Abstract
The complex gene expression mechanisms that occur in plant mitochondria, such as RNA editing and splicing, are not yet well understood. RNA editing in higher plant mitochondria is a highly specific process which modifies mRNA sequences by C-to-U conversions. It has been suggested that in some cases this process is required for splicing. Here, we use an experimental model based on the introduction of DNA into isolated mitochondria by electroporation to study organellar gene expression events. Our aim was to compare processing and editing of potato small ribosomal protein 10 gene (rps10) transcripts in heterologous (wheat mitochondria) and homologous (potato mitochondria) contexts. rps10 is a suitable model because it contains a group II intron, is absent in wheat mitochondria but is actively expressed in potato mitochondria, where transcripts are spliced and undergo five C-to-U editing events. For this purpose, conditions for electroporating isolated potato mitochondria were established. rps10 was placed under the control of either potato or wheat cox2 promoters. We found that rps10 was only transcribed under the control of a cognate promoter. In wheat mitochondria, rps10 transcripts were neither spliced nor edited while they are correctly processed in potato mitochondria. Interestingly, a wheat editing site grafted into rps10 was not recognized by wheat mitochondria but was correctly edited in potato mitochondria. Taken together, these results suggest that editing might occur only when the transcripts are engaged in processing and that they would not be available to editing factors outside of a putative RNA maturation machinery complex.
INTRODUCTION
Gene expression in plant mitochondria is a complex process involving multiple steps such as transcription, cis- and trans-splicing, RNA trimming and RNA editing on the way to translation (1). RNA editing has been found in a variety of organisms and occurs through different mechanisms such as insertion or deletion of nucleotides, or base conversions [for details see reference (2)]. In higher plant organelles, RNA editing is an important post-transcriptional event characterized by C-to-U changes via a deamination mechanism (3–5). In Arabidopsis thaliana 456 C-to-U editing events have been described (6). RNA editing occurs mostly in the coding regions which alter the identity of the encoded amino acid, but some editing events occur in highly structured regions of introns (7,8). Like other maturation processes, RNA editing is an essential post-transcriptional event in plant mitochondrial gene expression (9–11) required for the synthesis of functional proteins (12,13).
The introduction of foreign DNA into isolated mitochondria is a novel experimental model which has provided important information on RNA editing and RNA splicing enabling the use of a site-directed mutagenesis approach (14,15). Using a cognate cox2 chimeric gene construct, the cis-recognition elements required for plant mitochondria RNA editing have been determined (16,17). Using the same approach, Staudinger and Kempken (18) have reported that transcripts from A.thaliana cox2, but not Sorghum bicolor atp6, are edited when the genes are introduced into maize mitochondria.
Interestingly, higher plant mitochondrial genomes differ in their gene contents due to an evolutionary information transfer from the organelle to the nucleus (19). Of particular interest is the situation of the small ribosomal protein 10 gene (rps10), a group II intron-bearing gene which is encoded in Solanum tuberosum mitochondrial DNA but is absent from the wheat mitochondrial genome (20). Higher plant mitochondria contain group II introns either in cis- or trans- configuration (1) which can be folded in a characteristic secondary structure (21). Intron removal in plant mitochondrial mRNAs is not well documented because such introns are unable to self-splice. Previously, we described that electroporated cox2 constructs were a good model for the study of the splicing process. Using this model, we found that editing and splicing of cox2 transcripts were not linked in wheat mitochondria (15).
To challenge the ability of mitochondrial gene expression machinery to recognize genetic information which has been lost during evolution, we decided to introduce the S.tuberosum non-cognate rps10 gene into wheat mitochondria. Five C residues are changed to U in potato mitochondria rps10 transcripts by editing. Two of them have been postulated as being necessary for acquisition of a proper secondary and/or tertiary structure for splicing. To test this hypothesis, it was necessary to set up the conditions for electroporation of foreign DNA into S.tuberosum mitochondria. Here, we show that a potato rps10 construct is transcribed when introduced into wheat mitochondria, but transcripts are not recognized by the post-transcriptional processing machinery. In contrast, a rps10 construct is correctly expressed and processed in cognate potato mitochondria. Moreover, we present evidence that transcript editing might be linked to overall RNA processing. This is the first report on DNA electroporation into potato mitochondria.
MATERIALS AND METHODS
Plasmids
All plasmids used in this study are based on the previously described pCOXII vector (14). An NsiI restriction site was introduced at the initiation codon of the wheat cox2 open reading frame (ORF). Then, NsiI was used in combination with a SpeI restriction site present in the original vector after the stop codon, to produce the chimeric vectors.
S.tuberosum cox2 gene, including a 727 bp non-coding upstream region, was isolated by PCR from total DNA using the primer A designed from partial sequences reported by Löessl et al. (22), accession no. AF096321, containing the KpnI restriction sequence, and primer B derived from Triticum timopheevi cox2 (AF336134). A fragment of 2888 bp was obtained and cloned on pGEM-T vector. The complete sequence of the S.tuberosum cox2 was determined (accession no. DQ18064). To generate the plasmid pCOXIISt containing the S.tuberosum cox2 gene, a KpnI/SpeI fragment containing the 727 bp upstream region and the complete coding region was used to replace the wheat gene from pCOXII. As for pCOXII, a NsiI site was inserted at the ATG start codon and a 23 bp fragment was inserted at position −20. This 23 bp insertion provides a specific sequence allowing isolation of potato cox2 transcripts originating from the introduced DNA by RT–PCR. The chimeric wheat/potato cox2 gene was constructed by replacing 727 bp KpnI/NsiI promoter sequence from pCOXIISt with the 880 bp wheat upstream region from pCOXII.
