Abstract
Apolipoprotein (apo) E4, a 299-aa protein and a major risk factor for Alzheimer's disease, can be cleaved to generate C-terminal-truncated fragments that cause neurotoxicity in vitro and neurodegeneration and behavioral deficits in transgenic mice. To investigate this neurotoxicity, we expressed apoE4 with C- or N-terminal truncations or mutations in transfected Neuro-2a cells. ApoE4 (1-272) was neurotoxic, but full-length apoE4(1-299) and apoE4(1-240) were not, suggesting that the lipid-binding region (amino acids 241-272) mediates the neurotoxicity and that amino acids 273-299 are protective. A quadruple mutation in the lipid-binding region (I250A, F257A, W264R, and V269A) abolished the neurotoxicity of apoE4(1-272), and single mutations in the region of amino acids 273-299 (L279Q, K282A, or Q284A) made full-length apoE4 neurotoxic. Immunofluorescence staining showed that apoE4(1-272) formed filamentous inclusions containing phosphorylated tau in some cells and interacted with mitochondria in others, leading to mitochondrial dysfunction as determined by MitoTracker staining and flow cytometry. ApoE4(241-272) did not cause mitochondrial dysfunction or neurotoxicity, suggesting that the lipid-binding region alone is insufficient for neurotoxicity. Truncation of N-terminal sequences (amino acids 1-170) containing the receptor-binding region (amino acids 135-150) and triple mutations within that region (R142A, K146A, and R147A) abolished the mitochondrial interaction and neurotoxicity of apoE4(1-272). Further analysis showed that the receptor-binding region is required for escape from the secretory pathway and that the lipid-binding region mediates mitochondrial interaction. Thus, the lipid- and receptor-binding regions in apoE4 fragments act together to cause mitochondrial dysfunction and neurotoxicity, which may be important in Alzheimer's disease pathogenesis.
Keywords: Alzheimer's disease, mitochondria, proteolysis
Human apolipoprotein (apo) E, a 34-kDa protein with 299 aa, has three major isoforms, apoE2, apoE3, and apoE4 (1-4). ApoE4 is a major risk factor for Alzheimer's disease (AD) (5-7). The apoE4 allele, which is found in 40-65% of cases of sporadic and familial AD, increases the occurrence and lowers the age of onset of the disease (7, 8).
Biochemical, cell biological, transgenic animal, and human studies have suggested several potential mechanisms to explain the contribution of apoE4 to the pathogenesis of AD. These mechanisms include modulation of the deposition and clearance of amyloid β (Aβ) peptides and the formation of plaques (9-15), modulation of Aβ-caused synaptic and cholinergic deficits (16), acceleration of age- and excitotoxicity-related neurodegeneration (17), impairment of the antioxidative defense system and mitochondrial function (18-21), dysregulation of neuronal signaling pathways (22), altered phosphorylation of tau and neurofibrillary tangle formation (23-28), depletion of cytosolic androgen receptor levels in the brain (29, 30), potentiation of Aβ-induced lysosomal leakage and apoptosis in neuronal cells (31), and promotion of endosomal abnormalities linked to Aβ overproduction (32-34). The mechanisms of these apoE4-mediated detrimental effects are largely unknown.
We have shown that apoE can be cleaved by a neuron-specific chymotrypsin-like serine protease that generates bioactive C-terminal-truncated forms of apoE (25, 27, 28). The fragments are found at higher levels in the brains of AD patients than in age- and sex-matched controls (27), and apoE4 is more susceptible to cleavage than apoE3. When expressed in cultured neuronal cells or added exogenously to the cultures, apoE4 fragments are neurotoxic, leading to cell death (25). When expressed in transgenic mice, they cause AD-like neurodegeneration and behavioral deficits (27). Because apoE is synthesized by neurons under diverse pathophysiological conditions (35-49), we hypothesize that apoE4 produced in neurons in response to stress or injury (e.g., Aβ toxicity, brain trauma, or oxidative stress) is uniquely susceptible to proteolytic cleavage and that the resulting bioactive C-terminal-truncated fragments induce neuropathology and associated behavioral deficits. ApoE3 also undergoes proteolytic cleavage but to a lesser extent.
In this study, we investigated the cellular and molecular mechanisms of the neurotoxicity caused by apoE4 fragments in cultured neuronal cells. We also evaluated the roles of various regions [specifically, the receptor-binding region (amino acids 135-150) and the lipid-binding region (amino acids 241-272)] of apoE (1-4, 50).
Methods
Reagents. MEM, Opti-MEM, and FBS were obtained from Life Technologies (Rockville, MD). Polyclonal goat anti-human apoE was obtained from Calbiochem. Monoclonal antibodies that specifically recognize the lipid-binding region of apoE (3H1) were obtained from Karl H. Weisgraber (Gladstone Institutes, San Francisco). Anti-rabbit, anti-mouse, and anti-goat IgGs coupled to fluorescein or Texas red were obtained from Vector Laboratories. MitoTracker Deep Red 633 was obtained from Invitrogen. A cDNA construct encoding red fluorescent protein fused with a mitochondrial localization signal peptide (DsRed2-Mito) was obtained from BD Biosciences.
cDNA Constructs. PCR products encoding WT or N-terminal-truncated apoE4 with its signal peptide were subcloned into a pcDNA 3.1(+) vector (Invitrogen) containing the cytomegalo-virus promoter. A PCR product encoding a signal peptide-GFP-apoE4 fusion protein was also subcloned into the vector. cDNA constructs encoding apoE4 with various mutations or C-terminal truncations were made from the pcDNA-apoE4 or pcDNA-GFP-apoE4 construct with a QuikChange kit (Stratagene). All constructs were confirmed by sequence analysis.
