Abstract
MicroRNAs (miRs) are small noncoding RNAs that regulate gene expression primarily through translational repression. In erythropoietic (E) culture of cord blood CD34+ progenitor cells, the level of miR 221 and 222 is gradually and sharply down-modulated. Hypothetically, this decline could promote erythropoiesis by unblocking expression of key functional proteins. Indeed, (i) bioinformatic analysis suggested that miR 221 and 222 target the 3′ UTR of kit mRNA; (ii) the luciferase assay confirmed that both miRs directly interact with the kit mRNA target site; and (iii) in E culture undergoing exponential cell growth, miR down-modulation is inversely related to increasing kit protein expression, whereas the kit mRNA level is relatively stable. Functional studies show that treatment of CD34+ progenitors with miR 221 and 222, via oligonucleotide transfection or lentiviral vector infection, causes impaired proliferation and accelerated differentiation of E cells, coupled with down-modulation of kit protein: this phenomenon, observed in E culture releasing endogenous kit ligand, is magnified in E culture supplemented with kit ligand. Furthermore, transplantation experiments in NOD-SCID mice reveal that miR 221 and 222 treatment of CD34+ cells impairs their engraftment capacity and stem cell activity. Finally, miR 221 and 222 gene transfer impairs proliferation of the kit+ TF-1 erythroleukemic cell line. Altogether, our studies indicate that the decline of miR 221 and 222 during exponential E growth unblocks kit protein production at mRNA level, thus leading to expansion of early erythroblasts. Furthermore, the results on kit+ erythroleukemic cells suggest a potential role of these miRs in cancer therapy.
MicroRNAs (miRs) are a recently discovered class of small (≈22-nt) RNAs that play an important role in the negative regulation of gene expression by base-pairing to complementary sites on the target mRNAs (1). miRs, first transcribed as long primary transcripts (pri-miRs), are processed in the nucleus by the RNase III enzyme Drosha to generate the 60- to 120-nt-long precursor containing a stem–loop structure, known as pre-miR (2). This precursor, exported into the cytoplasm by the nuclear export factor Exportin-5 and the Ran-GTP cofactor, is finally cleaved by the RNase enzyme Dicer to release the mature miR (3).
miRs mostly bind to the 3′ UTRs of their target mRNAs (1). This process, requiring only partial matching, leads to translational repression. Target mRNAs having more stringent pairing requirement may be cleaved (4, 5).
Currently, >300 miRs have been identified in humans and other eukaryotic species (miR registry, www.sanger.ac.uk/Software/Rfam/mirna/index.shtml). Generally, miRs are phylogenetically conserved (6–9). Their expression pattern is often developmentally and/or tissue-specific, although some miRs are steadily expressed in the whole organism (10, 11). In lower species, miRs are involved in a variety of basic processes, e.g., cell proliferation and apoptosis (12, 13), neural development (14), fat metabolism (15), and stress response (16): in some studies, key target mRNAs have been identified (reviewed in ref. 17). In mammalian species, relatively little is known on their functional role: specifically, miR 181 is involved in the control of lymphopoiesis (18), miR 375 regulates insulin secretion by targeting myotrophin mRNA (19), and the miR-let7 family may play a role in oncogenesis via RAS oncogene mRNAs (20).
We have performed genomewide expression profiling of miRs (11) in the human hematopoietic lineages, as evaluated in unilineage differentiation/maturation culture of cord blood (CB) CD34+ hematopoietic progenitor cells (HPCs). This analysis indicated that miR 221 and 222, clustered on the X chromosome, are markedly down-modulated in erythropoietic (E) culture. Hypothetically, their decline may promote erythropoiesis by unblocking expression of key functional proteins. Our studies indicate that kit receptor mRNA is a major functional target of miR 221 and 222 in both normal erythropoiesis and TF-1 erythroleukemic cell line.
