Abstract
Recent outbreaks of highly pathogenic avian influenza (HPAI) viruses in poultry and their threatening zoonotic consequences emphasize the need for effective control measures. Although vaccination of poultry against avian influenza provides a potentially attractive control measure, little is known about the effect of vaccination on epidemiologically relevant parameters, such as transmissibility and the infectious period. We used transmission experiments to study the effect of vaccination on the transmission characteristics of HPAI A/Chicken/Netherlands/03 H7N7 in chickens. In the experiments, a number of infected and uninfected chickens is housed together and the infection chain is monitored by virus isolation and serology. Analysis is based on a stochastic susceptible, latently infected, infectious, recovered (SEIR) epidemic model. We found that vaccination is able to reduce the transmission level to such an extent that a major outbreak is prevented, important variables being the type of vaccine (H7N1 or H7N3) and the moment of challenge after vaccination. Two weeks after vaccination, both vaccines completely block transmission. One week after vaccination, the H7N1 vaccine is better than the H7N3 vaccine at reducing the spread of the H7N7 virus. We discuss the implications of these findings for the use of vaccination programs in poultry and the value of transmission experiments in the process of choosing vaccine.
Keywords: SEIR model, highly pathogenic avian influenza, final size, generalized linear model, reproduction ratio
Highly pathogenic avian influenza (HPAI) is a disease of poultry caused by H5 or H7 AI A strains, with mortality that ranges up to 100%. The number of outbreaks in the last few years has been unprecedented: Hong Kong (1997) (1), Italy (1999) (2), Chile (2002) (3), the Netherlands (2003) (4), Canada (2004) (5), and the continuing outbreaks in Southeast Asia (2003-2005) (6). Aside from causing havoc in poultry, it is becoming increasingly clear that certain HPAI viruses have the potential to directly cross the human-bird species barrier and may become a pandemic threat (6-8). To reduce the primary risk of human HPAI infection, it is crucial to prevent infection of poultry. Common methods to control outbreaks of HPAI are the killing and destruction of infected poultry, preemptive culling, biosecurity measures, and vaccination.
Several vaccines have been developed against H5 and H7 influenza viruses in poultry. Vaccination can protect chickens from overt disease and mortality (9). Although AI vaccines reduce the replication of HPAI viruses in the respiratory and gastrointestinal tracts, virus-shedding after vaccination is observed (10, 11), raising questions as to the effectiveness of vaccination in preventing transmission from animal to animal, and viral spread in the population is conceivable. Such silent transmission is very undesirable, because it increases the risk of new outbreaks and poses a threat to humans. To overcome this problem, sentinel chickens and differentiating infected from vaccinated animals (DIVA) vaccines have been used (10), but the best solution would be a vaccine that prevents transmission. An ideal vaccine against HPAI should reduce the spread of virus between animals in a flock and, subsequently, the spread of virus between flocks to such an extent that a major outbreak will not occur. Unfortunately, not much is known about the ability of AI vaccines to reduce transmission of HPAI viruses in chickens and the quantification of this reduction.
We studied the effect of vaccination on the spread of virus in a population of chickens by using so-called transmission experiments. In a transmission experiment, a number of infected chickens is housed together with a number of uninfected chickens, and the infection chain is monitored. Transmission experiments offer a way to look at the spread of virus under experimental conditions in a population of known composition, making it possible to quantify the effect of vaccination on transmission dynamics (12, 13). We focused on the transmission characteristics of A/Chicken/Netherlands/621557/03 H7N7 in chickens, using two different commercial inactivated oil-emulsion vaccines (H7N1 and H7N3) in different vaccination schemes. Our results show that vaccination not only protects chickens against mortality and morbidity but also reduces the spread of virus within a flock to such an extent that a major outbreak can be prevented.
Materials and Methods
Viruses. The virus used in this study was A/Chicken/Netherlands/621557/03 H7N7. This virus was isolated on the index farm of the outbreak in the Netherlands in 2003. The virus had an i.v. pathogenicity index of 2.93, as determined by the procedure described in ref. 14. Briefly, 10 chickens were injected i.v. with 0.1 ml of 10-fold diluted allantoic fluid. Birds were examined at 24-hour intervals for 10 days. At each observation, each chicken was recorded as 0, normal; 1, sick; 2, severely sick; or 3, dead. The index is calculated by adding up all scores and dividing the total by 100. When the index is >1.2, the AI virus is considered as highly pathogenic.
Animals. All animal experiments were undertaken in a high-containment unit under biosafety level 3+ conditions at the Central Institute for Animal Disease Control Lelystad. The experiments comply with Dutch law on animal experiments and were reviewed by an ethical committee. In all experiments, 6-wk-old specific-pathogen-free white leghorn chickens were used. The chickens were inoculated both intranasally and intratracheally with 0.1 ml of diluted allantoic fluid containing 106 median egg-infectious dose (EID50) per ml.
