Abstract
Many newly synthesized proteins have to become unfolded during translocation across biological membranes. We have analyzed the effects of various stabilization/destabilization mutations in the Ig-like module of the muscle protein titin upon its import from the N terminus or C terminus into mitochondria. The effects of mutations on the import of the titin module from the C terminus correlate well with those on forced mechanical unfolding in atomic-force microscopy (AFM) measurements. On the other hand, as long as turnover of the mitochondrial Hsp70 system is not rate-limiting for the import, import of the titin module from the N terminus is sensitive to mutations in the N-terminal region but not the ones in the C-terminal region that affect resistance to global unfolding in AFM experiments. We propose that the mitochondrial-import system can catalyze precursor-unfolding by reducing the stability of unfolding intermediates.
Keywords: mitochondria, titin, Hsp70, mechanical stability
A protein's function relies on its native folded conformation. However, normal cell functions also require transient unfolding of proteins in such processes as translocation across biological membranes and selective degradation by ATP-dependent proteases. In both cases, polypeptide chains have to translocate through narrow channels, the diameters of which are too small to accept folded protein domains. Therefore machineries for protein translocation and degradation have activities to induce unfolding of substrate proteins when they are folded (1).
The eukaryotic cell is subdivided into functionally distinct compartments or organelles that contain unique sets of proteins. Among them, mitochondria are essential organelles that are bounded by two membranes, the outer and inner membranes, and contain two aqueous compartments, the intermembrane space and the matrix. Most mitochondrial-matrix proteins and some inner mitochondrial-membrane proteins are synthesized in the cytosol as precursors with an N-terminal presequence that contains targeting information for mitochondria. They are translocated across the outer and inner membranes through the translocators in the two membranes, the TOM40 complex and TIM23 complex, respectively, with the aid of mitochondrial Hsp70 (mtHsp70) and mtHsp70-associated motor and chaperone proteins (2–5). Accumulated evidence suggests that precursor proteins that folded before import into mitochondria can be actively unfolded by the translocators to thread into their protein-conducting channels (6–9).
In vitro mitochondrial import of precursor proteins containing presequences of varying lengths showed that the import rates are higher for precursor proteins with a long presequence (>70 residues) than for those with a short presequence (<70 residues) (8), indicating that these two distinct situations are to be considered for unfolding of presequence-containing precursor proteins upon import into mitochondria. Because presequences do not take ordered structures in solution, the N terminus of a long presequence can reach the matrix without unfolding of the mature domain and engage mtHsp70 in the matrix. Then mtHsp70 pulls in the precursor protein by the mechanism, which is still under debate (10), and the import rates reflect the local structural stability around the N terminus of the mature domain (11–13). On the other hand, a mature domain with a short presequence needs to become unfolded before engagement of the presequence with mtHsp70, because the short presequence cannot reach the matrix without unfolding of the mature domain. Spontaneous and transient global unfolding of the mature domain would presumably allow the unfolded segment following the short presequence to move forward in the import channel by Brownian motion, so that the presequence can engage mtHsp70. Therefore, the import rates, in this case, would depend on the global, rather than local, structural stability of the mature domain (8, 11–13).
In this study, we systematically analyzed the effects of various stabilization/destabilization mutations in the mature passenger domain upon its import into mitochondria. We used the 27th Ig (I27) domain of the giant muscle protein titin as a model folded passenger domain, because unfolding of the I27 domain by atomic-force microscopy (AFM) (14–17) in combination with steered molecular-dynamics simulation (18, 19) and that by chemical denaturants (17, 20) has been extensively studied. Although the I27 domain is mechanically highly stable, the I27 domain attached to a mitochondrial-targeting signal can be efficiently unfolded by and imported into isolated mitochondria (21). We attached long or short mitochondrial-targeting sequences at the N terminus or C terminus of the I27 domain with various mutations, and the resultant fusion proteins were subjected to in vitro import into isolated mitochondria. The results show that mutations in the I27 domain affect the import rates for the N-terminal and C-terminal import differently, and, unexpectedly, the N-terminal import was not sensitive to the mutations that would affect the global stability. We propose that the mitochondrial-import system efficiently catalyzes precursor unfolding by changing the stability of unfolding intermediates differently from the forced mechanical unfolding by AFM and global unfolding by chemical denaturants.
