Abstract
DNA-PK is a protein complex that consists of a DNA-binding, regulatory subunit [Ku] and a larger ∼465 kDa catalytic subunit [DNA-PKcs], a serine/threonine protein kinase. The kinase activity of DNA-PKcs resides between residues 3745 and 4013, a PI3 kinase domain. Another recognized domain within this large protein is a leucine zipper (LZ) motif or perhaps more appropriately designated a leucine rich region (LRR) that spans residues 1503–1602. Whereas, DNA-PK's kinase activity has been shown to be absolutely indispensable for its function in non-homologous end joining (NHEJ), little is known about the functional relevance of the LRR. Here we show that DNA-PKcs with point mutations in the LRR can only partially reverse the radiosensitive phenotype and V(D)J recombination deficits of DNA-PKcs deficient cells. Disruption of the LRR motif affects the ability to purify DNA-PKcs via its binding to DNA-cellulose, but does not affect its interaction with Ku or its catalytic activity. These data suggest that the LRR region of DNA-PKcs may contribute to its intrinsic DNA affinity, and moreover, that intrinsic DNA binding is important for optimal function of DNA-PKcs in repairing double strand breaks in living cells.
INTRODUCTION
The DNA-dependent protein kinase (DNA-PK) plays an essential role in non-homologous DNA end joining (NHEJ) by initially localizing to DNA double strand breaks (DSBs). Its immense size (∼465 kDa) suggests that the catalytic subunit of DNA-PKcs (DNA-PKcs) may function (at least in part) as a scaffold to organize a DNA repair complex [reviewed in (1,2)]. Indeed, DNA-PKcs has been shown to interact with several other polypeptides including the Ku heterodimer, the XRCC4–DNA ligase IV complex, the Artemis endonuclease and the nuclear matrix protein C1D. An emerging consensus of DNA-PK's function is that after initial binding (one DNA-PK complex per DNA end), synapsis of two complexes activates DNA-PK's kinase activity (known to be requisite for NHEJ), and the active complex targets other repair factors to the site of DNA damage [reviewed in (1,2)]. Although numerous phosphorylation substrates of DNA-PK have been identified, to date the only DNA-PK phosphorylation event shown conclusively to be requisite in NHEJ are those targeting DNA-PKcs itself (3–13). Consistent with a model whereby DNA-PKcs serves a scaffolding role is data suggesting that DNA-PKcs regulates access of broken DNA ends to repair factors via autophosphorylation (4,7,8,14,15). Thus, autophosphorylation-induced conformational changes of DNA-PK may orchestrate an as yet undefined sequence of repair events.
Recent structural studies suggest that DNA-PKcs has three major domains, and that it shares this organization with the related kinase, ATM. The three domains have been termed as palm, arm and head (16,17). The head contains both the enzymatic (PI3 kinase) motifs as well as regions previously implicated in DNA-PKcs's interaction with Ku. The arm is a connecting domain and likely contains important regulatory autophosphorylation sites. The palm consists of two protruding regions termed claws. The distal claw most probably comprises the extreme N-terminus of DNA-PKcs; the proximal claw likely contains the leucine rich region (LRR). In the DNA-bound confirmation, a significant conformation change occurs such that the head and palm clamp together, possibly stabilizing the protein–DNA interaction.
In 1998, Jackson et al. (18) identified C1D as a factor that interacts specifically both in vitro and in vivo with the LRR of DNA-PKcs. C1D was originally identified (by expression cloning) as a 16 kDa polypeptide that is released from rigorously extracted and nuclease-digested DNA. The authors proposed that C1D is a nuclear matrix protein that might be involved in gene regulation (19). More recent studies have implicated C1D in promoting apoptosis in mammalian cells (20) and in contributing to both NHEJ and HR in yeast (21). Whether C1D functions in either NHEJ or HR in higher eukaryotes has not been addressed.
Here, using a mutagenic approach, we address the functional relevance of DNA-PK's LRR. We demonstrate that DNA-PKcs deficient cells expressing the LRR mutant (LRRm1) protein are considerably more radiosensitive and less proficient at V(D)J recombination than cells that express wild-type DNA-PKcs. Additionally, DNA-PKcs harboring a mutation within the LRR (LRRm1) fractionates poorly with DNA cellulose as compared to the wild-type protein. However, pull-down experiments demonstrate that the LRR mutant interacts with the DNA-binding protein, C1D analogous to wild-type DNA-PKcs. Similarly, in vitro LRRm1 interacts with Ku as well as wild-type DNA-PKcs. The poor fractionation onto DNA cellulose is therefore attributed to disruption of the innate DNA-binding capabilities of DNA-PKcs. Thus, we conclude that in addition to being recruited to DNA breaks by its DNA end-binding partner Ku, the intrinsic interaction of DNA-PKcs with DNA is also functionally important.
MATERIALS AND METHODS
Cell lines and culture conditions
The V3, DNA-PKcs deficient double strand break repair [DSBR] mutant CHO cell line (22) was the gift of Dr Martin Gellert. Cells were maintained in αMEM with 10% fetal calf serum (Gibco BRL, Gaithersburg, MD). Stable transfectants were maintained with 400 µg/ml G418. Sf9 cells were maintained in Sf-900 II SFM medium (Invitrogen, Carlsbad, CA) containing 1% antibiotic–antimycotic. The cells were grown at 27°C without CO2.