The vectors pRPS10W and the pRPS10St derivative, containing wheat and potato cox2 promoters, respectively, were constructed by inserting the 1178 bp rps10 sequence containing two exons separated by a 777 bp intronic sequence [(23), accession no. X74826]. The coding region was isolated from total S.tuberosum DNA by PCR using primers D1 and D2 containing the restriction sites NsiI and SpeI, respectively. The fragment NsiI/SpeI was purified and cloned into pCOXII and pCOXIISt, replacing the respective cox2 coding regions.
Since all vectors used here were based on pCOXII, they contain the downstream region from the wheat cob gene (Ir-cob) (accession no. AF337547) (14). This sequence, combined with the 23 bp upstream insert sequence served to distinguish, using primers 1a and 1b, foreign from endogenous transcripts. All mutants were obtained using QuickChange® Site-Directed Mutagenesis kit (Stratagene). PCR product purifications were carried out with the Wizard® Clean-Up System (Promega).
PCR primers used
Oligonucleotides sequences are in 5′ to 3′ orientation. (1a), GCGGTGCAGTCATACAGATCTGC; (1b), TATCCAGATTTGGTACCAAAC; (2a), GCAGTCATACAGATCTGCCAAAG; (2b), AGATTTGGTACCAAACCCGGG; (A), TATAGGTACCTCTCAGGTGTCAAAGTGTGGATTT; (B), TATAACTAGTTTAAGCTTCCCCG; (D1), TATAATGCATAGACAAAGGAGAGCACTTA; (D2), TATAACTAGTTCAGGAAAGGGTCAACGCAA. Restriction sites are underlined.
Mutagenesis primers (only sense primers in 5′–3′ orientation are indicated)
Single C2, C3 and C2+C3 double mutant plasmids were constructed with primers (C2) AAGAAGTTCTTTTGGTTAAAACGCC and (C3) CGCCGTGCGACTTGGAGGACATAAG. NsiI-pCOXII: GGAAATCCAATGCATCTTCGTTCATT and NsiI-pCOXIISt: CCAAACCAAATGCATGTTCTAGAATG. Construct pCOXIISt containing a 23 nt insertion in the promoter region, was carried out into two consecutive insertions using primers: TGGGGGGAGCAGAGCAGTGCGGTGCAGTCACAAAGAATGAACCAAACC and GCAGTGCGGTGCAGTCATACAGATCTGCCAAAGAATGAACCAAACC. For constructs MA and MAB containing the wheat cox2 C259 editing site, two consecutive insertions were carried out using primers: GGAAGATTGGATTACTATCGAAATTGCCCTGAATCA and TACTATCGAAATTATTCGGACCATGCCCTGAATCA. For ME6 and ME6b constructs, primers CCGCGAGGAATCAACTACTATCGAAATTATTGCCGGTGCTGAC and CAACTACTATCGAAATTATTCGGACCATATTGCCGGTGCTGAC were used. Inserted nucleotides are underlined and the modified residues are indicated in bold.
Mitochondria purification
S.tuberosum cv. Rosevalt tubers and T.aestivum var Fortal seeds were used. Potato mitochondria were prepared from 2 kg of tubers in batches of 200 g with 200 ml of a homogenization buffer containing 0.4 M mannitol, 25 mM MOPS (pH 7.8), 1 mM EGTA, 8 mM cysteine and 1 mg/ml fatty acid-free BSA. Homogenization was carried out for 15 s in a Waring blendor at full speed. Homogenate was centrifuged in a Sorvall GSA rotor at 1500 g for 10 min at 4°C. Supernatant was centrifuged in a GSA rotor at 12 000 g for 15 min. The mitochondrial pellet was resuspended in 50 ml of homogenization buffer and centrifuged at 1500 g. The supernatant was centrifuged in a Sorvall SS-34 rotor at 15 000 g for 10 min, the pellet was resuspended in 12 ml of homogenization buffer and mitochondria were purified by centrifugation on a sucrose gradient essentially as described for wheat embryo mitochondria (14).
Electroporation
Electrotransfer experiments were carried out with 1 mg of purified wheat embryo or potato tuber mitochondria in 50 µl of 0.33 M sucrose and 1 µg of recombinant plasmid as described (14). The electroporated mitochondria were incubated for 18 h at 25°C, then recovered by centrifugation in a Sigma N° 12024 rotor (Sigma 1K15 refrigerated centrifuge) at 15 000 g for 15 min at 4°C. In the case of potato mitochondria the incubation mixture was supplemented with 1 mg/ml of fatty acid-free BSA. RNA was purified with 200 µl of TRIzol™ reagent (Gibco-BRL) according to the supplier's protocol.
