Abstract
The nature of any long palindrome that might exist in the human genome is obscured by the instability of such sequences once cloned in Escherichia coli. We describe and validate a practical alternative to the analysis of naturally-occurring palindromes based upon cloning and propagation in Saccharomyces cerevisiae. With this approach we have investigated an intronic sequence in the human Neurofibromatosis 1 (NF1) locus that is represented by multiple conflicting versions in GenBank. We find that the site is highly polymorphic, exhibiting different degrees of palindromy in different individuals. A side-by-side comparison of the same plasmids in E.coli versus. S.cerevisiae demonstrated that the more palindromic alleles were inevitably corrupted upon cloning in E.coli, but could be propagated intact in yeast. The high quality sequence obtained from the yeast-based approach provides insight into the various mechanisms that destabilize a palindrome in E.coli, yeast and humans, into the diversification of a highly polymorphic site within the NF1 locus during primate evolution, and into the association between palindromy and chromosomal translocation.
INTRODUCTION
True DNA palindromes, where a sequence is immediately followed by its exact inverse complement (with no central asymmetry or spacer), can be extremely difficult to analyze by standard molecular genetic approaches. Palindromic sequences are able to self-pair, forming intrastrand hairpin structures and extensive stretches of palindromy can interfere with key aspects of DNA metabolism whether located chromosomally or on extrachromosomal replicons (1–3). For these reasons, pure palindromes longer than roughly 200 bp become impossible to clone in Escherichia coli, and no mutant has been described that fully overcomes the cloning block (4).
The word ‘palindrome’ is applied non-specifically to different types of inverted repeat in the literature, but for consistency we use the term only in its narrowest sense here. Palindromes and near-palindromes have overlapping properties, but a pure palindrome is significantly more susceptible to effects that drive cruciform extrusion than are inverted repeats separated by a central spacer or quasipalindromes with incompletely matched arms (5). The tendency to adopt a cruciform structure underlies at least some of the observed inviability and instability of palindromes in bacteria (2,6).
The suggestion that organisms other than E.coli tolerate long stretches of perfect palindromy to different extents (2,6) is supported by a handful of examples where introduced perfect palindromes (no central spacer) have been documented by DNA sequence analysis. In mice, a 15.7 kb perfect palindrome, transgenically introduced, could be transmitted in the germline (7,8). This huge palindrome, despite moderate instability, was inherited in a Mendelian ratio, and could be maintained in tissue culture cells without any apparent lethal effects. More recently a synthetic, pure palindrome with 900 bp arms was demonstrated to be stably maintained in Saccharomyces cerevisiae lacking SAE2 gene function (9). In both cases, the ultra-large palindromes were well above a size that might be propagated in E.coli, and both required special approaches in order to obtain DNA sequence information.
Suspected palindromes in higher eukaryotes have surfaced in diverse pathological situations. Natural palindromes or near-palindromes of about 200–800 bp exist at sites of some inherited chromosomal rearrangements in humans (10–12). Long inverted repeats that may be palindromic arise de novo at tumor-specific sites in the genome during oncogenesis (13). The origin of these structures may be mechanistically related to long palindromes or near-palindromes that can be experimentally generated in yeast (14–16). A recent observation of note is that yeast strains compromised for telomere maintenance can escape senescence concurrent with the generation of long palindrome-like structures near chromosome ends (17).
It is highly desirable to learn where perfect, or near-perfect palindromes exist naturally in the human genome, but this is difficult to determine because such sequences become corrupted or eliminated as soon as they are introduced into E.coli. It cannot be assumed, and as demonstrated in the present report, is in fact unlikely that the Human Genome Project database is representational with respect to palindromic features. At present most suspected palindromes in the human genome remain unverified and unrepresented as such in the consortium sequence (10–12). Alternatives to propagation of sequences in E.coli cloning have to be developed.
Here we focused on a site on human chromosome 17q11 for which conflicting evidence suggests the presence of a perfectly, or very nearly, palindromic sequence (18). At this position, a rare but recurrent translocation breakpoint in the intron between exons 31 and 32 of the Neurofibromatosis 1 (NF1) gene has been documented (18). The 17q11 palindrome is one of 67 perfect palindromes ≥90 bp retrieved in a search of the human genome database (S. M. Lewis, S. Chen, T. Alleyne, J. Cheung and R. Richard, manuscript in preparation). In the reference human genome sequence (NCBI Build 35), the 17q11 sequence is perfectly palindromic, but this site is significantly shorter than the sequence deduced from translocation breakpoint studies and other sequence analyses (18). Indeed multiple, inconsistent versions of this small region have been reported: disagreement exists between sources that include The Human Genome Project (chr17:26686385–26686474, NCBI Build 35), NF1 gene sequences deposited in GenBank (AC004526, L05367 and AY796305), data from breakpoint analysis of NF1 translocations (18,19) and additional full or partial BAC clones derived as part of the consortium sequence effort (AC134669, AC135274 and AC135183). All discrepancies involve the suspected palindrome and not all of the variants were fully palindromic. Consequently we were interested to learn which of these might be verified in human genomic DNA, whether the NF1 intronic site is polymorphic and if so, whether the longer or more perfectly palindromic alleles have an association with chromosomal translocation.
The undertaking was complicated at every turn by the structure-prone features of this particular NF1 intronic region. Each involved procedure, from the initial PCR amplification to the final DNA sequence determination had to be evaluated and modified. Presented here is a set of practical methods, suited to any standard molecular genetic laboratory, that circumvents difficulties inherent in the analysis of DNA palindromes. With these procedures we have been able to collect data that provide insight into the structure, evolution and pathological relevance of a naturally-arising palindrome in the human genome.
MATERIALS AND METHODS
DNA samples
Human genomic DNA was prepared from white blood cells isolated on Ficoll-Pacque (Amersham). Blood was collected from volunteers informed according to the protocol approved by the Ethics Review Board at the Hospital for Sick Children. White blood cells were isolated on Ficoll-Pacque (Amersham) and high molecular weight DNA was prepared by either a standard proteinase K digestion followed by successive multiple phenol and chloroform extractions and precipitation or with a GenomicPrep Blood DNA Isolation Kit (Amersham). CEPH DNA samples, as well as chimpanzee and gorilla DNA were analyzed in conjunction with the Center for Applied Genomics (TGAC) at the Hospital for Sick Children. The source of the gorilla DNA was cell line EB (JC) (ECACC cat# 89072703), the source of the chimpanzee DNA was the cell line: EB176(JC) (ECACC cat# 89072704). It is important to note that the chimpanzee cell line studied here is from a different animal than ‘Clint’ the donor for the database available on the UCSC browser [J. Clegg Weatherall (retired), Institute of Molecular Medicine, Oxford and E. Mardis, Co-Director Genome Sequencing Center, Washington University School of Medicine, personal communication].
