Abstract
Mammalian mitochondrial translational initiation factor 3 (IF3mt) promotes initiation complex formation on mitochondrial 55S ribosomes in the presence of IF2mt, fMet-tRNA and poly(A,U,G). The mature form of IF3mt is predicted to be 247 residues. Alignment of IF3mt with bacterial IF3 indicates that it has a central region with 20–30% identity to the bacterial factors. Both the N- and C-termini of IF3mt have extensions of ∼30 residues compared with bacterial IF3. To examine the role of the extensions on IF3mt, deletion constructs were prepared in which the N-terminal extension, the C-terminal extension or both extensions were deleted. These truncated derivatives were slightly more active in promoting initiation complex formation than the mature form of IF3mt. Mitochondrial 28S subunits have the ability to bind fMet-tRNA in the absence of mRNA. IF3mt promotes the dissociation of the fMet-tRNA bound in the absence of mRNA. This activity of IF3mt requires the C-terminal extension of this factor. Mitochondrial 28S subunits also bind mRNA independently of fMet-tRNA or added initiation factors. IF3mt has no effect on the formation of these complexes and cannot dissociate them once formed. These observations have lead to a new model for the function of IF3mt in mitochondrial translational initiation.
INTRODUCTION
The synthesis and assembly of the oligomeric complexes in mitochondria involved in electron transport and ATP synthesis require genetic information contained in both the nuclear and mitochondrial genomes. Limited information is available on the mechanism by which the mitochondrially-encoded components in these complexes are synthesized and assembled into the oligomeric complexes in the inner membrane of mitochondria. A number of interesting features distinguish the protein synthesizing system of mammalian mitochondria from other translational systems. Of particular interest is the observation that the mRNAs in this organelle have an almost complete lack of 5′- and 3′-untranslated nucleotides. The start codon is generally located within a few nucleotides of the 5′ end of the mRNA (1,2). Thus, a Shine/Dalgarno interaction between the mRNA and the 16S rRNA such as observed in prokaryotes is not used in mammalian mitochondrial protein synthesis. Mammalian mitochondrial ribosomes have low sedimentation coefficients (∼55S) and consist of 28S and 39S subunits (3). Animal mitochondrial ribosomes are 31% RNA and 69% protein. In contrast, bacterial ribosomes consists of ∼67% RNA and 33% protein (4,5).
In bacteria, three translational initiation factors, initiation factors 1, 2 and 3 (IF1, IF2, and IF3), are required for initiation (6–8). No homolog of IF1 has been detected in mammalian mitochondrial systems. However, mitochondrial initiation factor 2 (IF2mt), which promotes the binding of fMet-tRNA to the small subunit of mitochondrial ribosomes has been cloned and characterized (9–15). Recently, the mitochondrial homolog of initiation factor 3 (IF3mt) has been cloned and expressed. In bacterial protein synthesis initiation factor 3 has been assigned a number of discrete functions including (i) dissociation of ribosomes (7,16); (ii) increasing the forward rate constant for codon:anticodon interaction at the P-site (17); (iii) dissociation of fMet-tRNA at AUG codons at the 5′ end of leaderless mRNAs (18); (iv) proofreading the selection of the initiator tRNA and an AUG codon at the P-site (19–21) and (v) adjusting the position of the mRNA on the small subunit from a stand-by position to the decoding position (22).
The mature form of IF3mt, lacking the predicted mitochondrial import signal, is active in initiation complex formation on both mitochondrial 55S ribosomes and bacterial 70S ribosomes (23). IF3mt has a central region with weak homology (21–26% identity) to bacterial IF3 (Figure 1A). This homology region is divided into two domains (N- and C-domains) separated by a flexible linker as observed for the prokaryotic factors (23). The C-domain is thought to carry out the basic functions of IF3 with the N-domain increasing its affinity for the small subunit (24). In mammalian IF3mt, the central region with homology to the bacterial factors is preceded by an N-terminal extension of ∼30 residues, which is predicted to form a helical structure. The homology domain is followed by a hydrophilic C-terminal extension of ∼30 residues, which is also predicted to have significant helical content. The only organellar IF3 that has been studied at a detailed biochemical level to date is chloroplast IF3 (IF3chl) from Euglena gracilis (25–28). This factor has a long extension (∼150 amino acids) at the N-terminus and an acidic C-terminal extension of 63 residues. In IF3chl, sequences near the junction of the N-terminal extension and the homology domain and the C-terminal extension inhibit the activity of this factor (28) and may play a regulatory role in chloroplast protein synthesis. In the present work, we have examined some of the properties of human IF3mt and have tested the effects of the N-terminal and C-terminal extensions on the activity of this factor.
Figure 1.
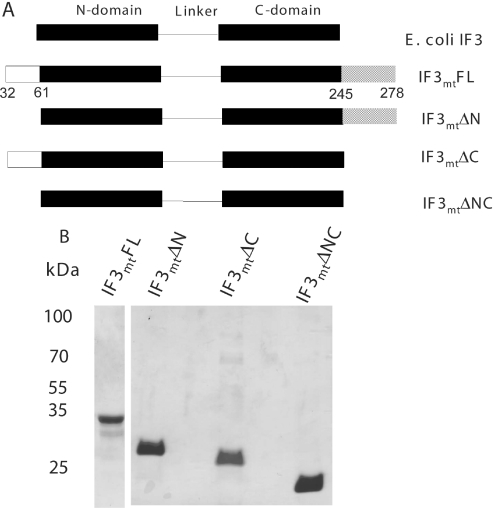
Domain organization of prokaryotic IF3 and mammalian IF3mt and deletion constructs for IF3mt. (A) Schematic diagram of the organization of E.coli IF3 and IF3mt and its deletion derivatives. IF3mt begins with an N-terminal import sequence that is not shown. The full-length version of the mature protein (IF3mtFL) encompasses residues 32–278. The region with homology to E.coli IF3 begins at residue 61 and goes through residue 245. Deletion of the N-terminal extension (IF3mtΔN) gives a derivative that includes amino acids 61–278. Deletion of the C-terminal extension (IF3mtΔC) includes amino acids 32 through and including residue 245. The deletion of both extensions (IF3mtΔNC) gives a construct that includes residues 61 through 245. (B) Analysis of the purity of the full-length and deletion derivatives by SDS–PAGE. Samples (1–2 µg) of IF3mtFL and its deletion derivatives were applied to a 12% SDS–PAGE gel and stained with Coosmassie blue.