DNAse I protection assay and DNA purification
After electroporation and centrifugation, the mitochondrial pellet was resuspended in 100 µl of buffer [10 mM Tris–HCl (pH 7.5), 2 mM magnesium acetate and 0.33 M sucrose] containing 60 U of DNase I (Gibco-BRL) and incubated for 1 h at room temperature. The DNase reaction was stopped by adding 4 µl of 0.5 M EDTA and then heating for 10 min at 65°C. Mitochondria were incubated with 100 µg of Proteinase K (Merck) for 4 h at 37°C. One microliter of 20% SDS was added to achieve mitochondrial lysis and the DNA was extracted with phenol/chloroform, precipitated with 0.1 vol of 3 M Sodium Acetate (pH 5.2), 3 vol of 100% ethanol and 100 ng of carrier yeast tRNA and left overnight at −20°C. After centrifugation, the DNA pellet was resuspended in TE buffer [10 mM Tris–HCl (pH 8) and 1 mM EDTA].
RT–PCR
RNA (1 µg) was treated with 2 U of Amplification grade DNase I (Promega). cDNA synthesis was performed with 200 U of Superscript II RT using 100 ng of random hexamers. The PCR were performed with primers 1a and 1b using Advantage® 2 Polymerase Mix (Clontech) as follows: 95°C, 1 min; 5 cycles at 95°C for 30 s and 68°C for 1 min; 30 cycles at 95°C for 30 s, 58°C for 30 s and 68°C for 30 s, and finally 68°C for 1 min. Primers 2a and 2b were used for nested PCR from 1 µl of the first PCR. No RT–PCR amplification products were obtained with RNA from non-electroporated mitochondria.
DNA sequencing
Sequence analyses were performed directly on the RT–PCR product using an automatic DNA sequencing equipment with the BigDye® Terminator Cycle Sequencing Kit (Applied Biosystem).
RESULTS
S.tuberosum rps10 transcripts are not processed in wheat mitochondria
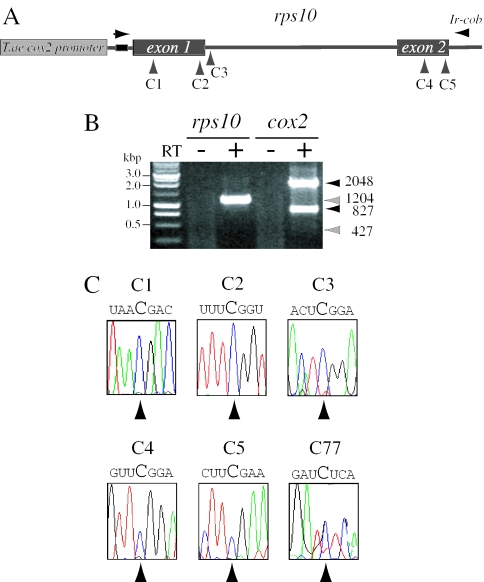
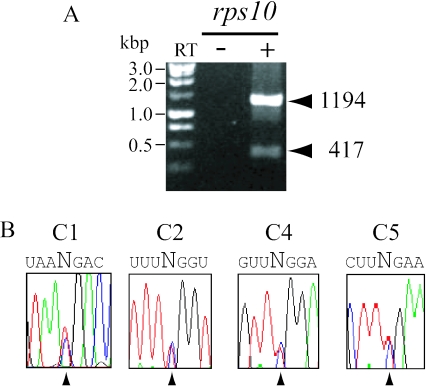
The S.tuberosum rps10 construct under control of a wheat cox2 promoter (Figure 1A) was incorporated into purified wheat mitochondria by electroporation as indicated in Materials and Methods. After electroporation and incubation in expression medium, mitochondrial RNA was extracted and analysed by RT–PCR. The primers used allowed detection of the transcripts generated by the constructs introduced and excluded any product from endogenous cox2 (or rps10 in the potato system). Two bands of 1204 and 427 bp were expected from precursor and mature rps10 mRNAs, respectively. Only precursor rps10 molecules were detected (Figure 1B). As a control, a cognate cox2 construct (16) was used. In this case, the 2048 and 827 bp RT–PCR bands representing the precursor and spliced cox2 products, respectively were observed (Figure 1B). In all cases, the PCR bands actually represent transcription products since when PCR was performed without the RT step we observed no amplification products.
Figure 1.
S.tuberosum rps10 gene expression in electroporated wheat mitochondria. (A) Schematic representation of the pRPS10W vector. Potato rps10 is under control of T.aestivum cox2 promoter. Horizontal arrows indicate the position of primers 1a and 1b used for specific PCR cDNA amplification. The same regions are present in all vectors used in this work. Vertical arrows indicate edited residues in potato rps10 RNA. (B) Agarose gel electrophoresis of RT–PCR products. Arrows indicate the position and numbers the size (in bp) of precursor and mature transcripts generated by rps10 and cox2 vectors. (C) Panels C1 to C5 represent the electropherograms of rps10 cDNA sequences in the regions normally edited in potato mitochondria. Panel C77 concerns one of the editing sites analysed in the T.aestivum cox2 transcript in a control experiment performed with pCOXII. Arrowheads signal editing target residues. In RT–PCR experiments, controls without the reverse transcription step were systematically performed and are indicated as RT(−) in all figures.
Previously, five C residues, two in exon 1, one in the intron and two in exon 2 have been reported to be changed to U by editing in potato mitochondria (23). To determine if the non-cognate transcript could be recognized by the wheat RNA editing machinery, the 1204 bp RT–PCR product was sequenced. While wheat cox2 editing sites were correctly edited in control as expected (Figure 1C, only site C77 is shown), all five editable residues in potato rps10 transcript remain unchanged. cox2 C77 editing sites are identical in potato and wheat.