Cell Culture and Transfection. Mouse neuroblastoma Neuro-2a cells (American Type Culture Collection) maintained at 37°C in MEM containing 10% FBS were transiently transfected with the apoE4 cDNA constructs by using Lipofectamine 2000 (Invitrogen) (25). ApoE4 expression levels were determined by anti-apoE Western blotting of cell lysates and media. The truncated and mutated forms of apoE4 that are neurotoxic were expressed at ≈15-30% lower levels than full-length apoE4. To exclude their potential weaker antibody responses, those forms of apoE4 were tagged with GFP, and their expression levels were determined by flow cytometry. Again, their expression levels were ≈15-30% lower than those of full-length apoE4. Thus, the results were not due to overexpression.
Immunocytochemistry and Confocal Microscopy. Neuro-2a cells transiently transfected with various apoE4 cDNA constructs were grown in serum-free MEM for 18-24 h, fixed in 3% paraformaldehyde, permeabilized for 45 min at room temperature with 500 units of Streptolysin-O (STP-O, Sigma) in BBII buffer (75 mM potassium acetate/25 mM Hepes, pH 7.2) (for plasma membranes) or 0.5% Tween 20 in PBS (for plasma and intracellular organelle membranes) (51), and stained with polyclonal anti-apoE (1:4,000 dilution) or monoclonal anti-apoE (3H1, 1:200 dilution) and a fluorescein-coupled secondary antibody (Vector Laboratories) (25). Labeled cells were mounted in VECTASHIELD (Vector Laboratories) and viewed with a Radiance 2000 laser-scanning confocal system (Bio-Rad) that was mounted on an Optiphot-2 microscope (Nikon). Neuro-2a cells transiently transfected with cDNA constructs encoding GFP-apoE4 with mutations or truncations were analyzed directly by confocal microscopy. Some Neuro-2a cells were cotransfected with various apoE cDNA constructs and a construct encoding red fluorescent protein fused with a mitochondrial localization signal peptide (DsRed2-Mito, BD Biosciences), stained with immunofluorescent polyclonal or monoclonal anti-apoE antibody, and analyzed by confocal microscopy.
Cell Survival. Neuro-2a cells grown in 24-well plates were transiently transfected with various apoE4 or GFP-apoE4 cDNA constructs in serum-free Opti-MEM. Cell survival was estimated with an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay (52) at 48 h after transfection.
Flow Cytometry Analysis of Mitochondrial Function and Integrity. Neuro-2a cells grown in six-well plates were transiently transfected with various GFP-apoE4 cDNA constructs. The culture medium was aspirated 48 h after transfection, and MitoTracker Deep Red 633 (100 nM in MEM containing 10% FBS) was added for 15 min at 37°C. After a wash with serum-free MEM, cells were trypsinized and suspended in 1 ml of PBS, washed twice with PBS by centrifugation (300 × g for 5 min), resuspended in 1 ml of PBS, and filtered through a mesh cap into a 5-ml tube. The fluorescence intensity of GFP, which represents apoE4 expression levels, and of MitoTracker Deep Red 633, which represents the levels of mitochondrial function and integrity (53), were analyzed by flow cytometry. Untransfected Neuro-2a cells served as a negative control.
Statistical Analysis. Results are reported as mean ± SD. Differences were evaluated by t test or analysis of variance.
Results
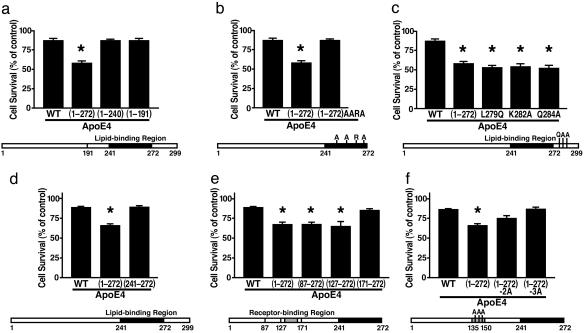
The Lipid-Binding Region Is Required for ApoE4 Fragment-Related Neurotoxicity. To assess the neurotoxicity of various apoE4 fragments in Neuro-2a cells, we used a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Expression of apoE4(1-272) caused 35% greater cell death than full-length apoE4; further C-terminal truncation to amino acids 240 or 191 to remove the lipid-binding region (amino acids 241-272) abolished the neurotoxicity (Fig. 1a). Four mutations of this region (I250A, F257A, W264R, and V269A) that are conserved across different species (54) also abolished the neurotoxicity (Fig. 1b).
Fig. 1.