Oligonucleotides
Oligonucleotides used in this study are as follows (5′ > 3′): KAM 110, GGAACAAGAGAATGGAGATGA; KAM 111, GCTTCTGTGCATGTGCTTGAC, (KAM 110, 111 flank the LRR); KAM 112 and its complement 113, GACTTCTGGACCCAGGCTTTGC, (used to generate LRRm1); KAM 281, TTGCGGCCGCGCACTTTTACTTTTTCCTTTATTGGC; KAM 282, TTAAGCTTCAGCCATAATGGCAGGTGAA, (Kam281 and 282 used to clone human C1D); KAM 390, CGCTCTAGATCTGCCATAATGCATCACCATCACCATCACGCAGGTGAAGAAATTAATGAAGAC; KAM 391, TCTAGACTCGAGCGGCCGCGCTTAACTTTTACTTTTTCC, (KAM 390 and 391 used to clone human C1D in baculovirus transfer vector); KAM 439, CGTGGGATCCCCAGGATGTCCACAGATTTG; KAM 440, GCGGCTCGAGTCCATTTTGGTATTATCCAC (used to subclone LRR into pGEX 5X-3); KAM 482 and its complement 483, CTCAGTTGAAGCAGCCGGCCAGCGGACTTCTG (used to generate LRRm2).
Construction and transfection of expression plasmids
Construction of the wild-type human DNA-PKcs expression vector was described previously (23). To generate the expression plasmids encoding the LRR mutant, duplex oligonucleotides KAM 112 and KAM 113 flanking the LRR were used to amplify a 5 kb fragment using human DNA-PKcs cDNA as the template DNA. The subsequent PCR fragment was cloned using the Topo-TA cloning kit according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). The resulting plasmid was utilized to generate the LRR mutant by Quik change mutagenesis according to the manufacturer's protocol (Stratagene, La Jolla, CA). This PCR fragment spans two EheI sites at positions 1770 and 6280 in the open reading frame of the human DNA-PKcs cDNA. This fragment was subsequently subcloned into the wild-type DNA-PKcs expression vector after sequencing analyses confirmed that no additional substitutions were introduced during PCR amplifications.
V3 transfectants expressing human DNA-PKcs were derived as described previously (4).
DNA cellulose pulldown of DNA-PK, measurement of protein kinase activity and nuclear matrix preparations
Whole cell extracts were prepared and kinase activity was measured as described previously (4). To assess fractionation of DNA-PKcs onto DNA-cellulose, whole cell extracts were absorbed onto DNA-cellulose in 500 µl buffer A [25 mM HEPES, pH 7.9; 50 mM KCl, 10 mM MgCl2, 10% (v/v) glycerol; 1 mM EDTA; 1 mM EGTA; 1 mM DTT and the indicated concentration of NaCl] for 1 h at 4°C. Beads were washed three times in the same buffer and washed beads were suspended in 2× SDS–PAGE buffer (25 mM Tris–HCl pH 6.8, 10% (v/v) glycerol, 1% SDS, 0.04% bromophenol blue and 25 µl 2-mercaptoethanol added fresh to 475 µl of the buffer). The beads were then heated for 5 min at 70°C, centrifuged at 1.5 r.p.m. for 2 min. The supernatant was then analyzed by immunoblotting as described previously (4).
To visualize mobilization of DNA-PKcs in response to DSBs, V3 transfectants were treated for 1 h with bleomycin (Sigma, St Louis, MO). Membrane-insoluble fractions were prepared as described previously (24).
V(D)J recombination assays, assessment of radiosensitivity and drug sensitivity
Extrachromosomal recombination assays were performed as described previously (4). To assess radiosensitivity, cells (3 × 103) were exposed to various amounts of ionizing radiation using a 60Co source and immediately seeded in complete medium containing 10% fetal bovine serum. After 7 days, cell colonies were fixed with ethanol, stained with crystal violet and colony numbers were assessed. To assess bleomycin sensitivity, cells (5000) were seeded in complete medium containing 10% fetal bovine serum in 24-well culture plates and exposed to various amounts of bleomycin sulfate. After 5 days, cell viability was assessed by MTT staining.
Expression of fusion proteins and pull-down experiments
A cDNA encoding full-length human C1D was amplified from human brain mRNA by RT–PCR using oligonucleotides KAM 281 and KAM 282. The termination codon was not included, and the amplified fragment was cloned into pcDNA-6V5/His expression vector (Invitrogen, Carlsbad, CA) that encodes a C-terminal V5-HIS tag. The above cDNA was then used as a template to amplify and subclone C1D into BamH1 and XhoI sites in pFastBac I (a baculovirus expression vector) using oligonucleotides KAM 390 and KAM 391. The oligonucleotides encode an N-terminal His tag. Human Ku70 and Ku80 cDNA cloned into pFastBac Dual was a gift from Dr Dale Ramsden.
Whole cell extracts from V3 transfectants (2 mg) and whole cell extracts from either C1D-infected, Ku-infected or control virus-infected Sf9 cells were co-incubated for 30 min in buffer A at 4°C. Subsequently, 30 µl of Ni+ agarose was added, and the extracts were absorbed in buffer A with 25 mM imidazole. After 1 h, the Ni+ agarose was washed three times with buffer A containing 50 mM imidazole. Proteins were eluted with SDS–PAGE buffer and analyzed by immunoblotting.
To express LRR fusion proteins in bacteria, the LRR (amino acids 1471–1592) was amplified using DNA-PKcs plasmids (either wild-type or LRR mutant) as the template using oligonucleotides KAM 439 and KAM 440. The PCR product was digested and subcloned into BamH1 and XhoI sites in pGEX-5X-3 (Amersham, Piscataway, NJ) to generate GST-LRR or GST-LRRm1 expression plasmids. The GST-LRRm1 was then utilized as a template to generate GST-LRRm2 (L1510P, E1516D, L1517P) by Quik change mutagenesis (Stratagene, LaJolla, CA) using oligonucleotides KAM 482 and KAM 483. GST-LRRdel was generated by deleting the Xba1-Nco1 fragment (amino acids 1504–1551) from GST-LRR. All plasmids were sequenced to confirm that no extra substitutions had been introduced by PCR amplification.