Failure of rps10 transcript splicing in wheat mitochondria is not linked to the absence of editing
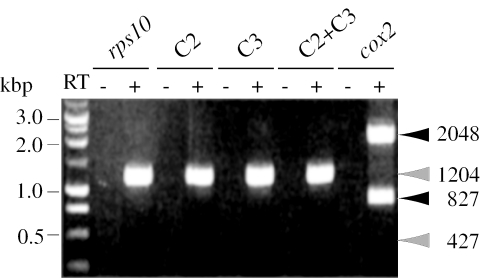
It has been suggested that residues C2 and C3 participate in the secondary structure of the intron necessary for splicing (23). A possible explanation to the failure observed in precursor rps10 splicing could be that the absence of edition of C2 and C3 prevent the intron from organizing itself in a catalytically competent conformation. To test this hypothesis, single C2 and C3 mutants and a C2+C3 double mutant were constructed by changing the C residues to T. Neither single nor double mutants were able to undergo splicing (Figure 2).
Figure 2.
Gene expression analysis of C2 and C3 single and double rps10 mutants introduced into wheat mitochondria. Mutants were obtained by changing to T two C residues that are edited in potato mitochondria (23). These mutants and the control rps10 construct (pRPS10W) were introduced into wheat mitochondria and analysed as described in Figure 1. The expected positions and sizes (in bp) of precursors and mature transcripts are shown for rps10 and cox2.
Electroporation of S.tuberosum mitochondria
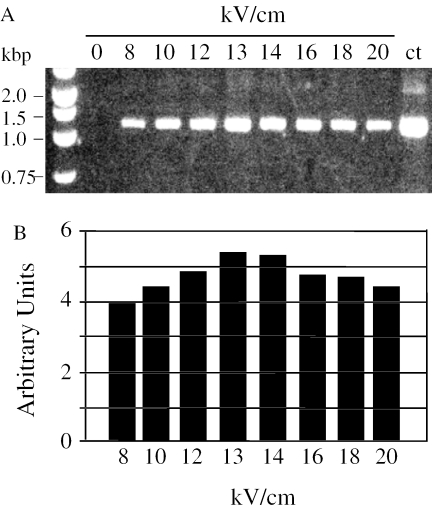
Since two important post-transcriptional processes, RNA editing and splicing, were inoperative when S.tuberosum rps10 was expressed in wheat mitochondria, we decided to verify whether the negative results were inherent to the rps10 chimeric constructs or due to the lack of trans-recognition elements in wheat mitochondria. For this purpose, it was necessary to set up an electroporation protocol adapted to S.tuberosum mitochondria. Purified organelles were prepared from potato tubers as indicated in Materials and Methods. Electric pulses in the range between 8 and 20 kV were performed. Internalization of exogenous DNA was measured by DNAse protection assays (14). Potato mitochondria show a broad range response with a maximum around 13 kV (Figure 3). This voltage setting was therefore used for further experiments.
Figure 3.
DNAse I protection experiments. Purified potato tuber mitochondria were electroporated in the presence of 1 µg of pCOXIISt plasmid at electric field strengths ranging from 0 to 20 kV/cm. To eliminate the non-internalized vector, mitochondria were treated with DNAse I. DNA was extracted and the target sequence was revealed by PCR using primers 1a and 1b. Panel A, agarose gel electrophoresis of PCR products. Panel B, the ethidium bromide UV fluorescence signal from the gel electrophoresis experiment in panel A was recorded with a CCD video camera and plotted as arbitrary units using the NIH Image software.
S.tuberosum mitochondria does not recognize the wheat cox2 promoter
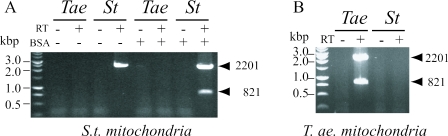
To ascertain the ability of electroporated mitochondria to perform expression of the exogenous gene construct, we used a plasmid containing the potato cox2 gene controlled either by T.aestivum or S.tuberosum promoters. As shown in Figure 4, the construct containing the wheat or potato promoters was transcribed only in cognate mitochondria. In potato, the mature product is barely detectable. In contrast to wheat mitochondria, the 821 nt mature transcript was only detected when the electroporated potato mitochondria were incubated in the presence of fatty acid-free BSA.
Figure 4.
Gene expression analysis of S.tuberosum cox2 under the control of T.aestivum (Tæ) and S.tuberosum (St) promoters. RT–PCR products were analysed by agarose gel electrophoresis and revealed with ethidium bromide stain. Panel A, electroporation of potato mitochondria. Panel B, electroporation of wheat mitochondria.
A cognate rps10 construct is correctly expressed, edited and processed in potato mitochondria
The construct expressing potato rps10 under the control of potato cox2 promoter was introduced into S.tuberosum isolated mitochondria. After incubation, the precursor and mature transcripts were amplified by RT–PCR (Figure 5A) and sequenced. As shown in Figure 5B, the four editing sites C1, C2, C4 and C5, described previously in endogenous transcripts, were found edited in mature mRNA. The presence of spliced molecules containing unedited residues (Figure 5B) indicates that rps10 transcripts could be spliced before editing. To confirm this observation, the PCR product was cloned and sequenced. One half of the individual clones were found edited at sites C1 and C2; 65% were edited at site C4 and 25% were edited at site C5.
Figure 5.