Vaccines. Two commercially available oil-emulsion vaccines were used: a H7N1 (A/Chicken/Italy/99) vaccine and a H7N3 (A/Chicken/Pakistan/95) vaccine. The dosage was 0.5 ml for the H7N1 vaccine and 0.3 ml for the H7N3 vaccine (as recommended by the manufacturer), and the vaccine was administered in the leg muscles. The hemagglutinin-antigen content was 45 μg/ml for the H7N1 vaccine and 13 μg/ml for the H7N3 vaccine (see Fig. 2, which is published as supporting information on the PNAS web site). The protein homology of the immunogenic part of the (HA1) between the challenge strain H7N7 and the H7N1 vaccine was 98% and for the H7N3 vaccine 92%.
Transmission Experiments. Group experiments were performed with unvaccinated chickens (experiment 1), with chickens challenged 1 wk after vaccination with H7N1 (experiment 2) or H7N3 (experiment 3) and with chickens challenged 2 wk after vaccination with H7N1 (experiment 4) or H7N3 (experiment 5). All group experiments were done in duplicate (see Table 1 for an overview). The design of the experiments was as follows: Five chickens were placed in a cage (1.2 × 1.2 m.) and inoculated with virus; 24 h later, five contact chickens were added. The chickens were monitored by taking tracheal and cloacal swabs daily for the first 10 days and twice a week for the next 11 days. A blood sample was taken weekly. The experiments were terminated 3 wk after the challenge.
Table 1. Overview of the experiments.
| Experiment no. | Type of experiment | Challenge after vaccination, weeks | Vaccine |
|---|---|---|---|
| 1 | Group | Unvaccinated | |
| 2 | Group | 1 | H7N1 |
| 3 | Group | 1 | H7N3 |
| 4 | Group | 2 | H7N1 |
| 5 | Group | 2 | H7N3 |
| 6 | Pair | 1 | H7N1 |
| 7 | Pair | 1 | H7N3 |
| 8 | Pair | 2 | H7N1 |
| 9 | Pair | 2 | H7N3 |
All experiments were done in duplicate.
Pair experiments were performed with vaccinated inoculated chickens and unvaccinated contact chickens (see Table 1 for an overview). All pair experiments were done with four pairs of chickens. Chickens were challenged 1 wk after vaccination with H7N1 (experiment 6) or H7N3 (experiment 7) or challenged 2 wk after vaccination with H7N1 (experiment 8) or H7N3 (experiment 9). In each experiment, a chicken was vaccinated and, 1 or 2 wk after vaccination, challenged with H7N7 virus. The vaccinated inoculated chicken was placed in a cage, and, 24 h later, an unvaccinated chicken was added. The chickens were monitored by taking tracheal and cloacal swabs daily for the first 10 days and at day 14 and by a weekly blood sample. As soon as a contact chicken showed signs of illness, the contact chicken was killed.
Vaccination-Response Experiment. The serological response after vaccination was studied by the hemagglutination inhibition (HI) assay. In total, 40 chickens were vaccinated with the H7N1 vaccine and 40 with the H7N3 vaccine. All animals were bled from the wing vein at days 6, 8, 10, 12, 14, 17, 21, 24, 28, 31, and 35.
Virus Isolation. Swabs were put in 2 ml of 2.95% tryptose phosphate buffer with 5 × 103 IU of penicillin-sodium and 5 mg of streptomycin per ml. The swabs were stored at -70°C until analyzed. Three embryonated chicken eggs incubated for 9 days were inoculated with 0.2 ml per egg. After 72 h, the allantoic fluid was harvested. A hemagglutination assay (HA) was performed following standard procedures. When at least one of the eggs was positive in the HA, the swab was considered to be positive.
HI Assay. This assay was performed by standard methods. Briefly, the test was performed in V-bottom 96-well microtiter plates with 8 hemagglutinating units of H7N7 challenge virus and 1% specific-pathogen-free chicken erythrocytes.
Sequencing of the Hemagglutinin. Before sequencing, the antigen was extracted from the vaccines as described in ref. 15: Vaccine (2 ml) was mixed with 8 ml of isopropylmyristate (Sigma). The mixture was centrifuged at 1,000 × g for 10 min, and the water phase was collected. Viral RNA was extracted by using the High Pure Viral Nucleic Acid kit (Roche Applied Science; Indianapolis). RT-PCR of the hemagglutinin was performed, and the PCR products were sequenced. The protein sequences of the HA1 were compared by using blastp 2.2.