Materials and Methods
Cell Culture. Unilineage E culture of CB CD34+ HPCs. Collection of CB, isolation of CD34+ cells, unilineage E culture, and morphology analysis were performed as described in refs. 21 and 22. The E culture was supplemented or not with kit ligand (KL, 100 ng/ml) (21, 22). KL level in E culture medium was evaluated by ELISA (R & D Systems).
TF-1 and HL60 cell culture. TF-1 and HL60 cell culture was performed by using standard methods. See Supporting Methods, which is published as supporting information on the PNAS web site, for further details.
miR 221 and 222 Expression. Microarray and bioinformatic analysis. Microarray and bioinformatic analysis was performed as described in ref. 11.
Northern blot. Total RNA isolation was performed as in ref. 23. RNA samples (25 μg each) were run as described in ref. 24. The expression levels were analyzed by using the program scion image (Scion, Frederick, MD). See Supporting Methods for further details.
kit Expression. Real-time PCR. Real-time PCR was performed according to standard procedures (25).
Western blot and FACS analysis. Total kit protein expression was analyzed by Western blotting (26) with an anti-kit antibody (R & D Systems) and a secondary anti-goat IgG antibody peroxidase conjugate (Chemicon). The expression levels were analyzed by using scion image. Membrane-bound kit was analyzed by FACS with a CyChrome conjugated anti-kit antibody (Pharmingen). See Supporting Methods for further details.
Plasmids and Constructs. PGL3–3′ UTR plasmid. The 3′ UTR from the kit gene was cloned by standard procedures in the pGL3-Promoter vector (Promega) XbaI site, downstream of the luciferase gene.
Lentiviral vectors. miR 221 and 222 precursors cDNA were PCR-amplified from a human BAC clone by using AccuPrime Taq DNA polymerase high fidelity (Invitrogen). miR 221 and 222 were first cloned in the pCR 2.1-TOPO vector (Invitrogen). Thereafter, they were inserted under CMV promoter into a variant third-generation lentiviral vector, pRRL-CMV-PGK-GFP-WPRE, called Tween (27, 28), to simultaneously transduce both the reporter GFP and the miR. See Supporting Methods for further details.
Luciferase Target Assay. K562 cells (5 × 104 cells per well) were cotransfected with 0.8 μg of pGL3–3′ UTR plasmid, 50 ng of Renilla, and 20 pmol of either a stability-enhanced nontargeting RNA control oligonucleotide (Dharmacon) or stability-enhanced miR 221 and/or 222 oligonucleotides (Dharmacon), all combined with Lipofectamine 2000 (Invitrogen). After 48 h, cells were washed and lysed with Passive Lysis Buffer (Promega), and their luciferase activity was measured by using the Femtomaster FB 12 (Zylux, Oak Ridge, TN). The relative reporter activity was obtained by normalization to the pGL3–3′ UTR/control oligonucleotide cotransfection.
Cell Transfection with miR 221 and 222 Oligonucleotides. Stability-enhanced miR 221 and 222 oligonucleotides and control nontargeting oligonucleotide, as well as their FITC-conjugated counterparts, were purchased from Dharmacon. On the day of transfection, cells were seeded in antibiotic-free media and transfected with miR and Lipofectamine 2000 (Invitrogen). CB progenitors cultured in E+KL culture were transfected on day 4.
Cell Infection with Lentiviral Vectors. Lentiviral supernatants preparation and infection were performed as described in refs. 27 and 28. See Supporting Methods for further details.
NOD-SCID Experiments. Seven 9-week-old NOD-SCID mice received a sublethal dose of whole-body irradiation (350 cGy). Within 24 h of irradiation, CB CD34+ cells were transfected with miR 221 or 222 oligonucleotides (see above), incubated overnight in presence of KL (1 ng/ml), and injected in the tail vein in a volume of 200 μl (5 × 104 cells per mouse), together with γ-irradiated (2,000 cGy) CB CD34-accessory cells (1 × 106 cells per mouse). Mice were killed 6 weeks after transplantation, and bone marrow cells were harvested and analyzed for human hematopoietic cell engraftment by standard procedures. Please see Supporting Methods for further details.