Materials and Methods
Fusion Proteins. The I27 fusion proteins used in this study were made as follows. The genes for pb2(35)-I27 and pb2(80)-I27 were derived from pb2(220)WT-dihydrofolate reductase (DHFR) (22). The codons for Arg30 and Leu62 of pb2(220)WT-DHFR were replaced by those for Gly30 and Pro62 by oligonucleotide-directed mutagenesis (23). The resulting genes were used as templates for PCR to generate DNA fragments encoding the first 35 residues and 80 residues of the precursor of cytochrome b2. The amplified fragments were cut with EcoRI and XhoI and ligated into the EcoRI/XhoI sites of the gene for pb2(220)WT-DHFR to generate DNA segments for pb2(35)-DHFR and pb2(80)-DHFR. The genes encoding the I27 domain and its variants Y9P, V11P, V13P, and V15P were amplified by PCR using pET AvaI, pET AvaI (Y9P), pET AvaI (V11P), pET AvaI (V13P), and pET AvaI (V15P), respectively, as templates (16). To construct pb2(35)-I27, pb2(35)-Y9P, pb2(35)-V11P, pb2(35)-V13P, pb2(35)-V15P, pb2(80)-I27, pb2(80)-Y9P, pb2(80)-V11P, pb2(80)-V13P, and pb2(80)-V15P, the amplified DNA fragments encoding I27, Y9P, V11P, V13P, and V15P were ligated into the XhoI/XbaI sites of the genes for pb2(35)-DHFR and pb2(80)-DHFR, respectively. Other point mutations (V4P, E5P, K6P, L60A, C47S/C63S, K6P/V11P, K6P/V13P, Y9P/V13P, and K6P/C47S/C63S) were introduced into the genes for pb2(35)-I27 and pb2(80)-I27 by PCR.
The DNA fragments encoding the C-terminal 37 residues (H37C) and 105 residues (H105C) of Hmi1p were amplified by PCR using the yeast genomic DNA as a template (24). The amplified DNA fragments were ligated into the BamHI/SalI sites of pGEM-4z. The DNA fragments encoding I27 and its variants were amplified by PCR using the genes for pb2(35)-I27 and pb2(35)-I27 variants as templates and ligated into the EcoRI/BamHI of pGEM-4z vectors containing H37C or H105C, respectively.
Import Assay. In vitro import of the I27-domain fusion proteins into isolated mitochondria was performed as described in ref. 25. Briefly, the fusion proteins were synthesized in rabbit reticulocyte lysate by coupled transcription/translation in the presence of [35S]methionine. Mitochondria were isolated from the yeast Saccharomyces cerevisiae strain D273-10B (26). The radiolabeled fusion proteins were incubated with isolated yeast mitochondria in import buffer (250 mM sucrose/10 mM Mops-KOH, pH 7.2/80 mM KCl/2 mM KPi/2 mM methionine/5 mM DTT/5 mM MgCl2/2 mM ATP/2 mM NADH/1% BSA). The import reactions were stopped by adding valinomycin to 10 μg/ml. The samples were halved, and one aliquot was treated with 100 μg/ml proteinase K for 30 min on ice. After addition of 1 mM phenylmethylsulfonyl fluoride, the mitochondria were isolated by centrifugation and washed once with SEM buffer (250 mM sucrose/5 mM EDTA/10 mM Mops-KOH, pH 7.2). Proteins were analyzed by SDS/PAGE and radioimaging with a Storm 860 image analyzer (Amersham Biosciences).
Results
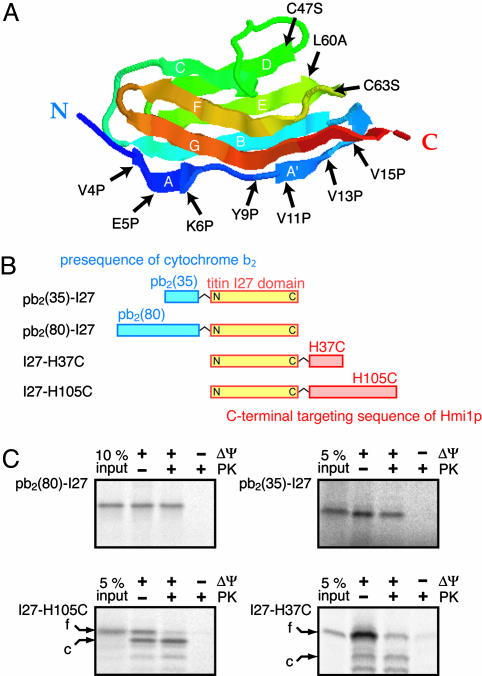
Mitochondrial-Targeting Sequences Can Direct the I27 Domain from Either the N Terminus or the C Terminus to the Mitochondrial Matrix. The titin I27 domain consists of 89 amino acid residues and forms seven β-strands (strands A–G), which fold into two face-to-face β-sheets through interactions, including backbone hydrogen bonds and hydrophobic core formation (Fig. 1A). The N-terminal β-strand A (residues 4–7) forms an antiparallel pair with the β-strand B (residues 18–25), whereas the C-terminal β-strand G (residues 78–88) forms a parallel pair with the β-strand A′ (residues 11–15). Structural elements that affect unfolding processes have been extensively studied for the titin I27 domain by AFM experiments and chemical denaturation (summarized in Table 1, which is published as supporting information on the PNAS web site). In the mechanical unfolding with extending forces applied by AFM at both the N and C termini (forced unfolding), the ability to withstand force depends on the interactions between the A′ and G strands (involving V11, V13, and V15 in the A′ strand), constituting a mechanical clamp but not on the ones bridging the A and B strands (involving V4, E5, and K6 in the A strand) (20). In unfolding by chemical denaturants (chemical unfolding), thermodynamic stability and kinetic stability of the entire I27 domain depend mainly on the core region and on the A′- and G-strand pair region, respectively (19), and the I27 mutant lacking the A strand is thermodynamically stable (27). The NMR H–D exchange experiments showed that backbone amide hydrogens in the A- and B-strand pair can exchange with the deuterium atom in the solvent, whereas those in the A′–G-strand pair and in the core region resist exchange, suggesting the frequent structural fluctuation in the A and B strands (28).
