Abstract
Topoisomerase II (Topo II) inhibitors are cell cycle–specific DNA-damaging agents and often correlate with secondary leukemia with chromosomal translocations involving the mixed-lineage leukemia/myeloid lymphoid leukemia (MLL) gene on chromosome 11 band q23 (11q23). In spite of the clinical importance, the molecular mechanism for this chromosomal translocation has yet to be elucidated. In this study, we employed 2-color FISH and detected intracellular chromosomal translocations induced by etoposide treatment. Cells such as ataxia-telangiectasia mutated–deficient fibroblasts and U2OS cells, in which the early G2/M checkpoint after treatment with low concentrations of etoposide has been lost, executed mitosis with etoposide-induced DNA double-strand breaks, and 2-color FISH signals located on either side of the MLL gene were segregated in the postmitotic G1 phase. Long-term culture of cells that had executed mitosis under etoposide treatment showed frequent structural abnormalities of chromosome 11. These findings provide convincing evidence for Topo II inhibitor–induced 11q23 translocation. Our study also suggests an important role of the early G2/M checkpoint in preventing fixation of chromosomal abnormalities and reveals environmental and genetic risk factors for the development of chromosome 11 translocations, namely, low concentrations of Topo II inhibitors and dysfunctional early G2/M checkpoint control.
Introduction
Interchromosomal translocations between 2 different chromosomes play critical roles in tumorigenesis by creating oncogenic fusion genes or deregulating gene expression. These cytogenetic aberrations are now considered to be of great importance as an initial step in tumorigenesis not only in hematological neoplasias but also in solid tumors (1). In spite of recent advances in our understanding of genomic instabilities such as microsatellite instability and chromosomal instability (2), little is known about the mechanism that underlies chromosomal structural aberrations.
The risk of malignancy reflects a complex interplay among the results of exposure to exogenous carcinogenic agents, inherited predisposition, and chance events. Topoisomerase II (Topo II) inhibitor–related secondary leukemia (3–6) and infant leukemia (7–9) are tumors significantly associated with chromosomal translocations caused by DNA-damaging agents. They are characterized by balanced chromosomal translocations involving the mixed-lineage leukemia/myeloid lymphoid leukemia (MLL) gene on chromosome 11 band q23 (11q23) (10). More than 30 different partner genes have been identified as involved in MLL gene translocations, and some MLL fusion genes reportedly have strong oncogenic effects (11–14). Topo II, which accumulates in a cell cycle–specific manner and peaks in the G2/M phase, forms a covalent linkage to both strands of the DNA helix, makes a transient double-strand break (DSB) in the helix, and re-ligates the cleaved DNA (15–19); it is essential for structural maintenance of genomic DNA in cell metabolism (17, 18). Agents with Topo II inhibitory activity, such as etoposide, doxorubicin, and mitoxantrone, maintain the DNA DSBs by stabilizing covalent Topo II–DNA complexes (15–17) and thereby exert their function as anticancer drugs. Previous studies using Southern hybridization showed that exposure to Topo II inhibitors induces DNA DSBs in the breakpoint cluster region (BCR) of the MLL gene that contains several Topo II affinity sites (20–22). These studies have suggested that Topo II inhibitors play an essential role in generating MLL gene rearrangement. Various substances contained in foods and the general environment are known to have Topo II inhibitory activity, and it has been hypothesized that maternal exposure to Topo II inhibitors is involved in the development of MLL-rearranged leukemia in infants (7–9).
In normal cells, the ataxia-telangiectasia mutated/ataxia-telangiectasia mutated and Rad3-related–regulated (ATM/ATR-regulated) DNA damage response network activates cell cycle checkpoints or apoptosis upon various endogenous or exogenous DNA damage and prevents cells from acquiring mutations (23, 24). Alternations of the components in this network increase the risk of cancer development (23, 24). Etoposide-induced DNA DSBs also activate this network during the S-to-G2 phase, such that cells accumulate in G2 (25–27) or more severely damaged cells undergo apoptosis (28). In cells arrested in the G2 phase, sister chromatids remain paired by cohesin (29), and cohesins are recruited to the damaged sites (30, 31). Thus, the undamaged sister chromatid tightly holds the etoposide-induced broken chromatid, supporting a coordinated repair of the DNA lesion. Additionally, Topo II inhibitors stabilize covalent Topo II–DNA complexes and keep the cleaved DNA ends adjacent to each other (17). In this context, Topo II inhibitor–induced DNA DSBs per se are not likely to produce chromosomal translocations, which require a spatial dynamic movement of chromosome segments generated by DNA DSBs (32). Based on these findings, we hypothesize that G2/M checkpoint failure due to a defect of the components in the DNA damage response could lead Topo II–induced DSBs to generate gross chromosomal translocations.
In the present study, we demonstrate that the failure of the early G2/M checkpoint to be activated by low concentrations of etoposide results in stable chromosomal aberrations, emphasizing a concomitant role of environmental and genetic risk factors for the development of chromosomal translocations.
Results
Segregation of 11q23 is not detected by 2-color FISH after etoposide treatment.
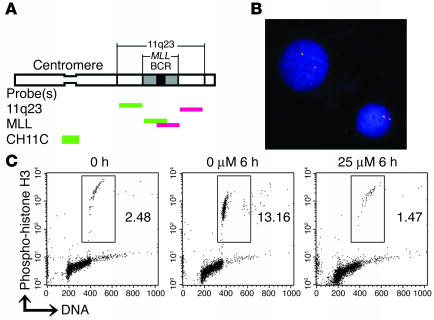
We first investigated whether 25 μM etoposide, which represents the mean plasma concentration in cancer patients during the first 6 hours after administration (22, 33), induced segregation of the centromeric and telomeric segments of 11q23 in BV173, a standard cell line commonly used for Topo II–DNA cleavage assays (20, 22). Cells were incubated with 25 μM etoposide for 6 hours and then hybridized with the 2-color FISH probes located on either side of the MLL gene (11q23 probes; Figure 1A). While previous studies indicated site-specific cleavage in the MLL gene using Southern hybridization (22), dynamic migration of the cleaved MLL gene in BV173 cells was minimal in FISH analyses (Figure 1B) (0.3% [solvent only] versus 0.7% [25 μM etoposide]; P = 0.34, Fisher’s exact test; n > 1,000 for each experiment).
Figure 1.
Broken chromosome ends caused by etoposide treatment are kept adjacent in BV173 cells. (A) Schematic diagram for the location of probes for FISH analysis. 11q23 probes are paired probes flanking centromeric (green) and telomeric (red) termini of the MLL gene. MLL probes are paired probes spanning the MLL gene with minimal overlap in the BCR. CH11C is specific for chromosome 11 centromere. (B) Representative image of BV173 cell line hybridized with the 11q23 probes. Cells were treated with 25 μM etoposide for 6 hours. Original magnification, ×600. (C) The percentages of M phase BV173 cells incubated with 0 or 25 μM of etoposide concomitantly with colcemid for 6 hours. M phase cells were simultaneously stained with anti–phospho-histone H3 antibody and PI and were determined by flow cytometry. Dots representing M phase cells are enclosed within the rectangles.