Results
miR 221, 222, and kit Expression in Unilineage E Culture. To investigate miR expression in E differentiation and maturation, we analyzed their level at discrete sequential stages of E culture of CB CD34+ HPCs (Fig. 1 A Top and Bottom). In some experiments, the culture was supplemented with KL (Fig. 1 A Middle and Bottom).
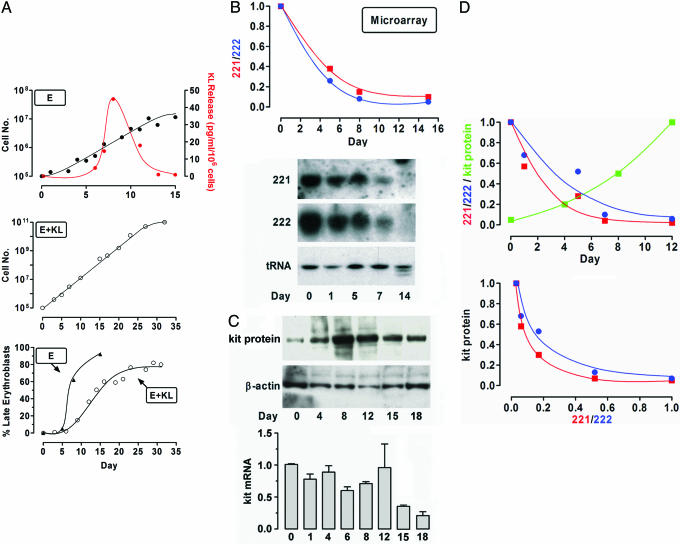
Fig. 1.
Expression of miR 221 and 222 and kit in unilineage E±KL culture. (A Top) Growth curve and KL release in HPC E culture. Shown are mean values from seven independent experiments. (A Middle) Growth curve of HPC E culture supplemented with KL. Shown are mean values from three separate experiments. (A Bottom) E maturation in E and E+KL culture: percentage of late (polychromatophilic + orthochromatic) erythroblasts is presented. (B) miR 221 and 222 expression in HPC E culture. (Upper) Microarray results, as compared with normalized day 0 level. (Lower) Northern blot results. Representative experiments are presented. (C) kit expression in HPC E culture. (Upper) Representative immunoblotting of kit protein. (Lower) Real-time PCR of kit mRNA level (mean ± SEM values from four separate experiments). (D) miR 221 and 222 expression versus kit protein level in E culture (Upper). (Lower) Inverse correlation of miR 221 and 222 vs. kit (r2 = 0.96, P < 0.01 in both cases).
miR 221 and 222 level is down-modulated in E culture. The analysis was performed by using a microarray chip containing as probes gene-specific 40-mer oligonucleotides, generated from 161 human and 84 mouse precursors miRs (11). The expression profile revealed that miR 221 and 222 are abundant in HPCs, but their level gradually and markedly declines during E differentiation-maturation (Fig. 1B Upper). Northern blot analysis confirmed the microarray data (Fig. 1B Lower).
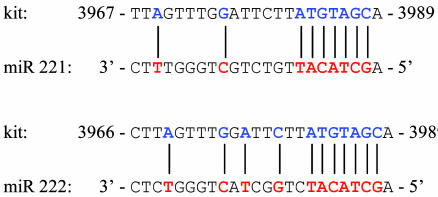
miR 221 and 222 may target kit mRNA. The miR 221 and 222 level decline may promote erythropoiesis by unblocking expression of key functional proteins. Bioinformatic analysis (1) suggested that in humans the kit 3′ UTR is a putative target of both miR 221 and 222. The “seed” sequence in miR 221 and 222 (5′-GCTACAT3-3′, nucleotides 2–8) matches nucleotides 3982–3988 in kit 3′ UTR (NM_000222) and is associated with additional flanking matches (Fig. 2). The seed sequence is conserved in mouse and rat (1).
