Mtr2p. As a consequence of these interactions, the mRNA export machinery could be targeted to the nuclear entrance of the pore channel. Upon binding of the mRNA exporter to FG repeat nucleoporins of the nuclear basket, dissociation of Sac3p from Mex67p–Mtr2p could be triggered, thereby allowing passage of the mRNP through the nuclear pore channel (Figure 9).
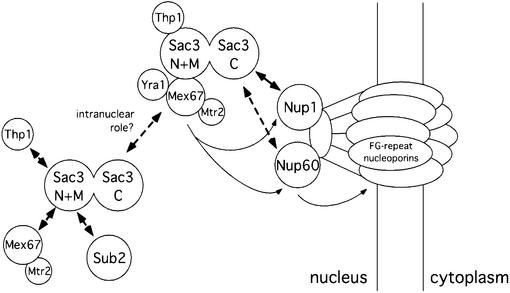
Fig. 9. Model of the function of the Sac3p–Thp1p complex in mRNA export. Already at the transcriptional level the Sac3p–Thp1p complex might be recruited to the intranuclear mRNP. Via its C-terminal domain, Sac3p docks the mRNP to the nuclear side of the NPC. At this point, the Mex67p–Mtr2p containing mRNP might leave the Sac3p–Thp1p complex and interact with different FG repeat nucleoporins on its way to the cytoplasm.
Materials and methods
Yeast strains, DNA recombinant work and microbiological techniques
Yeast strains used in this study are listed in Supplementary table I and plasmids in Supplementary table II. Double or triple knock-out strains were made by crossing single knock-out strains and tetrad dissection of derived sporulated diploids. Microbiological techniques, plasmid transformation and recovery, mating, sporulation of diploids, and tetrad analysis were done essentially as described (Santos-Rosa et al., 1998). Chromosomal integration of GFP (TRP1 marker), TAP (TRP1 marker) and HA (HIS3 marker) as C-terminal tags was performed as described (Gottschalk et al., 1998; Longtine et al., 1998; Gavin et al., 2002). The sacΔ3 THP1-GFP strain was made by disruption of SAC3 in the THP1-GFP strain. DNA recombinant work was performed according to Maniatis et al. (1982). The synthetic lethal screen with the yra1-ΔRRM allele was performed as described previously (Sträßer and Hurt, 2001).
Miscellaneous
Affinity purification of TAP-tagged or protein A-tagged proteins was performed as described (Gavin et al., 2002). TEV or EGTA eluates were analyzed by SDS–PAGE and Coomassie staining, using Novex 4–12% gradient gels (Invitrogen). MS using the tryptic digest from Coomassie-stained bands cut out from SDS–polyacrylamide gels was performed as described in Baßler et al. (2001). Proteins were identified using Mascot (Matrix Science) and the MSDB protein database. In situ hybridization of poly(A)+ RNA was performed according to Santos-Rosa et al. (1998). In situ localization of SSA1 mRNA was analyzed according to Hurt et al. (2000), and the localization of tRNA and SRP RNA according to Grosshans et al. (2000). Purification of GST-tagged proteins from Escherichia coli and in vitro binding studies to FG nucleoporins were performed as described (Sträßer et al., 2000).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Dr Susan Wente (Department of Cell and Developmental Biology, Nashville, TN) for providing GST–Nup116, Dr Michael Rexach (Department of Biological Sciences, Stanford, CA) for GST–Nup1p, Karin Bologa and Daniela Strauß for providing unpublished strains, and Dr Vikram Panse for helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB352 and Gottfried Wilhelm Leibniz Program), the Human Frontiers Science Program (HFSP) and Fonds der Chemischen Industrie.