In order to understand our contradictory findings in the context of cell cycle control and dynamic chromosome movements, BV173 cells were tested for G2/M checkpoint against etoposide exposure. When BV173 cells were incubated with the mitotic inhibitor colcemid for 6 hours, about 10% of the cells entered and accumulated in M phase, as detected by phospho-histone H3 positivity. In contrast, no cells entered mitosis during simultaneous treatment with 25 μM etoposide and colcemid (Figure 1C), indicating that etoposide-induced DNA damage triggers substantial G2 arrest in BV173 cells.
ATM-deficient cells are defective in early G2/M checkpoint upon etoposide treatment.
The findings in BV173 cells (Figure 1C) led us to hypothesize that disruption of G2/M checkpoints may permit damaged cells to enter mitosis, subsequently leading to the segregation of 11q23 probe signals in M or postmitotic G1 phase. In order to harvest cells at G2, M, and postmitotic G1 phases in the presence of etoposide, adherent cells in which M phase cells are easily enriched by shake-off were employed, even though MLL rearrangements have usually been observed in nonadherent hematopoietic cells.
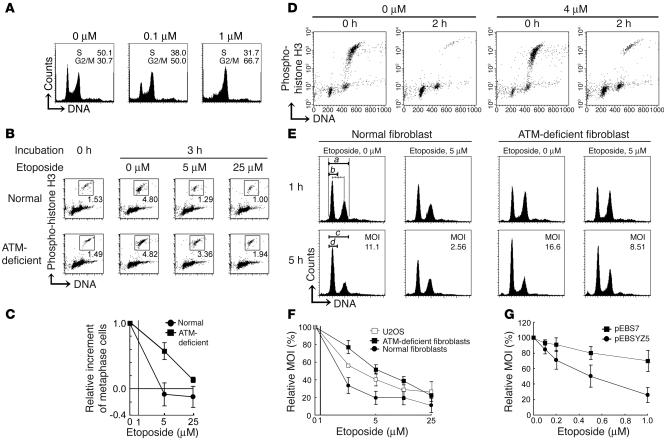
ATM-deficient cells are known to possess functional G2 and dysfunctional early G2/M checkpoints. ATM-deficient cells that have been damaged by ionizing radiation at S phase accumulate in the G2 phase, while they enter mitosis when DNA is damaged at the G2 phase (34). To confirm the cell cycle checkpoint activity on etoposide exposure, we compared the cell cycle distribution of SV-40–transformed ATM-deficient (GM05849C) and normal (GM00637Hmono1) skin fibroblasts in the presence of etoposide. In both lines, cells accumulated in the G2/M phase after treatment with 1 μM etoposide for 16 hours (Figure 2A and data not shown for normal fibroblasts). These data indicate that 1 μM etoposide apparently triggered the G2 accumulation checkpoint in response to DNA damage, irrespective of the presence or absence of ATM. To analyze the early response to etoposide treatment, cells were incubated with etoposide together with colcemid for up to 3 hours and then analyzed for the percentage of M phase cells. During the 3-hour incubation, about 3–5% of cells in either line entered and accumulated in M phase in the absence of etoposide (Figure 2B). When normal fibroblasts were exposed to 5 or 25 μM etoposide, the M phase population was significantly reduced (Figure 2, B and C). In contrast, a significant population of ATM-deficient fibroblasts entered mitosis under 5 μM or 25 μM etoposide treatment, though the latter was less prominent (Figure 2, B and C). These data indicate that ATM-deficient fibroblasts are defective in early G2/M checkpoint control in response to etoposide-induced DNA damage.
Figure 2.
ATM-deficient fibroblasts and U2OS cells execute mitosis in the presence of low-dose etoposide. (A) DNA content as analyzed in ATM-deficient fibroblasts after treatment with the indicated concentrations of etoposide for 16 hours. The percentage of cells at S and G2/M phases is indicated. (B and C) Percentages of M phase cells as analyzed before and after exposure to etoposide together with colcemid for 3 hours in normal and ATM-deficient fibroblasts. (B) Representative data analyzed by flow cytometry are shown. Dots representing M phase cells are enclosed within the squares. (C) The relative increment of the percentage of M phase cells under etoposide treatment was calculated against that under mock treatment. The abscissa is logarithmic (0 is shown separately). Data in the graph represent mean ± SD of 3 separate experiments. (D) Flow cytometric analysis for the M phase fraction of ATM-deficient fibroblasts. Cells were harvested soon after shake-off and incubated for an additional 2 hours with 0 or 4 μM etoposide. (E) The MOI, calculated as described in Methods. DNA content was analyzed in the BrdU-negative fraction of normal and ATM-deficient fibroblasts after 1- or 5-hour treatment. (F and G) The relative MOI in the presence of etoposide calculated against MOI in its absence. Cells were treated with the indicated concentrations of etoposide. Data represent mean ± SD of 3 separate experiments. (F) Relative MOI of normal ATM-deficient fibroblasts and U2OS cells. The abscissa is logarithmic (0 is shown separately). (G) Relative MOI of ATM-deficient fibroblasts with (pEBSYZ5) and without (pEBS7) ATM complementation.
ATM-deficient fibroblasts exit from M phase in the presence of etoposide.
There is some controversy as to whether DNA damage–associated mitotic arrest exists or not. We addressed this question using a mitotic arrest assay. M phase cells of ATM-deficient fibroblasts were enriched to 90% purity by shake-off after treatment with 0 or 4 μM of etoposide for 3 hours (indicated as 0 hours; Figure 2D). In the absence of etoposide, almost all M phase cells progressed to the postmitotic G1 phase during a subsequent 2 hours of incubation but, even under treatment with 4 μM etoposide, 60% also progressed to G1 (Figure 2D). These findings suggest that while mitotic exit is moderately delayed under treatment with 4 μM etoposide, a significant population of ATM-deficient fibroblasts overrides the M phase checkpoint in the presence of etoposide.
ATM-deficient fibroblasts execute mitosis in the presence of DNA DSBs induced by low concentrations of etoposide.
The above-described findings were validated by the M phase override assay, which enables direct analysis of whether G2 cells progress to the postmitotic G1 phase. Normal fibroblasts showed a relatively low M phase override index (MOI) under treatment with 5 μM etoposide compared with mock treatment (Figure 2E). This was in contrast to the MOI of etoposide-treated ATM-deficient fibroblasts, which was approximately half of that of the etoposide-free control. This difference was more prominent with low-dose etoposide and decreased with the high-dose etoposide (Figure 2F). The ATM dependence of these checkpoints was confirmed by experiments on the same cells with (GM05849-pEBSYZ5) or without (GM05849-pEBS7) ATM (Figure 2G). Thus, ATM-deficient fibroblasts could execute mitosis in the presence of low concentrations of etoposide due to a defect in DNA damage–induced early G2/M and M phase checkpoints.
Frequent segregation of etoposide-cleaved chromosome 11 in postmitotic G1 cells.
