Abstract
ATP-binding cassette (ABC) transporters couple ATP binding and hydrolysis to the movement of substances across the membrane; conformational changes clearly play an important role in the transporter mechanism. Previously, we have shown that a dimer of MalK, the ATPase subunit of the maltose transporter from Escherichia coli, undergoes a tweezers-like motion in a transport cycle. The MalK monomer consists of an N-terminal nucleotide binding domain and a C-terminal regulatory domain. The two nucleotide-binding domains in a dimer are either open or closed, depending on whether ATP is present, while the regulatory domains maintain contacts to hold the dimer together. In this work, the structure of MalK in a posthydrolysis state is presented, obtained by cocrystallizing MalK with ATP-Mg2+. ATP was hydrolyzed in the crystallization drop, and ADP-Mg2+ was found in the resulting crystal structure. In contrast to the ATP-bound form where two ATP molecules are buried in a closed interface between the nucleotide-binding domains, the two nucleotide-binding domains of the ADP-bound form are open, indicating that ADP, unlike ATP, cannot stabilize the closed form. This conclusion is further supported by oligomerization studies of MalK in solution. At low protein concentrations, ATP promotes dimerization of MalK, whereas ADP does not. The structures of dimeric MalK in the nucleotide-free, ATP-bound, and ADP-bound forms provide a framework for understanding the nature of the conformational changes that occur in an ATP-binding cassette transporter hydrolysis cycle, as well as how conformational changes in MalK are coupled to solute transport.
Keywords: ATP-binding cassette transporter, conformational change, posthydrolysis state, x-ray crystallography
Active transport is one of the basic mechanisms evolved to mediate traffic across cellular membranes. ATP-binding cassette (ABC) transporters are ubiquitous membrane protein complexes that use the energy generated from ATP binding and hydrolysis to selectively transport solutes across biological membranes (1). Genetic defects in these proteins have been implicated in more than a dozen disease states involving transport deficiencies (2), and many cancer cells become resistant to drugs during chemotherapy because of overexpression of a number of ABC transporters (3). Most ABC transporters contain two membrane-spanning domains or subunits and two highly conserved nucleotide-binding domains (NBDs) or subunits (also called ABCs) located at the cytosolic surface of the membrane. Crystallographic studies of isolated NBDs or subunits (4–15) show that almost all NBDs share a conserved fold, suggesting that the mechanism of coupling of transport to ATP hydrolysis may be conserved as well.
The maltose/maltodextrin transporter from Escherichia coli has long been a prototype for studies of ABC transporters. The membrane-associated components of this transporter form a complex containing one copy each of the membrane-spanning MalF and MalG proteins and two copies of the ATPase MalK. Bacterial uptake systems also require a periplasmic or cell surface-associated substrate-binding protein that binds nutrients with high affinity before translocation, and the periplasmically located maltose-binding protein (MBP or MalE) fills this role for the maltose transporter (16). The MalK subunit differs from most ABC ATPases in that it contains a C-terminal regulatory domain (RD) in addition to the highly conserved NBD. Recent structures of MalK reveal that the RD also mediates contact between two MalK monomers, stabilizing the MalK dimer and making it possible to directly compare the dimer structures of MalK in different conformational states (12). In nucleotide-free structures, the N-terminal NBDs are open, or separated, and the dimer is maintained solely through contacts with the C-terminal RDs. In the ATP-bound form, the NBDs are closed with two ATPs bound along the dimer interface, burying ≈2,000 Å2 of accessible surface per subunit.
In this work, we describe biochemical and x-ray crystallographic studies of the E. coli MalK dimer. Size-exclusion chromatography reveals that MalK forms a dimer in solution at concentrations of 20 μM or higher, indicating that the dimeric forms observed in crystal structures are not the result of crystal packing forces. In addition, we show that at 0.1 μM protein concentration, ATP promotes dimerization of MalK, whereas ADP does not. The structure of MalK in the posthydrolysis conformational state (ADP-bound) was determined at 2.3 Å by cocrystallizing MalK with ATP-Mg2+ and by crystallizing MalK directly with the hydrolysis product ADP-Mg2+. Both structures reveal ADP and Mg2+ at the nucleotide-binding sites, and the N-terminal NBDs are open with minimum intermolecular contacts. These studies support the hypothesis that ATP hydrolysis is required to reset the transporter to the resting state by opening the contact between the two NBDs, providing a possible mechanism of coupling ATP hydrolysis to maltose transport.
Methods
Oligomerization Assay. E. coli MalK with a C-terminal 10-aa extension, ASASHHHHHH, was overexpressed and purified as described in ref. 12. Protein samples of equivalent volume (500 μl) were loaded onto a QC-PAK GFC 200 gel filtration column (TOSOH Bioscience, Grove City, OH) at 22°C. Absorbances were monitored at both 280 and 215 nm because there was poor signal-to-noise ratio at 280 nm for runs containing nucleotide. The positions of the 280- and 215-nm elution peaks overlapped for all runs. Samples run in the presence of running buffer (150 mM NaCl/20% glycerol/5 mM 2-mercaptoethanol/0.1 mM EDTA/25 mM Tris·HCl, pH 8.0) containing nucleotides (0.5 mM ATP or 0.5 mM ADP + 10 mM MgCl2) were incubated at their injection concentration with the appropriate nucleotide solution for a minimum of 30 min before sample injection.