The lipid- and receptor-binding regions in apoE4 fragments act in concert to cause neurotoxicity, as determined with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. (a) Survival of cells transfected with WT apoE4, apoE4(1-272), apoE4(1-240), or apoE4(1-191). (b) Survival of cells transfected with WT apoE4, apoE4(1-272), or apoE4(1-272) with four mutations (I250A, F257A, W264R, and V269A). (c) Survival of cells transfected with WT apoE4, apoE4(1-272), or apoE4 with single mutations (L279Q, K282A, or Q284A). (d) Survival of cells transfected with WT apoE4, apoE4(1-272), or apoE(241-272). (e) Survival of cells transfected with WT apoE4, apoE4(1-272), apoE4(87-272), apoE(127-272), or apoE(171-272). (f) Survival of cells transfected with WT apoE4, apoE4(1-272), or apoE4(1-272) with double (K146A and R147A) or triple (R142A, K146A, and R147A) mutations. Values are given as mean ± SD of three to six assays at 48 h after transfection. *, P < 0.05 vs. WT apoE4.
Single C-Terminal Mutations Make Full-Length ApoE4 Neurotoxic. ApoE4(1-272) was more neurotoxic than full-length apoE4, suggesting that the 27 C-terminal amino acids protect against fragment-related neurotoxicity. Three amino acids in this region (L279, K282, and Q284) are highly conserved in 10 species (54). To assess their importance in this neuroprotective effect, we introduced mutations at each site (L279Q, K282A, or Q284A) into WT apoE4. Each mutation made full-length apoE4 as neurotoxic as apoE4(1-272) (Fig. 1c).
Neurotoxicity Requires Both Lipid- and Receptor-Binding Regions. To determine whether the lipid-binding region alone was neurotoxic, we analyzed Neuro-2a cells expressing only amino acids 241-272 of apoE4. No neurotoxicity was observed (Fig. 1d). To determine which region of the N terminus was also required for neurotoxicity, we transfected cells with cDNA constructs encoding apoE4(1-272) with progressively longer N-terminal truncations. Neurotoxicity was abolished only by a truncation that removed the receptor-binding region (amino acids 135-150) (Fig. 1e).
Positively Charged Amino Acids in the Receptor-Binding Region Are Critical for Neurotoxicity. The receptor-binding region contains a cluster of positively charged amino acids (arginine and lysine) (1-4). To test their importance in apoE4 fragment-related neurotoxicity, we introduced double (K146A and R147A) and triple (R142A, K146A, and R147A) mutations into apoE4(1-272). The triple mutation abolished the neurotoxic effect of apoE4(1-272), and the double mutation reduced it (Fig. 1f).
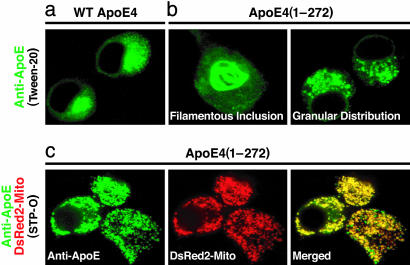
ApoE4 Fragments Escape the Secretory Pathway and Interact with Cytoskeletal Components and Mitochondria. To investigate the mechanisms of neurotoxicity, we assessed the intracellular localization of full-length or truncated apoE4 in Neuro-2a cells by immunofluorescence staining. Full-length apoE4 was typically located in the endoplasmic reticulum and Golgi apparatus (Fig. 2a), whereas apoE4(1-272) formed intracellular filamentous inclusions in some cells and had a granular distribution in others (Fig. 2b), suggesting mislocalization of the truncated apoE4 in Neuro-2a cells. Because intracellular filamentous inclusions contain phosphorylated tau and phosphorylated neurofilament proteins, as reported (25, 26), some of the fragments must have escaped the secretory pathway and interacted with cytoskeletal components. In cells expressing both apoE4(1-272) and DsRed2-Mito, the granule-associated apoE4 fragments were in the mitochondria (Fig. 2c).
Fig. 2.
Intracellular distribution of various forms of apoE4 as determined by immunocytochemistry and confocal microscopy. (a) Cells transfected with WT apoE4, permeabilized with Tween 20, and stained with anti-apoE (green). (b) Cells transfected with apoE4(1-272), permeabilized with Tween 20, and stained with anti-apoE (green). (c) Cells cotransfected with apoE4(1-272) and DsRed2-Mito (red), permeabilized with STP-O, and stained with anti-apoE (green). Yellow in the merged image indicates colocalization of apoE4(1-272) with mitochondria.
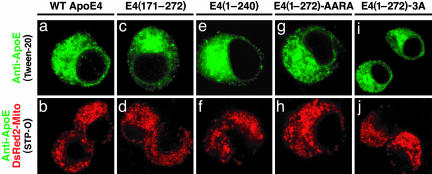
Mitochondrial Mislocalization Requires the Lipid- and Receptor-Binding Regions. Next, we investigated the intracellular location of apoE(171-272), containing only the lipid-binding region, and apoE4(1-240), containing only the receptor-binding region. Neither form was located in the mitochondria, and their intracellular distributions were similar to that of full-length apoE4 (Fig. 3 a-f). The mitochondrial mislocalization was also abolished by the quadruple mutation in the lipid-binding region [E4(1-272)-AARA] and the triple mutations in the receptor-binding region [E4(1-272)-3A] (Fig. 3 g-j).