Fig. 1.
The I27 domain can be imported from both the N terminus and C terminus. (A) Cartoon diagram, showing the β-sandwich structure of the I27 molecule and the amino acid residues that were substituted in this work. The β-strands are named A, A′, and B–G. (B) Fusion proteins with the I27 domain used in this study. Blue boxes, parts of the cytochrome b2 presequence; yellow boxes, the I27 domain; orange boxes, parts of the C-terminal targeting sequence of Hmi1p. (C) In vitro import of the radiolabeled I27 fusion proteins into mitochondria. The fusion proteins in B with the wild-type I27 domain were incubated with isolated mitochondria for 1 min (pb2(80)-I27), 5 min (pb2(35)-I27 and I27-H37C), or 20 min (I27-H105C) at 25°C. The mitochondria were treated with proteinase K and reisolated by centrifugation. The proteins were analyzed by SDS/PAGE and radioimaging. Valinomycin was added to 10 μg/ml to dissipate Δψ. f, full-length form; c, imported and processed form.
We made fusion proteins pb2(35)-I27 and pb2(80)-I27 by attaching the first 35 residues or entire 80 residues of the presequence of yeast cytochrome b2 (pb2) with R30G and L62P mutations (the R30G mutation eliminates the cleavage site for mitochondrial-processing peptidase (MPP), and L62P inactivates the intermembrane-space sorting signal), as short and long targeting sequences, respectively, to the N terminus of the I27 domain (Fig. 1B). We also made fusion proteins I27-H37C and I27-H105C by attaching residues 670–706 or residues 602–706 of yeast Hmi1p, which has a cleavable mitochondrial-targeting signal at the C terminus (24), as short and long targeting sequences, respectively, to the C terminus of the I27 domain (Fig. 1B).
The radiolabeled fusion proteins pb2(35)-I27, pb2(80)-I27, I27-H37C, and I27-H105C were synthesized with reticulocyte lysate in vitro and incubated with isolated yeast mitochondria. Treatment of the mitochondria with protease, which removed the fusion proteins outside the mitochondria, showed that the fusion proteins were imported into the mitochondria, depending on the membrane potential across the inner membrane (Δψ) (Fig. 1C). Because the presequence cleavage site for MPP was mutated (R30G), the targeting sequences of the imported pb2(35)-I27 and pb2(80)-I27 were not processed by MPP. On the other hand, the C-terminal targeting sequence of Hmi1p contains the MPP-processing site between residues 695 and 696, so that the targeting sequences of the imported I27-H37C and I27-H105C were proteolytically processed, although the cleavage efficiency of I27-H37C was not as high as that of I27-H105C. The fusion proteins with an Hmi1p-targeting sequence were previously shown to be imported into mitochondria by essentially the same mechanism as the N-terminal import guided by a mitochondrial presequence (24). Indeed, import of pb2(35)-I27, pb2(80)-I27, I27-H37C, and I27-H105C was significantly impaired by depletion of ATP, suggesting a requirement of mtHsp70 for their import (data not shown). Therefore, the I27 domain can be imported into the mitochondrial matrix from either the N terminus or the C terminus, depending on the positions of the attachment of the mitochondrial-targeting sequence.