Crystallization and Structure Determination. Protein stock (20 mg/ml MalK, 5 mM ATP-Mg2+, or 5 mM ADP-Mg2+ in buffer containing 150 mM NaCl/20% glycerol/5 mM 2-mercaptoethanol/25 mM Tris·HCl, pH 8.0) was mixed with crystallization buffer [0.2 M magnesium formate, 15∼20% poly(ethylene glycol) 3350/15% glycerol] at a 1:1 ratio. Crystals grew overnight at 20°C and were flash frozen in liquid nitrogen. Data were collected on beamline SBC19BM at the Advanced Photon Source and processed with hkl2000 (17).
The structure of MalK obtained by cocrystallization with ATP-Mg2+ was determined by the molecular replacement method using amore (18) and the coordinates of a monomeric MalK (Protein Data Bank ID code 1Q1B). Three solutions were found by a rotation and translation search, which yielded one MalK monomer and one MalK dimer per asymmetric unit cell. The coordinates of the dimeric solutions then were used as the search model in a second run of amore. Two dimers were found and displayed reasonable crystal packing. The model was improved by iterative model building in the program o (19) and rigid-body, simulated annealing, individual B factor and energy minimization refinement using the cns (20) program initially with noncrystallographic symmetry restraints, which were subsequently released in the final runs of refinement. Data collected from the ADP-Mg2+ cocrystallized crystal were used to refine the model obtained from ATP-Mg2+ cocrystallization. In both forms, there were four monomers of MalK per asymmetric unit, with each pair (A and B, C and D) arranged in two dimers. The higher-resolution structure (2.3 Å) contains residues 2–86, 108–121, and 143–371 of monomer A; residues 2–371 of monomer B; residues 2–10, 18–52, and 76–371 of monomer C; and residues 2–80 and 151–371 of monomer D.
Results
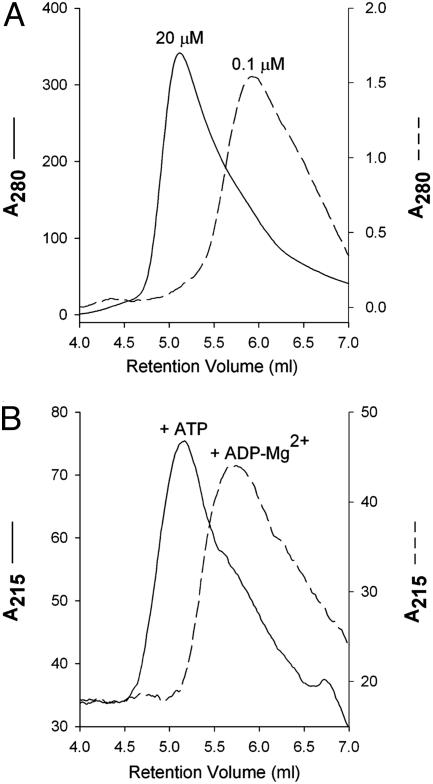
Oligomerization States of E. coli MalK in Solution. Despite the fact that NBDs function as dimers, many isolated ABC transporter ATPases, such as the histidine permease HisP (21) and the Thermococcus litoralis maltose transport MalK (22) purified as monomers. In contrast, E. coli MalK, at a concentration of 20 μM or higher, migrated as a dimer on a gel-filtration column (Fig. 1A). This result is consistent with a previously reported result that MalK forms a dimer independent of its assembly into the MalFGK2 complex (23). Although the isolated MalK hydrolyzes ATP slowly (turnover number of 4 per min), the intact MalFGK2 exhibits greatly enhanced rates, on the order of 10 per sec (24). At a lower protein concentration, 0.1 μM, MalK migrates as a monomer in the absence of nucleotides (Fig. 1A), suggesting that the Kd for the nucleotide-independent dimerization of MalK is in the low micro-molar range. At concentrations between 0.1 and 20 μM, MalK elutes from the size-exclusion column at intermediate positions between the monomer and dimer peaks, reflecting a monomer–dimer equilibrium at these concentrations (data not shown). To test whether nucleotides promote dimerization of MalK, protein samples at monomeric concentration were mixed with 0.5 mM ADP-Mg2+ or ATP-EDTA and then analyzed on a size-exclusion column preequilibrated with buffer containing 0.5 mM of the corresponding nucleotide solution. The elution profiles show that MalK elutes as a dimer in the presence of ATP and monomer in the presence of ADP-Mg2+ (Fig. 1B).
Fig. 1.
