Abstract
When oxygen supply is restricted, protein synthesis is rapidly abrogated owing to inhibition of global translation. However, HIF-1α protein expression can persist during hypoxia, owing to an internal ribosome entry site (IRES) in the 5′-untranslated region of its mRNA. Here, we report on the molecular mechanism of HIF-1α IRES-mediated translation during oxygen deprivation. Using RNA affinity chromatography and UV-crosslinking experiments, we show that the polypyrimidine tract binding protein (PTB) can specifically interact with the HIF-1α IRES, and that this interaction is enhanced in hypoxic conditions. Overexpression of PTB enhanced HIF-1α IRES activity, whereas RNA interference-mediated downregula-tion of PTB protein expression inhibited HIF-1α IRES activity. Furthermore, hypoxia-induced stimulation of the HIF-1α IRES was reduced in cells in which PTB function was downregulated. In agreement with these results, the IRES activity of HIF-1α IRES deletion mutants that are deficient in PTB-binding could not be stimulated by oxygen deprivation. All together, our data suggest that PTB plays a stimulatory role in the IRES-mediated translation of HIF-1α when oxygen supply is limited.
INTRODUCTION
Most higher organisms depend on an adequate oxygen supply for essential functions, such as ATP synthesis. In order to survive and to restore oxygen homeostasis when oxygen concentration becomes limiting, angiogenesis, erythropoiesis and anaerobic metabolism are stimulated. Most of these responses are mediated by the transcription factor hypoxia-inducible factor-1 (HIF-1), which enhances transcription of several genes, such as vascular endothelial growth factor (VEGF), erythropoietin, lactate dehydrogenase A and insulin-like growth factor 2 (IGF2) (1–7).
HIF-1 is a heterodimer composed of two basic helix–loop–helix–PAS-domain containing subunits HIF-1α and HIF-1β (8). In contrast to the β subunit, the HIF-1α subunit is unstable in normoxic conditions owing to oxygen-dependent ubiquitination and subsequent proteasomal degradation (9–11). In response to hypoxia, the HIF-1α subunit becomes stabilized and accumulates (8). Because the concentration of HIF-1α protein is very low during normoxia, de novo HIF-1α protein synthesis is required for the strong increase of the HIF-1α protein during hypoxia. This is supported by the observation that HIF-1α mRNA remains associated with the polysomal fraction whether cells are incubated in normoxic or hypoxic conditions (12,13). However, oxygen deprivation does not favor maximal rates of protein synthesis.
Interestingly, Lang et al. (12) identified an internal ribosome entry site (IRES) in the 5′-untranslated region (5′-UTR) of the HIF-1α mRNA that allows its efficient translation during hypoxic conditions and serum starvation, which agrees with our unpublished results. This mechanism involves direct binding of the small ribosomal subunit to the mRNA at an IRES independently of the cap structure (14,15). IRESs, originally discovered in picornaviruses, have been identified in a number of eukaryotic mRNAs (16–18). Several of these IRESs were shown to be active in stress situations when cap-dependent translation is repressed, including hypoxia (12,19).
The molecular mechanism of IRES-mediated translation is obscure, but it is clear that there is no single universal mechanism used by all IRES elements. For instance, the Cricket paralysis virus intergenic region IRES can recruit ribosomes independently of any transacting factor (20). In contrast, most other IRESs depend on canonical translation initiation factors and non-canonical IRES transacting factors (ITAFs) for efficient initiation of translation (15). It has been proposed that the major role of ITAFs is to act as RNA chaperones, either to maintain or to attain the correct 3D IRES structure required for efficient assembly of the 48S ribosomal initiator complex (21,22). For example, the binding of the polypyrimidine tract binding protein (PTB) to pyrimidine-rich regions within different IRES sequences modulates the activity of the IRESs. In this way PTB stimulates translation mediated by the polio, HRV, HAV, TMEV, EMCV, Apaf-1, Bag-1 and p27Kip1 IRESs, but inhibits the activity of the Bip and UNR IRESs (21–31). In this report we demonstrate that PTB binds to the HIF-1α IRES and is involved in the efficient translation of HIF-1α during hypoxia.
MATERIALS AND METHODS
Plasmid construction
The pSV-Sport firefly luciferase expression vector was constructed by inserting the firefly luciferase gene, obtained as a KpnI–XbaI fragment from the pGL3-basic vector (Promega, Madison, WI), in the pSV-Sport plasmid (Life technologies, Paisley, UK). To create the pSV-Sport Renilla luciferase expression vector, the Renilla luciferase gene was amplified by PCR from the pRL-SV40 vector (Promega, Madison, WI) using primers A and B (see below), and ligated as an EcoRI fragment in the pSV-Sport plasmid.
The mouse HIF-1α 5′-UTR was isolated by a 5′-RACE reaction on poly(A)+ mRNA from mouse Ba/F3 cells, using the SMART™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions. The gene-specific primers C and D (see below) were used to amplify the HIF-1α 5′-UTR. A product of 320 nt was cloned in the pT-Advantage vector, according to the manufacturer's instructions (Clontech, Palo Alto, CA). The HIF-1α 5′-UTR that we obtained contains the I.2 exon (gi 2821939) with the initiation codon at position 320. The HIF-1α 5′-UTR and the deletion mutants HIF-del1 and HIF-del2 were amplified by PCR with primer pairs E + F, I + F and E + J, respectively (see below). To generate the HIF-delPPT mutant, two fragments of the HIF-1α 5′-UTR were amplified using primer pairs E + K and L + F. These two fragments were subsequently fused by fusion-PCR using primer pair E + F. The PITSLRE IRES Di-4 fragment was amplified using primer pair M + N and the Di-4 plasmid as template (32).
The bicistronic pSV-Sport expression vectors Di-HIF, Di-HIFdel1, Di-HIFdel2, Di-HIFdelPPT and Di-PITSLRE were constructed by two successive three-point ligations as follows: (i) the PCR-amplified HIF-1α IRES fragments and PITSLRE IRES were digested with XbaI–NcoI and cloned together with Renilla luciferase (obtained as an NcoI–EcoRI fragment from pSV-Sport Renilla Luciferase) into the XbaI–EcoRI opened pUC18 vector; (ii) IRES-Renilla luciferase inserts were digested with XbaI–EcoRI and ligated to both the firefly luciferase gene (obtained as a KpnI–XbaI fragment from the pGL3-basic vector) and the KpnI–EcoRI opened pSV-Sport expression plasmid.
Making the monocistronic reporter construct pSV-HIF-Rluc was started by constructing plasmid pCAGGS-HIF-Rluc. This was done by a three-point ligation of the HIF-Renilla luciferase SalI–EcoRI fragment (obtained from the Di-HIF vector) and two fragments generated from the pCAGGS vector with XhoI–PvuI and EcoRI–PvuI. Subsequently the pSV-HIF-R vector was constructed by cloning HIF-Renilla luciferase, obtained as an EcoRI–EcoRI fragment from the pCAGGS-HIF-Rluc plasmid, into an EcoRI opened pSV-sport vector.