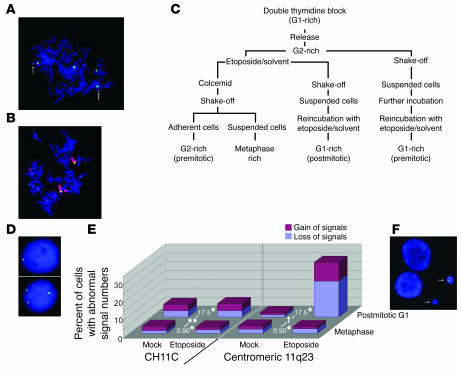
The background karyotypic features of the ATM-deficient fibroblasts were studied on metaphase spreads hybridized with a chromosome 11 centromere–specific probe (CH11C probe; Figure 1A) and a pair of probes spanning the MLL gene with minimal overlap in the BCR (MLL probes; Figure 1A). This cell line was found to have 4 copies of chromosome 11 as identified by CH11C signals, 2 of which have lost MLL signals (Figure 3A). 99.6% of MLL signals were associated with CH11C signals (n = 500), and all the centromeric and telomeric MLL signals were colocalized (n = 500). This cytogenetic feature was also confirmed by whole chromosome 11 painting together with hybridization with MLL probes (n = 500; Figure 3B). Having established these karyotypic characteristics, we extended our study on the ATM-deficient cells.
Figure 3.
FISH analysis for chromosome 11. (A) Representative image of metaphase ATM-deficient fibroblasts hybridized with the CH11C probe (green) and MLL probes (green and red overlap). Chromosomes were stained by DAPI. The arrows indicate overlapping MLL signals. (B) Representative image of metaphase ATM-deficient fibroblasts hybridized with probes for whole chromosome 11 (red) and MLL (green and red overlap). (C) Flow diagram of the cell fractionation procedure. (D) Etoposide-treated postmitotic G1 phase ATM-deficient fibroblasts with 1 (upper panel) and 3 (lower panel) centromeric 11q23 signals. (E) Proportion of metaphase and postmitotic G1 phase cells with gain (red) and loss (blue) of CH11C (left) or centromeric 11q23 (right) probe signals. n > 250 for each. *Odds ratio (OR) for etoposide treatment. **P = 0.77 and †P < 0.0001, differences in OR between postmitotic G1 and metaphase cells when cells were hybridized with CH11C and centromeric 11q23 probes, respectively (P for interaction term of cell cycle phase × treatment by a logistic regression model). (F) Micronuclei containing centromeric 11q23 signals (arrows). Cells were hybridized with centromeric 11q23 probes. Original magnification of FISH images, ×600.
G2, M, and postmitotic G1 phase cells of the ATM-deficient fibroblast were sequentially harvested by fractionation using double thymidine blockade and shake-off methods (Figure 3C and Methods). First, we counted the number of CH11C and centromeric 11q23 signals (Figure 1A) to confirm whether cells divided symmetrically when exposed to 2 μM etoposide. More than 96% of metaphase cells generated 4 CH11C signals, and parallel hybridization with the centromeric 11q23 probe revealed 2 replicated signals in 95% of metaphase cells either in the presence or absence of 2 μM etoposide, compatible with the original cytogenetic features. The majority of postmitotic G1 cells also showed 4 CH11C signals and 2 centromeric 11q23 signals in the absence of etoposide. These findings were in contrast to those in postmitotic G1 cells treated with etoposide, 30% of which manifested gain or loss of centromeric 11q23 signals (Figure 3D) in spite of the fact that 92% of these cells had 4 CH11C signals (Figure 3E). These observations indicate that etoposide induces DNA DSBs on chromosome 11 between the centromere and the centromeric terminus of band q23, and while the centromeric segments segregate symmetrically through mitosis, the telomeric segments result in asymmetrical distribution in daughter cells. When treated with 2 μM etoposide, micronuclei were occasionally seen in the postmitotic G1-rich fraction. Centromeric 11q23 signal lacking CH11C signal was observed in 8% of micronuclei (P < 0.0001, Fisher’s exact test; n = 250; Figure 3F). This finding is compatible with the interpretation that cleaved chromosomal fragments persist in M phase and telomeric segments of cleaved chromosomes distribute at random in micronuclei.
Splitting of the 11q23 signals in etoposide-treated postmitotic G1 cells.
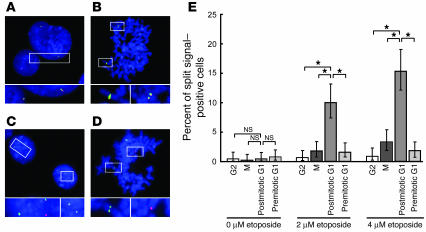
We next focused on the MLL gene locus of etoposide-treated ATM-deficient fibroblasts using 11q23 probes (Figure 1A). The cell cycle profile was determined based on the size of the nuclei, chromatin condensation, and probe signal localization (Figure 4, A and B). When ATM-deficient fibroblasts were cultured without etoposide, dissociation of centromeric (green) and telomeric (red) FISH signals was rarely seen in the G2 (Figure 4A), metaphase (Figure 4B), postmitotic G1, or premitotic G1 cells. When they were incubated with 2 or 4 μM etoposide, postmitotic G1 phase cells (Figure 4C) and metaphase cells (Figure 4D) showed frequent dissociation of the centromeric and telomeric 11q23 signals, though much less prominently in the latter (Table 1 and Figure 4E). In contrast, only a few cells remaining in the G2 or premitotic G1 phase during etoposide exposure showed split signals (Table 1). These results indicate that ATM-deficient fibroblasts acquire chromosomal segregation at 11q23 while undergoing cell cycle progression from the G2 to the postmitotic G1 phase in the presence of etoposide. Cleavage of the MLL gene in ATM-deficient fibroblasts was more directly confirmed in etoposide-treated postmitotic G1 cells using MLL probes (split signal–positive cells were present at 10.7% [2 μM] versus 2.4% [0 μM]; P < 0.0001, Fisher’s exact test; n > 300 cells for each; Figure 1A).
Figure 4.
Segregation of etoposide-cleaved DNA ends. (A and B) Mock-treated ATM-deficient fibroblasts were hybridized with centromeric (green) and telomeric (red) 11q23 probes. (A) G1 (a cell with small nucleus with 2 copies of 11q23 signals) and G2 (a cell with large nucleus with 2 sets of replicated 11q23 signals) phase cells. (B) A metaphase cell. (C and D) Segregation of 11q23 segments in 2 μM etoposide–treated ATM-deficient fibroblasts as identified by dissociation of centromeric and telomeric 11q23 signals. (C) Postmitotic G1 phase cells. (D) A metaphase cell. Original magnification of FISH images, ×600. The area enclosed by each rectangle is magnified in the bottom row. (E) Proportion of split 11q23 signal–positive cells in each fraction. All data from 3 independent experiments were combined for the analysis (n > 450 for each). Vertical lines indicate the 95% confidence intervals. Differences in the proportion of cells with dissociated signals were analyzed by Fisher’s exact test after adjustment for multiple comparisons by the Bonferroni-Holm method (3 comparisons at each drug concentration). *P < 0.0001.