Antigen Content of the Vaccines. Antigen was extracted from the vaccines as described above. A series of diluted BSA standard (Pierce) (600, 500, 400, 300, and 200 ng) and the vaccines (5, 4, and 2 μl) were run on a 12% denaturating Bis·Tris gel (NuPAGE, Invitrogen). The gel was stained for 60 min in SYPRO-orange dye (Molecular Dynamics) in 7.5% (vol/vol) acetic acid, washed for 1 minin7.5%(vol/vol) acetic acid, and scanned on a Storm 860 laser scanner (Molecular Dynamics) (16). Bands were quantified with imagequant 5.1 software (Molecular Dynamics).
Statistical Analysis. The analysis of the transmission experiments is based on a stochastic SEIR epidemic model in which individuals are susceptible (S), latently infected (i.e., infected but not yet infectious) (E), infected and infectious (I), and recovered and immune or dead (R). Throughout, the analyses are aimed at estimation of the (basic) reproduction ratio (R). The reproduction ratio is defined as the mean number of infections that would be caused by a single infected individual in a large population of susceptible animals. If R > 1, an infected animal infects, on average, >1 susceptible animal, and a chain reaction of infections may occur. If R < 1, a prolonged chain reaction of infections is not possible, and the epidemic comes to a halt. In our context, the reproduction ratio is given by the product of the mean infectious period E(TI) (dimension, time) and the transmission rate parameter β (dimension, time-1), R = βE(TI).
We used two different methods to estimate the reproduction ratio: (i) final-size methods and (ii) a generalized linear model (GLM). The appeal of final-size methods is that they are flexible and robust (17, 18). For instance, the final size does not depend on whether or not there is a period of latency, and different assumptions on the infectious-period distribution are easily incorporated. On the other hand, final-size methods do not make use of all of the information and do not allow separate estimation of the transmission-rate parameter and infectious period. For this purpose, the GLM is appropriate (13). The final size of an experiment is given by the number of contact animals that has been infected when the infection chain has ended. Central to our analysis is the fact that final-size distributions can be determined under a wide range of assumptions. Specifically, the probability p(k) of an outbreak of size k in a population where, initially, s0 uninfected and i0 infected animals are present is determined recursively from the equation
 |
[1] |
(17, 18), where L[z] is the Laplace transform of the infectious-period probability distribution. We focus on two extreme scenarios, one in which the infectious period is exponentially distributed and one in which the infectious period is of fixed duration. If the infectious period is exponentially distributed, L is given by L[z] = 1/(1 + (z/a), whereas, if the infectious period is of fixed duration, L is given by L[z] = e-z/a. By rescaling the time axis, we may measure time in units of the expected infectious period (17). As a consequence, E(TI) = 1 and R = βE(TI) = β. In other words, we may, without loss of generality, take α = 1 and equate β with R in the final-size equation (1).
With formulas for the final size at hand, it is possible to obtain estimates of the reproduction ratio by maximum likelihood (19, 20). Estimates of the reproduction ratio based on a final-size analysis are labeled by Rfix, in the case of an infectious period of fixed duration, and by Rexp, in the case of an exponentially distributed infectious period. Exact 95% confidence intervals are obtained by finding all r values for which the hypothesis H0:R = r is not rejected, i.e., by finding all values of r with a P value >0.05 (21).
In the same manner, exact tests of R against the threshold value 1 are performed (21). Furthermore, taking the difference in the number of contact infections between treatments as a natural test statistic, it is possible to make comparisons between treatments based on R, i.e., to test whether Rvaccine = Rcontrol. All calculations are carried out by using mathematica 5.1.
To take the time course of the experimental epidemics into account, two approaches are possible. One could take a Bayesian approach using methodology based on the likelihood of the data under the SEIR model (22). The Bayesian approach allows one to include prior information in the estimation procedure. Here, however, we take a standard approach based on a generalized linear regression. Specifically, assuming a fixed latent period, we estimated the transmission parameter β of the SEIR model by means of a GLM (13, 20, 23).
To this end, the data in Tables 2, 3, 4 are rendered into the format (S(t), I(t), C(t), Δt), where S(t) is the number of susceptible chickens at the beginning of a time period of length Δt, I(t) represents the average number of infectious chickens in this time period, and C(t) represents the number of new infections that have appeared. As in ref. 13, we assume a latent period of 2 days. The total number of chickens that are alive is also relevant and is denoted by N(t). By standard reasoning (13, 20), we accept that the number of cases is binomially distributed with parameter
 |
[2] |
(the probability of infection) and binomial totals S(t):
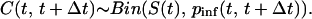 |
[3] |
The model specified by Eqs. 2 and 3 can be formulated as a GLM with a complementary log-log link function, taking log(I(t)/(N(t))) as offset variable. The intercept of this generalized regression estimates log(β). The analyses are carried out by using genstat 6.0.