For the construction of pcPTB see Cornelis et al. (30).
The coding sequence of the PTB fragment, ΔPTB, corresponding to amino acids 56–260 of the wild-type PTB, was amplified by PCR using the primer pair O + P and the pcPTB vector as template. The amplified fragment was cloned as an EcoRI–BglII fragment in a EcoRI–BglII opened pCAGGS vector.
Human Raf-1 cDNA was amplified using primer pair Q + R and the resultant amplicon was fused to an E-tag as a NotI–XhoI fragment in the PCAGGS expression vector.
For UV-crosslinking and RNA affinity chromatography assays, the HIF-1α IRES sequence, the different HIF-1α IRES mutants, the PITSLRE and EMCV IRES were inserted as XbaI–NcoI fragments into the corresponding sites of pUC19T7-fluc vector, described in Tinton et al. (32).
The following primers were used: A, 5′-TCCGGAATTCGCCATGGCTTCGAAAGTTTATGATCCAGAACAAAGG-3′; B, 5′-CCGGAATTCTTATTGTTCATTTTTGAGAACTCGCTCAA-3′; C, 5′-GTCAGGTAGACACGGAAGTAGAGTAG-3′; D, 5′-CGGCTCTAGAACGACGTAGAGATCTG-3′; E, 5′-TCTAGTCTAGAAAGCAGGGTGGTAACAACGCAGAGTAC-3′; F, 5′-TCTAGCCATGGCGAATCGGTGCCCGCGTTGTCTTCCCG-3′; G, 5′-TCCAGAGGCGGGGTGGGA-3′; H, 5′-CCTCCCACCCCGCCTCTGGAGCGCGCGCGGACAGAGCGGGCGCTTA-3′; I, 5′-CCGGAATTCCACCATGGTCCCCTCTAGAGTGATCCACA-3′; J, 5′-GAAGATCTCTAGAGCTTGGAAAAGTCGATGCGC-3′; K, 5′-CCTCCCACCCCGCCTCTGGCGCGCGCGGACAGAGCGGGCGCTTA-3′; L, 5′-ATCGATCTTGGTCCACAGAAGATGTTT-3′; M, 5′-CTAGTCTAGACATCACCGAACGATGAGAGAGG-3′; N, 5′-TTCTTCATCTTCACCCATGGCTTCCTCACTTAC-3′; O, 5′-CCGGAATTCCACCATGGTCCCCTCTAGAGTGATTCCACA-3′; P, 5′-GAAGATCTCTAGAGCTTGGAAAAGTCGATGCGC-3′; Q, 5′-ATAAGAAAGCGGCCGCTATGGAGCACATACAGGGAGCTTGGAAG-3′; R, 5′-AACCGCTCGAGCTAGAAGACAGGCAGCCTCGGGGAC-3′.
Cell culture and hypoxic conditions
Human embryonic kidney HEK293T cells (a gift from Dr M. Hall) were maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% penicillin, 1% streptomycin, 2 mM glutamine and 1 mM sodium pyruvate. African green monkey kidney BSC1 cells were maintained in DMEM supplemented with 10% heat-inactivated FCS, 1% penicillin, 1% streptomycin and 2 mM glutamine. For hypoxic stimulation, 6 h after transfection the cells were incubated in an atmosphere of 1% oxygen, 5% CO2 and 94% N2 at 37°C.
Transient DNA transfection and reporter gene assay
Human embryonic kidney HEK293T cells were transiently transfected by the calcium phosphate precipitation method (33). BSC1 cells were transiently transfected using lipofectamine plus reagent (Invitrogen) as specified by the manufacturer. For the RNA interference assays, HEK293T cells were seeded at a density of 1.5 × 105 cells/well in a 12-well plate. The following day they were transfected with 40 pmol of small interfering RNA (siRNA) duplex (Dharmacon) and 100 ng reporter plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The transfectants were split in wells of a 12-well plate 6 h later. Lysates were prepared 48 h afterwards using 1× Passive Lysis Buffer (Promega). Renilla and firefly luciferase activities were measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions, and light emission was detected by a Topcount scintillation counter (Perkin-Elmer).
UV-crosslinking assays and immunoprecipitation
For UV-crosslinking assays, DNA templates for synthesis of the respective RNA probes were generated by linearizing pUC19T7 plasmids containing the EMCV IRES, PITSLRE IRES or different fragments of the HIF-1α IRES with the appropriate restriction enzymes. Internally labeled RNA probes were synthesized by in vitro transcription with T7 polymerase (MAXIscript T7 RNA polymerase kit; Ambion) in the presence of 50 µCi (800 Ci/mmol) [α-32P]UTP (Amersham Pharmacia Biotech).
HEK293T cells were used to prepare cytoplasmic extracts for the UV-crosslinking assays. The cells were washed with cold phosphate-buffered saline and recovered by centrifugation at 2500 g for 5 min at 4°C. The pellets were dissolved in 100 µl lysis buffer A [10 mM HEPES–KOH (pH 7.4), 3 mM MgCl2, 40 mM KCl, 5% (v/v) glycerol, 1 mM DTT, 0.3% (v/v) Nonidet P40, 200 U/ml aprotinin, 0.1 mM phenylmethlysulfonyl fluoride (PMSF) and 10 µg/ml leupeptin]. The lysates were centrifuged at 20 000 g for 10 min at 4°C, the supernatants were recovered, the protein concentration was adjusted to 10 mg/ml, and the samples were stored at −80°C. UV-crosslinking assays were performed as described in Walker et al. (34). 32P-labeled RNA probes (±1 × 106 c.p.m.) were incubated with 400 ng recombinant PTB (gift from Dr Richard Jackson) or with 10 µl cytoplasmic extract (100 µg proteins) for 20 min at 30°C in a 25 µl reaction mixture containing 10 mM HEPES–KOH (pH 7.4), 3 mM MgCl2, 5% (v/v) glycerol, 1 mM DTT, 100 mM KCl, 40 U RNasin (Promega) and 6 µg tRNA. After RNA binding, the reaction mixtures were irradiated with UV light on ice for 30 min using a ‘GS gene pulser UV chamber’ (BioRad). The samples were then incubated with RNase A and RNase T1 for 60 min at 37°C. The RNA–protein complexes were resolved on 10% SDS–PAGE, the gels were dried, and the results were visualized with a Phosphorimager (Molecular Dynamics).
For immunoprecipitation, 1.5 µl of mouse monoclonal anti-E-tag antibody (Amersham Pharmacia Biotech) was added to each sample. After overnight incubation at 4°C in a rotary mixer, 30 µl of protein G–Sepharose beads (Amersham Pharmacia Biotech), equilibrated previously in lysis buffer A, were added and incubation was continued for 3 h. After washing six times with 1 ml buffer containing 50 mM HEPES–KOH (pH 7.4), 250 mM NaCl, 1% (v/v) NP-40, 5 mM EDTA, 200 U/ml aprotinin, 0.1 mM PMSF and 10 µg/ml leupeptin, resin-bound proteins were detached from the beads by adding 30 µl of Laemmli sample buffer and heating the mixture for 5 min at 95°C. The proteins were then resolved on 10% SDS–PAGE, the gels were dried, and the results were visualized with a Phosphorimager (Molecular Dynamics).