Dimerization assays of MalK by gel filtration. (A) Concentration-dependent dimerization in the absence of nucleotides. MalK at a 20 μM concentration (peak fraction) elutes at the expected position of a dimer, whereas at a concentration of 0.1 μM MalK elutes as a monomer. (B) Nucleotide-dependent dimerization at monomeric protein concentration. MalK stock at 1 μM concentration was mixed with 0.5 mM ATP (solid line) or 0.5 mM ADP and 10 mM MgCl2 (dashed line) and loaded to a size-exclusion column preequilibrated with buffer containing the corresponding nucleotide. The elution peak fraction contains MalK at ≈0.1 μM concentration. Note that in both A and B, for the purpose of comparison, the profiles do not use the same scale. Absorbance is shown in mAU. Figures were prepared with sigmaplot(Systat, Point Richmond, CA).
Structure of MalK in the Posthydrolysis State. Two methods were used to obtain crystals of MalK in the ADP-bound form. The posthydrolysis state of MalK was obtained by cocrystallizing MalK with ATP-Mg2+ in the absence of EDTA. Because isolated MalK shows ATPase activity in solution, we expected that MalK would hydrolyze most, if not all, of the ATP in the crystallization drop, resulting in ADP-bound MalK crystals. Alternatively, the product, ADP-Mg2+, was directly mixed with MalK, and crystals were obtained under the same conditions established for cocrystallization of MalK and ATP-Mg2+. The structure of MalK cocrystallized with ATP-Mg2+ was determined by molecular replacement method using nucleotide-free MalK monomer (12) as a search model (Table 1). The initial structure of MalK cocrystallized with ATP-Mg2+ was then used to determine the MalK structure cocrystallized with ADP-Mg2+. The conformation of MalK was essentially the same irrespective of the crystallization methods, with a rms deviation of 0.55 Å for all 698 Cα atoms in the dimer. The electron-density maps revealed that ADP and Mg2+ were bound in both crystal forms. In the remaining sections of this work, the structure obtained by means of ATP-Mg2+ cocrystallization will be considered as the representative of the product-bound form of MalK.
Table 1. Crystallographic and refinement statistics.
| Statistic | MalK* [ATP, Mg2+] | MalK† [ADP, Mg2+] |
|---|---|---|
| Data collection | ||
| Ligand | ADP, Mg2+ | ADP, Mg2+ |
| Wavelength, Å | 1.00 | 0.98 |
| Resolution range,‡ Å | 37-2.3 (2.4-2.3) | 50-2.8 (2.9-2.8) |
| No. of reflections | 70,224 | 40,629 |
| Completeness, % | 98.5 (89.1) | 92.0 (89.9) |
| Rsym | 0.074 (0.417) | 0.088 (0.374) |
| I/σI | 15.46 (2.55) | 17.52 (2.56) |
| Redundancy | 3.5 (3.0) | 3.6 (3.1) |
| Space group | P21 | P21 |
| a X b X c, Å | 70.3 × 102.7 × 130.8 | 70.2 × 102.0 × 131.5 |
| α X β X γ, o | 90 × 90.8 × 90 | 90 × 90.7 × 90 |
| No. of molecules per asymmetric unit | ||
| Refinement | ||
| Rwork,§ % | 23.0 | 23.9 |
| Rfree,¶ % | 27.1 | 27.7 |
| No. of reflections in the working set | 66,780 | 38,605 |
| No. of reflections in the test set∥ | 3,444 | 2,024 |
| No. of atoms | ||
| Protein | 10,355 | 10,897 |
| H2O | 206 | 0 |
| ADP | 108 (4 molecules) | 108 (4 molecules) |
| Mg2+ | 4 | 4 |
| Luzzati error, Å | 0.32 | 0.38 |
| rms deviations | 0.32 | 0.38 |
| Bond length, Å | 0.014 | 0.015 |
| Bond angle, o | 2.0 | 1.9 |
| % disallowed in | 0 | 0 |
| Ramachandran plot |
MalK was cocrystallized with ATP and Mg2+.
MalK was cocrystallized with ADP and Mg2+.
Numbers in parentheses refer to the highest resolution shell.
Rsym = ΣhΣi|Ij(h) - 〈I(h) 〉|/ΣhΣiI(h).
Rwork = Σ(|Fobs| - k|Fcal|)/Σ|Fobs|.
A random 5% of the reflection data was omitted in the refinement and used to calculate Rfree.
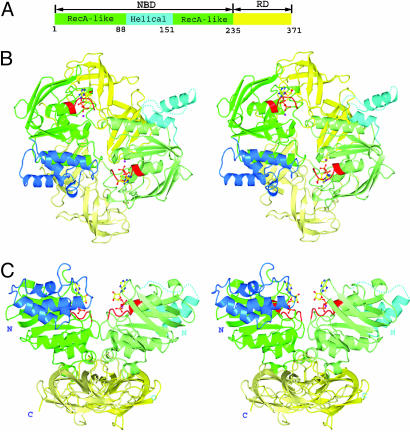
The MalK monomer consists of two domains, an N-terminal 235-residue NBD and a C-terminal 136-residue RD. The NBD can be further divided into two subdomains: a RecA-like subdomain (residues 1–87 and 152–235) containing Walker A and Walker B motifs involved in ATP-binding and hydrolysis (25) and a helical subdomain (residues 88–151) containing the ABC family signature motif LSGGQ (Fig. 2). The Q loop, which contains a highly conserved glutamine residue, joins the RecA-like subdomain to the helical subdomain. The two monomer structures in the ADP-Mg2+-bound dimer are very similar, with the exception of the relative orientation of the helical subdomain. The rms deviation is 0.52 Å over 162 Cα atoms out of 169 residues in the RecA-like subdomains and 0.36 Å over 104 Cα atoms out of 136 residues in the RDs (residues at crystal-packing contact regions are not included in the calculation). The relative orientations of the helical and RecA-like subdomains of the two monomers differ by ≈20°. The two helical subdomains of a dimer are involved in different contacts with neighboring molecules in the crystal lattice; therefore, this asymmetric orientation appears to be caused by the crystal-packing forces.