MATERIALS AND METHODS
Materials
Oligonucleotides used for mutagenesis were synthesized at Nucleic Acid Core Facility at the University of North Carolina, Chapel Hill. Bovine mitochondrial 55S ribosomes were prepared as described (29). Mitochondrial 28S and 39S subunits were purified on sucrose gradients (30). Escherichia coli ribosomes were prepared from E.coli W (31,32) and tight couples were collected from a sucrose gradient in the presence of 5 mM Mg2+ (33). Bovine IF2mt and E.coli initiation factors were prepared as described (13,23,32). E.coli IF2 was also prepared from an expression construct providing a mixture of the α and β forms of IF2 (A. C. Spencer and L. L. Spremulli, unpublished data). Yeast [35S]fMet-tRNA and [14C]Phe-tRNA were prepared and the [14C]Phe-tRNA was acetylated as described (32,34). A transcript encoding subunit 2 of bovine cytochrome oxidase was prepared by in vitro transcription (35–37).
Cloning and expression of IF3mt deletion derivatives
The construct carrying the N-terminal deletion was amplified by PCR using the mature IF3mt cDNA as template (23), the forward primer GGGAATTCCATATGACCCAGAATGAAGGAAAAAAGA and the reverse primer CGCGGATCCGCTCGAGCTGATGCAGAACAT. Deletion of the C-terminal extension was carried out using the forward primer CGCGGATCCAATTCATATGACAGCACCAGCACAG and the reverse primer CGCGGATCCGCTCGAGTTTGCTCAAAGCACG. The double deletion of the N- and C-terminal extensions was obtained using the forward primer GGGAATTCCATATGACCCAGAATGAAGGAAAAAAGA and the reverse primer CGCGGATCCGCTCGAGTTTGCTCAAAGCACG. These PCR products were digested with NdeI and XhoI and cloned into pET-21(+) (Novagen). This vector provides a sequence encoding six His residues (His-tag) at the C-terminus. The PCR products were transformed into E.coli ER2267 and the nucleotide sequence of the inserted DNA was confirmed. The plasmids were subsequently transformed into E.coli BL21(DE3) for expression.
Purification of IF3mtFL and deletion derivatives using Ni-NTA
The full-length mature form of human mitochondrial initiation factor 3 (IF3mtFL) was expressed in E.coli as described (23). The His-tagged protein was purified on Ni-NTA and on S-Sepharose. This later step separates the IF3mtFL from a 19 kDa degradation product. Ni-NTA preparations of the deletion derivatives did not contain this degradation product and did not require further purification. The N-terminus of the double truncated derivative (IF3mtΔNC) was sequenced using Edman degradation to ensure that the correct N-terminus was present on the expressed protein. Protein concentrations were determined using the Bradford assay with BSA as a standard (BioRad).
Initiation complex formation assay for IF3mtFL and its deletion derivatives
The activities of IF3mtFL and its deletion derivatives were determined by measuring their abilities to stimulate the binding of [35S]fMet-tRNA to either E.coli or mitochondrial ribosomes in filter-binding assays essentially as described previously (23). Reactions (100 µl) on E.coli ribosomes contained 50 mM Tris–HCl, pH 7.6, 1 mM DTT, 80 mM NH4Cl, 5 mM MgCl2, 0.25 mM GTP, 12.5 µg poly(A,U,G), 0.06 µM [35S]fMet-tRNA, 0.25 µM IF2mt, 0.24 µM E.coli 70S tight couples and varying amounts of IF3mt or its deletion derivatives as indicated. For initiation complex formation assays on mitochondrial ribosomes, reaction mixtures (100 µl) contained 50 mM Tris–HCl, pH 7.6, 1 mM DTT, 0.1 mM spermine, 35 mM KCl, 4.5 mM MgCl2, 0.25 mM GTP, 1 mM DTT, 12.5 µg poly(A,U,G), 0.42 µM IF2mt, 0.06 µM [35S]fMet-tRNA, 0.05 µM mitochondrial 55S ribosomes and varying amount of IF3mt or its deletion derivatives. Reaction mixtures were incubated for 15 min at 37°C then analyzed using a nitrocellulose filter-binding assay as described (9).
Proofreading assays
This assay has been modified from that described in (38,39) for E.coli IF3. A complex carrying [14C]AcPhe-tRNA bound to E.coli 30S subunits [AcPhe-tRNA:poly(U):30S] was formed by incubation of activated E.coli 30S subunits (0.08 µM), poly(U) (10 µg) and [14C]AcPhe-tRNA (0.3 µM) in a reaction mixture (50 µl) containing 50 mM Tris–HCl, pH 7.6, 0.1 mM spermine, 1 mM DTT, 50 mM NH4Cl and 15 mM MgCl2. After incubation at 37°C for 30 min, various amounts of IF3mt or buffer (50 µl) were added to the mixture and the incubation was continued for an additional 5 min at 37°C. The mixtures were then diluted 25-fold with pre-warmed dilution buffer (50 mM Tris–HCl, pH 7.6, 1 mM DTT, 50 mM NH4Cl and 15 mM MgCl2). The diluted reaction mixtures were incubated for 5 min at 37°C. The amount of initiation complex remaining was determined by a nitrocellulose filter-binding assay. A similar assay was carried out using a complex formed with E.coli 30S subunits (0.08 µM), poly(A,U,G) (12.5 µg) and [35S]fMet-tRNA (0.2 µM).