RNA affinity chromatography
Biotinylated RNA probes were synthesized from linearized pUC19 T7 plasmids by in vitro transcription with T7 polymerase (MEGAshortscript T7 RNA polymerase kit; Ambion) in the presence of biotinylated CTP (ratio 4:1, CTP/bioCTP) (Pierce). Biotinylated RNA (50 pmol) was incubated with 25 µl streptavidin beads (Pierce) in 200 µl binding buffer: 20 mM HEPES–KOH (pH 7.4), 50 mM KCl, 5% (v/v) glycerol, 1 mM DTT, 0.5 mM EDTA, 200 U/ml aprotinin, 0.1 mM PMSF, 10 µg/ml leupeptin and 25 µg/ml tRNA. After 1 h at 4°C in a rotary mixer, beads were washed two times with binding buffer and 200 µg cytoplasmic cell extract (prepared as described above) was added to a total volume of 500 µl; incubation was continued for 2 h. After washing the beads three times with binding buffer and two times with binding buffer in which KCl concentration was adjusted to 100 mM, RNA-bound proteins were eluted by addition of 200 µl Laemmli sample buffer and analyzed by western blot.
Western blot analysis
Cytoplasmic cell extracts were prepared as described above. For analysis of HIF-1α protein expression, total HEK293T cell extracts were prepared by direct lysis in Laemmli sample buffer. Protein concentration was determined by the BioRad RC/DC protein assay protocol (BioRad). Equal amounts of protein were separated by SDS–PAGE, and the proteins were transferred onto nitrocellulose membrane by electroblotting. The blots were probed with 1:1000 dilution of goat anti-PTB polyclonal antibody (Santa Cruz Biotechnology). Mouse anti-β-actin antibody was purchased from ICN Biomedicals and mouse anti-poly(ADP-ribose)polymerase antibody was purchased from Biomol. Monoclonal antibodies against HIF-1α were obtained from Becton Dickinson.
Membranes were incubated with a horseradish peroxidase-conjugated secondary antibody against mouse (Amersham Pharmacia Biotech) or goat (Santa Cruz Biotechnology) immunoglobulin. Proteins were revealed with an enhanced chemiluminescence kit (NEN Renaissance, Perkin-Elmer).
Cellular RNA purification and northern blotting
Total RNA was isolated from transfected HEK293T cells with RNAzol B Reagent (Wak Chemie Biomedical BmbH) according to the manufacturer's instructions. RNA was denatured in glyoxal/dimethylsulfoxide, separated on a 1% agarose gel and transferred by capillary blotting onto a nylon membrane. The filters were UV-crosslinked using a UV Stratalinker apparatus (Stratagene). They were subsequently hybridized with cDNA probes labeled with α-[32P]dCTP by randomly primed DNA synthesis using the Radprime DNA labeling system kit (Invitrogen). Results were visualized with the Phosphorimager (Molecular Dynamics).
RESULTS
The HIF-1α IRES mediates efficient translation during oxygen depletion
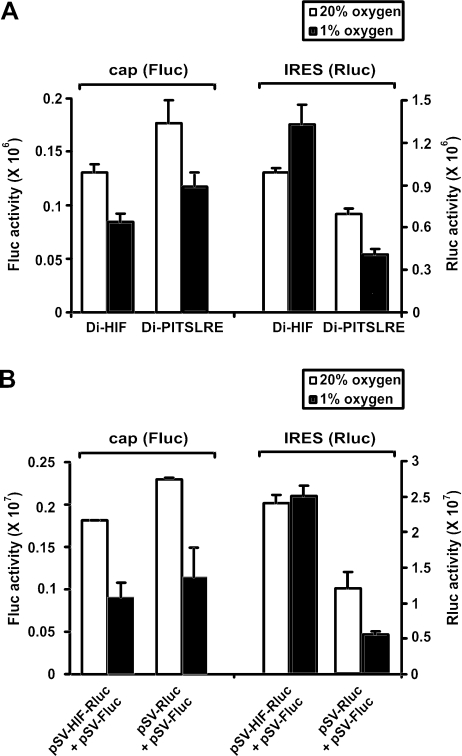
It has been shown that the HIF-1α 5′-UTR contains an IRES that is capable of mediating translation in conditions where cap-dependent translation is suppressed (12). Our results confirm this observation. Bicistronic reporter constructs harboring either the HIF-1α IRES (Di-HIF) or the PITSLRE IRES (Di-PITSLRE), as intercistronic sequences cloned between firefly (first cistron) and Renilla (second cistron) luciferase coding sequences, were transfected in HEK293T cells. The transfectants were grown in either normoxic or hypoxic conditions (Figure 1A). As expected, the cap-dependent cistron was expressed at a lower level in hypoxic than in normoxic conditions, in both Di-HIF and Di-PITSLRE transfectants. In transfectants in which the HIF-1α IRES drives translation of the second cistron, however, Renilla luciferase expression was sustained and even enhanced by oxygen depletion, while PITSLRE IRES-mediated translation was inhibited to the same extent as the cap-dependent translation.
Figure 1.
The HIF-1α IRES efficiently mediates translation during hypoxic conditions. Influence of oxygen depletion on HIF-1α and PITSLRE IRES activity. HEK293T cells were transfected with the bicistronic vectors Di-HIF and Di-PITSLRE (A), or with a combination of the monocistronic reporter plasmids pSV-Fluc + pSV-HIF-Rluc and pSV-Fluc + pSV-Rluc (B). Transfectants were then kept in either 20 or 1% oxygen for 24 h. The bars represent the averages (n = 3) ± SD of Firefly luciferase (Fluc) and Renilla luciferase (Rluc) activities in normoxic (open bars) or hypoxic (closed bars) conditions. Bars are representative of three independent transfection experiments.
In order to investigate whether the HIF-1α 5′-UTR can also promote translation during hypoxia in a monocistronic context, the HIF-1α 5′-UTR was cloned in the pSV-sport vector upstream of the Renilla luciferase coding sequence (pSV-HIF-Rluc). The pSV-Sport vectors containing the coding sequence for Renilla luciferase (pSV-Rluc) or the firefly luciferase (pSV-Fluc) were used as negative controls. HEK293T cells were transfected with a mixture of pSV-HIF-Rluc and pSV-Fluc, or a mixture of pSV-Rluc and pSV-Fluc, and then grown in normoxic or hypoxic conditions. As expected, luciferase expression by the vectors devoid of the HIF-1α IRES was inhibited by oxygen depletion. In contrast, the HIF-1α IRES could sustain Renilla luciferase expression in hypoxic conditions. All together, these data demonstrate that the HIF-1α IRES can guarantee efficient translation during oxygen deprivation.
PTB binds to the HIF-1α IRES
In order to investigate the molecular mechanism of HIF-1α IRES-mediated translation in hypoxic conditions, we examined its RNA sequence for the presence of ITAF consensus binding sites. This revealed a pyrimidine-rich sequence localized at nt 223–237, which is a potential binding site for the cellular RNA-binding protein PTB.