PCR amplification
PCR amplification was performed on a MJ thermal cycler PTC 2000, using a ‘forward’ oligonucleotide of the sequence 5′-GTCTAGTGCATGTCTCAGAG and a ‘return’ oligonucleotide of the sequence 5′-ACATCTACTGACACCTTTGG. Initial denaturation was at 94°C for 2 min and was followed by twelve cycles of 60.4°C for 30 s, 72°C for 40 s and 94°C for 30 s. Subsequently, 35 cycles were performed with the same annealing and extension conditions but using a lower denaturation temperature of 89°C. Oligonucleotides were at 100 nM, dNTP at 200 µM and MgCl2 at 1.5 µM in a final volume of 50 µl 20 mM Tris (pH 8.5), 50 mM KCl. 1 U of ‘Mercury’ Taq polymerase (Continental Lab Products) and ∼200 ng of genomic DNA was used per reaction. Taq polymerase from other suppliers was generally less suitable. PCR of bacterially-grown plasmid DNA (as used for ABI analysis) was performed on 1 µl of a 1/25 000 dilution. In the case of the BAC clone, dilution was 1/10 000. For yeast-grown plasmid clones, DNA was diluted 1/500.
Cloning of PCR products
PCR products were cloned after gel purification into pYES2.1/V5-his-TOPO (Invitrogen) with the pYES2.1 TOPO TA Expression Kit (Invitrogen catalog no. K4150-01).
Bacterial strains, transformation and minipreparations
TOP 10 cells (Invitrogen) are F-mcrA Δ(mrr-hsdRMS-mcrBC) f80lacZΔM15 ΔlacX74 deoR recA1 araΔ139 Δ(ara-leu)7697 galUΔgalK rpsL (StrR) endA1 nupG. Top 10 F′ cells are as TOP 10 but also F′[lacIqTn10(Tetr)]. SURE cells (Stratagene) are e14–(McrA–) Δ(mcrCB-hsdSMR-mrr)171 endA1 supE44 thi-1 gyrA96 relA1 lac recB recJ sbcC umuC::Tn5(Kanr) uvrC [F′ proAB lacIqZΔM15 Tn10 (Tetr)]. Transformation of all strains was as per the suppliers protocol. Plasmid DNA was prepared with the QIAprep Spin Miniprep Kit (Qiagen).
Colony PCR
Samples from fresh yeast or bacterial colonies were tooth picked directly without a wash step into 5 µl of Lyse-N-Go PCR reagent (Pierce) and lysed in the thermal cycler according to the manufacturer's specifications. For analysis of yeast colonies, results were improved by incorporating a freeze-thaw step either before or after lysis. PCR amplification was performed in a final volume of 50 µl.
Phi-29 amplification
Random-primed rolling circle amplification was performed as described in Dean et al (20) with the following modifications: denaturation of the template was not performed (21), yeast pyrophosphatase was omitted and we pre-treated samples with a restriction enzyme. Briefly, 2.5 µl of miniprep DNA was digested in a final volume of 10 µl with EcoR1 (New England Biolabs) in 1× buffer 4 from the same supplier [20 mM Tris-acetate, 10 mM magnesium acetate, 50 mM potassium acetate and 1 mM DTT (pH 7.9) at 25°C]. After incubation at 37°C, 20 µl of amplification mix was added directly to each 10 µl sample (without re-extraction or heat-inactivation of the restriction enzyme) and incubated at 32°C for 6 h to overnight as convenient. To minimize condensation, tubes were floated in a water bath in foam racks covered with aluminum foil. The amplification mix contained 50 µM exonuclease-resistant random hexamers (‘machine-mixed’ thiophosphate-modified random hexamers, 5′-NpNpNpNpsNpsN-3′; Integrated DNA Technologies), 600 µM dNTP (Pharmacia), 100 µg/ml BSA (New England Biolabs) and 3 U of phi-29 polymerase (New England BioLabs) in 1× buffer 4 (as above). After amplification, the reactions were heat-inactivated at 60°C for 20 min before further analysis or storage.
Transformation of S.cerevisiae
Small volumes (<5 µl of a topo-cloning reaction or of DNA minipreparations from yeast clones) were introduced into yeast with an EZ Transformation kit (Zymogen) as per the manufacturer's instructions. Transformants were selected on media lacking uracil.
DNA minipreparations from yeast
DNA minipreps were prepared as described (22) except that after ethanol precipitation, samples were further purified with a QIAprep spin miniprep kit. DNA was recovered from the column by two sequential 35 µl elutions.
Yeast strains
The initial transformation of yeast with pYES2.1 Topo ligation reactions was into a sae2 deletion derivative of strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YGL175c::KanMX4; Open Biosystems). The wild-type and sae2 mutant strains used for re-transformation experiments were GRY2565 (MATa ade2Δ::hisG can1Δ17 cyh2r his3 leu2 ura3Δ0) and GRY2566 (MATa ade2Δ::hisG can1Δ17 cyh2r his3 leu2 ura3Δ, sae2::NatMX)
DNA sequence analysis
Clones were sequenced at the NCI Laboratory of Molecular Technology (Frederick MD). Where automated sequencing failed, manual methods were employed in lab. Plasmid miniprep DNA was sequenced with a Sequenase version 2.0 sequencing kit (USB), and gel-purified yeast clone fragments were sequenced with Sequenase version 2.0 PCR Product Sequencing Kit (USB), following the manufacturer's protocols. Reactions were resolved on 6% acrylamide gels containing urea and 40% deionized formamide (23).
ABI fragment analysis
ABI analyses were performed by the Genetic Analysis Facility at the Centre for Applied Genomics (Toronto). Volumes of 1 µl of the PCR were suspended in a 10 µl mixture of 5 µl GeneScan™ 500[LIZ] size standard in 995 µl Hi-Di formamide (Applied Biosystems). The samples were run on an ABI3730XL genetic analyzer (Applied Biosystems) using the POP7 polymer and dye set G5. Results were analyzed using the software GeneMapper v. 3.5.
Database searches
Database searches were performed with a 1090 bp query sequence corresponding to nucleotides 26685885–26686974 of chromosome 17 of the NCBI May 2004 human genome assembly (24) as retrieved from the UCSC genome browser (http://genome.ucsc.edu/). GenBank sequences of the 17q11 region (accession nos AC004526, L05387, AB195814, AB195812, CTD-2538H15, CTD-2370N5, AY796305 AB195813 and AB195815) were obtained by BLAST (25) analysis of the non-redundant database, using the NCBI webserver (http://www.ncbi.nlm.nih.gov/blast/). The sequence designated ‘chimp NHGRI’ was obtained by a BLAT (26) search of the NHGRI alignment (November 2003) accessed through UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgBlat).
Sequence alignment
Alignments were performed with Clustal program accessed through the European Bioinformatics webserver (http://www.ebi.ac.uk/clustalw/index.html). Human alleles were aligned separately from chimpanzee and gorilla. By Clustal alignment, with default parameters, the human alleles assorted into the two groups shown. Hand adjustments were made by introducing gaps as required to have the conserved flanks on both sides in alignment for all sequences. Gaps were then arranged to highlight relationships.