A complex containing [14C]AcPhe-tRNA bound to 28S subunits [AcPhe-tRNA:poly(U):28S] was formed by incubation of mitochondrial 28S subunits (0.2 µM) with poly(U) (10 µg) and [14C]AcPhe-tRNA (0.3 µM) in a reaction mixture (50 µl) containing 50 mM HEPES–KOH, pH 7.8, 1 mM DTT, 0.1 mM spermine, 35 mM KCl and 25 mM MgCl2. After incubation at 27°C for 30 min, various amounts of IF3mt or compensating buffer (50 µl) were added to the mixture and incubated for 5 min at 27°C. The mixture was then diluted 50-fold with pre-warmed dilution buffer (50 mM HEPES–KOH, pH 7.8, 1 mM DTT, 0.1 mM spermine 35 mM KCl and 25 mM MgCl2). The reaction mixtures were incubated for an additional 15 min at 27°C. The amount of initiation complex remaining was determined by a nitrocellulose filter-binding assay. Attempts were also made to form a similar complex with mitochondrial 28S subunits (0.2 µM), poly(A,U,G) (12.5 µg) and [35S]fMet-tRNA (0.2 µM) and MgCl2 concentrations ranging from 15 to 50 mM (Results).
Effect of IF3mt on the binding of fMet-tRNA to 28S subunits in the presence or absence of mRNA
Reaction mixtures (100 µl) contained various amounts of IF3mt and 50 mM Tris–HCl, pH 7.6, 35 mM KCl, 0.1 mM spermine, 1 mM DTT, 7.5 mM MgCl2, 0.25 mM GTP, 1.25 mM phosphoenolpyruvate, 0.7 U pyruvate kinase, 0.06 µM [35S]fMet-tRNA, 0.14 µM IF2mt, 0.068 µM 28S subunits and, where indicated, 12.5 µg poly(A,U,G). Reaction mixtures were incubated for 20 min at 27°C and the amount of [35S]fMet-tRNA bound to the 28S subunit was determined using a filter-binding assay (9).
Sucrose gradient analysis of initiation complexes
Initiation complexes (200 µl) were assembled as described above containing 0.05 µM 28S subunits or 0.066 µM 39S subunits. Samples were incubated for 20 min at 27°C and then applied to a 5 ml sucrose gradient (10–30% sucrose in 50 mM Tris–HCl, pH 7.6, 40 mM KCl, 7.5 mM MgCl2 and 2 mM DTT). The gradients were subjected to centrifugation for 1 h 45 min at 48 000 r.p.m. in a Beckman SW50.1 rotor. Following centrifugation, gradients were fractionated on an Isco gradient fractionator at a flow rate of 0.8 ml/min. Fractions (0.2 ml) were collected and filtered through nitrocellulose membranes, dried and counted.
RESULTS
Role of the extensions on IF3mt in initiation complex formation
Mammalian IF3mt has N-terminal and C-terminal extensions just over 30 residues long surrounding the central region homologous to bacterial IF3. To assess the importance of these extensions on the activity of IF3mt, three deletion derivatives were constructed (Figure 1A). One derivative lacked the N-terminal extension (IF3mtΔN). A second lacked the C-terminal extension (IF3mtΔC) while the third lacked both extensions (IF3mtΔNC). These derivatives expressed well and were purified from E.coli (Figure 1B).
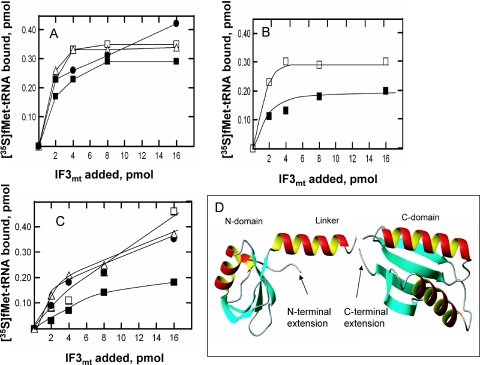
The activities of the full-length mature IF3mt (IF3mtFL) and the deletion derivatives were tested in initiation complex formation on bovine mitochondrial 55S ribosomes (Figure 2A). Interestingly, deletion of either the N-terminal or the C-terminal extension in IF3mt increased the activity of the factor slightly in promoting the binding of fMet-tRNA to mitochondrial ribosomes. This observation is reminiscent of the effects of removing the long extensions observed in E.gracilis IF3chl. However, removal of the extensions on IF3chl has a significantly larger positive effect (3.5- to 4-fold) on the activity of this factor in initiation complex formation. Removal of the extensions on IF3mt also increased the activity of this factor to a small extent in initiation complex formation with the mRNA for subunit 2 of cytochrome oxidase (Figure 2B). Finally, deleting the extensions on IF3mt had a slightly positive effect when this factor was tested in initiation complex formation on E.coli 70S ribosomes (Figure 2C) indicating that the effect is not dependent on the interaction of IF3mt with mitochondrial ribosomes. Structural information on bacterial IF3 (Figure 2D) suggests that the extensions in IF3mt could be positioned to interact with the linker region (Figure 1A). Since the linker is believed to play an important role in the binding of IF3mt to the ribosome (27), removal of these extensions may actually increase the binding of IF3mt to the small subunit slightly. In this context, it should be noted that the deletion of both extensions on IF3chl increases the affinity of IF3chl for chloroplast 30S subunits ∼100-fold (28). Alternatively, the extensions in IF3mt may be playing a different role in initiation complex formation.
Figure 2.