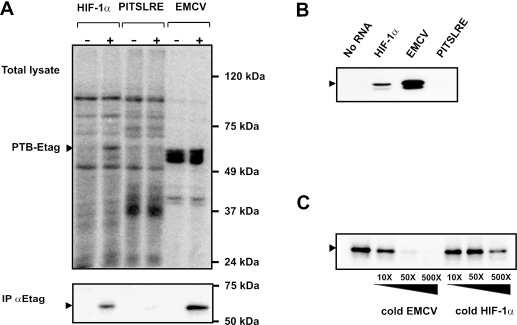
To examine whether PTB can interact with the HIF-1α IRES, UV-crosslinking experiments were performed. Radiolabeled RNA of HIF-1α IRES, EMCV IRES or PITSLRE IRES was incubated with cell extracts derived from HEK293T transfectants expressing either E-tagged PTB, or E-tagged c-Raf as an irrelevant protein. The EMCV IRES, which interacts strongly with PTB, served as a positive control, and the PITSLRE RNA, which is devoid of pyrimidine-rich sequences and does not bind PTB (S. Tinton, unpublished data), served as a negative control. After UV-crosslinking and RNAse treatment, the samples were analyzed by SDS–PAGE (Figure 2A, upper panel). A P32-labeled protein of 58 kDa, corresponding to the size of PTB, is crosslinked with the HIF-1α and EMCV IRES probes but not with the PITSLRE-specific probe. To confirm the identity of this protein, the UV-crosslinked complexes were immunoprecipitated with anti-E-tag antibody. Figure 2A (lower panel) illustrates the binding of E-tagged PTB to the HIF-1α and EMCV IRESs. As expected, none of the IRES probes revealed any binding to the irrelevant 74kDa E-tagged c-Raf protein.
Figure 2.
PTB binds to the HIF-1α IRES. (A) Cytoplasmic extracts from HEK293T cells overexpressing E-tagged c-Raf (−) or PTB (+) were prepared as described in the ‘Materials and Methods’ section. After incubation with the RNA probes corresponding to the HIF-1α IRES, the EMCV IRES, or the PITSLRE IRES, the samples were UV-irradiated and treated with an RNase cocktail. One part of the sample was analyzed by SDS–PAGE (upper panel), the other was mixed with anti-E-tag antibody for immunoprecipitation of labeled E-tagged PTB. Bound proteins were analyzed by SDS–PAGE (lower panel). The arrows depict the position of crosslinked E-tagged PTB. (B) Biotinylated HIF-1α, EMCV and PITSLRE IRES RNAs bound to streptavidin beads were incubated with cytoplasmic extracts from parental HEK293T cells as described in the ‘Materials and Methods’ section. After extensive washing, RNA-bound proteins were analyzed by western blotting using anti-PTB antibodies. The arrow depicts the position of endogenous PTB. (C) Recombinant PTB was UV-crosslinked to radiolabeled HIF-1α IRES RNA in the absence or presence of a 10–500 molar excess of unlabeled EMCV IRES or HIF-1α IRES RNA. After RNA binding and UV-irradiation, samples were treated with an RNase cocktail and resolved by SDS–PAGE. The arrow depicts the position of crosslinked His-tagged PTB.
To test the binding of endogenous PTB to the HIF-1α IRES, RNA affinity chromatography was performed. RNA corresponding to the HIF-1α, EMCV and the PITSLRE IRESs were synthesized in vitro in the presence of biotinylated CTP nucleotides. The biotinylated RNA probes were immobilized on streptavidin-coated beads and incubated with cell extracts from HEK293T cells. After intensive washing, the bound proteins were eluted and separated by SDS–PAGE. Western blot analysis using an anti-PTB antibody confirmed the binding of PTB to the HIF-1α IRES and to the EMCV IRES (Figure 2B). No PTB could be bound to PITSLRE IRES RNA or to beads not containing RNA.
In order to investigate the specificity of the observed interaction between PTB and the HIF-1α IRES, we examined whether the HIF-1α and the EMCV IRESs can compete for binding with recombinant PTB. To this end, UV-crosslinking experiments were performed in the presence or absence of excess unlabeled HIF-1α or EMCV IRES RNA. Incubation of recombinant PTB with a 10- to 500-fold excess of EMCV IRES RNA abolishes the interaction of PTB with radioactively labeled HIF-1α IRES RNA (Figure 2C). Similarly, addition of equal amounts of excess unlabeled HIF-1α IRES RNA also impedes the binding of PTB to radiolabeled HIF-1α IRES RNA, but to a lesser extent. These findings provide evidence for the binding of PTB to the HIF-1α IRES.
The polypyrimidine tract in the HIF-1α IRES is involved in the binding of PTB
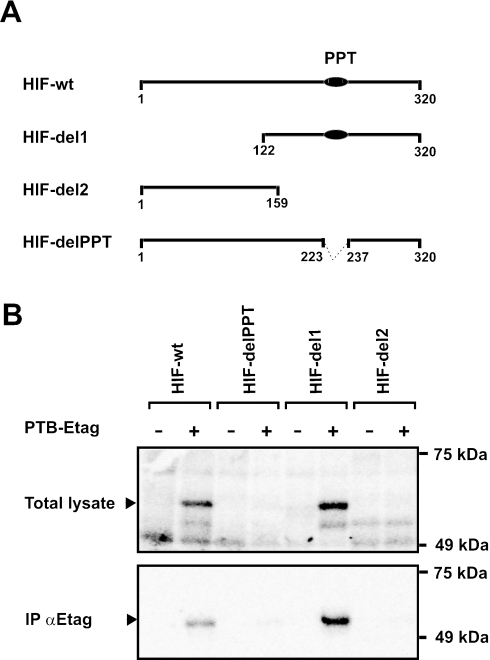
To investigate whether the pyrimidine-rich tract present within the HIF-1α IRES sequence is involved in the binding of PTB, UV-crosslinking experiments were performed with radiolabeled probes containing the wild-type HIF-1α IRES (HIF-wt) or particular fragments of the HIF-1α IRES. The HIF-del1 and HIF-del2 fragments contain the 3′-most half and the 5′-most half of the HIF-1α IRES, respectively, whereas the HIF-delPPT fragment differs from the wild-type HIF-1α IRES by a deletion of the 15 nt polypyrimidine tract (Figure 3A). The probes were incubated with cytoplasmic cell extracts derived from HEK293T cells overexpressing Raf-E-tag or PTB-E-tag, UV-crosslinked, and immunoprecipitated with the anti-E-Tag antibody (Figure 3B). A radioactive PTB-E-Tag band was clearly detected in the reactions with the HIF-wt and HIF-del1 probes, but not with the HIF-del2 or the HIF-delPPT probes. These results demonstrate that the pyrimidine-rich region is important for efficient interaction between PTB and the HIF-1α IRES. All together, these data indicate that the PTB-binding site is located at the 3′-most half of the HIF-1α sequence, and that the polypyrimidine tract is involved in this interaction.