RESULTS
PCR amplification
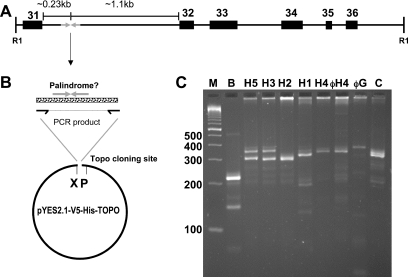
Analysis of the NF1 intron-derived sequence was initiated by PCR amplification of an ∼400 bp region encompassing the putative 100–200 bp palindrome from human genomic DNA (Figure 1). The NF1 gene itself covers 350 kb and has around 60 exons, the numbering of which varies. The site of interest lies on a 3.8 kb EcoR1 fragment as demonstrated by Southern blot (27) and occurs in the interval between exons 31 and 32 (numbered according to the Neurofibromatosis International Mutation Database; http://www.nfmutation.org/About.htm). Oligonucleotides were designed to hybridize to non-repeat sequences bracketing the palindromic area (Figure 1B). Thermophilic DNA polymerases from various suppliers were tested for their ability to generate a PCR product on genomic DNA templates. Consistent results were achieved with a ‘native’ version of Taq polymerase (Continental Lab Products). The CLP-supplied polymerase generated different bands ranging from about 280 to 340 bp for different human genomic DNA samples as well as from chimpanzee to gorilla templates (Figure 1C). Reproducibility of amplification products with the CLP enzyme was confirmed to 1 bp resolution by analyses on an ABI DNA analyzer (further details below).
Figure 1.
(A) Map (to scale) of the palindromic region within a 3.8 kb EcoR1 fragment of the NF1 gene. Exons are numbered according to L05367 (see text). ‘R1’ denotes EcoR1 sites according to Southern blot data (27) and the reference human genome sequence (May 2004). (B) Cloning strategy. Oligos used for PCR flank the palindromic site. The PCR product is ligated into a commercial vector (see Materials and Methods). ‘X’ and ‘P’ refer to the Xbal and PvuII sites used in diagnostic digests. (C) PCR amplification of various DNA templates. Lane ‘M’, markers (Trackit 100 bp ladder, Invitrogen). ‘B’ is a PCR with Bac clone CTD-2370N5; ‘H1’ to ‘H5’ are with human genomic DNA from the indicated individuals. ‘C’ is with chimpanzee DNA. ‘φH4’ and ‘φG’ are from an aliquot of H4 and gorilla genomic DNA that had first been amplified with phi-29 polymerase. The H4 samples demonstrate reproducibility of the PCR as well as the fidelity of phi-29 amplification.
To test the fidelity of the PCR on a known template, the BAC source clone used by the International Human Genome Sequencing Consortium was obtained (CTD-2370N5, Invitrogen). PCR amplification (see Materials and Methods) yielded the expected 215 bp product shown in Figure 1C. DNA sequence analysis confirmed this product was an exact replicate. Of note, the BAC-templated PCR product was significantly shorter than any of the 11 alleles we could differentiate among our collected human genomic DNAs (Figure 1C and data not shown).
Cloning PCR-amplified alleles into E.coli
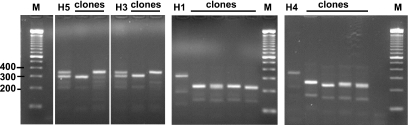
The PCR products from eight individuals as well as chimp and gorilla were gel-purified and topo-cloned into a 2 micron circle-based S.cerevisiae/E.coli shuttle plasmid, pYES2.1/V5-His-TOPO (Invitrogen) (Figure 1B). Consistently, certain alleles failed to give full-length E.coli clones when introduced into ‘TOP 10’ cells (Invitrogen), a recA-strain of E.coli. This was evident at the level of colony PCR (Figure 2) where clones derived from individuals H3 and H5 gave predicted amplification products, but those from H1 and H4 did not. Plasmid DNA preparations from the same colonies, cut with XbaI and PvuII (as shown in Figure 1B) confirmed the apparent deletions in H1 and H4 clones (data not shown). The close correspondence in the sizes of the deletion products and the structure of the BAC clone CTD-2370N5 (Figure 1C and data not shown), as well a second clone (CTD-2538H15, accession no. AC135724) is suggestive. As presented in more detail below, this site in the reference human genome (International Human Genome Sequencing Consortium) corresponds to a reproducible deletion artifact generated in E.coli.
Figure 2.
Colony PCR analysis of bacterial clones obtained from H1, H3, H4 and H5 genomic DNA as in Figure 1B. Ligation products were used to transform E.coli. Analytical colony PCR was with the oligonucleotides in Figure 1B. Examples of clones representing both alleles in H5 and H3 were obtained as shown. In contrast, no full-length H1 or H4 clones were recovered. Markers, ‘M’ are as in Figure 1C. For H3, 4 and 5, a single gel image has been cut with lanes removed. The H1 image is from a different gel. To simplify cross-comparisons here, as well as in Figures 3A and 4, images from different gels have been size-adjusted in the vertical dimension.
Cloning and analysis of difficult alleles in yeast
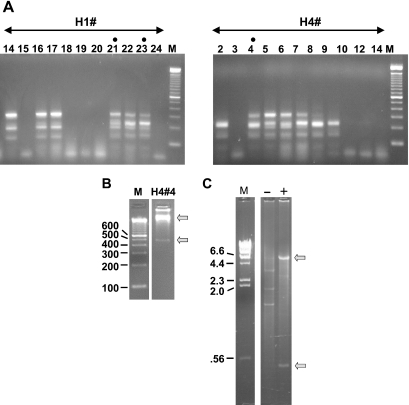
A different approach was attempted in order to recover representative clones from H1 and H4 DNA. Once the PCR products templated by these DNAs were ligated into the pYES vector, ligation reactions were transformed into S.cerevisiae (BY4742 YGL175c::kanMX4, a sae2 deletion mutant). In contrast to the results obtained with E.coli, colony PCR revealed that some of the yeast transformants had full-length inserts (Figure 3A). For confirmation, minipreparations of DNA from selected candidates were examined on gels after being subjected to isothermal multiply-primed rolling circle amplification with phi-29 DNA polymerase (20). Phi-29 amplification was a useful expedient because minipreparations of yeast plasmids give low yields, typically insufficient even for visualization on ethidium-stained gels. After Phi-29 amplification, the DNA samples were digested with XbaI and PvuII to liberate the insert as shown for one such clone, H4#4 in Figure 3B. The insert in H4#4 was about 450–480 bp, consistent with the size of the PCR product of H4, and significantly larger than any of the E.coli insertions (Figure 2) obtained from the same H4 PCR.
Figure 3.
(A) Colony PCR analysis of yeast clones obtained from H1 and H4 genomic DNA by the method shown in Figure 1B. Ligated samples (see legend, Figure 2) were used to transform S.cerevisiae. In contrast to results with E.coli (Figure 2), full-length candidate clones were obtained. Lanes with dots are from colonies used in further analyses (see text). The DNA ladder is as in Figures 1 and 2. (B) Verification of clone structure. To confirm the structure of the full-length candidates, yeast minipreps were treated with EcoR1 and subjected to random-primed rolling circle amplification with phi-29 DNA polymerase. As an example, H4#4 is displayed on a 2.5% agarose gel after diagnostic digestion with Xbal and PvuII. Upper and lower arrows indicate the vector backbone and insert bands, respectively. (C) Efficacy of the EcoR1 pre-digestion. The EcoR1 pre-treated sample in B (‘+’) is run on a 1% agarose gel alongside an untreated (‘−’) miniprep of H4#4. Treated and untreated samples were phi-29 amplified in parallel and digested with XbaI and PvuII. Bands evident in the untreated lane correspond in size to those predicted for 2 µ circle DNA. Bands observed with EcoR1 pre-treatment (arrows) correspond to the pYes2.1 vector and insert.