Activity of IF3mt and its deletion derivatives in initiation complex formation. (A) The activity of IF3mtFL (closed squares), IF3mtΔN (closed circles), IF3mtΔC (open triangles) and IF3mtΔNC (open squares) was tested in initiation complex formation on mitochondrial 55S ribosomes using poly(A,U,G) as the mRNA. A blank containing no IF3mt has been subtracted from each value (0.29 pmol). (B) Activities of IF3mtFL (closed squares) and IF3mtΔNC (open squares) were tested on mitochondrial 55S ribosomes using a transcript of the cytochrome oxidase subunit 2 gene as the mRNA. A blank containing no IF3mt (0.11 pmol) has been subtracted from each value. (C) The activities of IF3mtFL (closed squares), IF3mtΔN (closed circles), IF3mtΔC (open triangles) and IF3mtΔNC (open squares) were tested on E.coli 70S tight couples using poly(A,U,G) as mRNA. A blank containing no IF3mt (0.34 pmol) has been subtracted from each value. (D) Model for the N- and C-domains of Bacillus stearothermophilus IF3 created from the PDB coordinates (1TIF and 1TIG) using MolMol (55) indicating the location of the N- and C-terminal extensions. Homology modeling suggests that the N-domain of IF3mt has a similar fold to that observed with the B.stearothermophilus factor (23). The C-domain of IF3mt is not as highly conserved and cannot be modeled accurately. However, it is probable to have a similar overall fold.
Proofreading activity of IF3mt
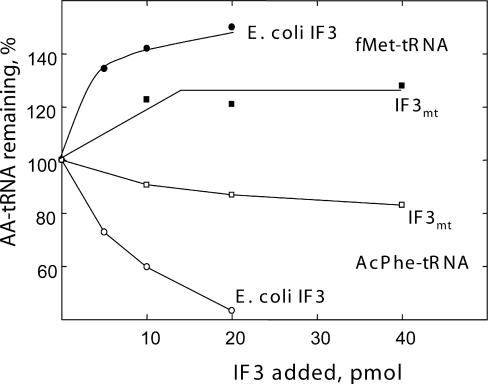
One of the roles assigned to IF3 in initiation is to proofread the selection of fMet-tRNA and the AUG codon in the P-site (20,38,40). This effect appears to occur through conformational changes in the subunit rather than by a direct interaction of IF3 with the fMet-tRNA bound at the P-site (41). One of the classical methods for measuring the proofreading function of IF3 is to test its ability to promote the dissociation of AcPhe-tRNA bound to the small subunit in response to poly(U). When these experiments are carried out with E.coli 30S subunits, the AcPhe-tRNA is bound non-enzymatically (in the absence of IF2) using an elevated concentration of Mg2+. As indicated in Figure 3, E.coli IF3 effectively dissociates AcPhe-tRNA bound to the 30S subunit but does not dissociate fMet-tRNA bound to the small subunit. When IF3mt is tested in this assay, it is quite anemic in dissociating the bound AcPhe-tRNA (Figure 3). As expected, it does not promote the release of fMet-tRNA. Deletions of both N- and C-terminal extensions had no effect on the response of IF3mt in this assay (data not shown). In contrast, deletion of these extensions improved that ability of IF3chl to dissociate the 30S:AcPhe-tRNA:poly(U) complex which correlated with the improved binding of this factor to chloroplast 30S subunits upon deletion of the extensions (28).
Figure 3.
Activity of E.coli IF3 and IF3mt in proofreading the initiation complex. The abilities of E.coli IF3 (circles) and IF3mt (squares) to promote the dissociation of a pre-formed complex [E.coli 30S:poly(U):[14C]AcPhe-tRNA] (open symbols) were measured in the presence of various concentrations of IF3 as described in Materials and Methods. The value for 100% complex remaining is 2.1 pmol. The effects of E.coli IF3 and IF3mt on a pre-formed complex [30S:poly(A,U,G):[35S]fMet-tRNA] (closed symbols) were tested under similar conditions. The 100% value for these experiments was 0.14 pmol.
Initial studies to examine the proofreading activity of IF3mt on mitochondrial 28S ribosomal subunits provided some surprises. Non-enzymatic binding of AcPhe-tRNA could be obtained on 28S subunits at 15–25 mM Mg2+ and IF3mt was active in destabilizing these complexes (Table 1). However, the positive control for these experiments should be the stability of complexes formed with fMet-tRNA and the AUG codon in the presence of IF3mt. Non-enzymatic binding of fMet-tRNA occurs readily in the E.coli system as the concentration of Mg2+ is raised. However, essentially no non-enzymatic binding of fMet-tRNA could be detected with 28S subunits even at Mg2+ ion concentrations as high as 40 mM (Table 1). The lack of non-enzymatic binding of fMet-tRNA prevented a true assessment of the ability of IF3mt to proofread initiation complex formation in the mitochondrial system.
Table 1.
Non-enzymatic binding of [14C]AcPhe-tRNA and [35S]fMet-tRNA to mitochondrial 28S subunits in the presence and absence of IF3
| Amino acid-tRNA | aminoacyl-tRNA bound to 28S (pmol) | |
|---|---|---|
| No IF3 added | With IF3 added | |
| [14C]AcPhe-tRNA | 0.83 | 0.40 |
| [35S]fMet-tRNA | 0.007 | 0.003 |
The 28S:poly(U):[14C]AcPhe-tRNA or 28S:poly(A,U,G):[35S]fMet-tRNA complexes were formed non-enzymatically and the effect of adding IF3mt was assessed as described in Materials and Methods. A blank representing the retention of label on the filters in the absence of subunits (∼0.16 pmol, 80–100 c.p.m., for [14C]AcPhe-tRNA and ∼0.02 pmol for [35S]fMet-tRNA) has been subtracted from each value.