Fig. 2.
Ribbon diagram of the structure of E. coli. MalK homodimer with bound ADP-Mg2+.(A) Schematic diagram showing the subunit structure of a monomer. The NBD (residues 1–235) is in green/cyan and RD (residues 236–371) in yellow. Different colors further distinguish the subdomains or segments in the NBD as follows: green, RecA-like subdomain (residues 1–87 and 152–235); cyan, helical subdomain (residues 88–151). (B) Stereoview of the homodimer viewed down the local twofold axis. ADP is represented in ball-and-stick model (O atoms, red; N atoms, blue; C atoms, yellow; P atoms, orange; and magnesium, purple). Walker A motif is colored in red. The color schemes for the domains of the two monomers are similar, except that one is rendered in lighter color. (C) Stereoview of the homodimer obtained by a 90° clockwise rotation of the structure shown in B about a horizontal axis. Missing residues are shown as dashed lines. Figures were prepared with pymol (www.pymol.org)
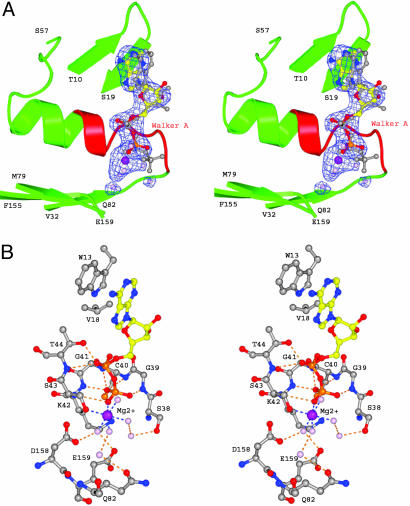
Nucleotide Binding. The electron-density map shows densities corresponding to an ADP and a Mg2+ molecule in the nucleotide-binding site next to the Walker A motif of each monomer (Fig. 3A). When the ATP- and ADP-bound structures of MalK are aligned based on the Walker A motifs, the α and β phosphates of ATP and ADP have identical conformation, whereas the ribose-base assemblies have slightly different orientations (Fig. 3A). This difference may result from additional contacts contributed by the second MalK subunit in the ATP-bound structure (Table 2). The adenosine ring and ribose of ADP are stabilized by ring-stacking interactions with the conserved aromatic residue Trp-13 and van der Waals interactions with Val-18. Several conserved residues in the Walker A motif, including Gly-39, Cys-40, Gly-41, Lys-42, and Thr-44, form hydrogen bonds with the oxygen atoms of the α or β phosphates (Fig. 3B and Table 2). The Mg2+ ion is coordinated by a total of six oxygen atoms: one from the side chain of Ser-43, one from the β phosphate of ADP, and those from four water molecules (Fig. 3). The angles and distances between the metal ion and the coordinating oxygen atoms agree well with those of an ideal octahedral coordination of an Mg–ligand complex (26). The position of the Mg2+ corresponding densities is very similar to those observed for Mg2+ in other high-resolution structures containing ADP-Mg2+ (8, 11). Similarly, the ATPase domain of TAP1 was crystallized in the presence of ATP and Mg2+, and the electron-density map showed density corresponding to ADP-Mg2+ (6) in a similar arrangement. Several conserved residues, including Asp-158 in Walker B and Glu-159, the putative catalytic base, form hydrogen bonds with the magnesium-coordinating waters. No density around the Walker A/B or LSGGQ motifs could be assigned to phosphate, indicating that the inorganic phosphate had already been released.
Fig. 3.
ADP binding. (A) Stereoview of the electron density (2σ contour level) of one of the bound ADP-Mg2+ obtained from a simulated annealing Fo – Fc map, with ADP-Mg2+ molecules omitted in the structure factor calculation. The ADP-Mg2+ is represented in ball-and-stick model. Water molecules are colored in pink. To illustrate that there is no density in the expected position of the γ phosphate, the ATP molecule also is shown in gray. The ATP model was obtained by aligning the Walker A motif of the ATP-bound form (12) with that of the ADP-bound form. (B) Atomic details of the interaction between MalK and ADP-Mg2+. Residues contacting the ADP and magnesium ion are labeled. Hydrogen-bonding and salt-bridge interactions are marked by dashed lines in orange and blue, respectively. Color identification for the residues is as follows: O atoms, red; N atoms, blue; C atoms, gray; and P and S atoms, orange.