Fig. 3.
The lipid and receptor-binding regions act in concert to cause mitochondrial mislocalization of apoE4 fragments. Cells transfected with WT apoE4 (a), apoE(171-272) (c), apoE4(1-240) (e), apoE4(1-272)-AARA with four mutations (I250A, F257A, W264R, and V269A) in the lipid-binding region (g), or apoE4(1-272)-3A with three mutations (R142A, K146A, and R147A) in the receptor-binding region (i) were permeabilized with 0.5% Tween 20 (a, c, e, g, and i) and stained with anti-apoE (green). Cells cotransfected with DsRed2-Mito (red) and various apoE4 constructs mentioned above were permeabilized with 500 units of STP-O (b, d, f, h, and j) and stained with anti-apoE (green). The cells were then analyzed by confocal microscopy for only green (a, c, e, g, and i) or both red and green (b, d, f, h, and j).
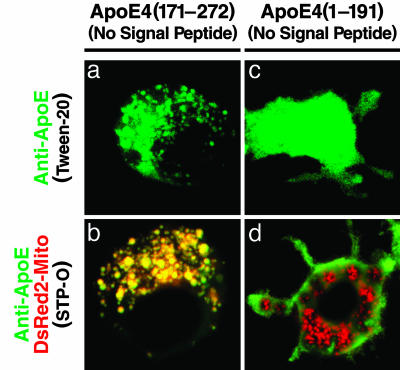
The Receptor-Binding Region Is Required to Escape the Secretory Pathway, and the Lipid-Binding Region Mediates Mitochondrial Interaction. To dissect the functions of the lipid- and receptor-binding regions, we assessed the effect of removing the N-terminal secretion signal peptide from fragments containing only one of the two regions. When expressed directly in the cytosol, apoE(171-272), containing only the lipid-binding region, interacted with the mitochondria (Fig. 4 a and b), although the same fragment with the signal peptide was retained in the secretory pathway and did not interact with the mitochondria (Fig. 3 c and d). Also, triple mutation of the receptor-binding region caused apoE4(1-272) with the signal peptide to be retained in the secretory pathway and, thus, no interaction with the mitochondria (Fig. 3 i and j). ApoE4(1-191), containing only the receptor-binding region, did not interact with the mitochondria, even when expressed directly in the cytosol (Fig. 4 c and d).
Fig. 4.
The receptor-binding region is required to escape the secretory pathway, and the lipid-binding region mediates mitochondrial interaction. Cells transfected with apoE(171-272) without signal peptide (a) or apoE4(1-191) without signal peptide (c) were permeabilized with 0.5% Tween 20 and stained with anti-apoE (green). Cells cotransfected with DsRed2-Mito (red) and either of those two apoE4 constructs were permeabilized with 500 units of STP-O (b and d) and stained with anti-apoE (green). The cells were analyzed as in Fig. 3.
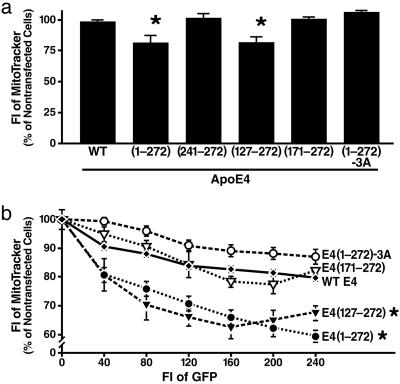
Lipid- and Receptor-Binding Regions Together Impair Mitochondrial Function and Integrity. To investigate the effect of apoE4 fragments on mitochondria, Neuro-2a cells transfected with various apoE4 constructs were incubated with MitoTracker Deep Red 633, and fluorescence intensity was analyzed by flow cytometry as a measure of mitochondrial function and integrity (53) (Fig. 5). Fluorescence intensity was 25% lower in cells expressing apoE4(1-272) or apoE(127-272) than in cells expressing full-length apoE4 (Fig. 5a). Because only functional mitochondria with a normal membrane potential can effectively take up and store MitoTracker Deep Red 633, this finding suggests that only apoE4 fragments with both the lipid- and receptor-binding regions can impair mitochondrial function and integrity. Importantly, this effect depended on the level of expression (Fig. 5b). Consistent with the immunocytochemical data, apoE4 fragments containing only one of the two regions and those fragments with the quadruple mutation in the lipid-binding region or the triple mutation in the receptor-binding region had no significant effect on mitochondrial function and integrity (Fig. 5).
Fig. 5.
The lipid- and receptor-binding regions in apoE4 fragments act in concert to cause mitochondrial dysfunction, as determined by MitoTracker Deep Red 633 staining and flow cytometry. (a) Effects of various forms of apoE4, expressed at similar levels, on mitochondrial function and integrity. (b) Effect on mitochondrial function and integrity depends on expression levels of apoE4 fragments, as measured by fluorescence intensity (FI) of GFP. Values are given as mean ± SD of three to six assays. *, P < 0.05 vs. WT apoE4, E4(171-272), and E4(1-272)-3A. E4(1-272)-3A, apoE4(1-272) with a triple mutation in the receptor-binding region.