Effect of IF3mt on the binding of fMet-tRNA to 28S subunits in the absence of mRNA
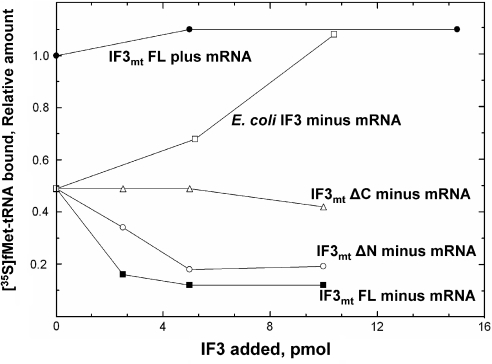
Control experiments used in analyzing the low numbers obtained in the proofreading experiments lead to the realization that a significant amount of fMet-tRNA binds to mitochondrial 28S subunits in the absence of mRNA. This binding is completely dependent on the presence of IF2mt (data not shown). Interestingly, the message-independent binding of fMet-tRNA to 28S subunits is destabilized by IF3mt (Figure 4). In contrast, IF3mt has no effect on the binding of fMet-tRNA to 28S subunits in the presence of mRNA. The destabilization of fMet-tRNA binding to mitochondrial 28S subunits is in contrast to observations made in E.coli in which IF3 is reported to stabilize the IF2-dependent binding of fMet-tRNA to 30S subunits in the absence of mRNA (42). We have retested this effect using E.coli 30S subunits and have observed that there is some mRNA-independent binding to these small subunits [∼10% of the level of binding observed in the presence of poly(A,U,G)]. And, as reported, E.coli IF3 stimulated this binding ∼2-fold (data not shown). Interestingly, E.coli IF3 also stimulates the mRNA-independent binding of fMet-tRNA to mitochondrial 28S subunits ∼2-fold. Thus, it behaves quite differently in this assay than does IF3mt.
Figure 4.
Effect of E.coli IF3, IF3mt and its derivatives on the binding of [35S]fMet-tRNA to mitochondrial 28S subunits in the presence and absence of mRNA. Initiation complexes were prepared containing [35S]fMet-tRNA, IF2mt and 28S subunits in the presence and absence of poly(A,U,G) as mRNA. After incubation with the indicated amounts of IF3mt or its derivatives, the amount of complex remaining was determined using a nitrocellulose filter-binding assay. The data are reported as the relative amount of binding obtained since data from different experiments were combined and the absolute numbers obtained depend on the percentage of active 28S subunits in different preparations. For these experiments, the value normalized to 1 for binding in the presence of mRNA represents 0.71 pmol while the value bound in the absence of mRNA was 0.35 pmol. For the experiments testing the effects of deletion of the N- and C-terminal extension, the level of binding obtained in the absence of mRNA and IF3mt was 0.12 pmol. IF3mtFL in the presence of mRNA (closed circles), IF3mtFL in the absence of mRNA (closed squares), IF3mtΔN (open circles) or IF3mtΔC (open triangles) or E.coli IF3 (open squares) were added as indicated. The amount of fMet-tRNA retained on the small subunit was measured as described in Materials and Methods.
The destabilization of message-independent binding to mitochondrial 28S subunits by IF3mt is quite rapid and is essentially complete within ∼20 s (data not shown). IF3mt was able to dissociate fMet-tRNA pre-bound to the 28S subunit in the absence of mRNA indicating that it does not have to be present on the small subunit prior to fMet-tRNA binding to carry out this activity.
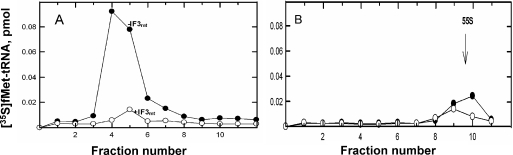
Sucrose gradient analysis was used to assess whether the fMet-tRNA bound to mitochondrial 28S subunits in the absence of mRNA could be incorporated into 55S monosomes. As indicated in Figure 5A, fMet-tRNA could be observed bound to 28S subunits when reaction mixtures were incubated in the absence of IF3mt. The presence of IF3mt resulted in a substantial decrease in the amount of fMet-tRNA bound as observed in the filter-binding assay. As expected, no fMet-tRNA binding was observed to mitochondrial 39S subunits in either the presence or absence of IF3mt (data not shown). About 25% of the fMet-tRNA bound in the absence of mRNA was observed in the 55S region of the gradient after 39S subunits were added indicating that at least a portion of this material could be chased into 55S complexes (Figure 5B). No fMet-tRNA was observed remaining in the 28S region of the gradient. Nitrocellulose filter-binding assays suggest that there is no loss of the fMet-tRNA bound in the absence of mRNA when 39S subunits are added suggesting that the lower yield of 55S complexes observed in the sucrose gradients arises from the reduced stability of these complexes compared with fMet-tRNA bound to 28S subunits directly. The loss of a portion of the fMet-tRNA bound to the subunit upon formation of 55S complexes probably reflects the release of IF2mt upon subunit joining.
Figure 5.
Effect of the addition of 39S subunits on the mRNA-independent binding of [35S]fMet-tRNA formed in the presence and absence of IF3mt. (A) The 28S subunits were incubated with [35S]fMet-tRNA and IF2mt in the absence (closed circles) or presence (open circles) of IF3mt. Reaction mixtures were analyzed by sucrose density gradient centrifugation and the position of the [35S]fMet-tRNA was located by filtering appropriate fractions as described in Materials and Methods. (B) [35S]fMet-tRNA binding was initially carried out with 28S subunits in the presence of IF2mt but in the absence of mRNA. Reactions mixtures were prepared in the absence (closed circles) or presence (open circles) of IF3mt. Following assembly of these complexes, 39S subunits (0.066 µM) were added and the incubation was continued for an additional 5 min at 27°C. The resulting complexes were then analyzed on sucrose gradients.