Table 2. Transmission of H7N7 in unvaccinated chickens.
| Day after challenge
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 14 | 16 | 21 |
| I | +/+ | +/+ | † | ||||||||||
| I | +/+ | +/+ | +/+ | † | |||||||||
| I | +/+ | +/+ | +/+ | † | |||||||||
| I | +/+ | +/+ | +/+ | +/+ | † | ||||||||
| I | +/+ | +/+ | +/+ | † | |||||||||
| S | nd | +/- | +/- | +/+ | +/+ | † | |||||||
| S | nd | -/- | +/- | +/+ | +/+ | +/+ | † | ||||||
| S | nd | +/- | +/- | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | -/+ | -/- | -/- | -/- |
| S | nd | -/- | +/- | +/+ | +/+ | +/+ | † | ||||||
| S | nd | +/- | +/- | +/- | +/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/- |
| I | +/+ | +/+ | +/+ | +/+ | † | ||||||||
| I | +/- | +/+ | +/+ | +/+ | † | ||||||||
| I | +/+ | +/+ | +/+ | +/+ | † | ||||||||
| I | +/- | +/+ | +/+ | +/+ | † | ||||||||
| I | +/+ | +/+ | +/+ | +/+ | +/+ | † | |||||||
| S | nd | -/- | +/+ | +/+ | +/+ | +/+ | † | ||||||
| S | nd | -/- | +/- | +/+ | +/+ | +/+ | † | ||||||
| S | nd | -/- | +/- | +/+ | +/+ | +/+ | † | ||||||
| S | nd | -/- | +/- | +/- | +/+ | +/+ | † | ||||||
| S | nd | -/- | +/- | +/- | +/- | +/+ | +/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/- |
Chickens were challenged with 0.2 ml of diluted allantoic fluid containing 106 EID50 per ml (0.1/ml intranasally and 0.1 ml intratracheally) of A/Chicken/Netherlands/621557/03. I, inoculated chicken; S, contact chicken; †, chicken died; nd, not determined; EID50, egg-infectious dose; +/+, positive trachael swab/positive cloacal swab; +/-, positive trachael swab/negative cloacal swab; -/+, negative trachael swab/positive cloacal swab; -/-, negative trachael swab/negative cloacal swab.
Table 3. Transmission of H7N7 in vaccinated chickens 7 days after vaccination.
| Day after challenge
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vaccine | Chicken | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 14 |
| H7N1 | I | +/- | -/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | nd/- |
| I | +/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | nd/- | |
| I | +/- | +/- | +/- | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/- | nd/- | |
| I | +/- | +/+ | -/+ | -/+ | -/- | -/+ | -/- | -/+ | -/- | -/- | nd/- | |
| I | +/- | -/- | -/+ | -/+ | -/+ | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/+ | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| I | +/- | +/- | -/+ | -/- | -/- | -/+ | -/- | -/- | -/- | -/- | nd/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| I | +/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/- | -/+ | -/- | nd/- | |
| I | +/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| I | +/+ | +/- | +/+ | +/+ | -/+ | -/+ | -/- | -/- | -/- | -/+ | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| H7N3 | I | +/+ | +/+ | +/+ | +/+ | +/+ | -/+ | -/+ | -/- | -/+ | -/+ | nd/- |
| I | +/- | +/+ | +/+ | +/+ | -/+ | -/+ | -/- | -/- | -/- | -/+ | nd/- | |
| I | +/- | +/+ | +/+ | -/+ | -/+ | +/+ | -/+ | -/- | -/- | -/- | nd/- | |
| I | +/- | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/- | -/- | -/- | nd/- | |
| I | +/+ | +/+ | +/+ | +/+ | +/+ | -/+ | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | +/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | +/- | -/- | +/- | -/- | -/+ | -/+ | -/- | -/- | nd/- | |
| S | nd | -/- | +/- | -/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/+ | +/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | +/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| I | +/- | +/- | -/+ | -/+ | -/+ | -/- | -/- | -/- | -/- | -/- | nd/- | |
| I | +/- | +/+ | -/+ | +/+ | -/+ | -/+ | -/- | -/- | -/- | -/- | nd/- | |
| I | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/+ | |
| I | +/+ | +/+ | +/+ | -/+ | -/+ | -/+ | -/+ | -/+ | -/- | -/- | nd/- | |
| I | +/- | +/+ | -/+ | +/+ | -/+ | -/+ | -/+ | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | +/- | -/- | -/- | -/- | +/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | nd/- | |
Chickens were challenged 7 days after vaccination with 0.2 ml of diluted allantoic fluid containing 106 EID50 per ml (0.1 ml intranasally and 0.1 ml intratracheally) of A/Chicken/Netherlands/621557/03. I, inoculated chicken; S, contact chicken; nd, not determined; EID50, egg-infectious dose; +/+, positive trachael swab/positive cloacal swab; +/-, positive trachael swab/negative cloacal swab; -/+, negative trachael swab/positive cloacal swab; -/-, negative trachael swab/ negative cloacal swab; nd/-, trachael swab not determined/negative cloacal swab.