Processing and editing of S.tuberosum rps10 transcripts in cognate mitochondria. (A) Agarose gel electrophoresis of RT–PCR products of electroporated potato mitochondria. The sizes of precursor and spliced products are indicated. (B) Sequence analysis of the regions containing the C1, C2, C4 and C5 editing sites in mature transcripts. Site C3, present in the intron, is lost after splicing. Arrowheads indicate the target residue. In sequences, N indicates a mixture of C and U residues corresponding to unedited and edited transcripts, respectively.
A cognate editing site in a non-native context is not recognized by wheat mitochondria but is recognized in heterologous mitochondria
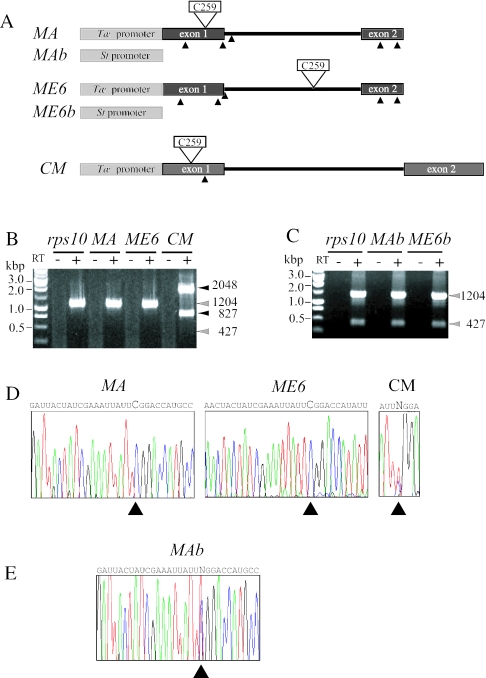
To test whether wheat mitochondria are able to edit a cognate site when placed in the context of a potato transcript, we introduced the C259 −16/+6 region from wheat cox2 into potato rps10 exon 1 or intron (Figure 6A, construct MA and ME6, respectively). As reported previously, the 23 nt region forming the C259 editing site from wheat cox2 was efficiently edited when grafted into a different context in a wheat transcript (17). Wild-type rps10 (pRPS10W) and the MA and ME6 mutants were expressed in wheat mitochondria but no splicing was observed (Figure 6B). Unexpectedly, wheat mitochondria appear to be unable to edit the cognate C259 site when the −16/+6 region is located on a potato rps10 precursor. The control CM construct, containing the C259 site grafted in a different context but in its own cox2 gene, was expressed, spliced and edited as expected [(17), Figure 6B and D]. To compare the editing status of RNAs at the same stage of processing, the editing status of the inserted C259 site in precursor cox2 transcripts is shown (Figure 6D). The analogous MAb and ME6b constructs, containing the S.tuberosum cox2 promoter and the C259 region inserted into rps10 exon 1 and intron were used to electroporate potato mitochondria. As shown in Figure 6C, MAb and ME6b were expressed and spliced at the same level as the control in the homologous context, indicating that insertion does not affect the splicing process. Interestingly, while C259 insertion was not recognized in wheat mitochondria (Figure 6D), the potato RNA editing machinery edited the inserted C259 region (Figure 6E). The C259 region inserted in the rps10 intron (construct ME6b) was also edited, although at a very low level, showing that editing may precede intron removal (not shown).
Figure 6.
Analysis of RNA editing in C259 insertion mutants. (A) Diagram of potato rps10 mutants bearing an inserted wheat editing site. The positions where the 23 nt editing site was inserted and the promoters used are indicated. CM is a wheat cox2 mutant containing the C259 editing site inserted at position 113 in cox2 exon 2 (17) (B) Gene expression analysis of rps10 wild-type, insertion mutants and wheat CM control after electroporation of wheat mitochondria. (C) Gene expression analysis of rps10 wild-type and insertion mutants after electroporation of potato mitochondria. (D) Sequence analysis of the inserted C259 region in RT–PCR products from MA and ME6 vectors electroporated in wheat mitochondria. To compare editing status of the inserted C259 site at the same maturation stage, only the sequence from CM precursor transcript is shown. (E) Sequence analysis of RT–PCR products of MAb transcript from potato electroporated mitochondria.
DISCUSSION
Plant mitochondria undergo complex expression mechanisms which are poorly understood. Most studies are based on analysis of in vivo mature or intermediate gene products giving clues on possible mechanisms and suggesting pathways operating in gene expression. Direct tests using in vitro approaches have been fruitful (4,5,24), but are hampered by the difficulty of obtaining active mitochondrial extracts. Previously, we devised an experimental model based on electroporation of isolated mitochondria that allows us to test gene expression of wild-type and mutant genes. This approach has been very useful in elucidating the process of RNA maturation in plant mitochondria, in particular splicing and editing (14,15). The aim of this work was to analyse different gene expression events when a foreign gene was expressed in a heterologous context. rps10 was chosen precisely because this genetic information is lacking in wheat mitochondria but is active in potato mitochondria (20,23,25). Moreover, rps10 transcripts undergo five C-to-U editing events in potato mitochondria and rps10 contains an intron, thus facilitating the analysis of post-transcriptional processing.