Phi-29 amplification is known to replicate genomic DNA with high fidelity (28), and this apparently holds true for the NF1 region as well (Figure 1C, and see ABI fragment analysis below). As applied to rolling circle amplification of yeast plasmids, improved results were obtained if we first linearized the resident 2 micron circle DNA. EcoR1 gave differential cleavage (i.e. cut the 2 micron circle and spared the plasmid clone) eliminating almost all amplification of the competing 2 micron circle DNA (Figure 3C). With EcoR1 pre-treatment, the amplified H4#4 miniprep gave two bands after a diagnostic digest with XbaI and PvuII: the expected pYES2.1 backbone fragment of 5.7 kb (Figure 4, left panel) and the full-length insert band of around 400 bp. Without pre-treatment, bands corresponding to the plasmid clone were almost invisible in the phi-29 amplified sample and instead diagnostic digests revealed the banding pattern expected for 2 µm circle DNA (3.8, 1.8 and 1.3 kb). (The strategy of differential digestion is generally applicable although of course in other cases, different candidates capable of cleaving the 2 micron circle might have to be tested in order to find an appropriate enzyme).
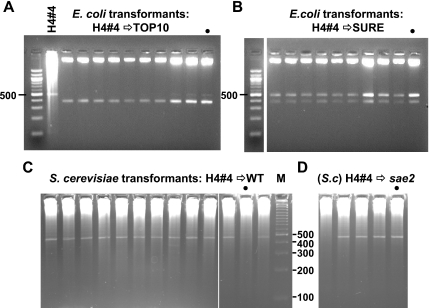
Figure 4.
Side-by-side comparison of microbial hosts for their ability to maintain the same plasmid. (A) Universal deletion in E.coli Top 10 cells. Plasmids re-isolated from TOP 10 clones transformed with the H4#4 plasmid are deleted. Each was XbaI and PvuII digested. H4#4 DNA (also cut with XbaI and PvuII after a phi-29 amplification) is loaded adjacent to the marker lane. Note, a different 100 bp ladder was used here (New England Biolabs) which has an intense 500 bp rather than 600 bp band as in previous figures. (B) Instability of the H4#4 plasmid in E.coli SURE cells. Plasmid DNA from individual SURE H4#4 transformants is a mixture of deleted and apparently non-deleted forms despite the lack of a functional SbcCD nuclease. The gel image was cut to remove one lane. (C) Stability of H4#4 plasmid in wild-type yeast. Phi-29 amplified minipreparations of DNA from random wild-type yeast clones transformed with H4#4 DNA were digested with XbaI and PvuII. Full-length inserts are observed. (D) Phi-29 amplified minipreparations of DNA from random sae2 yeast clones transformed and analyzed as in (C). In A–D, dots mark samples from colonies that were re-streaked as described in the text.
Side-by-side comparison of plasmid maintenance in E.coli versus S.cerevisiae
The ability to isolate apparently full-length clones in yeast provided the means by which to make a direct comparison of the stability of the same replicon after being introduced into E.coli or S.cerevisiae. Miniprepped DNA isolated from the yeast clones H4#4, H1#21 and H1#23 (Figure 3A, dots) were used without phi-29 amplification for these experiments.
Figure 4A shows results obtained when the H4#4 yeast plasmid was introduced into TOP 10 E.coli. All samples were digested as usual with Xbal and PvuII and, without exception, it was evident that all transformants had sustained deletions. A phi-29 amplified sample of the transforming H4#4 DNA, also cut, was included as a marker. Only a few of the transformants showed traces of an insert that co-migrated with the original DNA (see rightmost three lanes). Uniform deletion was again observed when miniprepped H1#21 and H1#23 plasmids were tested (data not shown). TOP 10 is a recA-strain, and these results were not unexpected given the known RecA independence of palindrome instability (3,29).
A second strain of E.coli, lacking expression of the SbcCD nuclease, was also tested, because mutation of SbcC or SbcD is known to reduce palindrome inviability and instability (30,31). Transformed sbcC mutants (‘SURE’ strain, Stratagene, see Materials and Methods) all contained some full-length plasmid but despite this, deletion products were apparent in every case (Figure 4B). The same result pertained when the two other yeast clones, H1#21 and #23, were used in the transformations (data not shown). On the one hand, the fact that each SURE transformant exhibited a full-sized band demonstrated that initially each must have taken up a full-length H4#4 molecule, and thus the yeast H4#4 preparation was largely homogenous. Moreover, the fact that each transformant contained deleted molecules implied that even without a functioning SbcCD nuclease, deletion was rapid and inevitable in E.coli.
To further investigate, an H4#4 SURE transformant (Figure 4B, lane with dot) was re-streaked, and DNA was prepared from 10 randomly picked single colonies. Each tested sample contained a mixture of full-length and deleted plasmid, just like the parental colony. The same test with an H1#23 transformant gave a similar result with nine mixed colonies and one lacking any remaining trace of full-length plasmid (data not shown). We conclude that deletion must be rapid in SURE cells. It either occurs soon after the transformation event—but after multiple plasmid copies are present in the initial transformant—and/or as the transformed colony was expanded to a saturated 3 ml liquid culture. There was no indication of a subpopulation of cells capable of sustained maintenance of full-sized plasmid. In other words, upon transformation with a full-length plasmid, a mixed population of plasmid molecules is rapidly elaborated and present in each individual E.coli cell: no ‘pure’ colony can be obtained by re-streaking.
Next, the three yeast minipreps were used to transform wild-type and sae2 mutant yeast cells (see Materials and Methods for genotypes). Altogether 22 wild-type and 10 sae2 transformants were examined; the results obtained with H4#4 are shown in Figure 4C and D. In contrast to the E.coli transformations, all but one of the wild-type yeast transformants was full-length (the deleted example was from an H1#21 transformation, data not shown). For the sae2 mutant, six transformants exhibited full-length insertions and four appeared to have lost the plasmid (a full-length PCR product was observed at the first screening, but plasmid was not detected once phi-29 polymerase amplified DNA was tested; data not shown).
To investigate whether or not plasmid stability in yeast was sustained, positive transformants were streaked out as for E.coli (above). One colony from each of the two strains (identified in Figure 4C) was selected and five random picks from each streak were miniprepped, amplified and cut with XbaI and PvuII. Nine of ten clones were full-length. The tenth, deleted, clone arose in the wild-type strain, but we know from other examples that deletions also occur in the sae2 strain, and we could not document any consistent difference between the two strains. Thus, whereas sae2 strains have been shown to enhance the stability of long palindromic sequences in plasmids and near-palindromic sequences in the yeast chromosome (9,16), the sae2 mutation did not augment the already significant stability of the shorter, endogenous sequences studied here. In sum, propagation of full-length plasmid clones from H1 and H4 is possible in yeast; only a low level instability was detected.
DNA sequence analysis of problematic alleles
Clones derived from H1 to H4 could not be sequenced by automated cycle-sequencing methods. Typically the traces became uninterpretable as they penetrated into the region of the suspected palindrome (data not shown). The block was not resolved with alternative chemistries nor with proprietary methods of one company (MTR Scientific, Ijamsville, MD; data not shown). Gel purification of PCR products or of inserts did not give improved results with automated methods. DNA sequence analysis was also problematic for some other alleles (such as those derived from chimp and gorilla) even though seemingly stable upon cloning in E.coli.