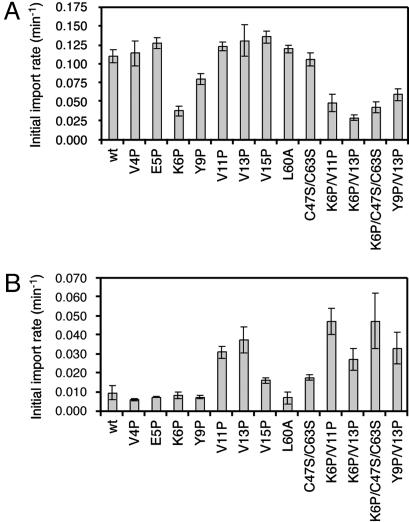
C-Terminal Import with a Long Targeting Sequence Reflects the Mechanical Stability of the I27 Domain. We made various I27-domain mutants with a single mutation in the A strand (V4P, E5P, and K6P), the linker between the A and A′ strands (Y9P), the A′ strand (V11P, V13P, and V15P), and the core region (L60A) and with double mutations in the core region (C47S/ C63S), the A and A′ strands (K6P/V11P and K6P/V13P), the linker and A′ strand (Y9P/V13P), and the A strand and core region (K6P/C47S/C63S) (Fig. 1 A). We then analyzed the effects of these I27 mutations on the rates of the N-terminal import (pb2(80)-I27) or of the C-terminal import (I27-H105C) with long targeting sequences (see Fig. 5 A and B, which is published as supporting information on the PNAS web site). Under the present experimental conditions with functional excess of mitochondria, binding of the fusion proteins to mitochondria is not rate-limiting (29) (data not shown), so that unfolding of the fusion proteins or their translocation across the membranes becomes rate-limiting for the entire import process. The mutations in the I27 domain do not affect exposure of the attached targeting sequences, because the targeting sequences were highly susceptible to externally added protease as compared with the I27 domain (data not shown).
The mutations affected the import rates quite differently between pb2(80)-I27 and I27-H105C. The mutations K6P, Y9P, K6P/V11P, K6P/V13P, K6P/C47S/C63S, and Y9P/V13P decreased the import rates of pb2(80)-I27, whereas the other mutations did not (Fig. 2A). On the other hand, the mutations V11P, V13P, V15P, C47S/C63S, K6P/V11P, K6P/V13P, K6P/ C47S/C63S, and Y9P/V13P increased the import rates of I27-H105C, whereas the other mutations did not (Fig. 2B). These results suggest that the unfolding pathways of the I27 domain differ between the N- and C-terminal import.
Fig. 2.
Effects of mutations in the I27 domain on the import of radiolabeled pb2(80)-I27 and I27-H105C. pb2(80)-I27 (A) or I27-H105C (B) fusion proteins with indicated mutations or without a mutation (wt) in the I27 domain were incubated with isolated mitochondria for various times at 25°C and imported. Proteinase-K-protected fractions were quantified, and import rates (initial slopes of the import reactions) were plotted. Values are means ± SD.
The effects of mutations in the I27 domain on its import from the C terminus (Fig. 2B) show close correlation with those on mechanical forced unfolding. Mutations in the A′ strand (V11P, V13P, K6P/V11P, K6P/V13P, and Y9P/V13P), but not in the A strand, increased the import rates of I27-H105C from the C terminus. The V15P mutation in the A′ strand and C47S/C63S mutation in the core region slightly increased the C-terminal import rates. In AFM experiments, the mutations V11P, V13P, and V15P (and C47S/C63S, slightly) decrease the force required to unfold the I27 domain (16, 17) (Table 1). In chemical denaturation, the mutations V11P, V13P, V15P, and C47S/C63S (and Y9P and L60A, slightly) accelerate the rate constants for global unfolding (17, 20) (Table 1). Mutations of L60 and C47/C63, which lie outside the C-terminal mechanical-clamp region, exhibit smaller effects on both the C-terminal import and forced mechanical unfolding, which may well require displacement of the C-terminal region, than on chemical unfolding, which involves cooperative disruption of the core and C-terminal regions. Therefore, upon import from the C terminus, disruption of the interactions between the A′- and G-strand pairs is immediately followed by or takes place together with the unfolding of the rest of the molecule containing the core region, as observed for mechanical unfolding (16, 17, 19). In other words, the rate-limiting step of the C-terminal import may well involve global unfolding triggered by the detachment of the A′ and G strands.
The effects of some, but not all, of the mutations in the I27 domain on the N-terminal import (Fig. 2 A) also show correlation with those in forced unfolding. The single mutations K6P in the A strand and Y9P in the linker segment between the A and A′ strands and double/triple mutations (K6P/V11P, K6P/V13P, K6P/C47S/C63S, and Y9P/V13P) containing the K6P or Y9P mutation decreased rates of the import of the I27 domain from the N terminus. The effect of Y9P on the import is consistent with the increased mechanical stability of Y9P observed by AFM (16) (Table 1). K6P may also increase the mechanical stability of the A- and B-strand pair comparable with that of the A′- and G-strand pair, because, in the AFM experiments, the K6P mutation led to loss of the unfolding intermediate before the global unfolding, resulting in the requirement of the higher force to unfold the mutant I27 domain completely (15) (Table 1). The mutations V4P and E5P in the A-strand residues that are not involved in the hydrogen bonds linking to the B strand did not affect the import from the N terminus.