Fig. 2.
kit mRNA 3′ UTR site targeted by miR 221 and 222
miR 221 and 222 level is inversely related to kit protein expression during exponential E cell growth. The bioinformatic analysis prompted us to investigate kit expression in E culture. kit protein level gradually increases up to day 12 (i.e., during E differentiation coupled with exponential growth) but then declines in terminal erythroblasts undergoing little proliferation (Fig. 1C Upper). Interestingly, kit mRNA is expressed at similar levels through the exponential growth phase but then is sharply down-modulated (Fig. 1C Bottom). It follows that the expression level of both miR 221 and 222 is inversely related to the amount of kit protein up to day 12 (Fig. 1D), thus strongly suggesting a posttranscriptional regulatory mechanism during expansion of early erythroblasts.
Exogenous KL enhances kit protein expression in E culture. kit protein was markedly up-regulated in E culture treated with KL (Fig. 6 Left, which is published as supporting information on the PNAS web site) as compared with standard E cultures releasing endogenous KL at a low level (see above). Both miR 221 and 222 were down-modulated during E differentiation in KL-treated culture (Fig. 6 Right) as observed in E culture not supplemented with KL. However, real-time PCR experiments revealed that kit mRNA was significantly increased upon KL treatment (data not shown). These results suggest that in E+KL culture, kit protein expression is up-regulated via both translational and transcriptional mechanisms.
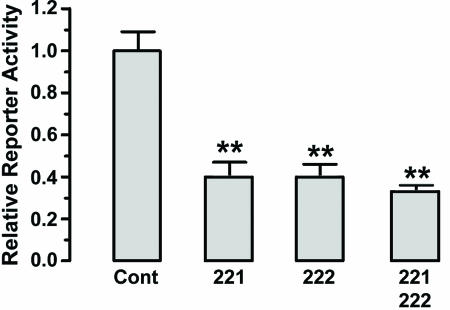
miR 221 and 222 Interact with the 3′ UTR of kit mRNA. To demonstrate the direct interaction between the miRs and kit mRNA, we inserted downstream of the luciferase ORF the 1,800-bp 3′ UTR of kit mRNA (Fig. 3). This reporter vector was cotransfected in the K562 cell line, which does not express miR 221 and 222, with (i) a control nontargeting RNA oligonucleotide or (ii) miR 221 and/or 222 oligonucleotides. The relative luciferase activity was markedly diminished after miR 221 and/or 222 cotransfection, as compared with the control RNA. These results indicate that the two miRs interfere with kit mRNA translation via direct interaction with the 3′ UTR.
Fig. 3.
miR 221 and 222 directly interact with kit 3′ UTR, as evaluated by luciferase targeting assay. Shown are mean ± SEM values from four separate experiments. **, P < 0.01 when compared with control.
miR 221 and 222 Oligonucleotides Down-Modulate kit Expression in TF-1 Erythroleukemic Line. To demonstrate that miR 221 and 222 modulate the expression of the kit protein, we analyzed the expression of kit in cells transfected with miR 221 and/or 222 (Fig. 7, which is published as supporting information on the PNAS web site). As a model system, we choose the erythroleukemia TF-1 cell line, which expresses high levels of kit protein (30) and, as expected, low levels of miR 221 and 222 by Northern blot (data not shown).
We transfected TF-1 cells with dsRNAs having the sequence of respectively the mature miR 221 and 222, as compared with the negative control oligonucleotide. In the initial experiments (Fig. 7 a and b), we transfected cells with FITC-conjugated miR 221 and carried out FACS analysis 16 h thereafter (Fig. 7a): 83% of cells were FITC-positive, indicating that dsRNA is efficiently transfected in these cells. Moreover, apoptosis analysis showed that the transfected miR was not toxic (data not shown). There was no difference in the transfection efficiency of different FITC-conjugated miRs (data not shown). In TF-1 cells transfected with miR 221 and/or 222, FACS analysis with anti-kit antibody revealed that the protein is strongly reduced, as compared with cells transfected with the control oligonucleotide (Fig. 7b). kit expression levels in nontransfected or Lipofectamine-treated cells were comparable with those observed in control oligonucleotide transfected cells (data not shown). In further experiments (Fig. 7 c–f), the inhibition of kit expression was time-dependent, showing a maximum inhibition at 48 h posttransfection (Fig. 7c). The down-modulation of kit expression was demonstrated by Western blot (Fig. 7d) and confirmed by FACS analysis (Fig. 7e) in TF-1 cells transfected with miR 221 and/or 222 at either 40 or 80 nM. In control studies, we analyzed kit mRNA expression in TF-1 cells transfected with miRs at 80 nM: real-time PCR indicated that kit mRNA expression levels were only moderately modified in miR-transfected cells, as compared with control cells treated with Lipofectamine or control oligonucleotide (Fig. 7f).