Figure 3.
Deletion of the polypyrimidine tract in the HIF-1α 5′-UTR RNA strongly reduces its binding to PTB. (A) Schematic representation of the radiolabeled HIF-1α fragments. The polypyrimidine tract (PPT) is indicated as a black ellipse. (B) Cytoplasmic extracts from HEK293T cells overexpressing E-tagged c-Raf (−) or PTB (+) were incubated with radiolabeled HIF-1α IRES RNA or radiolabeled RNA corresponding to the HIF-1α IRES fragments HIF-delPPT, HIF-del1 and HIF-del2. After UV-irradiation, samples were treated with an RNase cocktail. One part of the sample was analyzed by SDS–PAGE (upper panel), and the other was mixed with anti-E-tag antibody for immunoprecipitation of labeled E-tagged PTB. Bound proteins were analyzed by SDS–PAGE. The arrow depicts the position of crosslinked E-tagged PTB.
Hypoxia promotes the interaction between PTB and the HIF-1α IRES
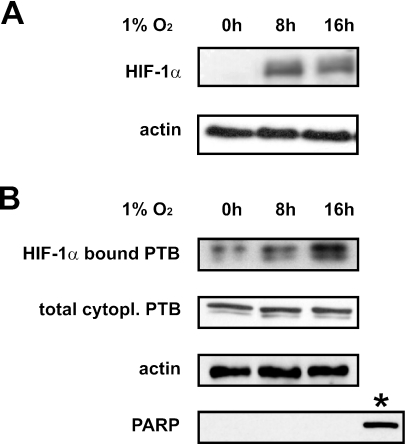
The HIF-1α IRES mediates efficient translation during hypoxic conditions. In order to investigate the physiological relevance of the interaction between PTB and the HIF-1α IRES, we examined their binding in hypoxic conditions. Cytoplasmic cell extracts were prepared from HEK293T cells grown in 1% oxygen for 0, 8 or 16 h, and induction of HIF-1α protein expression was verified by western blot analysis of total cell extracts (Figure 4A). Next, biotinylated HIF-1α IRES RNA immobilized on streptavidin-coated beads was incubated in the presence of normoxic (0 h) or hypoxic (8 and 16 h) cytoplasmic cell extracts. The bound proteins were separated by SDS–PAGE and analyzed with an anti-PTB antibody. Figure 3B (upper panel) illustrates that hypoxic treatment of the HEK293T cells coincides with a higher amount of bound PTB, indicating that hypoxia promotes the interaction between PTB and the HIF-1α IRES.
Figure 4.
The binding of PTB to the HIF-1α IRES is enhanced in hypoxic conditions. (A) Western blot analysis using an anti-HIF-1α antibody (upper panel), and an anti-β-actin antibody as loading control (lower panel) on blots containing total cell extracts derived from HEK293T cells grown in either 20% oxygen (0 h) or 1% oxygen for 8 or 16 h. (B) Biotinylated HIF-1α IRES RNA-bound to streptavidin beads was incubated with cytoplasmic extracts derived from the HEK293T cells described in (A). After extensive washing, RNA-bound proteins were analyzed by western blotting using an anti-PTB antibody (upper panel). Western blot analysis of the cytoplasmic cell extracts used for RNA affinity chromatography using an anti-PTB antibody (second panel), an anti-β-actin antibody as loading control (third panel) and an anti-poly(ADP-ribose)polymerase antibody, showed equal cytoplasmic PTB expression in all samples. As positive control for poly(ADP-ribose)polymerase expression whole cell extract from normal HEK293T cells (indicated by asterisk) was used.
Analysis of cytoplasmic extracts from hypoxic HEK293T cells for PTB protein expression shows that oxygen deprivation does not lead to accumulation of PTB protein in the cytoplasm (Figure 4B, lower panel), excluding that enhanced binding of PTB is due to enhanced expression of PTB in the hypoxic cytoplasmic cell extracts. Any nuclear contamination of the used cytoplasmic extracts was excluded by western blot analysis using an anti-poly(ADP-ribose)polymerase antibody. A sample of HEK293T total cell extract was used as positive control.
Overexpression of PTB stimulates the activity of the HIF-1α IRES
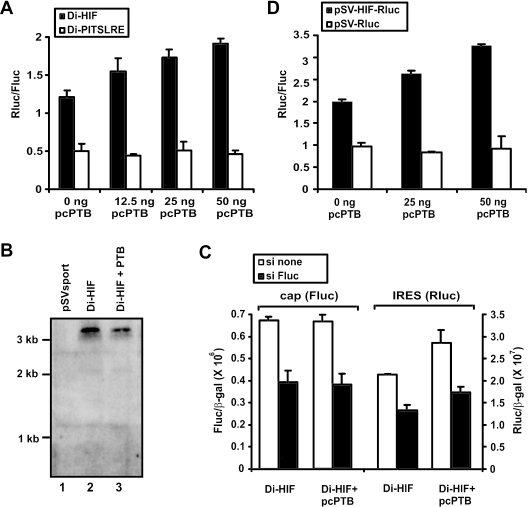
To further investigate a potential role for PTB in the regulation of HIF-1α IRES activity, we examined whether overexpression of PTB can modify the activity of the HIF-1α IRES. Bicistronic reporter constructs harboring either the HIF-1α IRES (Di-HIF) or the PITSLRE IRES (Di-PITSLRE) as intercistronic sequences were transfected in HEK293T cells, in the absence or presence of different amounts of an expression vector containing the PTB coding sequence (pcPTB). Figure 5A shows that overexpression of PTB, results in a concentration-dependent increase in activity of the HIF-1α IRES but not of the PITSLRE IRES. Increasing amounts of PTB were expressed in cells transfected with increasing amounts of pcPTB as confirmed by western blot analysis (data not shown). Similar results were obtained for BSC-1 cells (data not shown).
Figure 5.
PTB stimulates HIF-1α IRES-mediated translation. (A) Effect of PTB on HIF-1α IRES-driven expression in a bicistronic reporter assay. HEK293T cells were transfected with the bicistronic reporter plasmid Di-HIF or Di-PITSLRE, together with different amounts of the plasmid for PTB expression (pcPTB). The transfectants were lysed 24 h later and the luciferase activities were measured. The bars represent the average (n = 3) ± SD of the calculated ratio between Renilla and firefly luciferase activity. The results are representative of three independent experiments. (B) Overexpression of PTB does not affect the integrity of the bicistronic mRNA. HEK293T cells were transfected with the pSV-sport vector (lane 1) or with the Di-HIF vector in combination with either the empty pCAGGS vector (lane 2) or the PTB-containing pCAGGS expression vector pcPTB (lane 3). RNA was prepared 24 h later and analyzed by northern blotting using a radiolabeled probe directed against the coding sequence of Renilla luciferase. (C) Translation of the second cistron is initiated from an intact dicistronic mRNA. HEK293T cells were transfected with the bicistronic reporter plasmid Di-HIF, in combination with either an empty expression vector or pcPTB and in combination with siRNAs not directed against any known cellular RNA (si none, white bars), or against the Firefly luciferase mRNA (si Fluc, black bars). Cells were lysed 24 h later, and luciferase activities were measured. Bars represent the average (n = 2) ± SD of the calculated ratio between luciferase and β-galactosidase activities. (D) Effect of PTB on expression controlled by HIF-1α 5′-UTR in a monocistronic reporter assay. pSV-HIF-Rluc or pSV-Rluc were cotransfected with pSV-Fluc and different concentrations of pcPTB in HEK293T cells. The cells were lysed 24 h later and the luciferase activities were measured. The bars represent the average (n = 3) ± SD of the calculated ratio between Renilla luciferase and firefly luciferase. The results are representative of three independent experiments.