The expression plasmids for GST-fusion proteins were transformed into Escherchia coli BL-21 (DE3) (Stratagene, LaJolla, CA). To express GST-fusion proteins, a 5 ml overnight culture was used as a seed culture for 100 ml Luria–Bertani broth. The bacteria were grown at 37°C to reach the A600 of 0.6 and then induced with 1 mM IPTG at 27°C for 4 h. The cells were harvested and suspended in 5 ml lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X-100 and Protease inhibitor cocktail tablet) (Complete Mini; Roche, Indianapolis, IN) and sonicated for 15 min on ice. The lysate was then centrifuged in an Eppendorf centrifuge at maximum speed for 15 min at 4°C. The induction and solubility was assessed by SDS–PAGE followed by Coomassie staining.
Whole cell lysate from bacteria expressing either GST alone or equal amounts of GST-LRR, GST-LRRm1, GST-LRRm2 or GST-LRRdel and whole cell extracts from His tagged C1D-infected Sf9 cells were co-incubated in buffer A for 30 min at 4°C. Subsequently, 30 µl of pre-swollen glutathione–agarose (pre-equilibrated in buffer A) was added and incubated for 1 h at 4°C. After 1 h, the glutathione–agarose was washed three times with buffer A containing 2 mM glutathione (reduced form). Proteins were eluted with SDS–PAGE buffer and analyzed for C1D by immunoblotting using anti-His (Qiagen, Valencia, CA) as the primary antibody (1:5000 dilution).
RESULTS
LRRm1 only partially reverses V3's radiosensitive phenotype
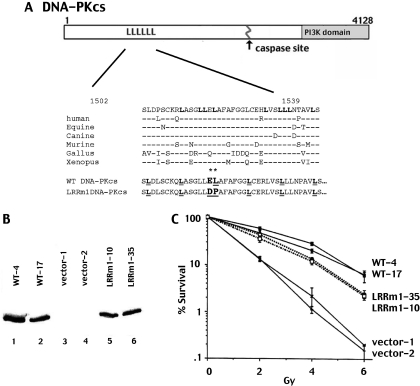
A potential leucine zipper (LZ) motif within the DNA-PKcs coding sequence spanning residues 1503–1538 was noted by Hartley et al. (25) in the original sequence analysis of the molecule. It consists of six leucine residues, each separated by six residues. More recently, other investigators consider this region (1503–1539) to be more appropriately designated as an LRR (33% leucine) since the first and fourth leucine residues (aa1502 and aa1524, respectively) are substituted in chicken and Xenopus DNA-PKcs. The other four leucine residues of the putative LZ are conserved among all vertebrates (26) (Figure 1A). A more extensive overlapping region within DNA-PKcs (1503–1602) is also relatively leucine rich (20%). DNA-PKcs is highly homologous (63–85%) in the six species with complete DNA-PKcs sequences available, (human, horse, dog, mouse, chicken and frog). The LRR is similarly well conserved.
Figure 1.
LRRm1 only partially reverses V3's radiosensitive phenotype. (A) Diagrammatic representation of mutations (*) in LRR region of DNA-PKcs. Bold ‘L’ indicate conserved leucine residues. Underlined ‘L’ indicate the six leucine residues assigned as a LZ motif by Hartley et al. (25). (B) Immunoblot analysis of whole cell extracts from two different V3 clonal transfectants expressing either full length DNA-PKcs (lanes 1 and 2), vector alone (lanes 3 and 4) or LRRm1 (lanes 5 and 6). (C) Radioresistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone or LRRm1 was assessed as described in Materials and Methods. Data are presented as percent survival of non-irradiated controls. Error bars depict standard error of the mean of three separate experiments.
In the report from Yavuzer et al. (18) demonstrating that C1D interacts with the LRR of DNA-PKcs, two site-specific mutants of a DNA-PKcs polypeptide that ablated its interaction with C1D were studied. One of the mutants contains two point mutations substituting aspartic acid and proline for the glutamic acid and leucine residues at amino acids 1516 and 1517 as indicated in Figure 1A. This mutant peptide was not able to interact with CID as assessed by yeast two-hybrid assays (18).
To begin to assess the functional relevance of the LRR of DNA-PKcs in living cells, these mutations were introduced into the complete DNA-PKcs cDNA. Wild-type and mutant constructs encoding DNA-PKcs (designated WT and LRRm1) were stably transfected into DSBR mutant V3 cells that lack DNA-PKcs and are thus defective in NHEJ. Clones with similar levels of DNA-PKcs expression were selected for further study (Figure 1B). To assess the ability of the wild-type and mutant constructs to reverse the known radiosensitive phenotype of V3 cells, cell irradiation assays were performed. As can be seen (Figure 1C), although the LRR mutant renders cells significantly more radioresistant than cells transfected with vector alone, cells expressing the mutant protein are still substantially more radiosensitive than cells expressing equivalent levels of wild-type DNA-PKcs. Thus, we conclude that the LRR mutant protein can only partially complement the radiosensitivity of V3 cells.
LRRm1 supports reduced levels of coding and signal end joining in V3 cells
The ability of the mutant DNA-PKcs to support RAG-induced V(D)J recombination in V3 cells was tested. As can be seen (Table 1), wild-type DNA-PKcs substantially complements the coding end joining deficit of V3 cells (assessed with substrate pJH290). In contrast LRRm1 supports only reduced levels of coding end joining as compared to wild-type DNA-PKcs (1.5- to 14-fold reduced). We and others have reported previously that V3 cells have a significant signal end joining deficit that is substantially reversed by transfecting wild-type DNA-PKcs (4,27,28). Consistent with those results, we find here that wild-type DNA-PKcs substantially increases the recovery of signal joints from V3 cells (assessed with substrate pJH201). Although co-transfection of LRRm1 also increases the recovery of signal joints from V3 cells, the numbers are consistently reduced as compared to transfections with wild-type DNA-PKcs (1.2- to 8-fold). As reported previously, co-transfection of an ATP binding site mutant (K>M) does not alter the levels of either coding or signal joints in transient assays as compared to transfections with no DNA-PKcs at all (29). These modest affects on coding and signal end resolution are consistent with the mild radiosensitivity observed in cells expressing LRRm1. Thus, we conclude that LRRm1 can only partially support either signal or coding end joining during V(D)J recombination.