The mutations V11P, V13P, and V15P in the A′ strand did not affect the import from the N terminus, suggesting that detachment of the A′- and G-strand pair is not rate-limiting for the N-terminal import. However, in forced unfolding of the I27 domain by AFM, even after the A strand is detached from the B strand, the rest of the domain resists unfolding until the A′ strand is detached from the G strand at a stronger force, and forced unfolding is, thus, sensitive to the destabilization mutations V11P, V13P, and V15P in the A′- and G-strand pair. Why is the import from the N terminus insensitive to these destabilization mutations? A possible explanation is that, because the import rates of pb2(80)-I27 are sufficiently fast, the import rate may be no longer limited by detachment of the destabilized A′–G-strand pair but, instead, determined by the turnover of the import machinery mtHsp70 (8, 12). If so, only the stabilization mutation that would decrease the unfolding rate, such as the one in the A–B-strand pair could limit the import rate; this result was, indeed, observed here for the K6P and Y9P mutations. However, on the other hand, K6P and Y9P mutations decreased the import rates, even in the presence of destabilization mutations in the A′ strand (K6P/V11P, K6P/V13P, and Y9P/K13P) or core region (K6P/C47S/C63S), suggesting that local unfolding around the A strand that is stabilized by K6P or Y9P mutation could likely represent the rate-limiting step of the import of the I27 domain, which is different from the case of forced unfolding. Possible reasons for uncoupling of the stability of the A′- and G-strand pair from the import rates will be discussed below, together with the N-terminal import directed by a short targeting sequence.
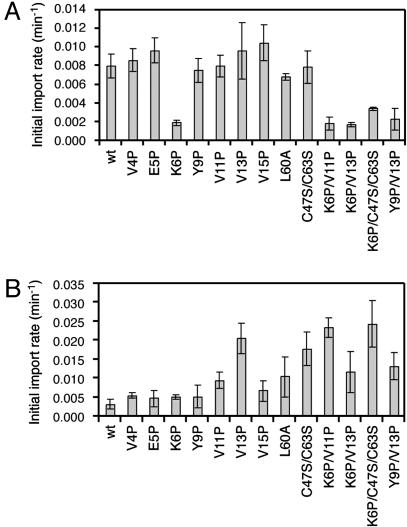
N-Terminal Import with a Short Targeting Sequence Does Not Reflect the Global Stability of the I27 Domain. Next, we tested the effects of mutations in the I27 domain on its import into mitochondria directed by short targeting sequences. The import of the I27 domain with the 35-residue pb2-targeting sequence from the N terminus (pb2(35)-I27) and with the 37-residue Hmi1p-targeting sequence from the C terminus (I27-H37C) (Fig. 3 A and B; and see Fig. 6 A and B, which is published as supporting information on the PNAS web site) was 10% and 50%, respectively, of that of the I27 domain with long targeting sequences (Fig. 2 A and B). The slower import rates of the I27 domain with a short targeting sequence compared with those with a long targeting sequence are because of the inability of the short targeting sequence to interact with mtHsp70 before unfolding of the I27 domain outside the mitochondria (8). In this case, for a short targeting signal, spontaneous global unfolding of the mature domain has been proposed to be essential for efficient import, because mutations affecting spontaneous global unfolding accelerate the import guided by a short presequence (8, 11–13). However, surprisingly, our observation with titin fusion proteins stands against this proposed model, as described below.
Fig. 3.
Effects of mutations in the I27 domain on the import of radiolabeled pb2(35)-I27 and I27-H37C. pb2(35)-I27 (A) or I27-H37C (B) fusion proteins with indicated mutations or without a mutation (wt) in the I27 domain were incubated with isolated mitochondria for various times at 25°C and imported. Proteinase-K-protected fractions were quantified, and import rates (initial slopes of the import reactions) were plotted. Values are means ± SD.