Table 2. Comparison of the contacts with ADP and ATP.
| Motif*/residues | ADP (Mg2+) | ATP (EDTA) |
|---|---|---|
| W13 | Adenine (vdW†) | Adenine (vdW) |
| V18 CG1 | Ribose (vdW) | Ribose (vdW) |
| Walker A | ||
| S38 OG | O2γ (H-bond‡) | |
| G39 N | O3β (H-bond) | O3β (H-bond) |
| N | O2γ (H-bond) | |
| C40 N | O1β (H-bond) | O1β (H-bond) |
| G41 N | O1β (H-bond) | O1β (H-bond) |
| K42 N | O1β (H-bond) | O1β (H-bond) |
| NZ | O3γ (H-bond) | |
| S43 N | O2β (H-bond) | O2β (H-bond) |
| OG | Mg2+ (salt bridge) | O2β (H-bond) |
| T44 N | O1α (H-bond) | O2α (H-bond) |
| OG1 | O1α (H-bond) | O2α (H-bond) |
| H192 NE2 | O3γ (H-bond) | |
| R129 NH2 | Ribose (vdW) | |
| K132 O | Adenine (vdW) | |
| A133 CB | Adenine (vdW) | |
| LSGGQ | ||
| S135 OG | O2γ (H-bond) | |
| G136 N | O1γ (H-bond) | |
| G137 N | O2γ (H-bond) | |
| Q138 OE1 | Ribose (vdW) | |
| NE2 | Ribose (vdW) |
The Walker A and LSGGQ motifs contacting one ATP molecule belong to the two opposite Malk monomers.
van der Waals (vdW) distance is ≤4 Å.
Hydrogen bond (H-bond) distance is <3.4 Å.
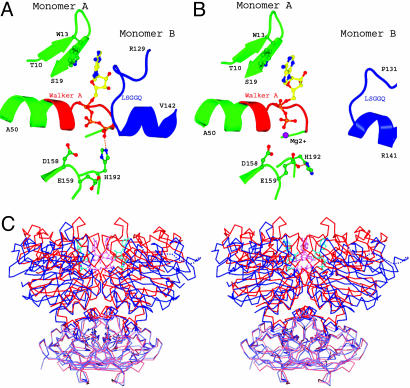
Conformational Change upon ATP Hydrolysis. Previously, we determined the dimeric structure of the MalK subunit in the ATP-bound form. EDTA was added in the crystallization drop to prevent ATP hydrolysis (12). The structure reveals two bound ATP molecules buried in the dimer interface, an architecture similar to those of Rad50 (27), an E171Q mutant of MJ0796 (9), and an H662A mutant of HlyB (28). In comparison with the ATP-bound dimer, major conformational changes occurred in the ADP-bound dimer that resulted in opening of the dimer interface between NBDs. In the ATP-bound form, each ATP molecule is sandwiched between the Walker A motif of one subunit and the LSGGQ motif of the other. The conserved residues Ser-135, Gly-136, and Gly-137 in the LSGGQ motif are engaged in extensive hydrogen bonds and van der Waals interactions with the ATP molecule (Table 2). Because most of the interactions are through the γ phosphate, it is clear that cleavage of the bond to the γ phosphate must destabilize the closed dimer, either through electrostatic repulsion (9) or release of inorganic phosphate. Indeed, in the posthydrolysis state of MalK, there is no interaction between the LSGGQ motif and ADP (Table 2), and the LSGGQ motif is withdrawn from the nucleotide by ≈10 Å in the binding site with unambiguous density for the LSGGQ motif (Fig. 4B). As a result of this opening at the nucleotide-binding interface, all dimer contacts mediated through bound nucleotide are completely lost in the ADP-bound form, leaving the two ADP molecules attached to the Walker A and B motifs of each monomer. Superimposition of both nucleotide-bound forms along the twofold symmetry axis reveals that the interactions between the RDs in both forms are essentially identical (Fig. 4C). The opening of the dimer interface is achieved by two steps of rigid-body rotation; the entire NBD is rotated outward ≈12° at residue 237 relative to the RD, and a second rotation of the helical subdomain relative to the RecA-like subdomain has occurred. Relative to the closed dimer structure, ≈8° and 28° bending angles of the helical subdomains generate the two asymmetric monomers in the ADP-bound form. The conformational change observed here fits well into our proposed tweezers-like motion of the NBDs, in which the two monomers of MalK are held together at the base by the RDs and the NBDs open and close like the tips of a pair of tweezers (12).
Fig. 4.
Conformational changes of MalK upon ATP hydrolysis. (A and B) The nucleotide-binding site of the ATP-bound dimer structure, with ATP sandwiched between the Walker A and B motifs of one monomer and the LSGGQ motif of the other monomer (A) and the posthydrolysis ADP-bound structure (B). The color scheme is similar to that of Fig. 2. (C) Stereoview of superimposed MalK structures in the ATP-bound (red) and ADP-bound (blue) forms. The RDs are rendered in lighter color than the NBDs. ATP (pink) and ADP (cyan) molecules are shown in ball-and-stick model.