Discussion
ApoE4 fragments found in cultured neuronal cells and in AD brains induce neurofibrillary tangle-like structures and cause neurotoxicity in vitro (25, 26, 55, 56) and neurodegeneration and behavioral deficits in transgenic mice (27, 28). This study demonstrates that both the lipid- and receptor-binding regions are required for neurotoxicity of apoE4 fragments in Neuro-2a cells and that, in addition to disrupting cytoskeletal structure and function (25, 27), the apoE4 fragments also interact with mitochondria and impair their function and integrity.
Our working model is that positively charged amino acids in the receptor-binding region enable apoE4 fragments to escape the secretory pathway and enter the cytosol, whereas the lipid-binding region mediates interactions with the mitochondria. When the secretion signal peptide was present, apoE(171-272), which contains only the lipid-binding region, could not escape the secretory pathway and did not interact with the mitochondria. However, when the signal peptide was removed and apoE(171-272) was directly expressed in the cytosol, it did interact with the mitochondria. Also, the triple mutation in the receptor-binding region caused apoE4(1-272) with the signal peptide to be retained in the secretory pathway, where it could not interact with the mitochondria. ApoE4(1-191), which contains only the receptor-binding region, did not interact with the mitochondria, even when expressed directly in the cytosol.
The receptor-binding region of apoE shares a feature [the enrichment of positively charged amino acids (arginine and lysine)] with the protein-translocation domain (PTD) of many viral proteins. PTD-containing proteins, such as HIV-1 Tat, penetrate the plasma membrane of cells in a receptor-independent, concentration-dependent fashion (57, 58). The Tat PTD, a short basic region of 10 aa, has been used as a carrier to deliver many peptides, proteins, and antisense oligodeoxynucleotides into cells (59-61). Likewise, the receptor-binding region of apoE has also been used to deliver antisense oligodeoxynucleotides into cells (62, 63), consistent with the membrane-penetrating ability that we observed in this study.
In vitro, the lipid-binding region is responsible for the interaction of apoE with Aβ peptides (9, 64), whereas the receptor-binding region is responsible for binding with tau (23). Thus, apoE4 fragments might also interact with Aβ or tau or both via two different regions and act synergistically to cause dysfunction of both the cytoskeleton and the mitochondria, leading to neuronal and behavioral deficits.
Mitochondrial dysfunction in AD (21, 65-67) varies with apoE genotype, being greater in apoE4 than in apoE3 carriers (19). ApoE4 is also associated with decreased cerebral glucose metabolism in both AD patients and nondemented subjects (68-71). Because normal cerebral glucose metabolism requires normal mitochondrial function, and because apoE4 fragments are found in AD brains (27), it is tempting to speculate that the impairment of mitochondrial function and integrity elicited by the expression of the truncated apoE4 in Neuro-2a cells relates to the mitochondrial dysfunction or damage observed in AD brains. Consequently, blocking the interaction of apoE4 fragments with the mitochondria is a potential strategy for inhibiting the detrimental effects of apoE4 in AD and other neurological diseases.
Acknowledgments
We thank Aubrey Bernardo for assistance in some cell-culture experiments; Drs. Karl Weisgraber, Lennart Mucke, and Luke Esposito for critical reading of the manuscript; Karina Fantillo and Sylvia Richmond for manuscript preparation; Stephen Ordway and Gary Howard for editorial assistance; John C. W. Carroll and Jack Hull for graphics; and Chris Goodfellow for photography. This work was supported in part by Program Project Grant P01 AG022074, National Institutes of Health Grant R01HL37063, and a research grant from GlaxoSmithKline.
Author contributions: S.C., R.W.M., and Y.H. designed research and wrote the paper; S.C., T.r.M., R.D.M., and M.E.B. performed research; and S.C., T.r.M., R.D.M., M.E.B., R.W.M., and Y.H. analyzed data.
Conflict of interest statement: No conflicts declared.
Abbreviations: Aβ, amyloid β; AD, Alzheimer's disease; apo, apolipoprotein; STP-O, Streptolysin-O.
References
- 1.Mahley, R. W. (1988) Science 240, 622-630. [DOI] [PubMed] [Google Scholar]
- 2.Mahley, R. W. & Huang, Y. (1999) Curr. Opin. Lipidol. 10, 207-217. [DOI] [PubMed] [Google Scholar]
- 3.Huang, Y. & Mahley, R. W. (1999) in Plasma Lipids and Their Role in Disease, eds. Barter, P. J. & Rye, K.-A. (Harwood, Amsterdam), pp. 257-284.