Table 4. Transmission of H7N7 in vaccinated chickens 14 days after vaccination.
| Day after challenge
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vaccine | Chicken | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| H7N1* | I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| H7N3* | I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| I | -/- | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
| S | nd | -/- | -/- | -/- | -/- | -/- | -/- | -/- | |
Chickens were challenged 14 days after vaccination with 0.2 ml of diluted allantoic fluid containing 106 EID50 per ml (0.1 ml intranasaly and 0.1 ml intratracheally) of A/Chicken/Netherlands/621557/03. I, inoculated chicken; S, contact chicken; nd: not determined; EID50, egg-infectious dose; -/-, negative trachael swab/negative cloacal swab.
Experiments were performed in duplicate. The results of the replicates were identical; therefore, only one replicate is shown
The infectious periods are directly observed from the infected contact animals. Hence, estimation of the infectious period and the construction of confidence interval is straightforward. An estimate of the reproduction ratio is given by the product of the estimates of the transmission parameter and infectious period (4, 13).
Results
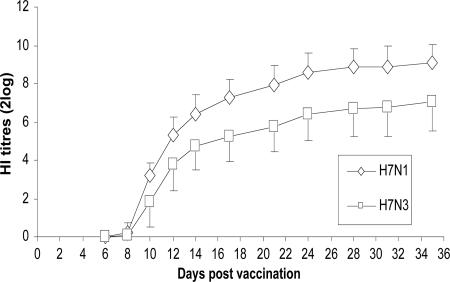
Kinetics of the Antibody Response. The antibody response in chickens vaccinated with H7N1 and H7N3 vaccine is given in Fig. 1 (and see Tables 7-8, which are published as supporting information on the PNAS web site). Antibodies were detectable by the HI assay from day 8 postvaccination. A significant difference between the geometric mean HI titers of the H7N1- and the H7N3-vaccinated groups developed: At day 35, the mean HI titer in the H7N1-vaccinated group is two 2 log steps higher than in the H7N3-vaccinated group. These differences in HI titers between the vaccines are also observed in the transmission experiments (see Tables 8-12, which are published as supporting information on the PNAS web site).
Fig. 1.
Serological response (HI titers) of chickens after vaccination. Groups of 40 chickens were vaccinated with H7N1 (0.5 ml per chicken i.m.) or with H7N3 (0.3 ml per chicken i.m.). Standard errors are given by vertical bars.
Transmission Experiments with Unvaccinated Chickens. To determine the transmission characteristics of H7N7 virus in a susceptible host population, we performed transmission experiments with unvaccinated chickens (experiment 1). All inoculated and contact chickens became positive by virus isolation (Table 2). However, although all inoculated chickens died within 2-5 days, two contact chickens in the first group and one contact chicken in the second group survived the infection. The estimate of the reproduction ratio based on the final-size method with a fixed infectious period is Rfix = ∞ [95% CI = (1.33-∞)] (Table 5). The estimate of the reproduction ratio based on the final-size method with an exponentially distributed infectious period gives Rexp = ∞ (1.30-∞), and the GLM is in good agreement with this estimate (Table 5). These experiments confirm that the H7N7 virus spreads easily in an unvaccinated population and can readily cause a major outbreak.
Table 5. Overview of the statistical analyses of the group experiments.
| Vaccine | Final size | Rfix (95% CI) | P H0:R ≥ 1 | P H0:Rv = Rc* | Infectious period (day) (95% CI) | Transmission parameter (day-1) (95% CI) | RGLM (95% CI) |
|---|---|---|---|---|---|---|---|
| Unvaccinated | 5.5 | ∞ (1.3-∞) | 1 | 6.3 (3.9–8.7) (n = 10) | 33 (n = 2) | 208 | |
| H7N1 (1 week)† | 1.0 | 0.2 (0.005–1.1) | 0.04 | <0.001 | 1 (n = 1) | 0.030 (0.01–0.09) (n = 18) | 0.03 |
| H7N3 (1 week)† | 5.1 | 1.4 (0.4–2.9) | 0.78 | 0.10 | 3.7 (0.7–6.7) (n = 6) | 0.30 (0.09–0.9) (n = 12) | 1.1 (0–3.1) |
| H7N1 (2 weeks)‡ | 0.0 | na§ | na§ | na§ | na§ | na§ | na§ |
| H7N3 (2 weeks)‡ | 0.0 | na§ | na§ | na§ | na§ | na§ | na§ |
CI, confidence interval; na, not applicable.