In the ATP-bound form, the highly conserved His-192 important for both transport and hydrolysis (29, 30) lies adjacent to the Walker A and forms a strong hydrogen bond with the γ phosphate (Fig. 4A). His-192 is located in the dimer interface and is one of the few amino acids that contact the opposite monomer directly (12). In the ADP-bound form where the γ phosphate has been released, the imidazole ring of His-192 bends away from the associated ADP and makes a hydrogen bond with the putative catalytic base, residue Glu-159, within the same monomer (Fig. 4B). The location of the histidine within the nucleotide-binding site and the dimeric interface suggests its role in ATP binding and closure of the NBDs in the formation of the ATPase active conformation. A recent study of HlyB suggests that the histidine residue plays a key role in the stabilization of the transition state of hydrolysis as part of a catalytic dyad (28).
Comparison with the Nucleotide-Free Forms. Two structures of the MalK dimer in the nucleotide-free state are described in ref. 12, showing identical subdomain structures but different relative orientations of the RecA-like and helical subdomains. Based on the extent of the separation between the NBDs in the dimer, these two forms were identified as semiopen and open forms. Compared with the two nucleotide-free forms, one monomer of the ADP-bound structure resembles the semiopen form (6° rotation of the NBD relative to the RD plus a 2° rotation of the helical subdomain relative to the RecA-like subdomain), whereas the other resembles the open form (a bending angle of 7° for the NBD and 8° for the helical subdomain). As a consequence, the intersubunit separation of the ADP-bound form falls in between the semiopen and open forms. The finding of two different nucleotide-free forms and the asymmetric arrangement of the ADP-bound form indicates considerable inherent interdomain flexibility in the MalK structure when the γ phosphate and transmembrane subunits are absent. The flexibility of the helical subdomain, reflected by differing orientations and high temperature factors in different crystal forms, may have important mechanistic implications as discussed in refs. 6–9.
Discussion
Data from both biochemical and crystallographic analyses demonstrate that MalK is a dimer in solution at concentrations of 20 μM or higher and that ATP binding and ATP hydrolysis drive conformational changes in the dimer. In the crystal structure of the nucleotide-free form, the N-terminal NBDs are open and the dimer is stabilized through interactions of the C-terminal RDs (12). The energy derived from ATP-binding, but not ADP-Mg2+-binding, to MalK along the dimer interface stabilizes the closure of the NBDs, suggesting that a cycle of ATP hydrolysis may involve the closing and opening of the N-terminal NBDs of the MalK dimer (Fig. 5). This hypothesis was originally proposed after determination of the structure of the catalytic domain of Rad50 and demonstration of ATP-dependent dimerization (27). In contrast to MalK, most isolated NBDs of ABC transporters are monomers in solution, and it has proved difficult to detect ATP-dependent dimerization in wild-type proteins. However, mutations of catalytic residues that prevent ATP hydrolysis do stabilize ATP-bound dimers, whereas ADP-bound NBDs remain monomers (31), as would be anticipated if ATP hydrolysis is responsible for opening the dimer interface. An open dimer with ADP-Mg2+ bound that bears similarity to the ADP-Mg2+-MalK dimer has also been reported for RNase-L inhibitor (32).
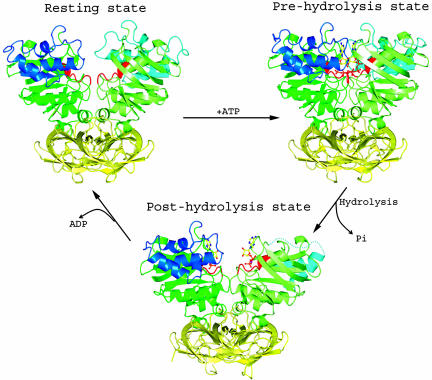
Fig. 5.
Three conformations of the MalK dimer. In the resting state, the two NBDs of MalK are separated from each other. In the ATP-bound state, the NBDs have closed, permitting ATP hydrolysis to occur. ATP hydrolysis and release of Pi open the dimer at the NBD interface. Finally, ADP is released from MalK.
In vivo, the MalK dimer is stably assembled into a complex with transmembrane proteins MalF and MalG, and several lines of evidence suggest that the conformational changes seen in the crystal structures of isolated MalK subunits also occur in the intact MalFGK2 transporter. When a Cys substitution is introduced to Ala-85 in the Q loop region, cross-linked MalK dimer is observed only upon addition of ATP (33). The failure of ADP to induce cross-linking suggests that ATP, but not ADP, induces closure of the NBDs in MalFGK2 (33). Photocleavage experiments using vanadate as a transition state analogue indicate that ATP hydrolysis occurs in the closed dimer (34), and the solvent accessibility of a fluorescent probe covalently attached to an amino acid in the Walker A motif is reduced in the catalytic transition state as compared with the resting state (35), consistent with the closure of the interface between the NBDs.