Not only mutations in the C-terminal mechanical-clamp region, V11P, V13P, K6P/V11P, K6P/V13P, and Y9P/V13P but also those in the core region, L60A, C47S/C63S, and K6P/ C47S/C63S, increased the rates of import from the C terminus of I27-H37C (Fig. 3B), consistent with the idea that the spontaneous global unfolding involving disruption of both the C-terminal and core regions of the I27 domain limits the C-terminal import rates. If spontaneous global unfolding of the I27 domain also limits the import rates for the I27 domains with the N-terminally attached targeting sequence pb2(35), which is too short to span both membranes (8, 11–13), mutations in the C-terminal clamp region and in the core region that destabilize the overall stability and increase the global unfolding rate of the I27 domain will affect the N-terminal import. However, only the single/double/ triple mutations with K6P in the A strand, not those in the A′ strand or in the core region without K6P, decreased the import rate of pb2(35)-I27 (Fig. 3A). In the Y9P/V13P mutation, Y9P mutation exhibits synergic effects with V13P mutation. The insensitivity of the N-terminal import to the destabilizing mutations in the A′ strand cannot be ascribed to the switch of the rate-limiting step from the unfolding of the I27 domain to turnover of the import machinery, because the import rates of pb2(35)-I27 derivatives are one-order-of-magnitude smaller than those of pb2(80)-I27, so that turnover of the import machinery should be too fast to limit the import rates. Therefore, in the case of pb2(35)-I27 (and likely the case of pb2(80)-I27 with K6P or Y9P mutation), unfolding around the A strand represents the rate-limiting step of the import; once the A strand is spontaneously detached from the B strand, unfolding of the rest of the molecule, including detachment of the A′ strand from the G strand and disruption of the core region, follows immediately. Apparently, trapping of the partially unfolded form of the I27 domain with the separated A and B strands by the mitochondrial import machinery destabilizes the rest of the molecule, including the C-terminal clamp region.
Discussion
In this study, we have systematically analyzed the effects of various stabilization/destabilization mutations in the model folded protein, the titin I27 domain, on its import from the N terminus and from the C terminus, guided by long or short targeting sequences. Generally, mitochondria can import folded proteins with a long targeting sequence much faster than these proteins unfold spontaneously in free solution, because mtHsp70 can tug at a long targeting sequence by directly exerting an iterative mechanical pulling force on or trapping local unfolding fluctuation in the segment downstream of the targeting signal and unravel the folded proteins from the targeting-sequence attachment site (usually the N terminus) at the expense of ATP hydrolysis. In this case, the resistance of the folded proteins to unfolding during import may well correlate with their mechanical stability against pulling force in AFM experiments. This study partly supports this model. The mutations around the C-terminal region that accelerate the C-terminal import of I27-H105C tend to lower the resistance of the I27 domain against the pulling force in AFM experiments (16). The mutations in the N-terminal region that lower the N-terminal import rates of pb2(80)-I27 tend to enhance resistance of the I27 domain against the applied force (15, 16). These results are consistent with the previous observation that mitochondrial protein-import rates correlate with the rates of transient local unfolding but not with the overall protein stability (10, 11). The destabilization mutations in the C-terminal region could not accelerate the N-terminal import of pb2(80)-I27, probably because turnover of the mtHsp70 system, not the unfolding of the I27 domain, is rate-limiting for the N-terminal import of pb2(80)-I27.
On the other hand, import of folded proteins with a short targeting sequence has been thought to require spontaneous global unfolding of the proteins, because the targeting sequence is too short to reach the mitochondrial matrix to engage the mtHsp70 system (8, 11–13). However, we found here that the N-terminal import of pb2(35)-I27 was not accelerated at all by mutations in the C-terminal A′–G-strand pair that can reduce the stability of the I27 domain, whether unfolded by force or by chemical denaturants. Because the N-terminal import of pb2(35)-I27 is sensitive to the stabilization mutation K6P in the N-terminal region, detachment of the A strand from the B strand is rate-limiting, whereas unfolding of the resultant intermediate with the detached A segment is not. N-terminal import of pb2(80)-I27 with K6P or Y9P mutation was not sensitive to mutations in the C-terminal A′–G-strand pair, likely for the same reason.
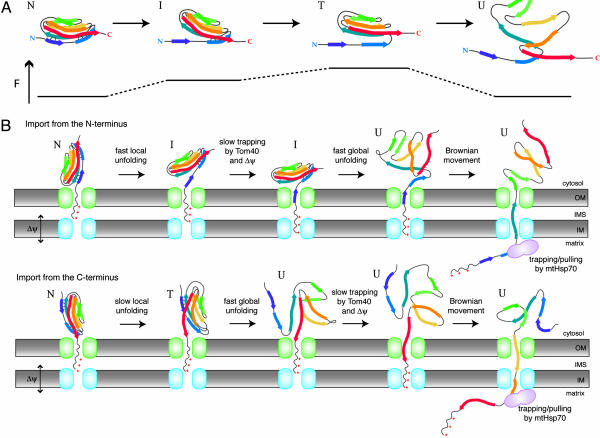
In AFM experiments, the A strand can be easily detached from the B strand without affecting the rest of the molecule, which unfolds only after detachment of the A′ strand from the G strand at stronger force (Fig. 4A). The mitochondrial import machinery should, therefore, change the stability of the partially unfolded intermediate with the detached A strand differently from the mechanical forced unfolding (Fig. 4B). What is the mechanism that changes the stability of the partially unfolded intermediate? In AFM experiments, the direction of the applied extension force, e.g., the orientation of β-strands relative to the force vector, changes the mechanical stability significantly (30, 31). Therefore, upon import, the geometrical constraints of the partially unfolded form, in which the unfolded segments are threaded in the import channel, whereas the remaining folded domain is placed at the opening of the channel, may well change the stability of the partially unfolded intermediate (Fig. 4B). Thus, the mitochondria-import system catalyzes precursor unfolding, most likely by lowering the stability of unfolding intermediates through specific geometrical constraints, thereby achieving global unfolding efficiently.