- 4.Mahley, R. W. & Rall, S. C., Jr., (2000) Annu. Rev. Genomics Hum. Genet. 1, 507-537. [DOI] [PubMed] [Google Scholar]
- 5.Strittmatter, W. J., Saunders, A. M., Schmechel, D., Pericak-Vance, M., Enghild, J., Salvesen, G. S. & Roses, A. D. (1993) Proc. Natl. Acad. Sci. USA 90, 1977-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roses, A. D. (1994) J. Neuropathol. Exp. Neurol. 53, 429-437. [DOI] [PubMed] [Google Scholar]
- 7.Corder, E. H., Saunders, A. M., Strittmatter, W. J., Schmechel, D. E., Gaskell, P. C., Small, G. W., Roses, A. D., Haines, J. L. & Pericak-Vance, M. A. (1993) Science 261, 921-923. [DOI] [PubMed] [Google Scholar]
- 8.Farrer, L. A., Cupples, L. A., Haines, J. L., Hyman, B., Kukull, W. A., Mayeux, R., Myers, R. H., Pericak-Vance, M. A., Risch, N. & Van Duijn, C. M. (1997) J. Am. Med. Assoc. 278, 1349-1356. [PubMed] [Google Scholar]
- 9.Strittmatter, W. J., Weisgraber, K. H., Huang, D. Y., Dong, L.-M., Salvesen, G. S., Pericak-Vance, M., Schmechel, D., Saunders, A. M., Goldgaber, D. & Roses, A. D. (1993) Proc. Natl. Acad. Sci. USA 90, 8098-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma, J., Yee, A., Brewer, H. B., Jr., Das, S. & Potter, H. (1994) Nature 372, 92-94. [DOI] [PubMed] [Google Scholar]
- 11.Wisniewski, T., Castaño, E. M., Golabek, A., Vogel, T. & Frangione, B. (1994) Am. J. Pathol. 145, 1030-1035. [PMC free article] [PubMed] [Google Scholar]
- 12.LaDu, M. J., Falduto, M. T., Manelli, A. M., Reardon, C. A., Getz, G. S. & Frail, D. E. (1994) J. Biol. Chem. 269, 23403-23406. [PubMed] [Google Scholar]
- 13.Holtzman, D. M., Bales, K. R., Tenkova, T., Fagan, A. M., Parsadanian, M., Sartorius, L. J., Mackey, B., Olney, J., McKeel, D., Wozniak, D. & Paul, S. M. (2000) Proc. Natl. Acad. Sci. USA 97, 2892-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bales, K. R., Verina, T., Cummins, D. J., Du, Y., Dodel, R. C., Saura, J., Fishman, C. E., DeLong, C. A., Piccardo, P., Petegnief, V., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 15233-15238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry, M. C., Cheung, B. S., Rebeck, G. W., Paul, S. M., Bales, K. R. & Hyman, B. T. (2000) Acta. Neuropathol. 100, 451-458. [DOI] [PubMed] [Google Scholar]
- 16.Buttini, M., Yu, G.-Q., Shockley, K., Huang, Y., Jones, B., Masliah, E., Mallory, M., Yeo, T., Longo, F. M. & Mucke, L. (2002) J. Neurosci. 22, 10539-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buttini, M., Orth, M., Bellosta, S., Akeefe, H., Pitas, R. E., Wyss-Coray, T., Mucke, L. & Mahley, R. W. (1999) J. Neurosci. 19, 4867-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyata, M. & Smith, J. D. (1996) Nat. Genet. 14, 55-61. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, G. E., Haroutunian, V., Zhang, H., Park, L. C. H., Shi, Q., Lesser, M., Mohs, R. C., Sheu, R. K.-F. & Blass, J. P. (2000) Ann. Neurol. 48, 297-303. [PubMed] [Google Scholar]
- 20.Ohta, S., Ohsawa, I., Kamino, K., Ando, F. & Shimokata, H. (2004) Ann. N.Y. Acad. Sci. 1011, 36-44. [DOI] [PubMed] [Google Scholar]
- 21.Kamino, K., Nagasaka, K., Imagawa, M., Yamamoto, H., Yoneda, H., Ueki, A., Kitamura, S., Namekata, K., Miki, T. & Ohta, S. (2000) Biochem. Biophys. Res. Commun. 273, 192-196. [DOI] [PubMed] [Google Scholar]
- 22.Herz, J. & Beffert, U. (2000) Nat. Rev. Neurosci. 1, 51-58. [DOI] [PubMed] [Google Scholar]
- 23.Strittmatter, W. J., Saunders, A. M., Goedert, M., Weisgraber, K. H., Dong, L.-M., Jakes, R., Huang, D. Y., Pericak-Vance, M., Schmechel, D. & Roses, A. D. (1994) Proc. Natl. Acad. Sci. USA 91, 11183-11186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tesseur, I., Van Dorpe, J., Spittaels, K., Van den Haute, C., Moechars, D. & Van Leuven, F. (2000) Am. J. Pathol. 156, 951-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Y., Liu, X. Q., Wyss-Coray, T., Brecht, W. J., Sanan, D. A. & Mahley, R. W. (2001) Proc. Natl. Acad. Sci. USA 98, 8838-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ljungberg, M. C., Dayanandan, R., Asuni, A., Rupniak, T. H., Anderton, B. H. & Lovestone, S. (2002) NeuroReport 13, 867-870. [DOI] [PubMed] [Google Scholar]