To best evaluate the expression of rps10 in the non-cognate mitochondria, it was necessary to set up electroporation conditions for introducing foreign DNA into S.tuberosum mitochondria. Potato tuber mitochondria were able to incorporate DNA essentially under the same conditions described for wheat embryo organelles (14), except that the optimal voltage range was larger. A major difference to wheat was that isolated potato mitochondria were viable for 3–4 h as measured by oxygen consumption (data not shown). Thus, to observe transcript maturation after electroporation the incubation mixture needed to be supplemented with fatty acid-free BSA (see Figure 4). This behaviour probably reflects the uncoupling of oxidative phosphorylation by fatty acids during incubation of potato mitochondria (26). In fact, previously we found that proper gene expression in isolated mitochondria requires a functional organelle able to generate ATP from ADP and succinate (14).
An interesting observation was that neither the wheat nor the potato cox2 promoters were recognized in a heterologous context (Figure 4A and B). This situation might by explained by the fact that potato and wheat promoter sequences have no recognizable homologous motifs. Plant mitochondria promoters are characterized by a conserved core CRTA sequence with differences in the extent and the composition of sequences around the consensus motif (27,28). While the transcription initiation site in wheat mitochondria is located at position −170 (29), the potato promoter has not yet been described. Transcription may be initiated at numerous sites suggesting a relaxed promoter recognition by the transcription machinery (28). However, the lack of crossed recognition of cox2 promoters in wheat and potato is not a general situation since the A.thaliana cox2 gene is expressed and spliced when introduced into maize mitochondria, indicating that the Arabidopsis gene shares some signals with maize cox2 promoter that are sufficient for transcription (18). The sites required for transcript initiation have been recently described in A.thaliana mitochondrial genes (28). Three regions were described as important for transcription initiation. We found no such homologous sequences in the potato cox2 upstream region indicating that the two dicot promoters do not have the same origin. This in turn may reflect the natural history of this particular mitochondrial gene evolving in its own context. It should be mentioned that the presence of a conserved sequence is not sufficient for expression since a region that acts as promoter in Arabidopsis, potato and Oenothera is inactive in vivo in pea (30). Further studies will be required to understand the transcriptional events in plant mitochondria. Electroporation of foreign DNA into isolated mitochondria provides an interesting functional model, complementary to in vitro transcription assay, for answering these questions.
The potato rps10 gene controlled by a T.aestivum promoter (Figure 1A) was transcribed as a 1204 nt precursor in wheat mitochondria, but no traces of mature RNA were observed. Moreover, the five C residues reported as RNA editing targets in vivo (23) remain unchanged, indicating that rps10 transcript was not recognized by the wheat splicing and editing machinery. A control using the cognate cox2 construct demonstrates that electroporated organelles were competent for splicing and editing (Figure 1B and C).
Some editing events occur in highly structured domains of introns. Because in some cases, editing corrects A-C mispairing improving conformation of the intron, it has been proposed that the C-to-U change might be necessary for efficient splicing (8,9,23,31). Based on the canonical structure of group II mitochondrial introns (21), two editing sites, C2 located at the Intron Binding Site 2 (IBS2) and C3 located in the intron, nine residues downstream from the end of exon 1 in S.tuberosum rps10 are of particular interest. It has been predicted that both edited residues participate in base-pair interactions in the putative secondary structure. Of particular interest is the site C3 located in intron domain I (23). This position may be crucial for splicing as inferred from mutants of a yeast mitochondrial intron (32). We addressed this question by introducing rps10 mutant genes presenting C2, C3 or both positions in the edited form into mitochondria. As shown in Figure 2, transcription was as efficient as for the cognate cox2 construct but neither wild-type rps10 nor the C-U mutants underwent splicing. These results clearly demonstrate that C2 and C3 editing is not sufficient for splicing of rps10 precursor in wheat mitochondria and lead us to conclude that splicing failure is probably linked to the lack of trans-recognition elements. One may speculate that these trans-acting factors, for instance nuclear-encoded maturases, were lost after rps10 was transferred to the nucleus in monocots (20,25). The C-to-U mutants were also tested in potato mitochondria; no significant differences were detected in splicing efficiency when compared to the unedited construct (D. Choury, unpublished data). It should be noted that the lack of splicing in wheat mitochondria is not a general feature of potato introns, since the potato cox2 intron is removed efficiently (Figure 4B). This is consistent with the hypothesis that potato and wheat mitochondria have similar trans-acting factors for cox2 intron removal.
A possible link between intron removal and editing has been proposed for splicing of nad1e and nad5III trans-introns where domain six may require editing to be structured in a catalytic competent secondary conformation (8). In rice, failure of RNA maturation and editing has been correlated with cytoplasmic male sterile phenotype. In this model, the absence of the nuclear gene Rf-1 affects B-atp6 RNA cleavage and editing (33). In organello studies have shown that S.bicolor atp6-1 gene was transcribed but not edited when introduced into maize mitochondria (18). However, partially edited molecules were detected in sorghum-maize atp6 chimeric transcripts when they included the maize atp6 5′-untranslated sequence, suggesting that the 5′ non-coding region provides a structural motif or binding site for a transcript-specific editing factor (34). Unfortunately, no data on atp6 processing was reported to determine whether this region is directly involved in editing or some other maturation event.