The solution was to revert to the older standard manual Sanger chain-termination method. Gel-purified inserts prepared after phi-29 amplification of the yeast clones H1#1, H1#23 and H4#4, gave easily interpretable ladders on formamide-containing acrylamide gels (data not shown). T4-DNA polymerase (Sequenase, USB) was suitable; BstN polymerase also worked well and could be used at elevated temperatures (32,33).
All clones of the NF1 intronic site exhibited substantial AT repeat content, and different clones from the same individual varied in AT tract length. Variation was due to ‘stutter’ at the dinucleotide repeat during the PCR (34,35). Thus, sequence analysis of individual cloned PCR products is unable to fix the actual number of AT repeats in the genomic template.
High-resolution fragment analysis
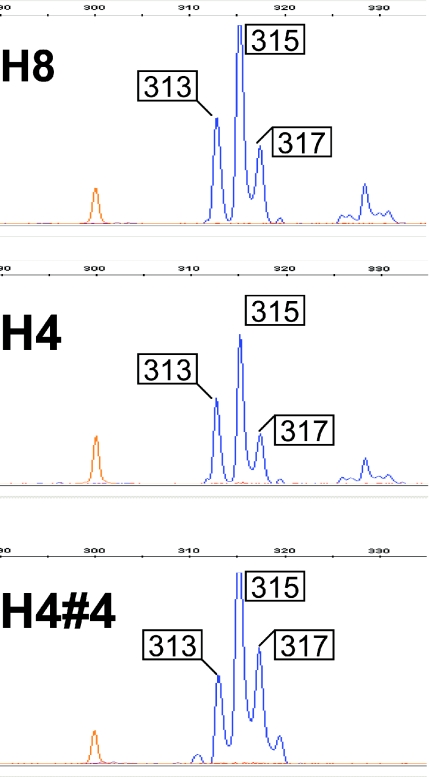
In order to precisely identify the number of AT repeats in a given allele, PCR from all genomic DNAs in this study were subjected to capillary gel electrophoresis on an ABI 3730XL fragment analyzer. In each case the major peak was flanked on either side by a series of lower peaks representing multiples of + and −2 bp stutter products. Consistent with expectation, ABI fragment analysis gives an invariant position for the highest peak (or peaks, in the case of a heterozygote), relative to standardized size markers from one run to the next. However without knowing in advance whether migration might be altered by sequence-specific features, it was not possible to get absolute values for fragment lengths. In practice, special allele-specific marker series are typically used for this purpose (36). Here we compared electropherograms of PCR products templated by candidate clones to that of the corresponding genomic template; thus identifying the clone that corresponded exactly to the genomic sequence (Figure 5). For example, the H1#21 clone matched the genomic H1 template, whereas H1#23 was evidently a ‘plus 2’ stutter product (data not shown). Sequence analysis showed that the H1 allele was a near-palindrome of 181 bp where all but the three central bp comprised a perfect inverted repeat. As suspected, DNA sequence analyses confirmed that PCR products from H1- and H4-derived yeast clones ran 7–8 bp smaller than their true sizes (Figure 5). In contrast to the clones derived from other alleles in this study all ran according to size in capillary gel electrophoresis, with not more than a 1–2 bp discrepancy relative to size markers.
Figure 5.
ABI fragment analysis of PCR templated by sibling H8 and H4 genomic DNA (top and middle) and the yeast clone H4#4 (bottom). Peaks occurring in 2 bp increments are PCR stutter products. For each sample, the main peak migrates at an apparent size of 315 bp. The coincidence of the H8 and H4 peaks demonstrates the reproducibility of both PCR amplification as well as capillary gel electrophoresis. The main peak from the H4#4 PCR products is at the same position as the PCR products from H4 genomic DNA, identifying H4#4 as a true clone of the genomic sequence. The H4#4 fragment is known by sequence to be 323 bp thus the H4 (and H8) allele migrates as though it were 8 bp shorter than its true size.
To summarize, the artifactual migration of the H1 and H4 PCR products, the presence of PCR stutter, and instability of E.coli clones means that virtually the only way to obtain a valid sequence of these two problematic alleles was by (i) cloning in yeast, (ii) identifying correct clones by high-resolution capillary gel electrophoresis and (iii) manual DNA sequence analysis.
DNA sequence comparisons
Twelve human sequences (all designated with ‘H’ in the left column), along with two chimp [‘chimp (a) and (b)’] and one gorilla sequence of the NF1 intronic region were determined in the present study and are given in Figure 6. Some individuals were related to one another as noted in the Figure legend. All of the sequences, including chimp and gorilla had an identical 38/39 bp inverted repeat at the outermost extremes (see arrows at bottom of Figure 6). There was considerable diversity among the alleles.
Figure 6.
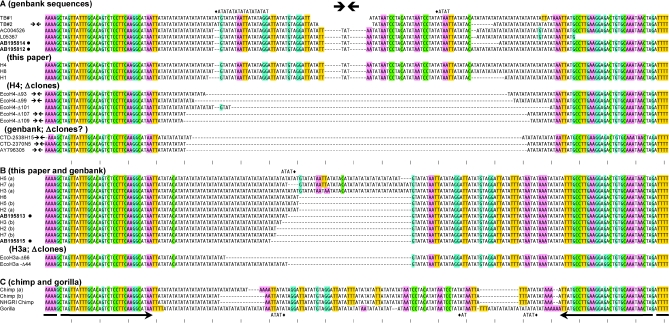
(A–C) Sequence comparisons of the NF1 site. All human sequences, as well as deletion derivatives fall into two groups, distinguished e.g. by the presence of ‘T’ or ‘C’ 46 bp from the left and TTAT or ATTT 34–37 bp from the right border of the sequence alignment. Chimp and gorilla sequences are given in (C). Where applicable, lower case ‘a’ and ‘b’ differentiate alleles. H7b and H2b were from a parent/child pair. H4 and H6 were from siblings. Other identical alleles such as H3a, H6 and H5b were from unrelated individuals. Known deletion artifacts generated in the TOP10 or TOP10F′ strain of E.coli are designated EcoH4-(#) and EcoH3-(#) in (A) or (B), respectively. All available versions of the 17q11 region in GenBank are given and identified by accession number. Some of these sequences match known deletion artifacts. ‘Chimp NHGRI’ is from the public chimpanzee genome database (see Materials and Methods), and originates from a different animal than the ‘chimp b’ allele sequenced here. Bulleted sequences (accession nos AB195814, AB195812, AB195813 and AB195815) were deposited while this manuscript was in preparation. TB#1 and TB#2 show the chr17 components of derivative 17:22 translocated chromosomes from (18) and (19), respectively. The gap (without dashes) indicates uncertainty because the chromosome 17 or 22 assignments close to the breakpoints of the derivative chromosomes is ambiguous. All sequences were first aligned using Clustal and then hand-adjusted. Presentation of the alignment was simplified by showing excess AT repeats for some sequences on the line above (TB#1, H5a) or below (Gorilla). The AT tract to which these belong is indicated by the diamond. G, C, AAA or TTT sequences are colored (G and C complementarity is indicated by blue and green; A and T are likewise red and yellow respectively). Perfectly palindromic sequences are marked by a double arrow in the left margin.