Table 1.
LRRm1 supports reduced levels of V(D)J recombination
| Transfected plasmidsa,b | Coding joints (pJH290) | Signal joints (pJH201) | ||
|---|---|---|---|---|
| #Amp Cam/#Amp | Recombination (%)c | #Amp Cam/#Amp | Recombination (%)c | |
| RAGS only | 1/2500 | 0.040 | 2/6750 | 0.030 |
| 1/33 250 | 0.003 | 0/18 750 | 0 | |
| 0/22 500 | 0 | 2/23 500 | 0.009 | |
| 0/33 750 | 0 | 2/11 750 | 0.017 | |
| 0/6500 | 0 | 3/3000 | 0.100 | |
| RAGS + wild type | 53/5000 | 1.060 | 184/25 750 | 0.715 |
| 108/103 000 | 0.105 | 106/17 000 | 0.624 | |
| 99/41 750 | 0.237 | 113/27 500 | 0.411 | |
| 96/51 250 | 0.187 | 157/13 750 | 1.142 | |
| 67/8750 | 0.766 | 95/4500 | 2.111 | |
| RAGS + LRR | 8/11 000 | 0.073 | 38/12 250 | 0.310 |
| 28/106 250 | 0.026 | 4/5250 | 0.076 | |
| 36/76 000 | 0.047 | 45/24 250 | 0.186 | |
| 30/23 250 | 0.129 | 440/54 000 | 0.815 | |
| 11/2250 | 0.489 | 77/4500 | 1.711 | |
| RAGS + K>M | 5/44 250 | 0.011 | 6/17 750 | 0.034 |
| 0/7500 | 0 | 0/10 750 | 0 | |
| 1/38 500 | 0.003 | 0/15 750 | 0 | |
| 0/6000 | 0 | 8/41 500 | 0.019 | |
| 0/5000 | 0 | 3/3750 | 0.080 | |
Transient V(D)J recombination assays were performed as described in Materials and Methods. RAG expression from plasmid vectors initiates recombination in the V3 cells, as assessed by the plasmid substrate pJH290 that detects coding joints or the pJH201 substrate that detects signal joints as indicated. Numbers of ampicillin (amp) and ampicillin/chloramphenicol (amp+cam) resistant colonies from five separate experiments are presented. Recombination rate (%R) is calculated as the number of chloramphenicol resistant colonies divided by ampicillin resistant colonies × 100.
aTransient V(D)J recombination assays were performed as described in Materials and Methods. RAG expression from plasmid vectors initiates recombination in V3 cells, as assessed by the plasmid substrate pJH 290, which detects coding joints, or the pJH 201 substrate, which detects signal joints, as indicated.
bResults from five independent experiments are presented.
cCalculated as (number of chloramphenicol-resistant colonies/number of ampicillin-resistant colonies) × 100.
Recently, our laboratory demonstrated that mutations in DNA-PKcs's numerous autophosphorylation sites dramatically affect end processing of coding joints (4,8). More specifically these data demonstrate that two conserved autophosphorylation site clusters reciprocally regulate end processing. Thus, blocking phosphorylation in one cluster (by mutagenesis) clocks end processing, whereas blocking phosphorylation in the second cluster dramatically enhances end processing. To determine whether mutations in the LRR affect end processing, DNA was prepared from recombined plasmids isolated from transient V(D)J assays. However, sequence analysis of coding joints mediated by the LRR mutant as compared to those mediated by the wild-type protein were comparable with regards to nucleotide loss and percent complete coding ends (Table 2). We conclude that coding joints mediated by the LRR mutant protein are structurally normal.
Table 2.
Coding joints mediated by LRRm1 have normal nucleotide loss from joined coding ends
| No. of sequences | Base loss/joint | % Complete ends | |
|---|---|---|---|
| Wild type | 72 | 4.8 | 29 (42/144) |
| LRR | 26 | 3.65 | 31 (16/52) |
| RAGS only | 16 | 14.69 | 41 (13/32) |
Average nucleotide losses per joint and percent intact coding ends were calculated from sequences obtained from isolated chloramphenicol-resistant pJH290 plasmids.
Both LRRm1 and wild-type DNA-PKcs interact with C1D
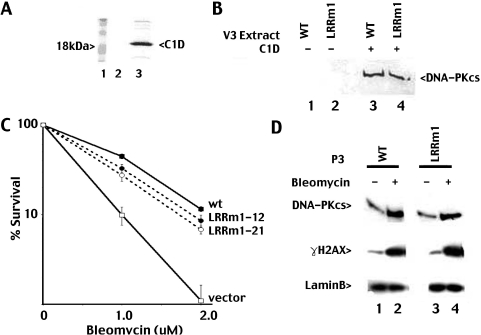
To confirm that the functional deficits of LRRm1 could be attributed to disruption of its interaction with C1D, a baculovirus vector expressing His-tagged C1D was constructed and expressed in Sf9 cells for pull down experiments (Figure 2A). Extracts from C1D-infected Sf9 or control virus-infected Sf9 cells were incubated with the cell extracts from V3 transfectants expressing either wild-type or LRRm1 DNA-PKcs and Ni+ agarose. To our surprise (and in contrast with the yeast two hybrid studies of Yavuzer et al.), both wild-type and mutant proteins fractionate similarly with immobilized C1D onto Ni+ agarose (Figure 2B).
Figure 2.