Since destabilization of mRNA-independent binding of fMet-tRNA to the small subunit is observed in the mammalian mitochondrial system but not in the prokaryotic system, the effects of the N- and C-terminal extensions on IF3mt on this activity were examined. As indicated in Figure 4, both the full-length factor and the derivative lacking the N-terminal extension were active in promoting the release of fMet-tRNA bound to the small subunit in the absence of mRNA. However, when the C-terminal extension was deleted, no destabilization of the fMet-tRNA bound to the small subunit in the absence of mRNA was observed (Figure 4). Deletion of both extensions gave a result identical to that observed when the C-terminal extension alone was deleted (data not shown). This observation suggests that this unusual activity of IF3mt requires the C-terminal extension and suggests that this extension developed on the mammalian mitochondrial factor in order to promote the dissociation of fMet-tRNA bound to the 28S subunit prior to mRNA binding.
The mitochondrial 28S subunit has the ability to bind mRNAs in a sequence independent manner in the absence of any added factors (35,43). The effect of IF3mt on this interaction was tested by monitoring the formation of the complex between 28S subunits and labeled mRNA for subunit 2 of cytochrome oxidase using a nitrocellulose filter-binding assay. IF3mt had no effect on the direct binding of mRNA to 28S subunits (data not shown) indicating that this complex can form in the presence of IF3mt and remains stable in its presence.
DISCUSSION
One of the classical features of bacterial IF3 is the ability to proofread the selection of fMet-tRNA and an AUG (or GUG) codon at the P-site during initiation. Detailed studies of the discrimination of IF3 against non-canonical initiation codons using tRNAs with the characteristic features of the initiator tRNA indicate that this factor recognizes primarily codon–anticodon interactions, at least at the second and third positions of the codon (20). The results reported here leave open the question of whether IF3mt has a proofreading function comparable with that observed with E.coli IF3. One might even question whether proofreading is important in the animal mitochondrial translational system with its limited repertoire of mRNAs to translate. Certain differences must apply to the mitochondrial system since both AUG and AUA serve as methionine codons in this organelle. In humans, 3 of the 13 translational start sites use AUA as the start codon. The basis for the ability of the mammalian fMet-tRNAMet to read the AUA codon is unclear although it has been postulated that the minor base 5-formyl cytidine has a critical role to play in decoding the AUA triplet (44). In humans, mice and presumably several other mammals, AUU also serves as a start codon for at least one mitochondrial mRNA (45,46). In addition, mutation of the AUG start codon to GUG in the ATPase 6 mRNA allows efficient initiation indicating that this codon can also serve as a start codon (47). These observations indicate that there is considerable tolerance for the start codon used in mammalian mitochondria with variations accepted in both position 1 and position 3 of the codon.
The same tRNAMet is used for both initiation and elongation in mammalian mitochondria. This tRNA has retained the classical set of three G:C base pairs at the bottom of the anticodon stem, a characteristic of initiator tRNAs (48,49). The three consecutive G:C pairs are critical for the binding of the initiator tRNA to the P-site during initiation and the anticodon stem is examined by IF3 during translational initiation (19,50). Interestingly, tRNAMet is the only mammalian mitochondrial tRNA characterized by three consecutive G:C base pairs at the bottom of the anticodon stem suggesting that this feature remains important for the selection of this tRNA for binding to the P-site during initiation (51).
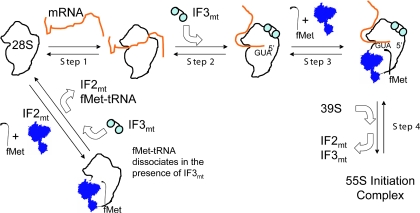
One of the most unusual observations emerging from these studies is that IF3mt has the ability to dissociate the IF2mt-dependent binding of fMet-tRNA to the 28S subunit in the absence of mRNA. Our current working hypothesis to account for this activity is illustrated by the model in Figure 6. In this model, premature binding of fMet-tRNA in the presence of IF2mt would lead to an unproductive complex. IF3mt dissociates this unproductive complex or prevents its formation. In the productive pathway, mRNA binds to the 28S subunit but the subunit is positioned randomly on the message (Step 1). The basis for this idea emerges from the observation that 28S ribosomal subunits bind mRNAs quite tightly (Kd of 25 nM at 50 mM KCl). This binding occurs randomly on the mRNA (35,43). IF3mt is postulated to alter the position of the mRNA, promoting the positioning of the 5′ start codon into the P-site. This idea has precedents in bacterial initiation in which the 5′-untranslated region near the Shine/Dalgarno sequence lies at the junction of the platform and the head of the small subunit (52) (Step 2). IF3 binding near the platform promotes the rearrangement of the mRNA facilitating the correct placement of the AUG start codon in the P-site (22,53). Following the correct positioning of the mRNA, IF2mt promotes the binding of fMet-tRNA (Step 3). Finally, the 39S subunit joins this complex leading to the release of the initiation factors and the formation of the 55S initiation complex (Step 4).
Figure 6.
Proposed model for the role of IF3mt in initiation complex formation in mammalian mitochondria. For a description of the steps see text.
The data presented here indicate that the N-terminal and C-terminal extensions on IF3mt are not essential for the activity of this factor in promoting initiation complex formation on mitochondrial 55S ribosomes. However, the C-terminal extension appears to be essential for allowing IF3mt to dissociate fMet-tRNA bound in the absence of mRNA. In the bacterial system, the order of binding of mRNA and fMet-tRNA to the small subunit appears to be random. Either will also bind to 30S subunits in the absence of the other although with less stability (7,54). We believe that the evolution of the C-terminal extension on mammalian IF2mt arose as a means to create an ordered pathway for the binding of mRNA prior to the binding of fMet-tRNA during initiation.