- 27.Harris, F. M., Brecht, W. J., Xu, Q., Tesseur, I., Kekonius, L., Wyss-Coray, T., Fish, J. D., Masliah, E., Hopkins, P. C., Scearce-Levie, K., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 10966-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brecht, W. J., Harris, F. M., Chang, S., Tesseur, I., Yu, G.-Q., Xu, Q., Fish, J. D., Wyss-Coray, T., Buttini, M., Mucke, L., Mahley, R. W. & Huang, Y. (2004) J. Neurosci. 24, 2527-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raber, J., Wong, D., Buttini, M., Orth, M., Bellosta, S., Pitas, R. E., Mahley, R. W. & Mucke, L. (1998) Proc. Natl. Acad. Sci. USA 95, 10914-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raber, J., Bongers, G., LeFevour, A., Buttini, M. & Mucke, L. (2002) J. Neurosci. 22, 5204-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji, Z.-S., Miranda, R. D., Newhouse, Y. M., Weisgraber, K. H., Huang, Y. & Mahley, R. W. (2002) J. Biol. Chem. 277, 21821-21828. [DOI] [PubMed] [Google Scholar]
- 32.Cataldo, A. M., Barnett, J. L., Pieroni, C. & Nixon, R. A. (1997) J. Neurosci. 17, 6142-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cataldo, A. M., Peterhoff, C. M., Troncoso, J. C., Gomez-Isla, T., Hyman, B. T. & Nixon, R. A. (2000) Am. J. Pathol. 157, 277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grbovic, O. M., Mathews, P. M., Jiang, Y., Schmidt, S. D., Dinakar, R., Summers-Terio, N. B., Ceresa, B. P., Nixon, R. A. & Cataldo, A. M. (2003) J. Biol. Chem. 278, 31261-31268. [DOI] [PubMed] [Google Scholar]
- 35.Beffert, U. & Poirier, J. (1996) Ann. N.Y. Acad. Sci. 777, 166-174. [DOI] [PubMed] [Google Scholar]
- 36.Beisiegel, U., Schneider, W. J., Goldstein, J. L., Anderson, R. G. W. & Brown, M. S. (1981) J. Biol. Chem. 256, 11923-11931. [PubMed] [Google Scholar]
- 37.Diedrich, J. F., Minnigan, H., Carp, R. I., Whitaker, J. N., Race, R., Frey, W., II & Haase, A. T. (1991) J. Virol. 65, 4759-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han, S.-H., Einstein, G., Weisgraber, K. H., Strittmatter, W. J., Saunders, A. M., Pericak-Vance, M., Roses, A. D. & Schmechel, D. E. (1994) J. Neuropathol. Exp. Neurol. 53, 535-544. [DOI] [PubMed] [Google Scholar]
- 39.Bao, F., Arai, H., Matsushita, S., Higuchi, S. & Sasaki, H. (1996) NeuroReport 7, 1733-1739. [DOI] [PubMed] [Google Scholar]
- 40.Metzger, R. E., LaDu, M. J., Pan, J. B., Getz, G. S., Frail, D. E. & Falduto, M. T. (1996) J. Neuropathol. Exp. Neurol. 55, 372-380. [DOI] [PubMed] [Google Scholar]
- 41.Xu, P.-T., Schmechel, D., Qiu, H.-L., Herbstreith, M., Rothrock-Christian, T., Eyster, M., Roses, A. D. & Gilbert, J. R. (1999) Neurobiol. Dis. 6, 63-75. [DOI] [PubMed] [Google Scholar]
- 42.Xu, P.-T., Gilbert, J. R., Qiu, H.-L., Ervin, J., Rothrock-Christian, T. R., Hulette, C. & Schmechel, D. E. (1999) Am. J. Pathol. 154, 601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, P.-T., Gilbert, J. R., Qiu, H.-L., Rothrock-Christian, T., Settles, D. L., Roses, A. D. & Schmechel, D. E. (1998) Neurosci. Lett. 246, 65-68. [DOI] [PubMed] [Google Scholar]
- 44.Xu, P.-T., Schmechel, D., Rothrock-Christian, T., Burkhart, D. S., Qiu, H.-L., Popko, B., Sullivan, P., Maeda, N., Saunders, A. M., Roses, A. D. & Gilbert, J. R. (1996) Neurobiol. Dis. 3, 229-245. [DOI] [PubMed] [Google Scholar]
- 45.Aoki, K., Uchihara, T., Sanjo, N., Nakamura, A., Ikeda, K., Tsuchiya, K. & Wakayama, Y. (2003) Stroke (Dallas) 34, 875-880. [DOI] [PubMed] [Google Scholar]
- 46.Dupont-Wallois, L., Soulié, C., Sergeant, N., Wavrant-de Wrieze, N., Chartier-Harlin, M.-C., Delacourte, A. & Caillet-Boudin, M.-L. (1997) Neurobiol. Dis. 4, 356-364. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira, S., Dupire, M.-J., Delacourte, A., Najib, J. & Caillet-Boudin, M.-L. (2000) Exp. Neurol. 166, 415-421. [DOI] [PubMed] [Google Scholar]