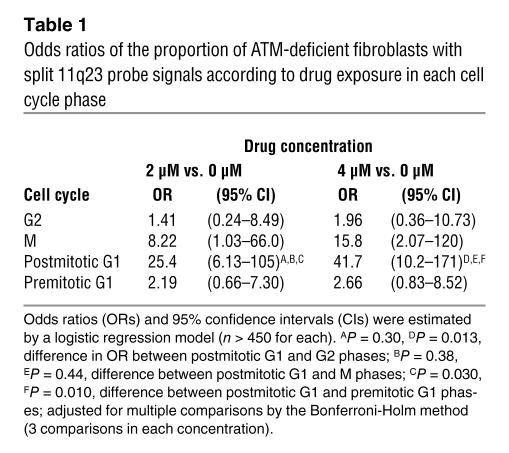
Table 1.
Odds ratios of the proportion of ATM-deficient fibroblasts with split 11q23 probe signals according to drug exposure in each cell cycle phase
The 11q23 locus also segregates in U2OS cells.
The U2OS osteosarcoma cell line has a high MOI under low-dose etoposide treatment in spite of normal function of ATM as analyzed by ionizing irradiation–induced phosphorylation of p53 Ser15 (35) (Figure 2F). Accordingly, U2OS cells were analyzed for split MLL probe signals in the same manner as were ATM-deficient cells (described above) and were found to have an increased frequency of split MLL signals in the etoposide-treated postmitotic G1–rich fraction (Table 2). These observations indicate that segregation of chromosomal breaks on 11q23 is in general attributable to the early G2/M checkpoint defect.
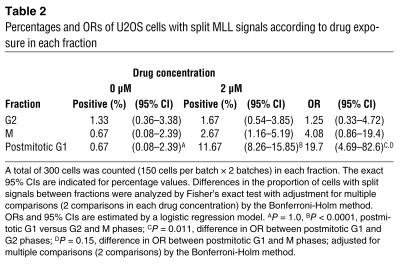
Table 2.
Percentages and ORs of U2OS cells with split MLL signals according to drug exposure in each fraction
DNA DSBs remaining unrepaired shortly after mitosis rejoin in the postmitotic G1 phase.
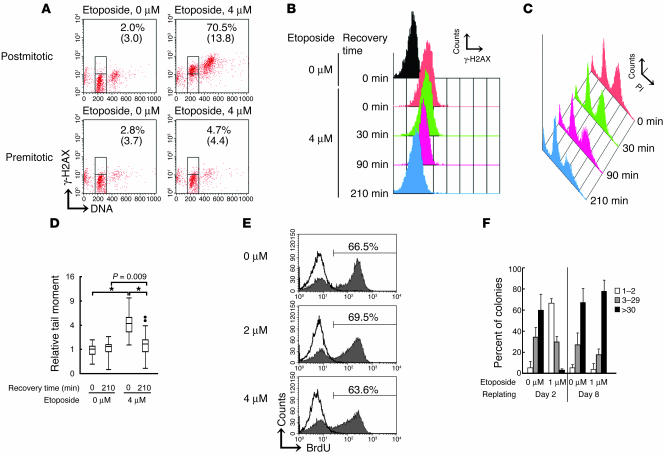
Phospho-histone H2AX (γ-H2AX) was sequentially monitored to determine the amount of DNA DSBs (36) in postmitotic G1 ATM-deficient fibroblasts. The M-rich fraction harvested following 4 μM etoposide treatment was reincubated with etoposide for 30 minutes to allow progression into the postmitotic G1 phase. They were then released from etoposide (Figure 5, B–D). Postmitotic G1 cells with 2N DNA content and cells remaining in mitosis with 4N DNA content stained strongly for γ-H2AX at time 0 (Figure 5A). In contrast, after 30 minutes etoposide treatment, cells previously synchronized at G1 (premitotic G1; Figure 3C) failed to increase γ-H2AX (Figure 5A). These data suggest that considerable amounts of DSBs in etoposide-treated postmitotic G1 cells were induced during the G2 and/or M phase and that these DSBs remained unrepaired shortly after mitosis. The high intensity of γ-H2AX staining in postmitotic G1 cells persisted for up to 30 minutes after removal of etoposide but started to decline from 90 minutes and reached the background level 210 minutes thereafter (Figure 5B). Reduced γ-H2AX staining was not due to death of the γ-H2AX–positive population, according to the results of subdiploid DNA content analysis for detection of apoptotic cells (Figure 5C). The neutral comet assay, another method for measuring cellular DNA DSBs, showed a significant comet tail moment in cells treated with 4 μM etoposide at time 0, but this disappeared by 210 minutes. (Figure 5D). These results are compatible with the interpretation that broken DNAs, including segregated chromosomes, are repaired while progressing through M to postmitotic G1 phases, leading to the formation of chromosomal translocations.
Figure 5.
DNA DSBs are repaired in the postmitotic G1 phase. (A) Flow cytometric analysis for γ-H2AX and DNA content. Cells were analyzed in mock- or 4 μM etoposide–treated postmitotic G1-rich and premitotic G1-rich fractions. The percentages of γ-H2AX–positive cells and geometric mean values of γ-H2AX intensity of G1 phase cells are indicated. (B) Sequential analysis of γ-H2AX intensity of postmitotic G1 cells. (C) DNA contents of the postmitotic G1-rich fraction at various recovery time points. Apoptotic cell population as identified by subdiploid DNA contents showed no significant difference. (D) Neutral comet assay for mock- or 4 μM etoposide–treated postmitotic G1-rich fraction at time 0 or 210 minutes after recovery. Relative tail moment (1 = the average of the tail moment of untreated cells at 0 minutes) was calculated, and data are shown in the box-and-whisker plot (n = 70 for each). Outliers are indicated as dots. The ordinate is a square-root scale. *P < 0.0001 for overall homogeneity by the Kruskal-Wallis test; P values are adjusted for multiple comparisons by the Bonferroni-Holm method. (E) BrdU incorporation of postmitotic G1 cells. Cells were pretreated with the indicated concentrations of etoposide. Filled histograms, cells incubated without blocking by thymidine; open histograms, cells incubated with blocking by excess thymidine. Percentages of BrdU-positive cells are indicated. (F) Distribution of colony-forming cells replated on days 2 and 8. One hundred randomly selected colonies were classified into 1 of 3 groups by the number of cells constituting the individual colonies. Percent of colonies composed of various cell numbers is shown on the y axis. The data represent mean ± SD of 3 separate experiments.
Postmitotic G1 cells have the ability to proliferate but are selected for stably growing clones.
Incorporation of BrdU by postmitotic G1 cells of ATM-deficient fibroblasts was measured at 24 hours. Flow cytometry showed that about 60% of the cells had incorporated BrdU whether or not they had been preincubated with etoposide (Figure 5E), and that this was blocked by addition of excess thymidine. These data indicate that postmitotic G1 ATM-deficient fibroblasts with etoposide-induced DNA DSBs nonetheless subsequently entered the cell cycle.
We next asked whether these postmitotic G1 cells could undertake multiple rounds of cell division. We replated the ATM-deficient fibroblasts 2 or 8 days after mitosis under 1 μM etoposide and then counted the number of cells constituting individual colonies in each assay. Cells replated on day 2 formed predominantly small colonies, more than 60% of which consisted of only a few cells (Figure 5F). This result is not consistent with data from the BrdU experiments described above, but rather indicates that the majority of etoposide-treated postmitotic G1 ATM-deficient fibroblasts either stop proliferating or die after only a couple of initial cell cycles. In contrast, cells replated on day 8 formed large colonies comparable to those subjected to mock treatment (Figure 5F). Together, these findings would suggest that many of the cells that acquire chromosomal translocations rapidly die or stop proliferating. This results in the selection of those clones with chromosomal abnormalities that have the advantage of stable cell growth.