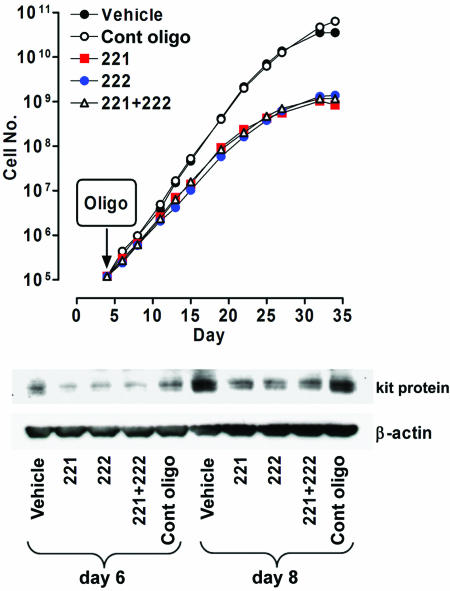
miR 221 and 222 Oligonucleotide Treatment of CD34+ Cells Inhibits E Growth and kit Protein Expression. CD34+ HPCs were grown in unilineage E liquid suspension culture in the presence of KL and transfected on day 4 with miR 221 and/or 222 oligonucleotides, or the control dsRNA at 160 nM (Fig. 4). Transfection efficiency, monitored with the FITC-conjugated miRs (see above), showed that 78% of cells were FITC-positive (data not shown). Furthermore, no apoptosis was associated with miR transfection (not presented).
Fig. 4.
miR 221 and 222 overexpression impairs cell growth in HPC E+KL culture. Growth curve (Upper) and kit protein expression (Lower) in E+KL culture transfected on day 4 with miR 221 and/or 222 oligonucleotide, as compared with vehicle and control oligonucleotide. A representative experiment of four independent experiments is presented.
Cells transfected with miR 221 and/or 222 showed a marked decrease in cell proliferation rate when compared with vehicle or control oligonucleotide transfected cells (Fig. 4 Upper). We also evaluated the miR effects on E differentiation and maturation: cells transfected with miR 221 and/or 222 undergo a more rapid maturation than control cells as indicated by the percentage of late (polychromatophilic and orthochromatic) erythroblasts at days 12–25 (data not shown). Finally, we analyzed kit expression in E culture overexpressing miR 221 and/or 222, or at 2 and 4 days after transfection. Western blotting analysis showed a marked decrease of kit protein expression in the transfected cells as compared with controls (Fig. 4 Bottom).
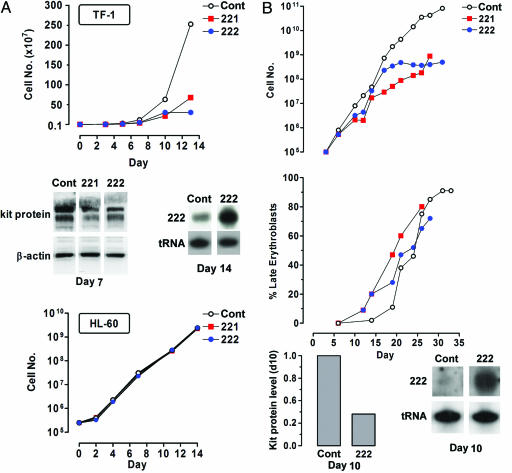
Lentiviral Gene Transfer of miR 221 and 222 Inhibits Cell Growth and Down-Modulates kit Protein Expression. TF-1 erythroleukemic cell line. We investigated the effects of miR 221 and 222 in the kit+ TF-1 erythroleukemic cell line (Fig. 5A). TF-1 cells were transduced with lentiviral vectors. The GFP+ sorted cells cultivated under standard conditions were analyzed at sequential times. TF-1 cells transduced with miR 221 or 222 showed a reduced proliferative rate as compared with control cells (Fig. 5A Top). The enforced expression of both miRs also induced a clear reduction of kit protein level (Fig. 5A Middle). Here again, no significant modulation of kit mRNA was observed by real-time PCR (data not shown).