Many reports illustrate that PTB is also involved in alternative splicing (35). Therefore, overexpression of PTB might result in aberrant mRNA species derived from the bicistronic vector, which could be responsible for the observed differences in luciferase expression. To exclude this possibility, we investigated the integrity of the bicistronic transcripts following transfection of HEK293T cells with Di-HIF alone (lane 2) or in combination with pcPTB (lane 3) by northern blot analysis (Figure 5B). HEK293T cells transfected with the empty pSV-Sport vector served as a negative control (lane 1). Bands corresponding to the expected bicistronic transcripts (3 kb) were detected with a probe against the second cistron (Renilla luciferase), but we could not detect any other transcripts, excluding a role for PTB-dependent splicing in the observed changes in HIF-1α IRES activity. The integrity of Di-HIF mRNA in cells transfected with either Di-HIF alone or in combination with pcPTB, was further investigated through RT–PCR. By this approach we could only amplify a fragment that corresponds to intact dicistronic mRNA (data not shown).
To more functionally demonstrate that translation of the second cistron, in both normal and PTB overexpressing cells, is initiated from an intact dicistronic mRNA, we analyzed the effect of siRNAs directed against the first cistron (si Fluc) on the expression of both cistrons (36). Figure 5C shows that in cells transfected with Di-HIF in combination with either an empty or a PTB expression vector, siRNA directed against the first cistron inhibits the expression of both the first and second cistron. These results demonstrate that translation of the second cistron is indeed initiated from an intact dicistronic mRNA and that PTB stimulated translation of this second cistron does not result from the possible generation of aberrant cryptic monocistronic mRNAs after PTB overexpression.
We also investigated whether PTB can influence translation initiated by the HIF-1α 5′-UTR in a monocistronic context. HEK293T cells were transfected either with a mixture of pSV-HIF-Rluc and pSV-Fluc, or with a mixture of pSV-Rluc and pSV-Fluc, in the absence or presence of different amounts of the pcPTB expression vector. Figure 5D shows that overexpression of PTB indeed enhances expression of a reporter from a monocistronic mRNA if its 5′ leader contains the HIF-1α 5′-UTR.
Downregulation of PTB reduces HIF-1α IRES activity
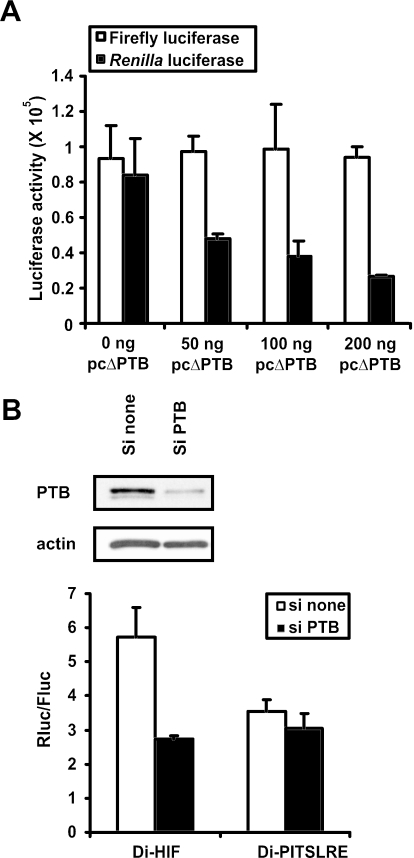
To further demonstrate that PTB is a positive regulator of the HIF-1α IRES, we set up an experiment in which we competed out the effect of endogenous PTB by overexpressing a PTB fragment (ΔPTB) that contains only the first two of the four RNA recognition motifs present within the wild-type PTB. Overexpression of this fragment significantly represses poliovirus IRES-mediated translation (37). HEK293T cells were cotransfected with Di-HIF and different concentrations of an expression vector containing the coding sequence of the ΔPTB polypeptide (pcΔPTB). Figure 6A shows that overexpressed ΔPTB leads to a concentration-dependent impairment of Renilla luciferase expression without affecting the expression of the first cistron. This observation indicates that inhibition of PTB function impedes the activity of the HIF-1α IRES.
Figure 6.
Downregulation of PTB reduces HIF-1α IRES activity. (A) Effect of the disruption of PTB function on the activity of the HIF-1α IRES. HEK293T cells were transfected with Di-HIF together with different amounts of the expression vector pcΔPTB. Luciferase activities were measured in the corresponding cell extracts 24 h later. The bars represent the average activities (n = 3) ± SD of firefly luciferase (open bars) and Renilla luciferase (black bars). Bars are representative of three independent experiments. (B) Knockdown of PTB by siRNAs inhibits HIF-1α IRES activity. HEK293T cells were transfected with Di-HIF or Di-PITSLRE together with siRNAs not directed against any known cellular RNA (si none), or against the PTB mRNA (si PTB). The cells were lysed 24 h later, and luciferase activities were measured. The bars represent the average (n = 3) ± SD of the calculated ratio between Renilla luciferase and firefly luciferase. Bars are representative of three independent experiments. Western blot analysis using anti-PTB and anti-actin antibodies confirmed the knockdown of PTB whereas the expression of actin was unchanged.
In order to further confirm the positive role of PTB in HIF-1α IRES-mediated translation, we also used siRNAs to suppress the expression of endogenous PTB. A mixture of four siRNA sequences complementary to the PTB mRNA, or a mixture not complementary to known cellular RNA, was cotransfected in HEK293T cells together with either Di-HIF or Di-PITSLRE. Western blot analysis with an anti-PTB antibody showed much lower levels of endogenous PTB 48 h after transfection of PTB siRNAs compared with PTB expression levels in cells transfected with non-specific siRNAs (Figure 6B). Expression of the unrelated β-actin protein was unchanged, indicating the specificity of the PTB siRNA. siRNA-mediated knockdown of PTB led to a 2-fold decrease of HIF-1α IRES activity. As expected, this was not seen with the PITSLRE IRES (Figure 6B). All together, these data indicate that PTB has a positive impact on HIF-1α IRES-mediated translation.