References
- Aguilera A. (2001) The connection between transcription and genomic instability. EMBO J., 21, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J., Grandi,P., Gadal,O., Leßmann,T., Tollervey,D., Lechner,J. and Hurt,E.C. (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell, 8, 517–529. [DOI] [PubMed] [Google Scholar]
- Bauer A. and Kölling,R. (1996) The SAC3 gene encodes a nuclear protein required for normal progression of mitosis. J. Cell Sci., 109, 1575–1583. [DOI] [PubMed] [Google Scholar]
- Bogerd A.M., Hoffmann,J.A., Amberg,D.C., Fink,G.R. and Davis,L.I. (1994) nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J. Cell Biol., 127, 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn,A. and Grosschedl,R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRa enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed] [Google Scholar]
- Clouse K.N., Luo,M.J., Zhou,Z. and Reed,R. (2001) A Ran-independent pathway for export of spliced mRNA. Nat. Cell Biol., 3, 97–99. [DOI] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Davis L.I. and Fink,G.R. (1990) The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell, 61, 965–978. [DOI] [PubMed] [Google Scholar]
- Denning D., Mykytka,B., Allen,N.P., Huang,L., Burlingame,A.L. and Rexach,M. (2001) The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol., 154, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth D.J., Suprapto,A., Padovan,J.C., Chait,B.T., Wozniak,R.W., Rout,M.P. and Aitchison,J.D. (2001) Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol., 153, 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Hurt,E.C., Aebi,U. and Panté,N. (1998) Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol., 143, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M. and Aguilera,A. (2001) A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics, 157, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.-C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Delahodde,A., Kodadek,T. and Johnston,S.A. (2002) Recruitment of a 19S proteasome subcomplex to an activated promoter. Science, 296, 548–550. [DOI] [PubMed] [Google Scholar]
- Gottschalk A. et al. (1998) A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA, 4, 374–393. [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Yates,J.R. and Workman,J.L. (1998) The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol. Cell, 2, 863–867. [DOI] [PubMed] [Google Scholar]
- Grant R.P., Hurt,E.C., Neuhaus,M. and Stewart.M. (2002) Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol., 9, 247–251. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Hurt,E. and Simos,G. (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev., 14, 830–840. [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Sträßer,K., Segref,A., Schlaich,N., Presutti,C., Tollervey,D. and Jansen,R. (2000) Mex67p mediates the nuclear export of a variety of Pol II transcripts. J. Biol. Chem., 275, 8361–8368. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Boulay,J., Rosbash,M. and Libri,D. (2001) The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol., 11, 1711–1715. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon,A.G., Luna,R. and Aguilera,A. (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J., 21, 3526–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.L., Quimby,B.B., Hood,J.K., Ferrigno,P., Keshava,P.H., Silver,P.A. and Corbett,A.H. (2000) SAC3 may link nuclear protein export to cell cycle progression. Proc. Natl Acad. Sci. USA, 97, 3224–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K. et al. (2001) Germinal center-associated nuclear protein (GANP) has a phosphorylation-dependent DNA-primase activity that is up-regulated in germinal center regions. Proc. Natl Acad. Sci. USA, 98, 10279–10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeHir H., Izaurralde,E., Maquat,L.E. and Moore,M.J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J., 19, 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeHir H., Gatfield,D., Izaurralde,E. and Moore,M.J. (2001) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J., 20, 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei E.P., Krebber,H. and Silver,P.A. (2001) Messenger RNAs are recruited for nuclear export during transcription. Genes Dev., 15, 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Ishigaki,Y., Li,X. and Maquat,L.E. (2002) The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J., 21, 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 10, 953–961. [DOI] [PubMed] [Google Scholar]
- Luo M.-J., Zhou,Z., Magni,K., Christoforides,C., Rappsilber,J., Mann,M. and Reed,R. (2001) Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature, 413, 644–647. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.T. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Novick P., Osmond,B.C. and Botstein,D. (1989) Suppressors of yeast actin mutations. Genetics, 121, 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. and Hurt,E.C. (2002) A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell, 108, 523–531. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.P., Rode,M., Gatfield,D., Blencowe,B., Carmo-Fonseca,M. and Izaurralde,E. (2001) REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA, 98, 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P. and Aitchison,J.D. (2001) The nuclear pore complex as a transport machine. J. Biol. Chem., 276, 16593–16596. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich N.L. and Hurt,E.C. (1995) Analysis of nucleocytoplasmic transport and nuclear envelope structure in yeast disrupted for the gene encoding the nuclear pore protein Nup1p. Eur. J. Cell Biol., 67, 8–14. [PubMed] [Google Scholar]
- Solsbacher J., Maurer,P., Vogel,F. and Schlenstedt,G. (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol., 20, 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D., Fahrenkrog,B. and Aebi,U. (1999) The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol., 11, 391–401. [DOI] [PubMed] [Google Scholar]
- Strahm Y., Fahrenkrog,B., Zenklusen,D., Rychner,E., Kantor,J., Rosbash,M. and Stutz,F. (1999) The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J., 18, 5761–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. and Hurt,E.C. (2000) Yra1p, a conserved nuclear RNA binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. and Hurt,E.C. (2001) Splicing factor Sub2p is required for nuclear export through its interaction with Yra1p. Nature, 413, 648–652. [DOI] [PubMed] [Google Scholar]
- Sträßer K., Baßler,J. and Hurt,E.C. (2000) Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol., 150, 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature, 417, 304–308. [DOI] [PubMed] [Google Scholar]
- Stutz F., Bachi,A., Doerks,T., Braun,I.C., Séraphin,B., Wilm,M., Bork,P. and Izaurralde,E. (2000) REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA, 6, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y., Swietlik,M., Tanoue,A., Tsujimoto,G., Kouzarides,T. and Laskey,R. (2001) MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep., 2, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Shah,S., Powers,M.A. and Forbes,D.J. (1999) The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell, 10, 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S.K. and Forbes,D.J. (2001) Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol., 13, 363–375. [DOI] [PubMed] [Google Scholar]
- Weis K. (2002) Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol., 14, 328–335. [DOI] [PubMed] [Google Scholar]
- Wente S.R. and Blobel,G. (1993) A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol., 123, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankulov K., Todorov,I., Romanowski,P., Licatalosi,D., Cilli,K., McCracken,S., Laskey,R. and Bentley,D.L. (1999) MCM proteins are associated with RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 6154–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra,P., Strahm,Y. and Stutz,F. (2001) The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol., 21, 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.L., Luo,M., Sträßer,K., Katahira,J., Hurt,E. and Reed,R. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature, 407, 401–405. [DOI] [PubMed] [Google Scholar]