Fig. 4.
Models of unfolding pathways of the I27 domain in forced mechanical unfolding (A) and upon mitochondrial import directed by a short targeting sequence (B). N, native state; I, unfolding intermediate with the detached A strand (19); T, transition state with the detached A′ strand for global unfolding (19); U, unfolded state; F, applied force.
Short targeting sequences cannot interact with mtHsp70 unless the I27 domain outside the mitochondria becomes unfolded. Besides, even after detachment of the A strand from the B strand, the N-terminal unraveled segment containing the 35-residue pb2(35) presequence (+5-residue linker) and the 7-residue A strand of the I27 domain cannot cross the two membranes to engage mtHsp70 in the matrix (8, 11). Then what element of the mitochondria-import system other than mtHsp70 can trap the partially unfolded I27 domain with its A segment detached? Because the import channel of the outer-membrane translocator, the TOM40 complex, has a chaperone-like function (32), the Tom40 channel may well trap the locally unfolded segment of the I27 domain, thereby preventing refolding of the unfolding intermediate (33, 34). Δψ across the inner membrane may also trap the unfolding intermediate or even directly act on the positively charged short presequences to promote further unfolding of the intermediate (13).
In conclusion, the implicated scenario of the import of the I27 domain is as follows. The I27 domain undergoes frequent detachment of the N-terminal A and B strands and (less frequently) that of the C-terminal A′ and G strands. The targeting sequence and the transiently unraveled N- or C-terminal segment of the I27 domain that thread into the mitochondrial import channels are trapped by Δψ and the Tom40 channel (for short targeting sequences) or mtHsp70 (for long targeting sequences). Trapping of the transiently unfolded species by the mitochondria-import system not only prevents refolding of the I27 domain but also destabilizes the unfolding intermediates, thereby efficiently leading to global unfolding. Then, translocation of the unfolded polypeptide through the import channels follows, with the aid of the hand-over-hand trapping or pulling by mtHsp70. More precise mechanisms for trapping or pulling of the unfolding intermediates by the mitochondria-import system are the important subject of future studies.
Supplementary Material
Acknowledgments
We thank members of the Endo laboratory for discussions and comments. This work was supported by Grants-in-Aid for Scientific Research from the Japan Ministry of Education, Culture, Sports, Science and Technology and a grant from the Japan Science and Technology Agency.
Author contributions: T.E. designed research; T.S. and M.E. performed research; J.M.F. contributed new reagents/analytic tools; T.S. analyzed data; and T.S. and T.E. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AFM, atomic force microscopy; DHFR, dihydrofolate reductase; I27, the 27th immunoglobulin; MPP, mitochondrial-processing peptidase; mtHsp70, mitochondrial Hsp70.
References
- 1.Prakash, S. & Matouschek, A. (2004) Trends Biochem. Sci. 29, 593–600. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann, J. M. & Neupert, W. (2000) Curr. Opin. Microbiol. 3, 210–214. [DOI] [PubMed] [Google Scholar]
- 3.Endo, T., Yamamoto, H. & Esaki, M. (2003) J. Cell Sci. 116, 3259–3267. [DOI] [PubMed] [Google Scholar]
- 4.Wiedemann, N., Frazier, A. E. & Pfanner, N. (2004) J. Biol. Chem. 279, 14473–14476. [DOI] [PubMed] [Google Scholar]
- 5.Koehler, C. M. (2004) Annu. Rev. Cell Dev. Biol. 20, 309–335. [DOI] [PubMed] [Google Scholar]
- 6.Eilers, M. & Schatz, G. (1986) Nature 322, 228–232. [DOI] [PubMed] [Google Scholar]
- 7.Rassow, J., Hartl, F. U., Guiard, B., Pfanner, N. & Neupert, W. (1990) FEBS Lett. 275, 190–194. [DOI] [PubMed] [Google Scholar]