Both LRRm1 and wild-type DNA-PKcs interact equivalently with C1D and are targeted similarly to nuclear matrix by DSBs. (A) Whole cell extract from Sf9 cells infected with baculovirus expressing His-tagged C1D was incubated with Ni+ agarose beads (lane 3). Beads were washed and analyzed for C1D expression by SDS–PAGE. (B) Ni+ agarose fractions of whole cell extracts (2 mg) from V3 transfectants expressing either wild-type DNA-PKcs (lanes 1 and 3) or LRRm1 (lanes 2 and 4) incubated with whole cell lysates from either control virus infected-Sf9 cells (lanes 1 and 2) or C1D-infected Sf9 cells (lanes 3 and 4) were immunoblotted for DNA-PKcs. (C) Bleomycin resistance of V3 transfectants expressing wild-type DNA-PKcs, vector alone or LRRm1 was assessed as described in Materials and Methods. Data are presented as percent survival of untreated controls. Error bars depict standard error of the mean of three separate experiments. (D) V3 transfectants expressing either wild-type DNA-PKcs (lane1–2) or LRRm1 (lanes 3–4) were left untreated or were treated with bleomycin (140 µM). Nuclear matrix fractions were prepared as described in Materials and Methods and then immunoblotted for DNA-PKcs, γH2AX and LaminB as indicated.
Both LRRm1 and wild-type DNA-PKcs mobilize to nuclear matrix in response to DSBs
There is a large body of evidence that DNA and RNA modifying enzymes are sequestered to particular nuclear compartments, in some cases by association with the nuclear matrix [reviewed in Ref. (30)]. Recently, Drouet et al. have shown that in response to DNA DSBs, DNA-PKcs is mobilized to a triton non-extractable fraction of the nuclear compartment (the nuclear matrix, 24). Yavuzer et al. (18) had previously hypothesized that C1D might be involved in targeting DNA-PKcs to the nuclear matrix in response to DNA damage. High doses of bleomycin were utilized to induce DSBs in the study of Drouet et al. As expected, cells expressing LRRm1 display intermediate sensitivity to bleomycin as compared to cells expressing wild-type DNA-PKcs or no DNA-PKcs at all (Figure 2C). We next examined the mobilization of LRRm1 to the nuclear matrix in response to DNA damage induced by bleomycin. V3 transfectants expressing either LRRm1 or wild-type DNA-PKcs were treated with bleomycin for 1 h to induce DSBs, and cells were fractionated as outlined by Drouet et al. The extracted fraction was then analyzed by immunoblotting for DNA-PKcs levels.
As shown in Figure 2D, there is a significant mobilization of DNA-PKcs and γ-H2AX to the nuclear matrix fraction in response to DSBs (laminB is a constitutive nuclear matrix protein). However, both LRRm1 and wild-type DNA-PKcs associate with the nuclear matrix equally well. Thus, we conclude that the NHEJ defect in the LRR mutant cannot be attributed to disruption of its interaction with either C1D or the nuclear matrix.
Both the LRR and regions outside the LRR in DNA-PKcs contribute to its interaction with C1D
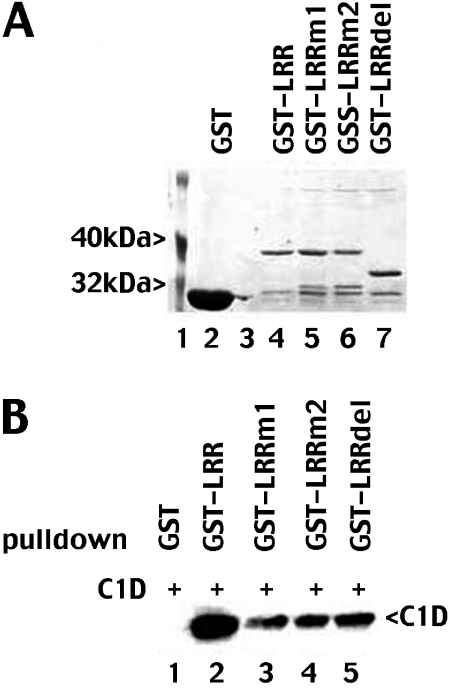
To understand the discrepancy between our results and the results published by Yavuzer et al., we considered that the LRR motif is not adequately disrupted in the full-length mutant protein. To address whether disruption of both L1510 and L1517 might more completely disrupt the LRR, we generated several GST fusion proteins spanning the LRR. These include wild-type DNA-PKcs, GST-LRR (spanning residues 1471–1592), GST-LRRm1 (with the same mutations already tested in the full length protein, E1516D and L1517P) GST-LRRm2 (including L1510P, E1516D, and L1517P) and GST-LRRdel (deleting residues 1504–1551) (Figure 3A). GST fusion proteins were co-incubated with extracts from C1D infected Sf9 cells and immobilized onto glutathione agarose beads and analyzed for C1D by immunoblotting (Figure 3B). GST-LRRm1 is substantially reduced in its ability to interact with C1D in agreement with the yeast two-hybrid results published by Yavuzer et al. (18) although the mutant retains weak affinity for C1D. Furthermore, GST-LRRm2 and GST-LRRdel also interact weakly with C1D. These data suggest that both the LRR and regions outside the LRR contribute to DNA-PKcs's interaction with C1D.
Figure 3.
Both the LRR and regions outside the LRR of DNA-PKcs contribute to its interaction with C1D. (A) Whole cell lysates from bacteria expressing GST (lane 2) or GST-LRR (lane 4), sGST-LRRm1 (lane 5), GST-LRRm2 (lane 6) or GST-LRRdel (lane 7) were incubated with glutathione-agarose beads. Beads were washed and analyzed for protein expression by SDS-PAGE followed by Coomassie blue staining. (B) Whole cell lysates from bacteria expressing GST (lane 1), GST-LRR (lane 2), GST-LRRm1 (lane 3), GST-LRRm2 (lane 4) or GST-LRRdel (lane 5) were co-incubated with whole cell lysates from Sf9 cells infected with baculovirus expressing His-C1D and absorbed onto glutathione-agarose. Glutathione beads were washed and immunoblotted for His-C1D.