PTB stimulates HIF-1α IRES activity during hypoxia
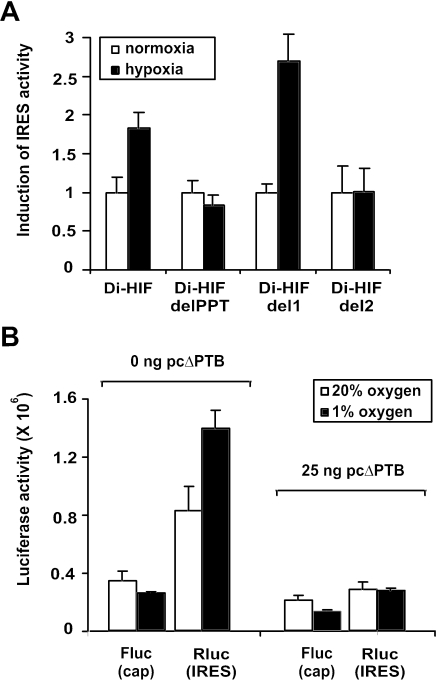
To evaluate the role of PTB in the induction of HIF-1α IRES activity during oxygen deprivation, we compared the IRES activities of different fragments of the HIF-1α IRES under normoxic and hypoxic conditions. The bicistronic reporter constructs Di-HIF, Di-HIFdelPPT, Di-HIFdel1 and Di-HIFdel2, containing the wild-type HIF-1α IRES and the fragments HIF-delPPT, HIF-del1 and HIF-del2 (see also Figure 3A) as intercistronic sequences, respectively, were transfected in HEK293T cells, and subsequently grown under normoxic or hypoxic conditions. Figure 7A shows that translation driven by the wild-type HIF-1α IRES and by the HIF-del1 fragment was induced upon oxygen deprivation, while translation driven by fragments missing the PPT-sequence remained unchanged. Thus, the ability of oxygen deprivation to induce the activity of the different IRES fragments correlates with their ability to bind PTB (also see Figure 3).
Figure 7.
PTB stimulates HIF-1α IRES activity during hypoxia. (A) The polypyrimidine tract is needed for the induction of HIF-1α IRES activity upon oxygen depletion. Di-HIF, Di-HIFdelPPT, Di-HIFdel1, Di-HIFdel2 or di-PITSLRE were transfected in HEK293T cells. Six hours later the transfectants were placed in either 20 or 1% oxygen for 24 h. Subsequently, the transfectants were lysed and luciferase activities were measured. The bars represent the average IRES activities (n = 3) ± SD calculated as the ratio between Renilla luciferase and firefly luciferase. The IRES activities measured in the transfectants grown in normoxic conditions were set as one. Bars are representative of two independent experiments. (B) Disruption of PTB function impairs the HIF-1α IRES activity induced by hypoxia. HEK293T cells were transfected with Di-HIF together with either the empty pCAGGS vector or pcΔPTB. Six hours later the transfectants were placed in either 20 or 1% oxygen for 24 h. The transfectants were then lysed and luciferase activities were measured. The bars represent the averages (n = 3) ± SD of Firefly luciferase (Fluc) and Renilla luciferase (Rluc) activities in normoxic (open bars) or hypoxic (closed bars) conditions. Bars are representative of three independent transfection experiments.
To further investigate the role of PTB in the induction of HIF-1α IRES activity during oxygen deprivation, we tested whether blocking the function of endogenous PTB by overexpressing the ΔPTB fragment can counteract the induction of IRES activity. The bicistronic reporter plasmid Di-HIF was cotransfected in HEK293T cells, together with either an empty pCAGGS vector or a pCAGGS vector harboring the coding sequence of the ΔPTB polypeptide (pcΔPTB). As expected, expression of the cap-dependent cistron was inhibited by hypoxia in pCAGGS-transfected and in pcΔPTB-transfected cells (Figure 7B). As shown before in Figure 6A, overexpression of ΔPTB impedes HIF-1α IRES-mediated translation in normoxic conditions. Interestingly, the hypoxia-induced stimulation of the HIF-1α IRES (pCAGGS transfectants) was completely blocked in the pcΔPTB transfectants, indicating that PTB positively modulates the HIF-1α IRES activity during oxygen deprivation.
DISCUSSION
The hypoxia-inducible factor-1α mRNA contains an IRES in its 5′-UTR that allows efficient translation during normoxia and hypoxia (12). In the present study, we set out to identify the mechanisms that regulate HIF-1α IRES-mediated translation in normoxic and hypoxic conditions. We began to survey the HIF-1α 5′-UTR for consensus binding sequences of known RNA-binding proteins. The presence of a pyrimidine-rich tract in the HIF-1α IRES suggested that PTB might be an HIF-1α IRES-binding factor. Using UV-crosslinking and RNA affinity, we showed that PTB specifically binds to the HIF-1α IRES. The interaction with PTB appears to be a common feature of many IRESs, but not all of them, as we could not detect binding of PTB to the PITSLRE IRES. For several of these IRESs, interaction with PTB seems to require a polypyrimidine tract usually located near the start codon (30,38,39). Also in the case of the HIF-1α IRES, a polypyrimidine tract sequence located at nt 223–237, is essential for PTB interaction.
Our results also demonstrate that PTB is a positive modulator of HIF-1α IRES-driven translation. Transient overexpression of the vector encoding PTB increased the HIF-1α IRES activity. In contrast, HIF-1α IRES activity was inhibited by disruption of PTB function, either by overexpression of a dominant-negative fragment of PTB or by knockdown of PTB protein expression by RNA interference. These results indicate that the status of PTB in the cell can potentially modulate translation initiated by the HIF-1α IRES during hypoxia.
Interestingly, the binding of the positive modulator PTB to the HIF-1α IRES is enhanced during oxygen depletion. Moreover, this observation parallels an increasing HIF-1α IRES activity, which could be counteracted by overexpression of dominant-negative PTB. In addition, HIF-1α IRES mutants incapable of binding PTB did not show any stimulatory effect during oxygen deprivation. All together, our results demonstrate that the induction of HIF-1α IRES activity during hypoxia depends on the binding of PTB to the HIF-1α IRES, and indicates that PTB plays an essential role in the up-regulation of HIF-1α protein expression during hypoxia.
Although PTB is found predominantly in the nucleus, it also shuttles between the nucleus and the cytoplasm (40). However, we could exclude that the enhanced binding of PTB to the HIF-1α IRES during hypoxia is due to increased translocation of nuclear PTB into the cytoplasm, since we did not observe any accumulation of PTB in the cytoplasm upon oxygen deprivation. Alternatively, enhanced binding of PTB to the HIF-1α IRES could result from secondary modifications of cytoplasmic PTB that alter its affinity for the HIF-1α IRES RNA. It has been shown that phosphorylation of hnRNPC1/C2 proteins upon TPA stimulation enhances their binding to the IRES present in the c-sis (PDGF-B) mRNA (41). It is also possible that the binding of other proteins to the IRES impact the binding of PTB. It has been illustrated that the binding of unr protein to the Apaf-1 IRES induces modifications in the secondary structure of the IRES that promote the binding of PTB to the IRES (38). Similarly, hypoxia may alter the binding of other proteins to the HIF-1α IRES to promote the binding of PTB.