One of the central questions in ABC transporter research is how ATP hydrolysis is coupled to substrate transport. Our current model for maltose transport, based in part on the observation that the periplasmic MBP becomes tightly bound to the transporter in the catalytic transition state for ATP hydrolysis (36), suggests that as the NBDs close to hydrolyze ATP, the ligand-binding site of the periplasmic MBP opens to release maltose into the transporter. Coupling of the closure of NBDs to the docking and opening of MBP on the opposing surface of the transporter ensures that maltose is delivered to the transporter during each catalytic cycle. These reciprocal changes could well be coupled by conformational changes in the transmembrane domains that simultaneously block access to a maltose translocation pathway from the cytoplasmic side of the membrane while opening the periplasmic side to receive the sugar from MBP. Electron paramagnetic resonance-based measurements (37) indicate that ATP-binding, but not ADP-binding, is able to trigger these major conformational changes in the intact transporter, increasing the affinity between MBP and the transporter and opening MBP. Remarkably, addition of Mg2+ to the ATP-bound conformation, permitting ATP hydrolysis to occur, restores the transporter to the resting state conformation in this experiment (37), just as it does in the crystals of MalK dimer.
Because of the presence of two nucleotide-binding sites along the dimer interface, several different mechanisms of ATP hydrolysis have been proposed that incorporate the hydrolysis of either one or both ATPs per cycle of substrate translocation (38, 39). The presence of positive cooperativity in ATP hydrolysis by MalFGK2 (40) suggests that both sites fill before hydrolysis, consistent with the structure of the closed, ATP-bound MalK dimer (12). If both ATPs are hydrolyzed in each cycle of transport, then the NBDs may dissociate fully between each transport event, as seen in the structure of ADP-bound MalK. Molecular dynamics simulations using the ATP-bound MJ0796 dimer with ATP in one site and ADP in the other site generated asymmetric changes that increased interactions with ATP while decreasing interactions with ADP (41). Although increased interactions with ATP may facilitate hydrolysis of the second ATP, leading to full dissociation of the NBD interface (41), the asymmetry also may indicate that only the ADP-bound site opens to exchange nucleotide whereas the second site remains closed with ATP bound. This concept is consistent with an “alternating catalytic sites” model proposed by Senior et al. (38), in which only one ATP is hydrolyzed per transport cycle. Such an intermediate, if it exists, would not have been captured in our experiments because most of the ATP in the drop is likely hydrolyzed before formation of the crystal.
Acknowledgments
We thank beamline staff at SBC19BM at the Advanced Photon Source and Dr. Dheeraj Khare for assistance with data collection and the Purdue Cancer Center for x-ray and DNA sequencing facilities. We also thank Drs. Jun-yong Choe, Matthew Clifton, Tomonori Kaneko, Dheeraj Khare, and Michael Oldham for critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 GM070515 (to J.C.) and R01 GM49261 (to A.L.D.) and the Pew Scholars Program in the Biomedical Sciences (to J.C). J.M.W. was supported in part by National Institutes of Health Institutional Training Award T32-GM009296.
Author contributions: G.L., J.M.W., A.L.D., and J.C. designed research; G.L. and J.M.W. performed research; and G.L., J.M.W., A.L.D., and J.C. analyzed data and wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ABC, ATP-binding cassette; MBP, maltose-binding protein; NBD, nucleotide-binding domain; RD, regulatory domain.
Data deposition: Atomic coordinates for the two structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2AWN and 2AWO).
References
- 1.Higgins, C. F. (1992) Annu. Rev. Cell Biol. 8, 67–113. [DOI] [PubMed] [Google Scholar]
- 2.Dean, M., Rzhetsky, A. & Allikmets, R. (2003) in ABC Proteins: From Bacteria to Man, eds. Holland, I. B., Cole, S. P. C., Kuchler, K. & Higgins, C. F. (Academic, London), pp. 47–61.