LRRm1 binds poorly to DNA-cellulose but has normal enzymatic activity
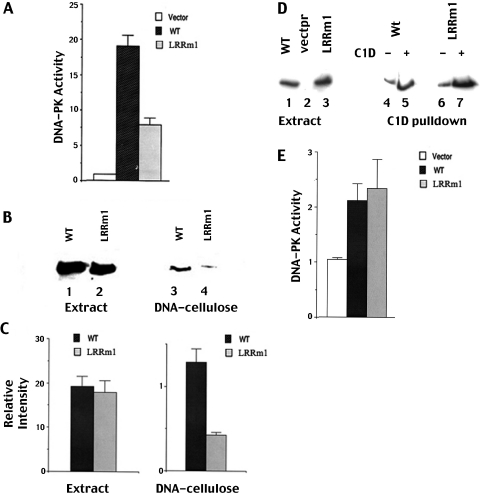
We next tested the mutant for its enzymatic activity utilizing standard DNA cellulose pull-down assays. Extracts from cells expressing LRRm1 consistently displayed approximately 2-fold reduced kinase activity as compared to extracts prepared from cells expressing similar amounts of wild-type DNA-PKcs (Figure 4A). However, immunoblotting of the DNA-cellulose fractions used for the kinase assays revealed that LRRm1 fractionates poorly onto DNA cellulose as compared to the wild-type protein (Figure 4B). This result was highly reproducible as revealed by densitometric quantification of three independent DNA-cellulose binding assays (Figure 4C). Furthermore, the reduction in DNA cellulose fractionation correlated well with the 2-fold reduction in kinase activity of the mutant suggesting that the reduced kinase activity observed in extracts from LRRm1 expressing cells is the result of reduced affinity for DNA and not an intrinsic enzymatic defect.
Figure 4.
LRRm1 binds poorly to DNA-cellulose but has normal enzymatic activity. (A) Whole-cell extracts (250 µg) from V3 cells transfected with either vector alone, wild-type DNA-PKcs or LRRm1 were assayed for DNA-PK activity as described in Materials and Methods. Each cell extract was tested in duplicate and three independent extracts were tested for each cell line. (B) Immunoblot analysis of whole cell extracts (500 µg) and DNA-cellulose fractions as indicated from V3 transfectants expressing wild-type DNA-PKcs (lanes 1 and 3) and LRRm1 (lanes 2 and 4). (C) Densitometric analyses of immunoblots from three independent experiments assessing DNA-cellulose fractionation of wild-type DNA-PKcs and LRRm1. Error bars, standard error of the mean. (D) Immunoblot analyses of Ni+ agarose fractions of whole cell extracts (2 mg) from V3 transfectants expressing either wild-type DNA-PKcs (lanes 1, 4 and 5) or LRRm1 (lanes 3, 6 and 7) incubated with whole cell lysates from either control virus infected-Sf9 cells or His-C1D-infected Sf9 cells as indicated. (E) DNA-PKcs absorbed onto C1D that was immobilized onto Ni+-agarose beads was assayed for enzymatic activity as described in Materials and Methods. Three independent extracts were tested for each cell line. Error bars, standard error of the mean.
To test this more formally, enzymatic activity was assessed by immobilizing LRRm1 or wild-type DNA-PKcs via Ni+-agarose immobilized C1D (Figure 4D). As can be seen, LRRm1 and wild-type DNA-PKcs partially purified by C1D affinity have similar enzymatic activity as measured by phosphorylation of the p53 peptide substrate (Figure 4E). We conclude that the LRRm1 mutation does not affect the enzyme's protein kinase activity.
The LRR contributes to the intrinsic affinity of DNA-PKcs for DNA
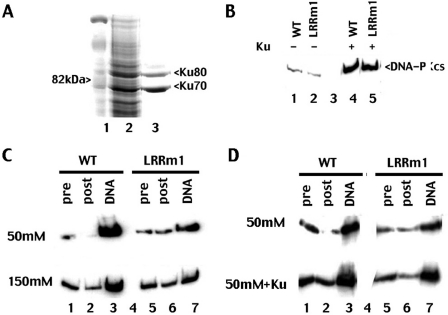
Previous studies have established that residues in the FAT domain of DNA-PKcs are involved in its interaction with Ku (31); thus, it seemed unlikely that mutation in LRRm1 disrupted the mutant's ability to interact with Ku. Still, since the mutant protein exhibits a defect in DNA-binding, we considered the possibility that the LRR facilitates the interaction of DNA-PKcs with Ku bound to DNA. To address this, His-tagged Ku70/80 was expressed in baculovirus (Figure 5A). Whole cell extracts from either Ku-infected or control-infected Sf9 cells were incubated with extracts from transfectants expressing wild-type or LRR mutant DNA-PKcs. The Ku-DNA-PKcs complex was then absorbed onto Ni+ agarose beads; as can be seen (Figure 5B) both LRRm1 and wild-type DNA-PKcs interact equivalently with Ni+agarose bound Ku.
Figure 5.
The LRR contributes to DNA-PKcs's intrinsic affinity for DNA but not its interaction with Ku. (A) Whole cell extract from Sf9 cells infected with baculovirus expressing His-tagged Ku was incubated with Ni+ agarose beads. Beads were washed and analyzed for Ku expression by SDS–PAGE followed by Coomassie blue staining. (B) Immunoblotting of Ni+ agarose fractions of whole cell extracts (2 mg) from V3 transfectants expressing either wild-type DNA-PKcs (lanes1 and 4) or LRRm1 (lanes 2 and 5) incubated with whole cell lysate from either control virus infected-Sf9 cells or Ku-infected Sf9 cells as indicated. (C) Immunoblot analysis of DNA-cellulose fractions of extracts (500 µg) from V3 transfectants expressing wild-type (WT) DNA-PKcs (lane 3) or LRRm1 (lane 7) in either 50 mM salt (upper panel) or 150 mM salt (lower panel) as indicated. Lysates, pre-absorption (lanes 1 and 5) and post-absorption (lanes 2 and 6) represents 2% of the total lysates input. (D) Immunoblot analysis of DNA-cellulose fractions of extracts (500 mg) from V3 transfectants expressing wild-type (WT) DNA-PKcs (lane 3) or LRRm1 (lane 7) in either 50 mM salt (upper panel) or 50 mM salt suspplemented with partially purified Ku (expressed in Baculovirus) (lower panel) as indicated. Lysates, pre-absorption (lanes 1 and 5) and post-absorption (lanes 2 and 6) represents 2% of the total lysates input.