Fig. 5.
Inhibition of cell growth in TF-1 cell line and unilineage E+KL culture upon infection with Tween-221 and Tween-222 vectors. (A Top) Growth curve of kit+ TF-1 cells infected with Tween-221 or Tween-222 vectors, as compared with empty Tween control vector. (A Middle) Western blot of kit protein and Northern blot of miR 222 in Tween-222-infected TF-1 cells. Similar results were obtained in Tween-221-infected cells (data not shown). (A Bottom) Growth curve of kit-HL-60 cells, infected with Tween-221 or Tween-222 vectors, as compared with empty Tween control vector. A representative experiment of six independent experiments is presented. (B) Growth curve (Top) and maturation of late erythroblasts (Middle) in E+KL culture of HPCs transduced with Tween-221 or -222 vectors, as compared with Tween control vector. (B Bottom) Western blot histogram of kit protein and Northern blot of miR 222 in Tween-222-infected cells at day 10 of culture, as compared with control value; similar results were obtained for Tween-221-infected cells (data not shown). A representative experiment of four independent experiments is presented.
To confirm the specificity of miR 221 and 222 activity on TF-1 cell lines, we investigated the effects of both miRs in HL-60 cell line, lacking the kit protein and expressing low levels of miR 221 and 222 (data not shown). HL-60 cells were transduced with lentivirus particles: as expected, miR-transduced cells did not show any difference in their proliferative rate, as compared with empty vector-treated cells (Fig. 5A Bottom), despite an increased level of miR 221 and 222 (data not shown).
CD34+ cells E culture. CD34+ cells were first incubated in E medium supplemented with KL for 24 h, and then transduced with a Tween lentivirus vector, either empty or containing either miR 221 or 222 (Fig. 5B). Two days later, the infection efficiency was controlled by FACS, and GFP+ cells were sorted. As expected, cells transduced with miR 221 or 222 showed a more elevated level of miR 221 or 222 RNA as compared with controls (Fig. 5B Bottom). HPCs transduced with either miR 221 or 222 exhibited a marked decrease of their growth rate as compared with the empty vector control group (Fig. 5B Top). The effect of miR 221 or 222 on E differentiation/maturation was also evaluated: cells transduced with either miR 221 or 222 show an accelerated E cell maturation compared with the empty vector (Fig. 5B Middle). Furthermore, miR 221- and 222-transduced cells showed an increased rate of cell death at late culture days (days 20–25), i.e., at terminal maturation (data not shown). Finally, down-modulation of kit expression was demonstrated by Western blot in E precursors (Fig. 5B Bottom).
miR 221 and 222 Impair CD34+ Engraftment upon Xenotransplantation into NOD-SCID Mice. As shown in a representative experiment, CB CD34+ cells treated with miR 221 or 222 show a marked decrease of stem cell repopulating activity in NOD-SCID mice as evaluated in terms of human CD45+ cell engraftment in the recipient bone marrow (Fig. 8, which is published as supporting information on the PNAS web site). Preliminary observations suggest that all hematopoietic lineages, as well as B lymphocyte production, are down-modulated upon miR 221 or 222 oligonucleotide transfection. Furthermore, control studies confirmed that miR 221 or 222 oligonucleotides transfection induced a significant down-modulation of kit protein in CD34+ cells maintained in E culture (data not shown), as observed in the other CD34+ cell transfection studies presented above.