Cells with fixed chromosomal translocations survive and grow.
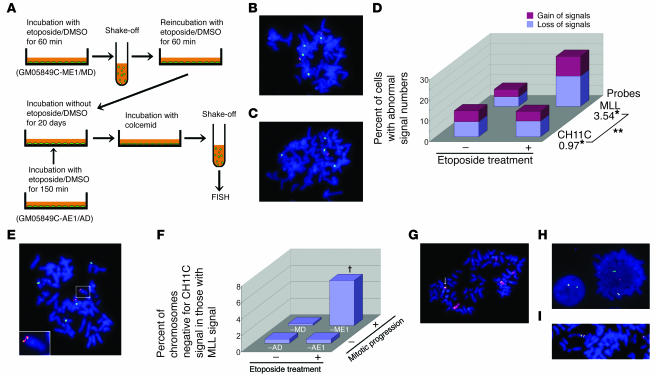
Postmitotic G1 phase ATM-deficient fibroblasts were collected after treatment with 1 μM etoposide (in the case of GM05849C-ME1 cells) or DMSO (in the case of GM05849C-MD cells) for 150 minutes. Viable cells were subsequently grown without etoposide for 20 days through several passages. Metaphase spreads of GM05849C-ME1 cells were hybridized with CH11C and MLL probes and the number of signals counted. Although the majority of GM05849C-ME1 cells yielded 4 CH11C signals as seen in GM05849C-MD cells (Figure 6, B and C), they showed frequent gain or loss of MLL signals (Figure 6, C and D). This was in contrast to GM05849C-MD cells, which showed almost no gain or loss of the MLL signals (Figure 6, B and D). The effect of etoposide treatment on the number of MLL signals but not CH11C signals was statistically significant (Figure 6D). These results were consistent with the analyses undertaken soon after mitosis in the presence of etoposide (Figure 3E). We next focused on the MLL signal–positive chromosomes and counted those with or without CH11C signals. Notably, 5.5% of MLL signal–positive chromosomes had lost CH11C signals (Figure 6, E and F) in GM05849C-ME1 cells. In contrast, GM05849C-MD and long-term-cultured cells (GM05849C-AE1 and-AD) that had been exposed to 1 μM etoposide or DMSO for 150 minutes under asynchronous conditions (Figure 6A) showed no loss of CH11C signals in MLL signal–positive chromosomes (Figure 6F). Similar results were obtained when cells were simultaneously hybridized with a chromosome 11 painting probe and with MLL probes. The translocation of chromosome 11 including the MLL locus to another chromosome (Figure 6G) was frequently observed in GM05849C-ME1 cells (2.4% versus 0.4% in GM05849C-MD cells; P = 0.004, Fisher’s exact test; n = 500 for each). Finally, metaphase spreads were hybridized with overlapping MLL probes to focus on the MLL gene rearrangement. MLL gene translocation (2/700; Figure 6H) and MLL tandem gene duplications (3/700; Figure 6I) were occasionally seen in GM05849C-ME1 but not in GM05849C-MD cells (0/700). These observations imply that a fraction of cells that acquired chromosomal translocations survived and could grow. In these selected cells, chromosome 11 abnormality was one of the prominent karyotypic features.
Figure 6.
Chromosomal aberrations in a mixture of stable clones of ATM-deficient fibroblasts, which had executed mitosis under etoposide treatment. (A) Flow diagram for long-term culture procedure. (B, C, and E) Metaphase spreads hybridized with the CH11C probe (green) and MLL probes (green and red overlap). (B) One GM05849C-MD cell showing 4 CH11C signals and 2 pairs of MLL signals. (C) One GM05849C-ME1 cell showing 4 CH11C signals with only 1 of them bearing MLL signals. (D) Proportion of cells with gain and loss of CH11C or MLL signals. n = 300 for each. *OR for etoposide treatment. **P = 0.0002, difference in OR for interaction term of probe × treatment by a logistic regression model. (E) One GM05849C-ME1 cell with the MLL gene translocated to another chromosome. The inset is a magnified image of the enclosed area. (F) Percent of chromosomes negative for CH11C signal among those positive for MLL signals. Data were analyzed by multiple logistic regression. n = 400 for each. †P = 0.028 for interaction term of etoposide treatment × mitotic progression. Neither of the pairwise comparisons for etoposide treatment in asynchronous cells nor for mitotic progression in etoposide-untreated cells was statistically significant (P = 1.0). (G) Representative image of chromosome 11 translocation (arrow) in GM05849C-ME1 cells hybridized with chromosome 11 painting (red) and MLL probes (green and red overlap). (H and I) Abnormal MLL gene configuration. GM05849C-ME1 cells were hybridized with MLL probes. Chromosomal translocation of MLL BCR (H) and tandem duplication of the MLL gene (I). Original magnification for FISH images, ×600.
Discussion
Taking advantage of cells with early G2/M checkpoint failure, we demonstrated that etoposide induced chromosomal translocations in cycling cells. In ATM-deficient fibroblasts, etoposide-induced DNA DSBs on chromosome 11 were visualized as chromosomal aberrations in the postmitotic G1 phase (Figures 3E and 4E and Table 1), but DNA DSBs on 11q23 were kept adjacent in the premitotic G1 and G2 cells (Figure 4E). A fraction of cells, which acquired chromosomal translocations during the G2 to postmitotic G1 phases, retained sufficient functional integrity for continued proliferation, which results in a selection of stable chromosome 11 translocations, as observed in long-term cultures (Figures 5F and 6). In light of the existence of 11q23 segments in micronuclei without centromeric regions as well as persistent DNA DSBs throughout the mitosis in the presence of etoposide, we favor for the idea that the chromosomal breaks persist during M phase and translocation is completed in postmitotic G1 phase. Our assay, however, does not exclude the possibility that some of the translocations have been generated during G2 as a concomitant rare event, and this will be an issue for future investigation.
The G2/M checkpoint prevents cells from initiating mitosis when they are damaged during G2 phase (early G2/M checkpoint) or when they progress into G2 with unrepaired damage inflicted during previous S or G1 phases (G2 accumulation). Of these 2 distinct G2/M checkpoints, ATM is reportedly responsible only for the early G2/M checkpoint after irradiation (34). Here, we demonstrated that ATM was also responsible for the early G2/M checkpoint following exposure to low-dose etoposide (Figure 2B). Although the early G2/M checkpoint is reportedly dose independent when triggered by ionizing radiation (34), it was clearly dose dependent for etoposide exposure (Figure 2, B, C, F, and G).