Acknowledgments
This work was supported in part by funds provided by the National Institutes of Health (Grant GM32734). Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Anderson S., de Brujin M., Coulson A., Eperon I., Sanger F., Young I. Complete sequence of bovine mitochondrial DNA: Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 2.Montoya J., Ojala D., Attardi G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien T.W., Denslow N.D., Faunce W., Anders J., Liu J., O'Brien B. Structure and function of mammalian mitochondrial ribosomes. In: Nierhaus K., Franceschi F., Subramanian A., Erdmann V., Wittmann-Liebold B., editors. The Translational Apparatus: Structure, function Regulation and Evolution. NY: Plenum Press; 1993. pp. 575–586. [Google Scholar]
- 4.van Holde K., Hill W. General physical properties of ribosomes. In: Nomura M., Tissieres A., Lengyel P., editors. Ribosomes. Cold Spring Harbor NY: Cold Spring Harbor Laboratory; 1974. pp. 53–91. [Google Scholar]
- 5.Wittmann H. Structure of ribosomes. In: Hardesty B., Kramer G., editors. Structure, Function and Genetics of Ribosomes. NY: Springer-Verlag; 1986. pp. 1–27. [Google Scholar]
- 6.Van Knippenberg P. Aspects of translation initiation in Escherichia coli. In: Hill W., Dahlberg A., Garrett R., Moore P., Schlessinger D., Warner J., editors. The Ribosome: Structure, Function and Evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 265–274. [Google Scholar]
- 7.Gualerzi C., Pon C. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 8.Gualerzi C.O., Brandi L., Caserta E., Teana A., Spurio R., Tomsic J., Pon C.L. Translation initiation in bacteria. In: Garrett R.A., Douthwaite S.R., Liljas A., Matheson A.T., Moore P.B., Noller H.F., editors. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Washington, D.C.: ASM Press; 2000. pp. 477–494. [Google Scholar]
- 9.Liao H.-X., Spremulli L.L. Initiation of protein synthesis in animal mitochondria: Purification and characterization of translational initiation factor 2. J. Biol. Chem. 1991;266:20714–20719. [PubMed] [Google Scholar]
- 10.Liao H.-X., Spremulli L.L. Identification and initial characterization of translational initiation factor 2 from bovine mitochondria. J. Biol. Chem. 1990;265:13618–13622. [PubMed] [Google Scholar]
- 11.Ma L., Spremulli L.L. Cloning and sequence analysis of the human mitochondrial translational initiation factor 2 cDNA. J. Biol. Chem. 1995;270:1859–1865. doi: 10.1074/jbc.270.4.1859. [DOI] [PubMed] [Google Scholar]
- 12.Ma J., Farwell M., Burkhart W., Spremulli L.L. Cloning and sequence analysis of the cDNA for bovine mitochondrial translational initiation factor 2. Biochim. Biophys. Acta. 1995;1261:321–324. doi: 10.1016/0167-4781(95)00041-e. [DOI] [PubMed] [Google Scholar]
- 13.Ma J., Spremulli L.L. Expression, purification and mechanistic studies of bovine mitochondrial translational initiation factor 2. J. Biol. Chem. 1996;271:5805–5811. doi: 10.1074/jbc.271.10.5805. [DOI] [PubMed] [Google Scholar]
- 14.Spencer A.C., Spremulli L.L. Interaction of mitochondrial initiation factor 2 with mitochondrial (f)Met-tRNA. Nucleic Acids Res. 2004;32:5464–5470. doi: 10.1093/nar/gkh886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer A.C., Spremulli L.L. The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim. Biophys. Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Dottavio-Martin D., Suttle D.P., Ravel J.M. The effects of initiation factors IF-1 and IF-3 on the dissociation of Escherichia coli 70 S ribosomes. FEBS Lett. 1979;97:105–110. doi: 10.1016/0014-5793(79)80062-9. [DOI] [PubMed] [Google Scholar]
- 17.Wintermeyer W., Gualerzi C. Effect of Escherichia coli initiation factors on the kinetics of N-AcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry. 1983;22:690–694. doi: 10.1021/bi00272a025. [DOI] [PubMed] [Google Scholar]
- 18.Tedin K., Moll I., Grill S., Resch A., Graschopf A., Gualerzi C.O., Bläsi U. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- 19.Hartz D., Binkley J., Hollingsworth T., Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by E.coli IF-3. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 20.Meinnel T., Sacerdot C., Graffe M., Blanquet S., Springer M. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 1999;290:825–837. doi: 10.1006/jmbi.1999.2881. [DOI] [PubMed] [Google Scholar]
- 21.Sussman J., Simons E., Simons R. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 22.La Teana A., Gualerzi C., Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30 S subunit under the influence of the initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- 23.Koc E.C., Spremulli L.L. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 24.Petrelli D., LaTeana A., Garofalo C., Spurio R., Pon C.L., Gualerzi C.O. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Q., Yu N.-J., Spremulli L.L. Expression and functional analysis of Euglena gracilis chloroplast initiation factor 3. Plant Mol. Biol. 1996;32:937–945. doi: 10.1007/BF00020490. [DOI] [PubMed] [Google Scholar]
- 26.Lin Q., Ma L., Burkhart W., Spremulli L.L. Isolation and characterization of cDNA clones for chloroplast translational initiation factor-3 from Euglena gracilis. J. Biol. Chem. 1994;269:9436–9444. [PubMed] [Google Scholar]
- 27.Yu N.-J., Spremulli L.L. Structural and mechanistic studies on chloroplast translational initiation factor 3 from Euglena gracilis. Biochemistry. 1997;36:14827–14835. doi: 10.1021/bi971185y. [DOI] [PubMed] [Google Scholar]