Discussion
Despite extensive studies carried out so far on miR genes (reviewed in ref. 4), relatively little is known on their functional role and even less on their targets in mammalian species (18–20).
Our approach involved expression profiling of miRs in the different hematopoietic lineages, as evaluated in unilineage differentiation/maturation culture of CB CD34+ HPCs (21, 22). Particularly, we observed that miR 221 and 222, clustered on the X chromosome, are markedly down-modulated through the E pathway. This decline may unblock the translation of key functional proteins underlying erythropoiesis. In this regard, we focused on the kit receptor, based on two observations: (i) the bioinformatic analysis and the luciferase assay indicated that miR 221 and 222 interact with the 3′ UTR of kit mRNA; and (ii) in E culture undergoing exponential cell proliferation, the miR 221 and 222 level is inversely related to that of kit protein, whereas the abundance of kit mRNA is relatively stable.
Functional studies indicated that miR 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth at least in part via kit receptor down-modulation, whereas they do not affect kit-hematopoietic cell lines (e.g., HL-60). Specifically, treatment of CD34+ HPCs with miR 221 and 222, via oligonucleotide transfection or lentiviral vector infection, causes impaired proliferation and accelerated differentiation of E cells, coupled with down-modulation of kit protein. This phenomenon, observed in E cultures releasing endogenous KL, is magnified by exogenous KL treatment. Furthermore, transplantation experiments in NOD-SCID mice reveal that miR 221 or 222 treatment of CD34+ cells impairs their engraftment capacity, thus indicating that both miRs also hamper early hematopoiesis.
Our studies indicate that in erythropoiesis, the decline of miR 221 and 222 unblocks kit protein production at translational level, thus leading to expansion of early E cells. Our results do not rule out the possibility that miR 221 and 222 hamper erythropoiesis by blocking the translation of not only kit mRNA but also other target mRNAs. On the other hand, we cannot exclude the possibility that other miRs cooperate with miR 221 and 222 to block kit mRNA translation.
Our studies further suggest that in erythropoiesis kit protein expression is regulated not only at translational level via miR 221 and 222, but also through transcriptional mechanisms. Thus, (i) the final phase of E culture, coupled with erythroblast maturation and declining proliferation, is characterized by a drop of kit mRNA and protein levels indicating transcriptional repression of the kit gene. This process may be mediated by GATA-1 transcription factor, which exerts a suppressive action on kit gene promoter in late erythropoiesis (31). (ii) Addition of KL to E culture causes an enhanced proliferation, coupled with a sharp increase of kit mRNA and protein levels: this finding suggests that in this culture system, the unblocking of kit mRNA translation via miR 221 and 222 is potentiated by activation of kit gene transcription.
Modulation of kit protein by miR 221 and 222 treatment may provide an important tool in biotechnology and therapy. Indeed, KL is a key factor controlling proliferation of primitive hematopoietic and E cells (22, 32). Furthermore, constitutive activation of kit underlies diverse neoplasias, e.g., gastrointestinal stromal tumors (GIST) (33) and selected acute leukemias (34). In this regard, we observed that miR 221 and 222 gene transfer blocks proliferation of the kit+ TF-1 erythroleukemic cell line. Hypothetically, modulation of kit protein level by miR 221 and 222 treatment may contribute to studies on early hematoerythropoiesis (e.g., ex vivo expansion of stem cells), as well as to therapy in oncology patients.
It is also noteworthy that kit plays a key functional role in nonhematopoietic tissues, e.g., in smooth muscle progenitors termed Cajal cells (35), neural progenitors (36), melanocytes (35), etc. Therefore, the miR 221- and 222-mediated inhibition of kit mRNA translation may not be restricted to early hematopoiesis and erythropoiesis, but may also operate in a variety of other tissues.