Rv, reproduction ratio among vaccinated chickens; Rc, reproduction ratio among unvaccinated chickens
Birds were challenged 7 days after vaccination
Birds were challenged 14 days after vaccination
No virus was detected from the inoculated or the contact chickens; therefore, no statistical analysis was performed
Transmission Experiments with Vaccinated Chickens. Does vaccination reduce transmission of HPAI viruses? To answer this question, experiments were done with chickens challenged 1 or 2 weeks after vaccination (Experiments 2-5). When challenged 1 week after vaccination with the H7N1 vaccine, all but one of the chickens in the inoculated groups became positive by virus isolation (Table 3). In the contact groups, one and none of the five chickens became positive. The chickens showed no signs of illness. Analysis of these data shows that the estimate of the reproduction ratio based on the final-size method is Rfix = 0.2 (0.005-1.1), which is significantly below 1 (P = 0.04, Table 5). The results based on the final-size method with an exponentially distributed infectious period [Rexp = 0.2 (0.005-1.4)] and based on the GLM, again, are in good agreement with this estimate. When challenged 1 wk after vaccination with the H7N3 vaccine, all challenged chickens became positive. In the contact group, five and one of the five chickens became positive. These chickens also showed no signs of illness. When analyzed, the estimate of the reproduction ratio based on the final-size method is Rfix = 1.4 (0.4-2.9) (Table 5). The results from the final-size method with an exponentially distributed infectious period [Rexp = 1.7 (0.4-4.3)] and the GLM yield similar results.
The transmission characteristics were also studied when chickens were challenged 2 wk after vaccination (experiments 4-5). All inoculated and contact chickens in the H7N1- and H7N3-vaccinated groups remained negative in the tracheal and cloacal swabs (Table 4). In these experiments, the SEIR model was not applied, because no virus could be detected, even from the inoculated chickens.
To decide whether transmission is significantly reduced by vaccination, the reproduction ratios of the vaccinated groups and the unvaccinated groups were compared. A significant difference is found for the H7N1 vaccine (P < 0.001) 1 wk after vaccination but not for the H7N3 vaccine (P = 0.1) (Table 5). In the groups challenged 2 wk after vaccination, the SEIR model is not applied, but it is obvious that no transmission occurs.
Transmission from Vaccinated to Unvaccinated Chickens. Do vaccinated chickens still excrete virus and pose a threat of infection to unvaccinated chickens? To answer this question, we carried out so-called pair experiments with one inoculated vaccinated chicken and one unvaccinated contact chicken (experiments 6-9; Table 1).
For both vaccines, transmission was still observed when the experiments were carried out 1 wk after vaccination (experiments 6-7; and see Table 13, which is published as supporting information on the PNAS web site). For the H7N1 vaccine, all four inoculated animals became positive by virus isolation, and three of four unvaccinated contact chickens were infected. For the H7N3 vaccine, all of the vaccinated and unvaccinated chickens became positive by virus isolation. In both experiments, the contact animals became positive at day 3 or 4, indicating efficient transmission from the vaccinated to unvaccinated chickens. The infected contact animals died from the infection, whereas the inoculated chickens showed no signs of illness and survived.
When the pair experiments were carried out 2 wk after vaccination (experiments 8-9), none of the inoculated and contact chickens became positive by virus isolation, and all chickens survived. These experiments show that no transmission is possible from vaccinated to fully susceptible contact chickens 2 wk after vaccination.
Discussion
Our transmission experiments demonstrate that vaccination not only protects chickens against disease symptoms and mortality but is also an effective strategy to reduce transmission. Specifically, when challenged 2 wk after vaccination, transmission of the virus is completely halted, and a major outbreak can be prevented. In our experiments, 1 wk after vaccination with the H7N1 vaccine, some transmission is still observed, but the reproduction ratio is already significantly <1. This finding implies that an introduction of the virus may cause a small number of secondary infections, but the virus probably cannot spread extensively. In contrast, we were unable to disprove the hypothesis that R ≥ 1 for the H7N3 vaccine 1 wk after vaccination (P = 0.78). This finding may indicate that the H7N3 vaccine does not sufficiently reduce transmission 1 wk after vaccination (i.e., R > 1), but it could also indicate that the number of replicates was too small (i.e., the power of the experimental setup was not sufficiently high). A (two-sided) test of the hypothesis RH7N3 = RH7N1 against the alternative RH7N3 ≠ RH7N1 yields P = 0.10, indicating that there is marginal evidence that the H7N1 vaccine performs better than the H7N3 vaccine. Summarizing, we have shown that vaccination can be an attractive tool to prevent outbreaks of highly pathogenic AI viruses in poultry, thereby achieving the aim of eliminating the source of human infections.