- 28.Yu N.-J., Spremulli L.L. Regulation of the activity of chloroplast translational initiation factor 3 by NH2- and COOH-terminal extensions. J. Biol. Chem. 1998;273:3871–3877. doi: 10.1074/jbc.273.7.3871. [DOI] [PubMed] [Google Scholar]
- 29.Matthews D.E., Hessler R.A., Denslow N.D., Edwards J.S., O'Brien T.W. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- 30.Sharma M.R., Koc E.C., Datta P.P., Booth T.M., Spremulli L.L., Agrawal R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 31.Graves M., Breitenberger C., Spremulli L.L. Euglena gracilis chloroplast ribosomes: Improved isolation procedure and comparison of elongation factor specificity with prokaryotic and eukaryotic ribosomes. Arch. Biochem. Biophys. 1980;204:444–454. doi: 10.1016/0003-9861(80)90055-7. [DOI] [PubMed] [Google Scholar]
- 32.Graves M., Spremulli L.L. Activity of Euglena gracilis chloroplast ribosomes with prokaryotic and eukaryotic initiation factors. Arch. Biochem. Biophys. 1983;222:192–199. doi: 10.1016/0003-9861(83)90516-7. [DOI] [PubMed] [Google Scholar]
- 33.Hapke B., Noll H. Structural dynamics of bacterial ribosomes. IV. Classification of ribosomes by subunit interactions. J. Mol. Biol. 1976;105:97–109. doi: 10.1016/0022-2836(76)90196-0. [DOI] [PubMed] [Google Scholar]
- 34.Pon C.L., Gualerzi C. Qualitative and semiquantitative assay of Escherichia coli translational initiation factor IF-3. Methods Enzymol. 1979;60:230–239. doi: 10.1016/s0076-6879(79)60020-4. [DOI] [PubMed] [Google Scholar]
- 35.Liao H.-X., Spremulli L.L. Interaction of bovine mitochondrial ribosomes with messenger RNA. J. Biol. Chem. 1989;264:7518–7522. [PubMed] [Google Scholar]
- 36.Wang C.C., Roney W., Alston R., Spremulli L.L. Initiation complex formation on chloroplast 30 S subunits in the presence of natural mRNAs. Nucleic Acids Res. 1989;17:9735–9747. doi: 10.1093/nar/17.23.9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betts L., Spremulli L.L. Analysis of the role of the Shine–Dalgarno sequence and mRNA secondary structure on the efficiency of translational initiation in the Euglena gracilis chloroplast aptH mRNA. J. Biol. Chem. 1994;269:26456–26463. [PubMed] [Google Scholar]
- 38.Risuleo G., Gualerzi C., Pon C. Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30-S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur. J. Biochem. 1976;67:603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 39.Gualerzi C., Risuleo G., Pon C. Mechanism of the spontaneous and initiation factor 3-induced dissociation of 30 S:aminoacyl-tRNA:polynucleotide ternary complexes. J. Biol. Chem. 1979;254:44–49. [PubMed] [Google Scholar]
- 40.Meinnel T., Sacerdot C., Graffe M., Blanquet S., Springer M. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 1999;290:825–837. doi: 10.1006/jmbi.1999.2881. [DOI] [PubMed] [Google Scholar]
- 41.Hartz D., McPheeters D., Gold L. Selection of the initiator tRNA by E.coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- 42.Dallas A., Noller H.F. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 43.Noll M., Noll H. Translation of R17 RNA by Escherichia coli ribosomes. Initiator transfer RNA-directed binding of 30 S subunits to the starting codon of the coat protein gene. J. Mol. Biol. 1974;89:477–494. doi: 10.1016/0022-2836(74)90477-x. [DOI] [PubMed] [Google Scholar]
- 44.Liao H.-X., Spremulli L.L. Effects of length and mRNA secondary structure on the interaction of bovine mitochondrial ribosomes with messenger RNA. J. Biol. Chem. 1990;265:11761–11765. [PubMed] [Google Scholar]
- 45.Takemoto C., Ueda T., Miura K., Watanabe K. Nucleotide sequences of animal mitochondrial tRNAs(Met) possibly recognizing both AUG and AUA codons. Nucleic Acids Symp. Ser. 1999;42:77–78. doi: 10.1093/nass/42.1.77. [DOI] [PubMed] [Google Scholar]
- 46.Anderson S., Bankier A.T., Barrell B.G., Debruijn M.H.L., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 47.Chomyn A., Mariottini P., Cleeter M., Ragan C., Matsuno-Uagi A., Hatefi Y., Doolittle R., Attardi G. Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase. Nature. 1985;314:596–602. doi: 10.1038/314592a0. [DOI] [PubMed] [Google Scholar]
- 48.Dubot A., Godinot C., Dumur V., Sablonniere B., Stojkovic T., Cuisset J.M., Vojtiskova A., Pecina P., Jesina P., Houstek J. GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene. Biochem. Biophys. Res. Commun. 2004;313:687–693. doi: 10.1016/j.bbrc.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 49.Mandal N., Mangroo D., Dalluge J., McCloskey J., RajBhandary U. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherchia coli. RNA. 1996;2:473–482. [PMC free article] [PubMed] [Google Scholar]
- 50.RajBhandary U., Chow C. Initiator tRNAs and initiation of protein synthesis. In: RajBhandary U., Soll D., editors. tRNA: Structure, Biosynthesis and Function. Washington, D.C.: ASM Press; 1995. pp. 511–528. [Google Scholar]
- 51.Seong B.L., RajBhandary U.L. Escherichia coli formylmethionine transfer-RNA—mutations in GGG-CCC sequence conserved in anticodon stem of initiator transfer-RNAs affect initiation of protein-synthesis and conformation of anticodon loop. Proc. Natl Acad. Sci. USA. 1987;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clemons W.M.J., May J.L., Wimberly B.T., McCutcheon J.P., Capel M.S., Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 53.Canonaco M., Gualerzi C., Pon C. Alternative occupancy of a dual ribosomal binding site by mRNA affected by translation initiation factors. Eur. J. Biochem. 1989;182:501–506. doi: 10.1111/j.1432-1033.1989.tb14856.x. [DOI] [PubMed] [Google Scholar]
- 54.McCarthy J., Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 55.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]