Our findings would predict that an inherent dysfunction of the cell cycle checkpoint is 1 of the conceivable risk factors in MLL-rearranged leukemia. Although familial clustering is not seen and no predisposing constitutional alleles have been identified in these cases (37), it was noted that 2 young children diagnosed with MLL-rearranged leukemia were reported to have a constitutional dysfunction of ATM, and 1 of them had a heterozygous germline missense mutation with dominant negative effect (35). The biological significance of germline missense changes of the ATM gene has also been demonstrated in breast cancer patients (38). In an animal model, mice heterozygous for the mutation, an in-frame deletion that was previously found to cause ataxia-telangiectasia in humans, are predisposed to various cancers (39). Although we mainly focused on ATM deficiency in this report, the analysis using the U2OS cell line (Table 2) implies that dysfunction of any other genes critical for the early G2/M checkpoint pathway could act as a predisposing condition for acquiring chromosome translocations.
In ATM-deficient cells, treatment with a high concentration of etoposide results in activation of the early G2/M checkpoint, and cells accumulate in G2 (Figure 2F). A large number of DNA DSBs caused by higher concentrations of etoposide may trigger additional DNA damage responses other than ATM, either leading to direct apoptosis or G2 arrest even in ATM-deficient cells. Based on these findings, a few chromosomal breaks induced by low-dose Topo II inhibitors could conceivably act as initiators of chromosomal translocations. This is consistent with the cytogenetic features of infant and secondary leukemias characterized by a reciprocal chromosomal translocation and are also supported by several epidemiological studies (7, 8). When Topo II inhibitors are used at high doses either intravenously or in the diet, it is still possible that cells are temporarily exposed to a lower concentration of the agent before or after the maximum concentration is achieved. Mitosis may take just an hour and could be completed during the period in which plasma concentration of Topo II inhibitor remains at a low level. Together with clinical data, where the cumulative dose of epipodophyllotoxin does not seem to be of primary importance in determining the risk of secondary leukemia (6), our findings are compatible with the interpretation that a limited window of Topo II inhibitor concentration triggers chromosomal translocations.
Our data also indicate that etoposide-induced chromosomal translocations are not restricted to the MLL locus and would be induced on any other loci on chromosome 11. This is not unexpected given the function of Topo II (17). Topo II inhibitors have been reported to cause secondary leukemia with chromosomal translocations involving genes other than MLL, such as t(8;21), t(15;17), t(9;22), t(11;20), and others (40–42). Recently, mitoxantrone-related acute promyelocytic leukemia with the t(15;17) translocation was reported to be 1 of the most frequent secondary cancers that arise after breast cancer treatment (43–45). In vitro Topo II–DNA cleavage assays also revealed that mitoxantrone cleaves the PML gene at the cryptic Topo II–sensitive site (46). It is not known how the specificity of chromosomal translocations is determined, but it would involve a complex interplay among types of Topo II inhibitor, additional anticancer drugs, or environmental stress and degree of active cycling of tissue-specific target cells. In cells with acquired chromosomal abnormalities, the clone(s) with a growth advantage will be selected depending on the translocation and fusion of the critical genes transcribed in tissue- and differentiation-specific manners. Cells thus selected would eventually progress to malignant tumors by acquiring additional critical mutations. In this context it should be noted that the hematopoietic system involves cycling precursor cells where the MLL gene is actively transcribed, which underscores a pivotal role of MLL translocation for leukemogenesis (11–14).
Together, our findings emphasize the important role of the G2/M checkpoints in preventing chromosome aberrations and predict a possible intrinsic genetic background predisposing to etoposide-induced chromosomal structural abnormalities. Further genetic analysis of patients with infant leukemia and secondary leukemia should provide new insights into how and why these leukemias arise under the complex interplay of host and environmental risk factors.
Methods
Cells.
GM00637Hmono1 cells (normal fibroblasts) were subcloned from GM00637H. GM05849C cells (ATM-deficient fibroblasts) and GM00637H cells were obtained from Coriell Cell Repositories (Coriell Institute for Medical Research). The osteosarcoma cell line U2OS was obtained from the Health Science Research Resources Bank. GM05849C, GM00637Hmono1, and U2OS cells were cultured in DMEM supplemented with 10% FBS. Philadelphia chromosome–positive acute leukemia cell line BV173 was cultured in RPMI supplemented with 10% FBS. GM05849-pEBYZ5 and GM05849-pEBS7 cells were obtained as reported previously (47).
G2/M accumulation assay, early G2/M checkpoint assay, mitotic arrest assay, and M phase override assay.
For G2/M accumulation assays, cells were exposed to various concentrations of etoposide (Sigma-Aldrich) for 16 hours. Cells were fixed with 70% ethanol and treated with RNase (QIAGEN) and propidium iodide (PI) (Sigma-Aldrich), and then more than 10,000 cells were analyzed by flow cytometry (BD). Cell cycle distribution was analyzed using ModFit LT (BD). For the early G2/M checkpoint assay, cells were incubated with different concentrations of etoposide in the presence of 25 ng/ml colcemid (Sigma-Aldrich) for 3 hours. For the mitotic arrest assay, M phase cells were purified by shake-off after treatment with etoposide or DMSO (Sigma-Aldrich) for 3 hours and reincubated on collagen-coated dishes for 2 hours with or without etoposide, respectively. The cells were harvested and fixed with 0.34% formaldehyde and then permeabilized with 90% methanol. Cells were stained with phospho-histone H3 monoclonal antibody (Cell Signaling Technology), followed by FITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories Inc.) and PI staining, and then more than 10,000 cells were analyzed by flow cytometry. For the M phase override assay, the cells were synchronized at the G1/S border by double thymidine block using 2.5 mM thymidine (MP Biomedicals LLC) and then released. At the time point when the cells accumulated at late S/G2 phase, they were labeled with 10 μM BrdU (BD Biosciences — Pharmingen) for 60 minutes and then washed and incubated with different concentrations of etoposide for 1 or 5 hours. At the indicated time points, the cells were harvested, fixed, and stained with FITC-conjugated anti-BrdU antibody (BD Biosciences — Pharmingen) after incubation with DNase I (BD Biosciences — Pharmingen). Cells were stained with 7-amino-actinomycin D (7-AAD) (BD Biosciences — Pharmingen), and BrdU-negative cells were subjected to cell cycle analysis by FACScan. To avoid counting postmitotic G1 fraction twice, because mitotic cells are doubled by undergoing mitosis, we calculated MOI as follows. Algebras (a, b, c, d) are indicated in Figure 2E (a, percentage of G1, S, and G2/M phase cells at 1 hour of treatment; b, percentage of G1 and early S phase cells at 1 hour of treatment; c, percentage of G1, S, and G2/M phase cells at 5 hours of treatment; d, percentage of G1 and early S phase at 5 hours of treatment). The right-hand borders of b and d correspond to the average of the mean values of 7-AAD intensity of 2N and 4N cells. On the assumption that X (= MOI) of total cells after 1 hour of treatment underwent mitosis during treatment (b + 2X) : (a + 2X – X) = d : c. MOI is calculated as X = (da – bc)/(2c – d). The relative MOI percentage was calculated against the MOI of the etoposide-free control as percent of control MOI.
Fractionation of cells.