Since wheat mitochondria do not have rps10 encoded in the mitochondrial genome (20), it was interesting to test whether wheat mitochondria were able to recognize editing sites which had been lost during evolution. In other words, whether the mitochondrial trans-recognition elements are specific for each site or whether they are operating on a subset of editing sites. This question is important since to date the factors responsible for RNA editing in plant mitochondria remain unknown. Solving this issue may provide clues to uncover such factors. As described above, the rps10 transcript was not processed in wheat but it was correctly spliced in cognate mitochondria (Figure 5A). Indeed, the mature transcript had identical exon1 and exon2 junction as found in endogenous rps10 mRNA (data not shown). More importantly, all editing sites, C1, C2, C4 and C5 were significantly converted to Us (Figure 5B). The fact that the four editing sites were found either edited or unedited in spliced rps10 mRNA is an indication that editing does not precede splicing as was previously found for cox2 in wheat (15).
It may be argued that the absence of splicing precluded editing. This possibility can be discarded since potato rps10 precursor mRNA was found to be edited in vivo (23). These data clearly show that splicing is not required for rps10 editing, similar to previous findings for cox2 mRNA and also in cox2 mutant derivatives unable to remove the intron (15). This led us to postulate that the inability of wheat mitochondria to recognize rps10 editing sites is likely due to the fact that wheat mitochondria have lost the editing trans-recognition elements which become dispensable after transfer of rps10 to the nucleus. To test this possibility, rps10 chimeric plasmids containing editing site C259 from wheat cox2 inserted either in exon1 or the intron were constructed. Site C259 is formed by 23 nt corresponding to the −16/+6 sequence embedding the target C. Previously we found that this small region could be recognized by the RNA editing machinery when placed outside of its natural context (16,17). The wheat C259 editing site in the chimeric construct was correctly edited by potato mitochondria. This result is not unexpected since the corresponding region in endogenous potato cox2 mRNA, which presents two differences at positions −3 (C instead of A) and −7 (G instead of A), is edited. Furthermore, these positions were not crucial for editing of C259 (17). Surprisingly, the C259 editing site grafted in rps10 was not recognized by the wheat editing machinery. We cannot exclude the possibility that editing efficiency in precursor molecules is very low and so is undetectable when sequencing RT–PCR products representing a pool of transcripts. However, analysis of the same region in cox2 precursors indicates that this was not the case since significant C-to-U conversion was observed (Figure 6D). These observations lead us to postulate that editing is occurring only when the transcript is engaged in post-transcriptional processing, suggesting that rps10 transcripts are not available to editing factors independently of the RNA maturation machinery. The fact that the wheat editing site C259 inserted in chimeric rps10 transcripts was not recognized by wheat mitochondria is a strong argument for this hypothesis. One might speculate that in plant mitochondria, transcripts have to be engaged in a kind of multiprotein processing complex. A failure to be recognized at some early stage will lead to their accumulation as unmodified precursors.
Acknowledgments
We thank Ms Evelyne Sargos for excellent technical assistance. This research was supported in part by the French Ministère de l'Enseignement Supérieur et de la Recherche, the Université Victor Segalen Bordeaux 2, the research grants 1020930 and 7020930 (Cooperación Internacional) from Fondecyt-Chile and PICS no. 2179 grant from CNRS. Funding to pay the Open Access publication charges for this article was provided by Laboratoire REGER, Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Giegé P., Brennicke A. From gene to protein in higher plant mitochondria. C R Acad Sci III. 2001;324:209–217. doi: 10.1016/s0764-4469(00)01293-2. [DOI] [PubMed] [Google Scholar]
- 2.Brennicke A., Marchfelder A., Binder S. RNA editing. FEMS Microbiol. Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 3.Blanc V., Jordana X., Litvak S., Araya A. Control of gene expression by base deamination: the case of RNA editing in wheat mitochondria. Biochimie. 1996;78:511–517. doi: 10.1016/0300-9084(96)84757-2. [DOI] [PubMed] [Google Scholar]
- 4.Blanc V., Litvak S., Araya A. RNA editing in wheat mitochondria proceeds by a deamination mechanism. FEBS Lett. 1995;373:56–60. doi: 10.1016/0014-5793(95)00991-h. [DOI] [PubMed] [Google Scholar]
- 5.Yu W., Schuster W. Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J. Biol. Chem. 1995;270:18227–18233. doi: 10.1074/jbc.270.31.18227. [DOI] [PubMed] [Google Scholar]
- 6.Giegé P., Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder S., Marchfelder A., Brennicke A. Regulation of gene expression in plant mitochondria. Plant Mol. Biol. 1996;32:303–314. doi: 10.1007/BF00039387. [DOI] [PubMed] [Google Scholar]
- 8.Farré J.C., Araya A. The mat-r open reading frame is transcribed from a non-canonical promoter and contains an internal promoter to co-transcribe exons nad1e and nad5III in wheat mitochondria. Plant Mol. Biol. 1999;40:959–967. doi: 10.1023/a:1006296422485. [DOI] [PubMed] [Google Scholar]
- 9.Börner G.V., Mörl M., Wissinger B., Brennicke A., Schmelzer C. RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol. Gen. Genet. 1995;246:739–744. doi: 10.1007/BF00290721. [DOI] [PubMed] [Google Scholar]