HIF-1 acts as an oxygen sensor and activates the transcription of several genes (e.g. VEGF, FGF) upon oxygen depletion (1–7). Interestingly, an IRES has been shown to be involved in the translation of the mRNA of several of the HIF-1 targets (VEGF-A, FGF-2, PDGF-2, IGF-2) (19,42–44). It would be very interesting to investigate a role for PTB as a modulator for these IRESs. Binding of PTB to a polypyrimidine tract within the VEGF IRES has already been reported by Huez et al. (45). IRESs have been identified not only in HIF-1 target genes, but also in the mRNA of genes whose expression is induced by hypoxia independently of the HIF-1 transcription factor (ODC, p27, PKCδ, Rbm3) (46–49). Similar to the HIF-1α IRES, the IRES present in the VEGF mRNA is also stimulated by oxygen deprivation (12,19). The observation that PTB can also bind to a polypyrimidine tract within the VEGF and p27 IRESs suggests that its binding might be involved in their activity during hypoxia (25,45). IRES-mediated translation, orchestrated by PTB, might therefore be a general theme in the cellular response to hypoxia.
Acknowledgments
We thank Dr R. Jackson (Department of Biochemistry, University of Cambridge, UK) for providing recombinant PTB. We are grateful to Dr A. Bredan for critical reading of the manuscript, and to A. Meeus and W. Burm for their technical assistance. This work was supported by grants from the ‘Belgische Federatie tegen Kanker’, the ‘Interuniversitaire Attractiepolen’ and the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen (FWO). B.S. is a predoctoral research fellow with the ‘Vlaams Instituut voor de Bevordering van het Wetenschappelijk-technologisch Onderzoek in de Industrie’. S.C. is a postdoctoral research associate with the FWO. Funding to pay the Open Access publication charges for this article was provided by the FWO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shweiki D., Itin A., Soffer D., Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P., Dor Y., Herbert J.M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 3.Ryan H.E., Lo J., Johnson R.S. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer N.V., Kotch L.E., Agani F., Leung S.W., Laughner E., Wenger R.H., Gassmann M., Gearhart J.D., Lawler A.M., Yu A.Y., et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang B.H., Rue E., Wang G.L., Roe R., Semenza G.L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 6.Semenza G.L., Wang G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldser D., Agani F., Iyer N.V., Pak B., Ferreira G., Semenza G.L. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 1999;59:3915–3918. [PubMed] [Google Scholar]
- 8.Wang G.L., Semenza G.L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 9.Huang L.E., Arany Z., Livingston D.M., Bunn H.F. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 10.Huang L.E., Gu J., Schau M., Bunn H.F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl Acad. Sci. USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salceda S., Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 12.Lang K.J., Kappel A., Goodall G.J. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlach A., Camenisch G., Kvietikova I., Vogt L., Wenger R.H., Gassmann M. Efficient translation of mouse hypoxia-inducible factor-1alpha under normoxic and hypoxic conditions. Biochim. Biophys. Acta. 2000;1493:125–134. doi: 10.1016/s0167-4781(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 14.Pestova T.V., Hellen C.U., Shatsky I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang S.K., Krausslich H.G., Nicklin M.J., Duke G.M., Palmenberg A.C., Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelletier J., Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 18.Macejak D.G., Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 19.Stein I., Itin A., Einat P., Skaliter R., Grossman Z., Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson J.E., Powell M.J., Hoover S.E., Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilipenko E.V., Pestova T.V., Kolupaeva V.G., Khitrina E.V., Poperechnaya A.N., Agol V.I., Hellen C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 22.Borovjagin A., Pestova T., Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 23.Hellen C.U., Witherell G.W., Schmid M., Shin S.H., Pestova T.V., Gil A., Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell S.A., Brown E.C., Coldwell M.J., Jackson R.J., Willis A.E. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol. 2001;21:3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho S., Kim J.H., Back S.H., Jang S.K. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickering B.M., Mitchell S.A., Evans J.R., Willis A.E. Polypyrimidine tract binding protein and poly r(C) binding protein 1 interact with the BAG-1 IRES and stimulate its activity in vitro and in vivo. Nucleic. Acids Res. 2003;31:639–646. doi: 10.1093/nar/gkg146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 1996;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- 28.Hunt S.L., Jackson R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi M., Schultz D.E., Lemon S.M. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 2000;74:6459–6468. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornelis S., Tinton S.A., Schepens B., Bruynooghe Y., Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic. Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y.K., Hahm B., Jang S.K. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- 32.Tinton S.A., Schepens B., Bruynooghe Y., Beyaert R., Cornelis S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2alpha. Biochem. J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Mahoney J.V., Adams T.E. Optimization of experimental variables influencing reporter gene expression in hepatoma cells following calcium phosphate transfection. DNA Cell Biol. 1994;13:1227–1232. doi: 10.1089/dna.1994.13.1227. [DOI] [PubMed] [Google Scholar]
- 34.Walker J., de Melo Neto O., Standart N. Gel retardation and UV-crosslinking assays to detect specific RNA-protein interactions in the 5′ or 3′ UTRs of translationally regulated mRNAs. Methods Mol. Biol. 1998;77:365–378. doi: 10.1385/0-89603-397-X:365. [DOI] [PubMed] [Google Scholar]
- 35.Valcarcel J., Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 1997;7:R705–708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 36.Van Eden M.E., Byrd M.P., Sherrill K.W., Lloyd R.E. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Back S.H., Kim Y.K., Kim W.J., Cho S., Oh H.R., Kim J.E., Jang S.K. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) J. Virol. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell S.A., Spriggs K.A., Coldwell M.J., Jackson R.J., Willis A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 39.Vagner S., Waysbort A., Marenda M., Gensac M.C., Amalric F., Prats A.C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 40.Xie J., Lee J.A., Kress T.L., Mowry K.L., Black D.L. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sella O., Gerlitz G., Le S.Y., Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell. Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagner S., Gensac M.C., Maret A., Bayard F., Amalric F., Prats H., Prats A.C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol. Cell. Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein J., Sella O., Le S.Y., Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J. Biol. Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 44.Pedersen S.K., Christiansen J., Hansen T.O., Larsen M.R., Nielsen F.C. Human insulin-like growth factor II leader 2 mediates internal initiation of translation. Biochem. J. 2002;363:37–44. doi: 10.1042/0264-6021:3630037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huez I., Creancier L., Audigier S., Gensac M.C., Prats A.C., Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pyronnet S., Pradayrol L., Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol. Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 47.Kullmann M., Gopfert U., Siewe B., Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrish B.C., Rumsby M.G. The 5′ untranslated region of protein kinase Cdelta directs translation by an internal ribosome entry segment that is most active in densely growing cells and during apoptosis. Mol. Cell. Biol. 2002;22:6089–6099. doi: 10.1128/MCB.22.17.6089-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chappell S.A., Owens G.C., Mauro V.P. A 5′ leader of Rbm3, a cold-stress induced mRNA, mediates internal initiation of translation with increased efficiency under conditions of mild hypothermia. J. Biol. Chem. 2001;24:24. doi: 10.1074/jbc.M106008200. [DOI] [PubMed] [Google Scholar]