- 3.Chen, C.-J., Chin, J. E., Ueda, K., Clark, D. P., Pastan, I., Gottesman, M. M. & Roninson, I. G. (1986) Cell 47, 381–389. [DOI] [PubMed] [Google Scholar]
- 4.Hung, L. W., Wang, I. X., Nikaido, K., Liu, P. Q., Ames, G. F. & Kim, S. H. (1998) Nature 396, 703–707. [DOI] [PubMed] [Google Scholar]
- 5.Diederichs, K., Diez, J., Greller, G., Muller, C., Breed, J., Schnell, C., Vonrhein, C., Boos, W. & Welte, W. (2000) EMBO J. 19, 5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudet, R. & Wiley, D. C. (2001) EMBO J. 20, 4964–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan, Y. R., Blecker, S., Martsinkevich, O., Millen, L., Thomas, P. J. & Hunt, J. F. (2001) J. Biol. Chem. 276, 32313–32321. [DOI] [PubMed] [Google Scholar]
- 8.Karpowich, N., Martsinkevich, O., Millen, L., Yuan, Y., Dai, P. L., MacVey, K., Thomas, P. J. & Hunt, J. F. (2001) Structure (London) 9, 571–586. [DOI] [PubMed] [Google Scholar]
- 9.Smith, P. C., Karpowich, N., Millen, L., Moody, J. E., Rosen, J., Thomas, P. J. & Hunt, J. F. (2002) Mol. Cell 10, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt, L., Benabdelhak, H., Blight, M. A., Holland, I. B. & Stubbs, M. T. (2003) J. Mol. Biol. 330, 333–342. [DOI] [PubMed] [Google Scholar]
- 11.Verdon, G., Albers, S. V., Dijkstra, B. W., Driessen, A. J. & Thunnissen, A. M. (2003) J. Mol. Biol. 330, 343–358. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., Lu, G., Lin, J., Davidson, A. L. & Quiocho, F. A. (2003) Mol. Cell 12, 651–661. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, H. A., Buchanan, S. G., Burley, S. K., Conners, K., Dickey, M., Dorwart, M., Fowler, R., Gao, X., Guggino, W. B., Hendrickson, W. A., et al. (2004) EMBO J. 23, 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ose, T., Fujie, T., Yao, M., Watanabe, N. & Tanaka, I. (2004) Proteins 57, 635–638. [DOI] [PubMed] [Google Scholar]
- 15.Scheffel, F., Demmer, U., Warkentin, E., Hulsmann, A., Schneider, E. & Ermler, U. (2005) FEBS Lett. 579, 2953–2958. [DOI] [PubMed] [Google Scholar]
- 16.Hor, L. I. & Shuman, H. A. (1993) J. Mol. Biol. 233, 659–670. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 18.Navaza, J. (2001) Acta Crystallogr. D 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 20.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido, K., Liu, P. Q. & Ames, G. F. (1997) J. Biol. Chem. 272, 27745–27752. [DOI] [PubMed] [Google Scholar]
- 22.Greller, G., Horlacher, R., DiRuggiero, J. & Boos, W. (1999) J. Biol. Chem. 274, 20259–20264. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy, K. A. & Traxler, B. (1999) J. Biol. Chem. 274, 6259–6264. [DOI] [PubMed] [Google Scholar]
- 24.Sharma, S., Davis, J. A., Ayvaz, T., Traxler, B. & Davidson, A. L. (2005) J. Bacteriol. 187, 2908–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. (1982) EMBO J. 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding, M. M. (2000) Acta Crystallogr. D 56, 857–867. [DOI] [PubMed] [Google Scholar]
- 27.Hopfner, K. P., Karcher, A., Shin, D. S., Craig, L., Arthur, L. M., Carney, J. P. & Tainer, J. A. (2000) Cell 101, 789–800. [DOI] [PubMed] [Google Scholar]
- 28.Zaitseva, J., Jenewein, S., Jumpertz, T., Holland, I. B. & Schmitt, L. (2005) EMBO J. 24, 1901–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson, A. L. & Sharma, S. (1997) J. Bacteriol. 179, 5458–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shyamala, V., Baichwal, V., Beall, E. & Ames, G. F.-L. (1991) J. Biol. Chem. 266, 18714–18719. [PubMed] [Google Scholar]
- 31.Moody, J. E., Millen, L., Binns, D., Hunt, J. F. & Thomas, P. J. (2002) J. Biol. Chem. 277, 21111–21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karcher, A., Buttner, K., Martens, B., Jansen, R. P. & Hopfner, K. P. (2005) Structure (London) 13, 649–659. [DOI] [PubMed] [Google Scholar]
- 33.Hunke, S., Mourez, M., Jehanno, M., Dassa, E. & Schneider, E. (2000) J. Biol. Chem. 275, 15526–15534. [DOI] [PubMed] [Google Scholar]
- 34.Fetsch, E. E. & Davidson, A. L. (2002) Proc. Natl. Acad. Sci. USA 99, 9685–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannering, D. E., Sharma, S. & Davidson, A. L. (2001) J. Biol. Chem. 376, 12362–12368. [DOI] [PubMed] [Google Scholar]
- 36.Chen, J., Sharma, S., Quiocho, F. A. & Davidson, A. L. (2001) Proc. Natl. Acad. Sci. USA 98, 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Austermuhle, M. I., Hall, J. A., Klug, C. S. & Davidson, A. L. (2004) J. Biol. Chem. 279, 28243–28250. [DOI] [PubMed] [Google Scholar]
- 38.Senior, A. E., Al-Shawi, M. K. & Urbatsch, I. L. (1995) FEBS Lett. 377, 285–289. [DOI] [PubMed] [Google Scholar]
- 39.Sauna, Z. E. & Ambudkar, S. V. (2001) J. Biol. Chem. 276, 11653–11661. [DOI] [PubMed] [Google Scholar]
- 40.Davidson, A. L., Laghaeian, S. S. & Mannering, D. E. (1996) J. Biol. Chem. 271, 4858–4863. [PubMed] [Google Scholar]
- 41.Campbell, J. D. & Sansom, M. S. (2005) FEBS Lett. 579, 4193–4199. [DOI] [PubMed] [Google Scholar]