The fractionation procedure flow chart is shown in Figure 3C. The cells were synchronized by thymidine block and then released. At the time point when the cells accumulated at the G2/M phase, they were exposed to different concentrations of etoposide. Cells that had entered the M phase shortly after etoposide exposure were removed by shake-off. The remaining cells were placed in fresh medium with the same concentration of etoposide. After etoposide treatment for 3 hours, together with colcemid for the last hour, the metaphase-rich fraction was collected by shake-off, enabling us to harvest cells overriding the early G2/M checkpoint. The adherent cells remaining on the flasks were harvested as the G2-rich fraction. To harvest postmitotic G1 cells, M phase cells that had overridden the early G2/M checkpoint were collected by shake-off after exposure to different concentrations of etoposide without colcemid for 3 hours and then reincubated on collagen-coated dishes with the same concentration of etoposide. The postmitotic G1 cells were harvested after an additional 2-hour reincubation. For the premitotic G1-rich fraction, the M phase cells obtained by shake-off without etoposide were incubated again to allow them to proceed to G1.
Generation of long-term cultured cells having passed through mitosis under etoposide treatment.
GM05849C cells were synchronized at G2 and exposed to 0 or 1 μM etoposide. After 60 minutes, M phase cells were collected by shake-off and then reincubated on collagen-coated dishes. After 90 minutes, cells were released from etoposide and cultured for 20 days without etoposide (several passages). As controls, GM05849C cells were treated with 0 or 1 μM etoposide under asynchronous condition for 150 minutes, then released and grown without etoposide for 20 days.
FISH and statistics.
Bacterial artificial chromosome probes (RP11-30E1, RP11-59N1) flanking centromeric and telomeric termini of the MLL gene (11q23 probes) were isolated based on the database provided by the University of California–Santa Cruz (Human [Homo sapiens] Genome Browser Gateway; http://genome.ucsc.edu/cgi-bin/hgGateway). Each probe was labeled by nick-translation with biotin-16-dUTP or digoxigenin-11-dUTP (Roche Diagnostics Corp.) and used for hybridization. Specific signals for biotin- and digoxigenin-labeled probes were detected with FITC-avidin (Roche Diagnostics Corp.) and anti–digoxigenin-rhodamine (Roche Diagnostics Corp.), respectively (48). CH11C probes, specific for chromosome 11 centromere, labeled with SpectrumGreen (CH11C probe), paired probes spanning the MLL gene with minimal overlap in the BCR (MLL probes), and a whole chromosome painting probe specific for chromosome 11 were obtained from VYSIS and processed according to the protocols provided by the manufacturer. Images were acquired on a Nikon Optiphot-2 microscope at ×600 magnification with a Photometrics CH250 camera. For counting split signals in ATM-deficient fibroblasts, at least 150 cells that did not show gain or loss of signals were examined at each cell cycle stage for each fraction in each experiment (165 cells on average). Data were obtained from 3 separate experiments. Since the frequencies of split signals in some groups were very sparse and the proportion of cells with split signals among the 3 experimental batches was not significantly different (P = 0.16, by Cochran-Mantel-Haenszel test, after controlling for the drug concentration levels and cell cycles), all data from these 3 independent experiments were combined for the analysis. Data were obtained from 3 separate experiments and the combined data were analyzed for statistical significance using Fisher’s exact test, after adjustment for multiple comparisons using the Bonferroni-Holm method (3 comparisons at each drug concentration). Differences were considered statistically significant at P < 0.05. Odds ratios and 95% confidence intervals were estimated by a logistic regression model. U2OS cells occasionally have 3 MLL locus signals in normal culture as determined by MLL probes. Thus, determining the cell cycle phase of each U2OS cell was impossible. For this reason, the 150 U2OS cells were counted for split signals of MLL in fractions. Two independent experiments were combined for statistical analysis for the reason mentioned above. Statistical significance of the differences among the proportions of cells with split signals between fractions was analyzed using Fisher’s exact test, after adjustment for multiple comparisons using the Bonferroni-Holm method. Differences were considered statistically significant at P < 0.05. Odds ratios and 95% confidence intervals were estimated by a logistic regression model.
γ-H2AX analysis. M phase cells were harvested after exposure to 4 μM etoposide for 3 hours and then reincubated with the same concentration of etoposide for 30 minutes. They were subsequently incubated in etoposide-free media for 0, 30, 90, or 120 minutes and processed for flow cytometric analysis. The cells were stained with FITC-conjugated anti–γ-H2AX antibodies (Upstate) according to the protocol provided by the manufacturer, followed by RNaseA treatment and PI staining. More than 10,000 cells were analyzed by flow cytometry.
Neutral comet assay.
Cells were processed using Comet Assay kits (Trevigen Inc.) according to the protocols provided by the manufacturer. Slides were stained with PI (2.5 μg/ml), and comets were analyzed using an inverted epifluorescent microscope at ×20 magnification. Finally, 70 cells were analyzed per slide using Scion Image (Scion Corp.) with the comet assay macro scion_comet1.3 (49) for tail moment.
BrdU incorporation assay.
Postmitotic G1 ATM-deficient fibroblasts were incubated with 10 μM BrdU with or without 2.5 mM thymidine for 24 hours. Cells were harvested and processed using BrdU Flow kits (BD Biosciences — Pharmingen) according to the protocol provided by the manufacturer. More than 10,000 cells were analyzed by flow cytometry.
Colony component enumerating assay.
Postmitotic G1 ATM-deficient fibroblasts pretreated with or without 1 μM etoposide were further cultured in etoposide-free medium. After 2 or 8 days, cells were harvested and 1 × 104 cells were replated on collagen-coated dishes. After 7 days, cells were subjected to Giemsa staining. Numbers of cells in 100 randomly selected colonies were determined under microscopic observation at magnification ×100 and then classified into 3 categories, colonies composed of 1 or 2 cells, 3–29 cells, or 30 or more cells.
Acknowledgments
We are grateful to Y. Shiloh for the kind gift of pEBSYZ5. We thank L. Wiedemann for the critical comments and discussion. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, and Culture, Japan, and by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare, Japan.