- 10.Marchfelder A., Brennicke A., Binder S. RNA editing is required for efficient excision of tRNA(Phe) from precursors in plant mitochondria. J. Biol. Chem. 1996;271:1898–1903. doi: 10.1074/jbc.271.4.1898. [DOI] [PubMed] [Google Scholar]
- 11.Maréchal-Drouard L., Cosset A., Remacle C., Ramamonjisoa D., Dietrich A. A single editing event is a prerequisite for efficient processing of potato mitochondrial phenylalanine tRNA. Mol. Cell. Biol. 1996;16:3504–3510. doi: 10.1128/mcb.16.7.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bégu D., Graves P.V., Domec C., Arselin G., Litvak S., Araya A. RNA editing of wheat mitochondrial ATP synthase subunit 9: direct protein and cDNA sequencing. Plant Cell. 1990;2:1283–1290. doi: 10.1105/tpc.2.12.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernould M., Suharsono S., Litvak S., Araya A., Mouras A. Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc. Natl Acad. Sci. USA. 1993;90:2370–2374. doi: 10.1073/pnas.90.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farré J.C., Araya A. Gene expression in isolated plant mitochondria: high fidelity of transcription, splicing and editing of a transgene product in electroporated organelles. Nucleic Acids Res. 2001;29:2484–2491. doi: 10.1093/nar/29.12.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farré J.C., Araya A. RNA splicing in higher plant mitochondria: determination of functional elements in group II intron from a chimeric cox II gene in electroporated wheat mitochondria. Plant J. 2002;29:203–213. doi: 10.1046/j.1365-313x.2002.01207.x. [DOI] [PubMed] [Google Scholar]
- 16.Farré J.C., Leon G., Jordana X., Araya A. cis-Recognition elements in plant mitochondrion RNA editing. Mol. Cell. Biol. 2001;21:6731–6737. doi: 10.1128/MCB.21.20.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choury D., Farré J.C., Jordana X., Araya A. Different patterns in the recognition of editing sites in plant mitochondria. Nucleic Acids Res. 2004;32:6397–6406. doi: 10.1093/nar/gkh969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staudinger M., Kempken F. Electroporation of isolated higher-plant mitochondria: transcripts of an introduced cox2 gene, but not an atp6 gene, are edited in organello. Mol. Genet. Genomics. 2003;269:553–561. doi: 10.1007/s00438-003-0863-x. [DOI] [PubMed] [Google Scholar]
- 19.Adams K.L., Qiu Y-L., Stoutemyer M., Palmer J.D. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl Acad. Sci. USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanlungo S., Quiñones V., Moenne A., Holuigue L., Jordana X. A ribosomal protein S10 gene is found in the mitochondrial genome in Solanum tuberosum. Plant Mol. Biol. 1994;25:743–749. doi: 10.1007/BF00029612. [DOI] [PubMed] [Google Scholar]
- 21.Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns–a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 22.Löessl A., Adler N., Horn R., Frei U., Wenzel G. Chondriome-type characterization of potato: mt alpha, beta, gamma, delta, epsilon and novel plastid-mitochondrial configurations in somatic hybrids. Theor. Appl. Genet. 1999;99:1–10. [Google Scholar]
- 23.Zanlungo S., Quiñones V., Moenne A., Holuigue L., Jordana X. Splicing and editing of rps10 transcripts in potato mitochondria. Curr. Genet. 1995;27:565–571. doi: 10.1007/BF00314449. [DOI] [PubMed] [Google Scholar]
- 24.Neuwirt J., Takaneka M., Van Der Merwe J.A., Brennicke A. An in vitro RNA editing system from cauliflower mitochondria: editing site recognition parameters can vary in different plant species. RNA. 2005;11:1563–1570. doi: 10.1261/rna.2740905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams K.L., Daley D.O., Qiu Y.L., Whelan J., Palmer J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408:354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 26.Douce R. Mitochondria in Higher Plants: Structure, Function, and Biogenesis. American Society of Plant Physiologist Monograph Series. Orlando Florida. USA: Academic Press, Inc.; 1985. [Google Scholar]
- 27.Lupold D.S., Caoile A.G., Stern D.B. The maize mitochondrial cox2 gene has five promoters in two genomic regions, including a complex promoter consisting of seven overlapping units. J. Biol. Chem. 1999;274:3897–3903. doi: 10.1074/jbc.274.6.3897. [DOI] [PubMed] [Google Scholar]
- 28.Kühn K., Weihe A., Börner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanic-Joyce P.J., Gray M.W. Accurate transcription of a plant mitochondrial gene in vitro. Mol. Cell. Biol. 1991;11:2035–2039. doi: 10.1128/mcb.11.4.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giese A., Thalheim C., Brennicke A., Binder S. Correlation of nonanucleotide motifs with transcript initiation of 18S rRNA genes in mitochondria of pea, potato and Arabidopsis. Mol. Gen. Genet. 1996;252:429–436. doi: 10.1007/BF02173008. [DOI] [PubMed] [Google Scholar]
- 31.Wissinger B., Schuster W., Brennicke A. Trans-splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell. 1991;65:473–482. doi: 10.1016/0092-8674(91)90465-b. [DOI] [PubMed] [Google Scholar]
- 32.Schmelzer C., Schweyen R.J. Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell. 1986;46:557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- 33.Iwabuchi M., Kyozuka J., Shimamoto K. Processing followed by complete editing of an altered mitochondrial atp6 RNA restores fertility of cytoplasmic male sterile rice. EMBO J. 1993;12:1437–1446. doi: 10.1002/j.1460-2075.1993.tb05787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staudinger M., Bolle N., Kempken F. Mitochondrial electroporation and in organello RNA editing of chimeric atp6 transcripts. Mol. Genet. Genomics. 2005;273:130–136. doi: 10.1007/s00438-005-1117-x. [DOI] [PubMed] [Google Scholar]