- 8.Matouschek, A., Azem, A., Ratliff, K., Glick, B. S., Schmid, K. & Schatz, G. (1997) EMBO J. 16, 6727–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voisine, C., Craig, E. A., Zufall, N., von Ahsen, O., Pfanner, N. & Voos, W. (1999) Cell 97, 565–574. [DOI] [PubMed] [Google Scholar]
- 10.Neupert, W. & Brunner, M. (2002) Nat. Rev. Mol. Cell Biol. 3, 555–565. [DOI] [PubMed] [Google Scholar]
- 11.Gaume, B., Klaus, C., Ungermann, C., Guiard, B., Neupert, W. & Brunner, M. (1998) EMBO J. 17, 6497–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, S., Ratliff, K. S., Schwartz, M. P., Spenner, J. M. & Matouschek, A. (1999) Nat. Struct. Biol. 6, 1132–1138. [DOI] [PubMed] [Google Scholar]
- 13.Huang, S., Ratliff, K. S. & Matouschek, A. (2002) Nat. Struct. Biol. 9, 301–307. [DOI] [PubMed] [Google Scholar]
- 14.Carrion-Vazquez, M., Oberhauser, A. F., Fowler, S. B., Marszalek, P. E., Broedel, S. E., Clarke, J. & Fernandez, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3694–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marszalek, P. E., Lu, H., Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Shulten, K. & Fernandez, J. M. (1999) Nature 402, 100–103. [DOI] [PubMed] [Google Scholar]
- 16.Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Marszalek, P. E. & Fernandez, J. M. (2000) Nat. Struct. Biol. 7, 1117–1120. [DOI] [PubMed] [Google Scholar]
- 17.Brockwell, D. J., Beddard, G. S., Clarkson, J., Zinober, R. C., Blake, A. W., Trinick, J., Olmsted, P. D., Smith, D. A. & Radford, S. E. (2002) Biophys. J. 83, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, H., Isralewitz, B., Krammer, A., Vogel, V. & Schulten, K. (1998) Biophys. J. 75, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best, R. B., Fowler, S. B., Herrera, J. L., Steward, A., Paci, E. & Clarke, J. (2003) J. Mol. Biol. 330, 867–877. [DOI] [PubMed] [Google Scholar]
- 20.Kenniston, J. A., Baker, T. A., Fernandez, J. M. & Sauer, R. T. (2003) Cell 114, 511–520. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto, K., Brinker, A., Paschen, S. A., Moarefi, I., Hayer-Hartl, M., Neupert, W. & Brunner, M. (2002) EMBO J. 21, 3659–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esaki, M., Kanamori, T., Nishikawa, S. & Endo, T. (1999) Proc. Natl. Acad. Sci. USA 96, 11770–11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel, T. A., Roberts, J. D. & Zakour, R. A. (1987) Methods Enzymol. 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. M., Sedman, J., Neupert, W. & Stuart, R. A. (1999) J. Biol. Chem. 274, 20937–20942. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto, H., Esaki, M., Kanamori, T., Tamura, Y., Nishikawa, S. & Endo, T. (2002) Cell 111, 519–528. [DOI] [PubMed] [Google Scholar]
- 26.Daum, G., Bohni, P. C. & Schatz, G. (1982) J. Biol. Chem. 257, 13028–13033. [PubMed] [Google Scholar]
- 27.Fowler, S. B., Best, R. B., Toca Herrera, J. L., Rutherford, T. J., Steward, A., Paci, E., Karplus, M. & Clarke, J. (2002) J. Mol. Biol. 322, 841–849. [DOI] [PubMed] [Google Scholar]
- 28.Improta, S., Politou, A. S. & Pastore, A. (1996) Structure 4, 323–337. [DOI] [PubMed] [Google Scholar]
- 29.Lithgow, T. & Schatz, G. (1995) J. Biol. Chem. 270, 14267–14269. [DOI] [PubMed] [Google Scholar]
- 30.Brockwell, D. J., Paci, E., Zinober, R. C., Beddard, G. S., Olmsted, P. D., Smith, D. A., Perham, R. N. & Radford, S. E. (2003) Nat. Struct. Biol. 10, 731–737. [DOI] [PubMed] [Google Scholar]
- 31.Carrion-Vazquez, M., Li, H., Lu, H., Marszalek, P. E., Oberhauser, A. F. & Fernandez, J. M. (2003) Nat. Struct. Biol. 10, 738–743. [DOI] [PubMed] [Google Scholar]
- 32.Esaki, M., Kanamori, T., Nishikawa, S., Shin, I., Schultz, P. G. & Endo, T. (2003) Nat. Struct. Biol. 10, 988–994. [DOI] [PubMed] [Google Scholar]
- 33.Mayer, A., Neupert, W. & Lill, R. (1995) Cell 80, 127–137. [DOI] [PubMed] [Google Scholar]
- 34.Kanamori, T., Nishikawa, S., Nakai, M., Shin, I., Schultz, P. G. & Endo, T. (1999) Proc. Natl. Acad. Sci. USA 96, 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.