Whether transmission between two chickens occurs depends on the infectiousness of the infected chicken and the susceptibility of the uninfected chicken. The reproduction ratio is a composite measure that incorporates both factors (24). An indication for infectiousness is the amount of virus shed, which is reduced by vaccination (10, 11). This finding was confirmed by our experiments: When chickens were challenged 2 wk after vaccination, no transmission occurred, not even to unvaccinated chickens in close contact, suggesting that vaccination reduces infectiousness. The minimum infective dose can be used as a proxy for susceptibility. For example, in vaccinated turkeys, the minimum infective dose was higher than in unvaccinated turkeys (25). Our experiments confirm this finding: When the contact chickens were vaccinated (experiments 2-3), the number of contact infections was lower than expected from the experiments where the contact chickens were not vaccinated (experiments 6-7). This decrease in transmission can be attributed to the decrease in susceptibility of the contact chickens, because the infectiousness in both experiments is the same. We conclude that vaccination reduces the infectiousness of infected chickens as well as the susceptibility of uninfected chickens.
The two vaccines differed in their effect on transmission. Two important factors that determine the effectiveness of a vaccine are antigen content (11, 26) and antigenic differences between the vaccine and the challenge virus (11, 27). In the H7N1 vaccine, the hemagglutinin-antigen content per dose was higher than in the H7N3 vaccine (H7N1, 22 μg per dose and H7N3, 4 μg per dose). The other factor is the sequence similarity of the hemagglutinin, which is correlated with the reduction of virus-shedding from the oropharynx or trachea (9, 27, 28). The H7N1 vaccine had higher homology with the challenge virus (98%) than did the H7N3 vaccine (92%). These differences in antigen content and sequence homology are in agreement with the observed differences between the two vaccines. To what extent antigen content and homology contribute to transmission reduction is a question that merits further investigation.
Is there a relationship between the immune response that develops after vaccination and the reduction of transmission? Antibodies against the hemagglutinin evoke the major protection against infection with HPAI (29). HI titers after vaccination with H7N1 and H7N3 are detectable from day 8 (Fig. 1). These results are in good agreement with HI titers from our transmission experiments (see Table 6). Remarkably, despite the low titers at days 8-10 after vaccination (days 1-3 after challenge), there is already a considerable reduction in transmission, suggesting that other immune mechanisms might contribute to protection or that low HI titers are already effective in preventing infection.
The choice of a vaccine is an important issue. In poultry, a number of vaccines are available for use against highly pathogenic H5 and H7 influenza A viruses. Given the choice, which of these vaccines should be used? In our opinion, besides safety aspects and side-effects, the most important requirement is that the vaccine is effective in preventing viral spread. As we have shown, transmission experiments are eminently suited to address this question. Our experiments indicate that vaccines against highly pathogenic H7N7 influenza virus can completely block transmission 2 wk after vaccination. We conclude that vaccination of poultry can be an effective tool to prevent the spread of highly pathogenic AI viruses.
Supplementary Material
Acknowledgments
We thank Marieke Tijms, Marlies Kolkman, and John Voermans for technical assistance. Two reviewers are gratefully acknowledged for their helpful comments. This work was supported by European Union Grant QLRT-CT2001-01454 (AVIFLU Project) and by the Dutch Ministry of Agriculture, Nature, and Food Quality.
Author contributions: J.A.v.d.G., G.K., M.C.M.d.J. \tand M.v.B. designed research; J.A.v.d.G. and G.K. performed research; M.C.M.d.J. and M.v.B. analyzed data; and J.A.v.d.G. and M.v.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AI, avian influenza; GLM, generalized linear model; HI, hemagglutination inhibition; HPAI, highly pathogenic AI; R, reproduction ratio; SEIR, susceptible, latently infected, infectious, recovered.