Just internal to the conserved repeats are differences that define two groups of human alleles (‘group-A’ and ‘-B’, Figure 6). Notably, the unstable alleles (as defined by uniform deletion when propagated in E.coli) are together and separate from the other alleles, all of which belong to group-B; Figure 6A). Only one group-B allele (designated H3a) showed a tendency to delete when cloned, such that three of five randomly picked TOP 10 clones were deleted. In this case, full-length plasmids were nevertheless recovered and were analyzable by automated sequencing methods. Group-A and -B, are distinguished not only by SNPs adjoining a common 39 bp inverted repeat, but also by a difference in palindromy. The distinction between the two groups in this regard can be appreciated by visual inspection, by noting the vertical striping to either side of the central double arrow in Figure 6. In group-A stripes to the left are mirrored on the right, a feature that is lacking in group-B. Thus, instability and palindromy are well correlated.
When all available versions of the NF1 intronic sequence (either published or from GenBank) are retrieved and included in the alignment, they assort unambiguously into either group-A or -B (Figure 6A and B). An indication that symmetry and instability of NF1 sequences in the human genome have a physiological impact is provided by the two examples of chromosome 17 translocation involving the site (18,19). The translocated sequences have been determined, so that even though the pre-existing chr17 sequences are not available, it is possible to piece together the chr17 contribution to the sequence of the derivative chromosomes for each case. These have been designated TB#1 and TB#2 as based upon information published by other groups (18,19). Although the exact sequence at the central crossover site is uncertain (see legend Figure 6), in both examples, the sequences adjacent to the 39 bp inverted repeat clearly identify these alleles as belonging to group-A. The TB#2 sequence inferred in Figure 6 is a perfect palindrome.
Deletion artifacts
As mentioned, the PCR product from the source clone for the human consortium sequence, CTD230N5, was also cloned and sequenced. The sequence of the clone identified it as having arisen from a group-A allele and was in full agreement with the consortium sequence (chr17: chr17:26686380–26686479 on the UCSC browser, May 2004 assembly). However, there were no human alleles similar in size to the BAC clone. In addition to the eight distinct alleles defined in our sequence analyses, an ABI analysis of four additional CEPH DNA samples (1362-1, 1362-2, 884-1 66-1 and 66-2) indicated the presence of three other human polymorphisms, all of which were longer than CTD-2370N5.
To investigate the possible origin of the consortium sequence, and to further analyze the mechanism of deletion in our E.coli clones, we sequenced deleted clones derived from the group-A allele H4 and group-B allele, H3a (EcoH4 Δ93, 99, 101, 107, 109 and EcoH3a Δ44, Δ66). Sequences of the deletion products are given in Figure 6A and B, respectively. Significantly, the BAC clone CTD230N5 is identical in sequence to one of the spontaneous deletions arising from the group-A allele, H4 (EcoH4 Δ107).
DISCUSSION
Methods for analysis of palindromic sequences in the human genome
Standard molecular genetic techniques are ineffective in the analysis of palindromic DNA longer than several 100 bp. This would be of no concern if palindromes were uniformly excluded from all genomes. However long perfect palindromes do occur naturally in the human genome [e.g. (12), and S. M. Lewis, S. Chen, T. Alleyne and J. Cheung, manuscript in preparation]. Moreover, palindromes in human DNA appear to be involved in provoking rearrangement (10,12,18,19,37–40) and amplification (13). As demonstrated here, cloning artifacts in E.coli will obliterate the sequence of regions with significant palindromic character. Clearly any relationship between palindromic alleles and pathological conditions will be difficult, or more likely, impossible to establish without employing alternative approaches.
We find that palindromic inserts can be maintained in yeast plasmids. Although the low DNA yields when plasmids are isolated from yeast has been a deterrent to the use of this microbe as a cloning vehicle in the past, it is possible to compensate for this deficit. Isothermal amplification with phi-29 DNA polymerase as described easily generates sufficient quantities of DNA from a small aliquot of miniprep sample for restriction enzyme and DNA sequence analyses.
Dinucleotide repeats are a component of palindromes at other sites in the human genome (unpublished data) as well as of other suspected palindromes corresponding to translocation breakpoints (10,18,37). We have shown that ‘stutter’ induced by such sequences during PCR need not obviate the ability to get the accurate sequence. Uncertainty as to the exact dinucleotide repeat count can be resolved by comparative ABI analysis of amplicons produced by cloned versus genomic DNA.
Finally, palindromic DNA sequences are notoriously difficult to ascertain. We show that at least for palindromes in the size-range studied here, the problem can be circumvented by use of manual Sanger sequencing methods and resolving products on 40% formamide gels.
Sources of palindrome corruption
Two separable origins for the cloning difficulties presented by palindromic sequences in E.coli have been defined. One is non-propagation (‘inviability’) the other, instability (6). Inviability has been traced to the destruction of palindrome-bearing replicons by the hairpin-cleaving nuclease SbcCD (2,6,30). Instability is attributed to faulty replication, where a hairpin structure in the template strand is bypassed by discontinuous copying (41,42). Both inviability and instability are proposed to occur if (and only if) hairpin structures form as a result of intrastrand self-annealing of the palindrome. The reproducible deletion products generated here in E.coli (Figure 6) can be attributed to the second, replication-based, scenario; they are seen whether or not the SbcCD nuclease is active.
While the machinery exists in S.cerevisiae to cause instability and inviability, it is possible that in general, the conditions causing a palindrome to buckle into an intrastrand-base paired structure do not. Torsional strain drives cruciform extrusion at palindromic sites, and extrusion, which reduces the interwinding of DNA strands, relaxes the molecule (43). DNA must be torsionally strained in E.coli for essential functions, such as the coordinate transcription of certain genes, and the proper level of strain is actively maintained by topoisomerases (44). The topoisomerase, gyrase, is key in this process because it is able to negatively supercoil DNA. There is no gyrase equivalent in yeast or mammals. Thus a unique situation exists in E.coli: any negative supercoiling relieved by a spontaneous extrusion event is likely to be quickly re-established by gyrase. That is, once cruciform extrusion occurs at a palindromic sequence, there may be no going back. In yeast, DNA wrapped around nucleosomes is relaxed. Fluxes in levels of negative superhelicity can occur in transiently-generated domains (45,46) but there is no equivalent homeostatic mechanism for actively maintaining negative superhelical stress. Extrusion events in yeast are likely more fleeting, and reversible, as changes in torque are generated by various metabolic activities. Lower global stress as well as an inability to re-instate negative supercoiling after extrusion, can quite simply account for the superior ability of yeast to stably maintain palindromic sequences. The insignificant level of deletion in yeast, which is not suppressed by mutation of Sae2, suggests that cruciform formation, in the case of the near-palindrome in the present study, is rare in this environment.
We can rule out the alternative possibility that AT dinucleotide repeats at the NF1 site, rather than palindromy, is what makes the H1 and H4 clones highly unstable in E.coli. With the identical vector and strains, we obtain pure AT repeat clones from another polymorphic region of the genome of around 64 bp in length. This is a far more extensive uninterrupted repeat than the two 18 bp AT tracts in H1#21 e.g. yet the pure AT clones are both stable and sequenceable by automated methods (S. M. Lewis and S. Chen, unpublished data). Also, some clones of group-B alleles have longer uninterrupted AT runs than those in either H1#21 or H4#4 (as in the case of H6a, H5b and H2a; Figure 6B) but these were stable. Apparently, AT dinucleotide tracts contribute to instability only in the context of a larger palindromic sequence. Likely, the AT repeat provides a large target for replication bypass events, allowing the nascent strand, if obstructed by hairpin formation and disengaged from its template, to reanneal at the homologous sequence beyond the obstruction.