Footnotes
Nonstandard abbreviations used: ATM, ataxia-telangiectasia mutated; BCR, breakpoint cluster region; CH11C probe, chromosome 11 centromere–specific probe; DSB, double-strand break; γ-H2AX, phospho-histone H2AX; MLL, mixed-lineage leukemia/myeloid lymphoid leukemia; MOI, M phase override index; PI, propidium iodide; 11q23, chromosome 11 band q23; Topo II, topoisomerase II.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 2.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, et al. Secondary acute myeloid leukemia in children treated for acute lymphoid leukemia. N. Engl. J. Med. 1989;321:136–142. doi: 10.1056/NEJM198907203210302. [DOI] [PubMed] [Google Scholar]
- 4.Felix CA, et al. Common region of ALL-1 gene disrupted in epipodophyllotoxin-related secondary acute myeloid leukemia. Cancer Res. 1993;53:2954–2956. [PubMed] [Google Scholar]
- 5.Kumar L. Epipodophyllotoxins and secondary leukaemia. Lancet. 1993;342:819–820. doi: 10.1016/0140-6736(93)92692-m. [DOI] [PubMed] [Google Scholar]
- 6.Smith MA, et al. Secondary leukemia or myelodysplastic syndrome after treatment with epipodophyllotoxins. J. Clin. Oncol. 1999;17:569–577. doi: 10.1200/JCO.1999.17.2.569. [DOI] [PubMed] [Google Scholar]
- 7.Ross JA, Potter JD, Reaman GH, Pendergrass TW, Robison LL. Maternal exposure to potential inhibitors of DNA topoisomerase II and infant leukemia (United States): a report from the Children’s Cancer Group. Cancer Causes Control. 1996;7:581–590. doi: 10.1007/BF00051700. [DOI] [PubMed] [Google Scholar]
- 8.Alexander FE, et al. Transplacental chemical exposure and risk of infant leukemia with MLL gene fusion. Cancer Res. 2001;61:2542–2546. [PubMed] [Google Scholar]
- 9.Felix CA, Lange BJ. Leukemia in infants. Oncologist. 1999;4:225–240. [PubMed] [Google Scholar]
- 10.Pui CH, Relling MV. Topoisomerase II inhibitor-related acute myeloid leukaemia. Br. J. Haematol. 2000;109:13–23. doi: 10.1046/j.1365-2141.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 11.Corral J, et al. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 12.Dobson CL, et al. The Mll-AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J. 1999;18:3564–3574. doi: 10.1093/emboj/18.13.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster A, et al. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer. Cancer Cell. 2003;3:449–458. doi: 10.1016/s1535-6108(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 14.So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen GL, et al. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J. Biol. Chem. 1984;259:13560–13566. [PubMed] [Google Scholar]
- 16.Burden DA, et al. Topoisomerase II.etoposide interactions direct the formation of drug-induced enzyme-DNA cleavage complexes. J. Biol. Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 17.Kellner U, Sehested M, Jensen PB, Gieseler F, Rudolph P. Culprit and victim - DNA topoisomerase II. Lancet Oncol. 2002;3:235–243. doi: 10.1016/s1470-2045(02)00715-5. [DOI] [PubMed] [Google Scholar]
- 18.Berger JM. Type II DNA topoisomerases. Curr. Opin. Struct. Biol. 1998;8:26–32. doi: 10.1016/s0959-440x(98)80006-7. [DOI] [PubMed] [Google Scholar]
- 19.Heck MM, Hittelman WN, Earnshaw WC. Differential expression of DNA topoisomerase I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broeker PL, et al. Distribution of 11q23 breakpoints within the MLL breakpoint cluster region in de novo acute leukemia and in treatment-related acute myeloid leukemia: correlation with scaffold attachment regions and topoisomerase II consensus binding sites. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 21.Aplan PD, Chervinsky DS, Stanulla M, Burhans WC. Site-specific DNA cleavage within the MLL breakpoint cluster region induced by topoisomerase II inhibitors. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 22.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4790–4795. doi: 10.1073/pnas.070061297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 24.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 25.Grieder A, Maurer R, Stähelin H. Effect of an epipodophyllotoxin derivative (VP 16-213) on macromolecular synthesis and mitosis in mastocytoma cells. in vitro. Cancer Res. 1974;34:1788–1793. [PubMed] [Google Scholar]
- 26.Krishan A, Paika K, Frei E., III Cytofluorometric studies on the action of podophyllotoxin and epipodophyllotoxins (VM-26, VP-16-213) on the cell cycle traverse of human lymphoblasts. J. Cell Biol. 1975;66:521–530. doi: 10.1083/jcb.66.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow KC, Ross WE. Topoisomerase-specific drug sensitivity in relation to cell cycle progression. Mol. Cell. Biol. 1987;7:3119–3123. doi: 10.1128/mcb.7.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sordet WO, Khan QA, Kohn KW, Pommier Y. Apoptosis induced by topoisomerase inhibitors. Curr. Med. Chem. Anti-Canc. Agents. 2003;3:271–290. doi: 10.2174/1568011033482378. [DOI] [PubMed] [Google Scholar]
- 29.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 30.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 31.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 32.Aten JA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 33.Relling MV, et al. Etoposide and antimetabolite pharmacology in patients who develop secondary acute myeloid leukemia. Leukemia. 1998;12:346–352. doi: 10.1038/sj.leu.2400928. [DOI] [PubMed] [Google Scholar]
- 34.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 2002;22:1049–1059. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oguchi K, et al. Missense mutation and defective function of ATM in a childhood acute leukemia patient with MLL gene rearrangement. Blood. 2003;101:3622–3627. doi: 10.1182/blood-2002-02-0570. [DOI] [PubMed] [Google Scholar]
- 36.Rothkamm K, Löbrich M. Evidence for lack of DNA double-strand break repair in human cells exposed to very low X-ray doses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. [PubMed] [Google Scholar]
- 38.Scott SP, et al. Missense mutations but not allelic variants alter the function of ATM by dominant interference in patients with breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2002;99:925–930. doi: 10.1073/pnas.012329699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spring K, et al. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat. Genet. 2002;32:185–190. doi: 10.1038/ng958. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen-Bjergaard J, Andersen MK, Christiansen DH, Nerlov C. Genetic pathways in therapy-related myelodysplasia and acute myeloid leukemia. Blood. 2002;99:1909–1912. doi: 10.1182/blood.v99.6.1909. [DOI] [PubMed] [Google Scholar]
- 41.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Med. Pediatr. Oncol. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 42.Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: overview report. Genes Chromosomes Cancer. 2002;33:331–345. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- 43.Beaumont M, et al. Therapy-related acute promyelocytic leukemia. J. Clin. Oncol. 2003;21:2123–2137. doi: 10.1200/JCO.2003.09.072. [DOI] [PubMed] [Google Scholar]
- 44.Andersen MK, et al. Balanced chromosome abnormalities inv(16) and t(15;17) in therapy-related myelodysplastic syndromes and acute leukemia: report from an international workshop. Genes Chromosomes Cancer. 2002;33:395–400. doi: 10.1002/gcc.10043. [DOI] [PubMed] [Google Scholar]
- 45.Carli PM, et al. Increase therapy-related leukemia secondary to breast cancer. Leukemia. 2000;14:1014–1017. doi: 10.1038/sj.leu.2401787. [DOI] [PubMed] [Google Scholar]
- 46.Mistry AR, et al. DNA topoisomerase II in therapy-related acute promyelocytic leukemia. N. Engl. J. Med. 2005;352:1529–1538. doi: 10.1056/NEJMoa042715. [DOI] [PubMed] [Google Scholar]
- 47.Takagi M, et al. Identification and characterization of polymorphic variations of the ataxia telangiectasia mutated (ATM) gene in childhood Hodgkin disease. Blood. 2004;103:283–290. doi: 10.1182/blood-2003-01-0094. [DOI] [PubMed] [Google Scholar]
- 48.Nakakuki K, et al. Novel targets for the 18p11.3 amplification frequently observed in esophageal squamous cell carcinomas. Carcinogenesis. 2002;23:19–24. doi: 10.1093/carcin/23.1.19. [DOI] [PubMed] [Google Scholar]
- 49.Helma C, Uhl M. A public domain image-analysis program for the single-cell gel-electrophoresis (comet) assay. Mutat. Res. 2000;466:9–15. doi: 10.1016/s1383-5718(99)00232-6. [DOI] [PubMed] [Google Scholar]