Ku binds directly to DNA ends. When Ku recruits DNA-PKcs, Ku translocates to internal sites in the DNA and DNA-PKcs interacts directly with the DNA end (32,33). Previous studies have established that DNA-PKcs can bind DNA ends in the absence of Ku (34–36), but direct DNA binding by DNA-PKcs is salt labile. DNA-PKcs only binds DNA ends in Ku's absence in low salt concentrations. We considered that LRRm1 might have a deficit in its intrinsic affinity for DNA. Thus, we tested the ability of LRR mutant to bind DNA in various salt conditions by incubating whole cell extracts from stable transfectants with DNA cellulose (Figure 5C). As can be seen, the LRR mutant protein fractionates poorly onto DNA cellulose in both salt concentrations (50 and 150 mM). However, fractionation of wild-type DNA-PKcs onto DNA cellulose is markedly enhanced in low salt concentrations. In contrast, reducing the salt concentration has little affect on DNA-cellulose fractionation of LRRm1. Finally, if the LRRm1 mutant depends primarily on Ku for its interaction with DNA, we reasoned that addition of excess Ku might enhance the fractionation of the mutant onto DNA cellulose. Thus, DNA cellulose binding was assessed in the presence or absence of excess recombinant Ku. As can be seen, fractionation of LRRm1 is substantially increased by the addition of excess Ku. These data are consistent with a model whereby the LRR of DNA-PKcs contributes to its intrinsic affinity for DNA.
DISCUSSION
C1D is a nuclear matrix associated protein and binds DNA with very high affinity (19). Previous work from Jackson and colleagues had shown that DNA-PKcs interacts directly with C1D both in vitro and in vivo (18). The authors inferred that this interaction requires an intact LRR region because in a yeast two-hybrid experiment, a DNA-PKcs fragment with the LRR mutated failed to interact with C1D. Thus, at the onset of our studies, our working model was that DNA-PKcs is targeted to the nuclear matrix or chromatin network via its interaction with C1D and after DNA damage, Ku targets DNA-PKcs to DNA ends. To test this model, we introduced two mutations in the LRR region of DNA-PKcs (E1516D, L1517P) that are identical to the construct used in the previous yeast two-hybrid studies. When expressed in DNA-PKcs deficient cells, the LRR mutant could only partially reverse their NHEJ deficits. However (to our surprise), when tested for its interaction with C1D, the LRR mutant was found not only to interact normally with C1D in vitro, but was also efficiently mobilized to nuclear matrix in response to DNA damage in living cells. Thus, the NHEJ deficits in cells expressing the mutant protein could not be attributed to defective targeting of DNA-PKcs to nuclear matrix via its interaction with C1D.
Several additional in vitro assays were performed to try to delineate the functional deficit of the LRR mutant. Whereas the mutant was found to have normal catalytic activity as a protein kinase, and was able to interact as well as wild-type DNA-PKcs with its DNA end-binding partner, Ku, the mutant did not fractionate onto DNA cellulose as well as wild-type DNA-PKcs. The difference in DNA cellulose binding was consistently most dramatic in low salt conditions when DNA-PKcs can interact with DNA both via its interaction with Ku and via its own intrinsic affinity for DNA (34–36). Thus, we propose that the LRR of DNA-PKcs contributes to the intrinsic ability of DNA-PKcs to bind DNA, and that its intrinsic DNA affinity is functionally important.
Our hypothesis is consistent with recent structural studies of DNA-PKcs (17,18). Llorca and colleagues have demonstrated that the palm region of DNA-PKcs undergoes a conformational change when bound to a DSB. The palm domain contains two ‘claws’ (proximal and distal) that likely facilitate DNA binding. The LRR resides in the proximal claw possibly explaining the DNA-binding deficits of the LRR mutant. These data, in concert with emerging data from our laboratory and others (4,7,8,14), suggest that after targeting to sites of DNA damage by Ku (and possibly C1D), DNA-PKcs regulates DNA end access via its direct interaction with DNA termini, and then regulates end access by a series of autophosphorylation-induced conformational changes.