In brief, yeast ought to be more fully exploited in closing physical gaps in the human genome sequence. This of course is not a new suggestion. Methods for capturing genomic sequences in yeast have been described (47). Here we contribute a practical set of tools allowing one to proceed after clones are identified. If captured on a large low copy yeast vector, subsequent shotgun cloning and preparation of DNA for sequence analysis can be performed without passing the DNA through E.coli. The manipulation of plasmid clones in yeast, demonstrated here to provide faithful replicates of difficult genomic sequences (Figure 5), make S.cerevisiae a truly workable molecular genetic alternative.
Evolution of the NF1 intronic palindrome
Our survey of human genomic DNA samples revealed that the NF1 region is highly polymorphic. Two groups of alleles emerged on the basis of SNPs within a common shared region (Figure 6A and B). The same two groups could be independently distinguished from one another according to the degree of palindromy they displayed. Features suggestive of special biophysical properties were seen for the more palindromic group-A alleles. The group-A derived sequences migrated anomalously in an ABI fragment analyzer and could not be sequenced by automated methods. Further, group-A alleles were impossible to stably propagate in E.coli. Notably two alleles clearly belonging to group-A are interrupted by translocation in humans (18,19). Our findings support the suggestion that naturally-occurring palindromes in the mammalian genomes can be hotspots for rearrangement (7,48–51) and will contribute to chromosomal instability in humans due to an inherent tendency to assume a secondary structure (10,18,37,39).
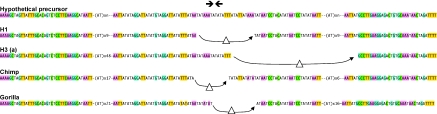
The variation in the 17q11 sequence (Figure 6) constrains possible models for its evolution. Replication errors that vary the length of the AT tracts and cause deletions between them can account for many of the variations seen. However the differences between group-A and -B as well as the differences between chimp, gorilla and human alleles are not so simply explained. Group-A cannot be derived from group-B by deletions or expansions at AT tracts. Instead, differences between the two groups of human alleles qualitatively resemble a process seen in mice and other eukaryotes called ‘center-break’ palindrome modification.
Center-break palindrome modifications are central asymmetric deletions that can be distinguished from replication errors because they bear hallmarks of non-homologous end-joining (7,48–50,52). The scheme in Figure 7 illustrates how four independent center-break revisions operating on an ancestral palindrome could have created the variant sequences seen in chimp, gorilla and in the two human groups today. The putative ancestral palindrome cannot be unambiguously reconstructed, but the relationships between the sequences studied here suggest its existence. In this view, at least one center-break event took place since human radiation, and neither human group-A nor group-B is ancestral to the other. Experimentally, center-break revision of palindromes are observed in both germline and somatic contexts in mice (7,48,52,53). It follows that center-break activity could drive rapid evolution of any true palindrome as seen here (Figures 6 and 7).
Figure 7.
Center-break events in the evolution of the 17q11 palindrome. A precursor sequence is postulated to have undergone four independent center-break events to give rise to the structures in human group-A and -B, chimp and gorilla. AT repeat expansion/contractions and SNPs that subsequently accumulate within the repetitive AT tracts in each lineage are not shown. The mechanism by which the central asymmetric deletions are generated is described more in detail elsewhere (48,50).
While this manuscript was in preparation an analysis of the same NF1 target was published (54). The study by Inagaki et al. (54) provides alternatives to the PCR and DNA sequence techniques described here, but did not address the central problem of unstable propagation in E.coli. As the authors note, the strategy of directly sequencing PCR products amplified from genomic DNA fails if a heterozygous individual alleles aren't well-resolved on gels. Where cloning of PCR products had to be performed, apparently full-length inserts were excised from mixed DNA preparations (similar to the SURE clones shown in Figure 4B) for DNA sequence analysis, and a consensus was deduced from multiple determinations. The data provided in Inagaki et al. (54) shows by its consistency with the present report, that representative sequences were indeed derived. However our methods are generally applicable and more importantly, will yield a definitive determination of the sequences in a given individual. We show e.g. that we can resolve alleles that do not separate well on gels (see data for individual H2 in Figures 1C and 6B). Shared alleles such as those in siblings H4 and H8 or in the parent–child couple (H2 and H7) had the identical sequence in fully independent determinations. The component steps in our approach—PCR amplification from genomic DNA, cloning of amplicons in a yeast vector, preparation of DNA after phi-29 amplification, identification of true clones by ABI analysis and DNA sequencing—all can be carried out from different genomic DNA samples, with a reproducible result.
A final point to be emphasized is that, among endogenous palindromes, the ∼200 bp near-palindrome at the NF1 locus is marginally stable: longer and clearly more unstable palindromes exist in the human genome (10,18,39). The methods we describe can be extended to palindromes that are much too long to be propagated at all in SURE cells or BAC clones. For example, a plasmid with a 600 bp palindrome is propagated in sae2 yeast mutants (9), and we have seen that even 12 kb fully-palindrome DNA circle is maintainable in yeast (A. G. Coté and S. M. Lewis, unpublished data).
Palindromic variation in the human population is likely to be a risk factor for mutation in the NF1 gene and elsewhere (10,54). By cloning a near-palindrome from the human genome without use of E.coli, we can prove that systematic corruption of the sequence occurs. At the present time, the number of palindromes that are similarly corrupted or even completely eradicated from the human reference sequence is unknown. To fully explore the implications of palindromy with respect to genome stability, it is essential to have the means by which to faithfully propagate and test the impact of DNA palindromes in relevant model systems. Our other studies in mice (7,48–51) and work by ourselves and others with S.cerevisiae (9,14–16,55,56)(A. G. Coté unpublished data) are beginning to shed light on the molecular details of how DNA palindromes effect illegitimate recombination in eukaryotes.
Acknowledgments
This study was initiated while S.M.L. was on sabbatical in the laboratory of J.N.S. The support and guidance received from members the Strathern group, as well others in the Genome Regulation and Chromosome Biology Laboratory is gratefully acknowledged. We thank Brenda Shafer for technical assistance. In addition we thank Dr Tara Patton and Zhanqin Liu of the Center for Applied Genomics (Toronto) for the ABI analysis. Dr Knut Madden (Invitrogen) generously donated two samples pYES2.1 TOPO TA expression kits. This work was supported in part by the Intramural Research Program of the NIH, Genome Regulation and Chromosome Biology Laboratory, NCI Frederick and by the National Cancer Institute of Canada with funds from the Canadian Cancer Society. Funding to pay the Open Access publication charges for this article was provided by The Canadian Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pinder D.J., Blake C.E., Lindsey J.C., Leach D.R. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 1998;28:719–727. doi: 10.1046/j.1365-2958.1998.00831.x. [DOI] [PubMed] [Google Scholar]
- 2.Leach D.R. Cloning and characterization of DNAs with palindromic sequences. Genet Eng (N Y) 1996;18:1–11. doi: 10.1007/978-1-4899-1766-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Bzymek M., Lovett S.T. Evidence for Two mechanisms of palindrome-stimulated deletion in Escherichia coli. Single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty J.P., Lindeman R., Trent R.J., Graham M.W., Woodcock D.M. Escherichia coli host strains SURE and SRB fail to preserve a palindrome cloned in lambda phage: improved alternate host strains. Gene. 1993;124:29–35. doi: 10.1016/0378-1119(93)90758-u. [DOI] [PubMed] [Google Scholar]
- 5.Benham C.J., Savitt A.G., Bauer W.R. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 2002;316:563–581. doi: 10.1006/jmbi.2001.5361. [DOI] [PubMed] [Google Scholar]
- 6.Leach D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 7.Akgün E., Zahn J., Baumes S., Brown G., Liang F., Romanienko P.J., Lewis S., Jasin M. Palindrome resolution and recombination in the mammalian germline. Mol. Cell. Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham L.A. Characterization of palindrome instability in mammalian cell lines. 2002. MSc thesis, University of Toronto.