- 48.Harris, F. M., Tesseur, I., Brecht, W. J., Xu, Q., Mullendorff, K., Chang, S., Wyss-Coray, T., Mahley, R. W. & Huang, Y. (2004) J. Biol. Chem. 279, 3862-3868. [DOI] [PubMed] [Google Scholar]
- 49.Huang, Y., Weisgraber, K. H., Mucke, L. & Mahley, R. W. (2004) J. Mol. Neurosci. 23, 189-204. [DOI] [PubMed] [Google Scholar]
- 50.Narayanaswami, V. & Ryan, R. O. (2000) Biochim. Biophys. Acta 1483, 15-36. [DOI] [PubMed] [Google Scholar]
- 51.Du, X., Stoops, J. D., Mertz, J. R., Stanley, C. M. & Dixon, J. L. (1998) J. Cell Biol. 141, 585-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berridge, M. V. & Tan, A. S. (1993) Arch. Biochem. Biophys. 303, 474-482. [DOI] [PubMed] [Google Scholar]
- 53.Kalbácová, M., Vrbacky, M., Drahota, Z. & Melková, Z. (2003) Cytometry 52A, 110-116. [DOI] [PubMed] [Google Scholar]
- 54.Weisgraber, K. H. (1994) Adv. Protein Chem. 45, 249-302. [DOI] [PubMed] [Google Scholar]
- 55.Tolar, M., Marques, M. A., Harmony, J. A. K. & Crutcher, K. A. (1997) J. Neurosci. 17, 5678-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolar, M., Keller, J. N., Chan, S., Mattson, M. P., Marques, M. A. & Crutcher, K. A. (1999) J. Neurosci. 19, 7100-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frankel, A. D. & Pabo, C. O. (1988) Cell 55, 1189-1193. [DOI] [PubMed] [Google Scholar]
- 58.Green, M. & Loewenstein, P. M. (1988) Cell 55, 1179-1188. [DOI] [PubMed] [Google Scholar]
- 59.Schwarze, S. R., Ho, A., Vocero-Akbani, A. & Dowdy, S. F. (1999) Science 285, 1569-1572. [DOI] [PubMed] [Google Scholar]
- 60.Cao, G., Pei, W., Ge, H., Liang, Q., Luo, Y., Sharp, F. R., Lu, A., Ran, R., Graham, S. H. & Chen, J. (2002) J. Neurosci. 22, 5423-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fawell, S., Seery, J., Daikh, Y., Moore, C., Chen, L. L., Pepinsky, B. & Barsoum, J. (1994) Proc. Natl. Acad. Sci. USA 91, 664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miura, S.-I., Okamoto, T., Via, D. P. & Saku, K. (2002) Circ. J. 66, 1054-1056. [DOI] [PubMed] [Google Scholar]
- 63.Liu, K., Ou, J., Saku, K., Jimi, S., Via, D. P., Sparrow, J. T., Zhang, B., Pownall, H. J., Smith, L. C. & Arakawa, K. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 2207-2213. [DOI] [PubMed] [Google Scholar]
- 64.Cho, H. S., Hyman, B. T., Greenberg, S. M. & Rebeck, G. W. (2001) J. Neuropathol. Exp. Neurol. 60, 342-349. [DOI] [PubMed] [Google Scholar]
- 65.Ghosh, S. S., Swerdlow, R. H., Miller, S. W., Sheeman, B., Parker, W. D., Jr. & Davis, R. E. (1999) Ann. N.Y. Acad. Sci. 893, 176-191. [DOI] [PubMed] [Google Scholar]
- 66.Trimmer, P. A., Swerdlow, R. H., Parks, J. K., Keeney, P., Bennett, J. P., Jr., Miller, S. W., Davis, R. E. & Parker, W. D., Jr. (2000) Exp. Neurol. 162, 37-50. [DOI] [PubMed] [Google Scholar]
- 67.Hirai, K., Aliev, G., Nunomura, A., Fujioka, H., Russell, R. L., Atwood, C. S., Johnson, A. B., Kress, Y., Vinters, H. V., Tabaton, M., et al. (2001) J. Neurosci. 21, 3017-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Small, G. W., Mazziotta, J. C., Collins, M. T., Baxter, L. R., Phelps, M. E., Mandelkern, M. A., Kaplan, A., La Rue, A., Adamson, C. F. & Chang, L. (1995) J. Am. Med. Assoc. 273, 942-947. [PubMed] [Google Scholar]
- 69.Small, G. W., Ercoli, L. M., Silverman, D. H. S., Huang, S.-C., Komo, S., Bookheimer, S. Y., Lavretsky, H., Miller, K., Siddarth, P., Rasgon, N. L., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 6037-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reiman, E. M., Caselli, R. J., Chen, K., Alexander, G. E., Bandy, D. & Frost, J. (2001) Proc. Natl. Acad. Sci. USA 98, 3334-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reiman, E. M., Chen, K., Alexander, G. E., Caselli, R. J., Bandy, D., Osborne, D., Saunders, A. M. & Hardy, J. (2004) Proc. Natl. Acad. Sci. USA 101, 284-289. [DOI] [PMC free article] [PubMed] [Google Scholar]