References
- 1.Claas, E. C. J., Osterhaus, A. D. M. E., Van Beek, R., De Jong, J. C., Rimmelzwaan, G. F., Senne, D. A., Krauss, S., Shortridge K. F. & Webster R. G. (1998) Lancet 351, 472-477. [DOI] [PubMed] [Google Scholar]
- 2.Capua, I. & Marangon, S. (2000) Avian Pathol. 29, 289-294. [DOI] [PubMed] [Google Scholar]
- 3.Suarez, D. L., Senne, D. A., Banks, J., Brown, I. H., Essen, S. C., Lee, C. W., Manvell, R. J., Mathieu-Benson, C., Mareno, V., Pedersen, J., et al. (2004) Emerg. Infect. Dis. 10, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stegeman, A., Bouma, A., Elbers, A. R. W., de Jong, M. C. M., Nodelijk, G., De Klerk, F., Koch, G. & van Boven, M. (2004) J. Infect. Dis. 190, 2088-2095. [DOI] [PubMed] [Google Scholar]
- 5.Bowes, V. A., Ritchie, S. J., Byrne, S., Sojonky, K., Bidulka, J. J. & Robinson, J. H. (2004) Avian Dis. 48, 928-934. [DOI] [PubMed] [Google Scholar]
- 6.Li, K. S., Guan, Y., Wang, J., Smith, G. J., Xu, K. M., Duan, L., Rahardjo, A. P., Puthavathana, P., Buranathai, C., Nguyen, T. D., et al. (2004) Nature 430, 209-213. [DOI] [PubMed] [Google Scholar]
- 7.De Jong, J. C., Claas, E. C. J., Osterhaus, A. D. M. E., Webster, R. G. & Lim, W. L. (1997) Nature 389, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier, R. A. M., Schneeberger, P. M., Rozendaal, F. W., Broekman, J. M., Kemink, S. A. G., Munster, V. Kuiken, T., Rimmelzwaan, G. F., Schutten, M., Van Doornum, G. J. J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swayne, D. E., Perdue, M. L., Beck, J. R., Garcia, M. & Suarez, D. L. (2000) Vet. Microbiol. 74, 165-172. [DOI] [PubMed] [Google Scholar]
- 10.Capua, I., Terregino, C., Cattoli, G., Mutinelli, F. & Rodriguez, J. F. (2002) Avian Pathol. 32, 47-55. [DOI] [PubMed] [Google Scholar]
- 11.Swayne, D. E., Beck, J. R., Garcia, M. & Stone, H. D. (1999) Avian Pathol. 28, 245-255. [DOI] [PubMed] [Google Scholar]
- 12.de Jong, M. C. M. & Kimman T. G. (1994) Vaccine 12, 761-766. [DOI] [PubMed] [Google Scholar]
- 13.van der Goot, J. A., de Jong, M. C. M., Koch, G. & van Boven, M. (2003) Epidemiol. Infect. 31, 1003-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Council Directive 92/40/EEC (1992) Off. J. Eur. Communities L 167, 1-16. [Google Scholar]
- 15.Maas, R. A., Komen, M., Van Diepen, M., Oei, H. L. & Claassen, I. J. T. M. (2003) Vaccine 21, 3137-3142. [DOI] [PubMed] [Google Scholar]
- 16.Steinberg, T. H., Jones, L. J., Haughland, R. P. & Singer, V. L. (1996) Anal. Biochem. 239, 223-237. [DOI] [PubMed] [Google Scholar]
- 17.Ball, F. (1986) Adv. Appl. Prob. 18, 289-310. [Google Scholar]
- 18.Ball, F. (1995) in Epidemic Models: Their Structure and Relation to Data, ed. Mollison, D. (Cambridge Univ. Press, Cambridge, U.K.), pp. 34-52.
- 19.Bailey, N.T.J. (1975) The Mathematical Theory of Infectious Diseases and Its Applications (Griffin, London).
- 20.Becker, N. G. (1989) Analysis of Infectious Disease Data (Chapman and Hall, London).
- 21.Kroese, A. H. & de Jong, M. C. M. (2001) in Proc. Soc. Vet. Epidemiol. Prevent. Med., eds. Menzies, F. D. & Reid, S. W. J. (SVEPM Proceedings series, Noorwijkerhout, The Netherlands), pp. 21-37.
- 22.Streftaris, G. & Gibson, G. J. (2004) Proc. R. Soc. London B 271, 1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCullagh, P. & Nelder, J. A. (1989) Generalized Linear Models (Chapman and Hall, London).
- 24.Diekmann, O. & Heesterbeek, J. A. P. (2000) Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation, ed. Levin, S. (Wiley, Chichester, U.K.).
- 25.Capua, I., Terregino, C., Cattoli, G. & Toffan, A. (2004) Avian Pathol. 33, 158-163. [DOI] [PubMed] [Google Scholar]
- 26.Brugh, M., Beard, C. W. & Stone, H. D. (1979) Am. J. Vet. Res. 40, 165-169. [PubMed] [Google Scholar]
- 27.Lee, C. W., Senne, D. A. & Suarez, D. L. (2004) J. Virol. 78, 8372-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swayne, D. E., Garcia, M., Beck, J. R., Kinney, N. & Suarez, D. L. (2000) Vaccine 18, 1088-1095. [DOI] [PubMed] [Google Scholar]
- 29.Webster, R. G., Kawaoka, Y., Weinberg, R. & Paoletti, E. (1991) Vaccine 9, 303-308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.