- 9.Rattray A.J. A method for cloning and sequencing long palindromic DNA junctions. Nucleic Acids Res. 2004;32:e155. doi: 10.1093/nar/gnh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurahashi H., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 11.Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henthorn P.S., Mager D.L., Huisman T.H., Smithies O. A gene deletion ending within a complex array of repeated sequences 3′ to the human b-globin gene cluster. Proc. Natl Acad. Sci. USA. 1986;83:5194–5198. doi: 10.1073/pnas.83.14.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka H., Bergstrom D.A., Yao M.C., Tapscott S.J. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nature Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 14.Rattray A.J., Shafer B.K., Neelam B., Strathern J.N. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler D.K., Gillespie D., Steele B. Formation of large palindromic DNA by homologous recombination of short inverted repeat sequences in Saccharomyces cerevisiae. Genetics. 2002;161:1065–1075. doi: 10.1093/genetics/161.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 17.Maringele L., Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurahashi H., Shaikh T., Takata M., Toda T., Emanuel B.S. The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehrer-Sawatzki H., Haussler J., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- 20.Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman C.S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 23.Slatko B.E., Albright L.M. Denaturing gel electrophoresis for sequencing. In: Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Albright L.M., Coen D.M., Varki A., editors. Current Protocols in Molecular Biology. NY: John Wiley & Sons, Inc.; 2003. [DOI] [PubMed] [Google Scholar]
- 24.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. A basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viskochil D., Buchberg A.M., Xu G., Cawthon R.M., Stevens J., Wolff R.K., Culver M., Carey J.C., Copeland N.G., Jenkins N.A., et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- 28.Paez J.G., Lin M., Beroukhim R., Lee J.C., Zhao X., Richter D.J., Gabriel S., Herman P., Sasaki H., Altshuler D., et al. Genome coverage and sequence fidelity of phi29 polymerase-based multiple strand displacement whole genome amplification. Nucleic Acids Res. 2004;32:e71. doi: 10.1093/nar/gnh069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins J., Volckaert G., Nevers P. Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene. 1982;19:139–146. doi: 10.1016/0378-1119(82)90198-6. [DOI] [PubMed] [Google Scholar]
- 30.Connelly J.C., Leach D.R. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 31.Connelly J.C., Kirkham L.A., Leach D.R. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClary J., Ye S.Y., Hong G.F., Witney F. Sequencing with the large fragment of DNA polymerase I from Bacillus stearothermophilus. DNA Seq. 1991;1:173–180. doi: 10.3109/10425179109020768. [DOI] [PubMed] [Google Scholar]
- 33.Shengyu Y., Guofan H. Heat-stable DNA polymerase I large fragment resolves hairpin structure in DNA sequencing. Sci. sin. 1987;30:503–506. [PubMed] [Google Scholar]
- 34.Hauge X.Y., Litt M. A study of the origin of ‘shadow bands’ seen when typing dinucleotide repeat polymorphisms by the PCR. Hum. Mol. Genet. 1993;2:411–415. doi: 10.1093/hmg/2.4.411. [DOI] [PubMed] [Google Scholar]
- 35.Murray V., Monchawin C., England P.R. The determination of the sequences present in the shadow bands of a dinucleotide repeat PCR. Nucleic Acids Res. 1993;21:2395–2398. doi: 10.1093/nar/21.10.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler J.M., Buel E., Crivellente F., McCord B.R. Forensic DNA typing by capillary electrophoresis using the ABI Prism 310 and 3100 genetic analyzers for STR analysis. Electrophoresis. 2004;25:1397–1412. doi: 10.1002/elps.200305822. [DOI] [PubMed] [Google Scholar]
- 37.Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fodde R., Losekoot M., Casula L., Bernini L.F. Nucleotide sequence of the Belgian G gamma+(A gamma delta beta)0- thalassemia deletion breakpoint suggests a common mechanism for a number of such recombination events. Genomics. 1990;8:732–735. doi: 10.1016/0888-7543(90)90263-t. [DOI] [PubMed] [Google Scholar]
- 39.Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2003;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Game L., Bergounioux J., Close J.P., Marzouka B.E., Thein S.L. A novel deletion causing (epsilon,gamma,delta,beta)deg thalassaemia in a Chilean family. Br J. Haematol. 2003;123:154–159. doi: 10.1046/j.1365-2141.2003.04564.x. [DOI] [PubMed] [Google Scholar]
- 41.Lovett S.T. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004;52:1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 42.van Noort V., Worning P., Ussery D.W., Rosche W.A., Sinden R.R. Strand misalignments lead to quasipalindrome correction. Trends Genet. 2003;19:365–369. doi: 10.1016/s0168-9525(03)00136-7. [DOI] [PubMed] [Google Scholar]
- 43.Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 1982;156:229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- 44.Snoep J.L., van der Weijden C.C., Andersen H.W., Westerhoff H.V., Jensen P.R. DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur. J. Biochem. 2002;269:1662–1669. doi: 10.1046/j.1432-1327.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 45.Brill S.J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988;54:403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Droge P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996;15:581–589. [PMC free article] [PubMed] [Google Scholar]
- 47.Noskov V.N., Kouprina N., Leem S.H., Ouspenski I., Barrett J.C., Larionov V. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics. 2003;4:16. doi: 10.1186/1471-2164-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cunningham L.A., Coté A.G., Cam-Ozdemir C., Lewis S.M. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis S.M. P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis S.M. Palindromy is eliminated through a structure-specific recombination process in rodent cells. Nucleic Acids Res. 1999;27:2521–2528. doi: 10.1093/nar/27.12.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis S.M., Akgün E., Jasin M. Palindromic DNA and genomic stability: further studies. Ann. N Y Acad. Sci. 1999;870:45–57. doi: 10.1111/j.1749-6632.1999.tb08864.x. [DOI] [PubMed] [Google Scholar]
- 52.Collick A., Drew J., Penberth J., Bois P., Luckett J., Scaerou F., Jeffreys A., Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 53.Honchel R., Rosenzweig B.A., Thompson K.L., Blanchard K.T., Furst S.M., Stoll R.E., Sistare F.D. Loss of palindromic symmetry in Tg.AC mice with a nonresponder phenotype. Mol. Carcinog. 2001;30:99–110. [PubMed] [Google Scholar]
- 54.Inagaki H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H. Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rattray A.J., McGill C.B., Shafer B.K., Strathern J.N. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nasar F., Jankowski C., Nag D.K. Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol. Cell Biol. 2000;20:3449–3458. doi: 10.1128/mcb.20.10.3449-3458.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]