Acknowledgments
This work was supported by Public Health Service grant AI048758 (K.M.). Funding to pay the Open Access publication charges for this article was provided by Public Health Service grant AIO48758 (K.M.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Meek K., Gupta S., Ramsden D.A., Lees-Miller S.P. DNA-PK: Activity director at the end. Immunol. Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 3.Chan D.W., Chen B.P., Prithivirajsingh S., Kurimasa A., Story M.D., Qin J., Chen D.J. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Q., Reddy Y.V., Wang W., Woods T., Douglas P., Ramsden D.A., Lees-Miller S.P., Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soubeyrand S., Pope L., Pakuts B., Hache R.J. Threonines 2638/2647 in DNA-PK Are Essential for Cellular Resistance to Ionizing Radiation. Cancer Res. 2003;63:1198–1201. [PubMed] [Google Scholar]
- 6.Chen B.P., Chan D.W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D.J. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 7.Block W.D., Yu Y., Merkle D., Gifford J.L., Ding Q., Meek K., Lees-Miller S.P. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui X., Yu Y., Gupta S., Lees-Miller S.P., Meek K. Separate clusters of autophosphorylation sites in DNA-PKcs regulate distinct steps in DNA end processing. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas P., Gupta S., Morrice N., Meek K., Lees-Miller S.P. Ku is phosphorylated by a staurosporine sensitive protein kinase but not DNA-PK in vivo. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y., Wang W., Ding Z., Ye R., Merkle D., Douglas P., Chen D., Schriemer D., Meek K., Lees-Miller S.P. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair. 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 11.Douglas P., Sapkota G.P., Morrice N., Yu Y., Goodarzi A.A., Merkle D., Meek K., Alessi D.R., Lees-Miller S.P. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002;369:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wechsler T., Chen B., Harper R., Morotomi-Yano K., Huagn B.C.B., Meek K., Cleaver J.E., Chen D.J., Wabl M. DNA-PKcs function regulated specifically by protein phosphatase PP5. Proc. Natl Acad. Sci. USA. 2004;101:1247–1252. doi: 10.1073/pnas.0307765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee K.J., Jovanovic M., Udayakumar D., Bladen C.L., Dynan W.S. Identification of DNA-PKcs phosphorylation sites in XRCC4 and effects of mutations at these sites on DNA end joining in a cell-free system. DNA Repair. 2004;3:267–276. doi: 10.1016/j.dnarep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Reddy Y.V., Ding Q., Lees-Miller S.P., Meek K., Ramsden D.A. Nonhomologous end-joining requires the DNA-PK complex to undergo remodeling at DNA ends. J. Biol. Chem. 2004;279:39408–39413. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- 15.Weterings E., Verkaik N.S., Bruggenwirth H.T., Hoeijmakers J.H.J., vanGent D.C. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boskovic J., Rivera-Calzada A., Maman J.D., Chacon P., Willison K.R., Pearl L.H., Llorca O. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J. 2003;22:5875–5882. doi: 10.1093/emboj/cdg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Calzada A., Maman J.D., Spagnolo L., Pearl L.H., Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243–255. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Yavuzer U., Smith G.C., Bliss T., Werner D., Jackson S.P. DNA end-independent activation of DNA-PK mediated via association with the DNA-binding protein C1D. Genes Dev. 1998;12:2188–2199. doi: 10.1101/gad.12.14.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nehls P., Keck T., Greferath R., Spiess E., Glaser T., Rothbarth K., Stammer H., Werner D. cDNA cloning, recombinant expression and characterization of polypeptides with exceptional DNA affinity. Nucleic Acids Res. 1998;26:1160–1166. doi: 10.1093/nar/26.5.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothbarth K., Spiess E., Juodka B., Yavuzer U., Nehls P., Stammer H., Werner D. Induction of apoptosis by overexpression of the DNA-binding and DNA-PK-activating protein C1D. J. Cell Sci. 1999;112:2223–2232. doi: 10.1242/jcs.112.13.2223. [DOI] [PubMed] [Google Scholar]
- 21.Erdemir T., Bilican B., Cagatay T., Goding C.R., Yavuzer U. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 2002;46:947–957. doi: 10.1046/j.1365-2958.2002.03224.x. [DOI] [PubMed] [Google Scholar]
- 22.Whitmore G.F., Varghese A.J., Gulyas S. Cell cycle responses of two X-ray sensitive mutants defective in DNA repair. Int. J. Radiat. Biol. 1989;56:657–665. doi: 10.1080/09553008914551881. [DOI] [PubMed] [Google Scholar]
- 23.Shin E.K., Rijkers T., Pastink A., Meek K. Analyses of TCRB rearrangements substantiate a profound deficit in recombination signal sequence joining in SCID Foals: Implications for the role of DNA-dependent protein kinase in V(D)J recombination. J. Immunol. 2000;164:1416–1424. doi: 10.4049/jimmunol.164.3.1416. [DOI] [PubMed] [Google Scholar]
- 24.Drouet J., Delteil C., Lefrancois J., Concannon P., Salles B., Calsou P. DNA-PK and XRCC4/DNA ligase IV mobilization in the cell in response to DNA double-strand breaks. J. Biol. Chem. 2005;280:7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 25.Hartley K.O., Gell D., Smith G.C., Zhang H., Divecha N., Connelly M.A., Admon A., Lees-Miller S.P., Anderson C.W., Jackson S.P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 26.Fujimori A., Araki R., Fukumura R., Ohhata T., Takahashi H., Kawahara A., Tatsumi K., Abe M. Identification of four highly conserved regions in DNA-PKcs. Immunogenetics. 2000;51:965–973. doi: 10.1007/s002510000227. [DOI] [PubMed] [Google Scholar]
- 27.Kurimasa A., Kumano S., Boubnov N.V., Story M.D., Tung C.S., Peterson S.R., Chen D.J. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods T., Wang W., Convery E., Errami A., Zdzienicka M.Z., Meek K. A single amino acid substitution in DNA-PKcs explains the novel phenotype of the CHO mutant, XR-C2. Nucleic Acids Res. 2002;30:5120–5128. doi: 10.1093/nar/gkf625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kienker L.J., Shin E.K., Meek K. Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000;28:2752–2761. doi: 10.1093/nar/28.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Adv. Enzyme Regul. 2002;42:39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- 31.Jin S., Kharbanda S., Mayer B., Kufe D., Weaver D.T. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J. Biol. Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 32.Yoo S., Kimzey A., Dynan W.S. Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 1999;274:20034–20039. doi: 10.1074/jbc.274.28.20034. [DOI] [PubMed] [Google Scholar]
- 33.Yoo S., Dynan W.S. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dynan W.S., Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammarsten O., Chu G. DNA-dependent protein kinase - DNA binding and activation in the absence of Ku. Proc. Natl Acad. Sci. USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan D.W., Mody C.H., Ting N.S., Lees-Miller S.P. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem. Cell. Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]