Altogether, these findings indicate that miR 221 and 222 play a key functional role in early hematopoiesis and E differentiation, at least in part via unblocking of kit receptor mRNA translation. The results further suggest that miR 221 and 222 may modulate the growth of kit+ leukemic cells. Hypothetically, the functional role of miR 221 and 222 may be extended to other kit+ nonhematopoietic tissues of either normal or abnormal type, e.g., Cajal cells and GIST tumors.
Supplementary Material
Acknowledgments
We thank A. Addario and S. Chicarella for technical assistance, M. Blasi and M. Fontana for editorial assistance, A. Zito for graphics, and M. Fontana for statistical analysis. This work was supported by the Italy–USA Oncology Program, Istituto Superiore di Sanità (Rome), and National Institutes of Health Grant 1R01 HL63168 (to C.P.).
Author contributions: N.F., C.M.C., and C.P. designed research; N.F., L.F., E.P., R.B., D.B., F.F., F.L., V.L., O.M., S.S., M.V., G.A.C., C.-G.L., and A.S. performed research; N.F., C.M.C., and C.P. analyzed data; and N.F., C.M.C., and C.P. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CB, cord blood; E, erythropoietic; HPC, hematopoietic progenitor cell; KL, kit ligand; miR, microRNA.
References
- 1.Bartel, D. P. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 2.Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. (2003) Nature 425, 415-419. [DOI] [PubMed] [Google Scholar]
- 3.Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. (2003) Genes Dev. 17, 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros, V. (2004) Nature 431, 350-355. [DOI] [PubMed] [Google Scholar]
- 5.Yekta, S., Shih, I. H. & Bartel, D. P. (2004) Science 304, 594-596. [DOI] [PubMed] [Google Scholar]
- 6.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A. & Tuschl, T. (2003) RNA 9, 175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houbaviy, H. B., Murray, M. F. & Sharp, P. A. (2003) Dev. Cell 5, 351-358. [DOI] [PubMed] [Google Scholar]
- 9.Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. (2003) Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 10.Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrowsky, E. & Ambros, V. (2004) Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, C. G., Calin, G. A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, C. D., Shimizu, M., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9740-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, P., Guo, M. & Hay, B. A. (2004) Trends Genet. 20, 617-624. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, A. M., Byrom, M. W., Shelton, J. & Ford, L. P. (2005) Nucleic Acids Res. 33, 1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnova, L., Grafe, A., Seiler, A., Schumacher, S., Nitsch, R. & Wulczyn, F. G. (2005) Eur. J. Neurosci. 6, 1469-1477. [DOI] [PubMed] [Google Scholar]
- 15.Xu, P., Vernooy, S. Y., Guo, M. & Hay, B. A. (2003) Curr. Biol. 13, 790-795. [DOI] [PubMed] [Google Scholar]
- 16.Dresios, J., Aschrafi, A., Owens, G. C., Vanderklish, P. W., Edelman, G. M. & Mauro, V. P. (2005) Proc. Natl. Acad. Sci. USA 102, 1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrington, J. C. & Ambros, V. (2003) Science 301, 336-338. [DOI] [PubMed] [Google Scholar]
- 18.Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. (2004) Science 303, 83-86. [DOI] [PubMed] [Google Scholar]
- 19.Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., MacDonald, P. E., Pfeffer, S., Tuschl, T., Rajewski, N., Rorsman, P., et al. (2004) Nature 432, 226-230. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K. L., Brown, D. & Slack, F. J. (2005) Cell 120, 635-647. [DOI] [PubMed] [Google Scholar]
- 21.Sposi, N. M., Zon, L. I., Carè, A., Valtieri, M., Testa, U., Gabbianelli, M., Mariani, G., Bottero, L., Mather, C., Orkin, S. H. & Peschle, C. (1992) Proc. Natl. Acad. Sci. USA 89, 6353-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabbianelli, M., Testa, U., Massa, A., Pelosi, E., Sposi, N. M., Riccioni, R., Luchetti, L. & Peschle, C. (2000) Blood 95, 3555-3561. [PubMed] [Google Scholar]
- 23.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 24.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak, K. J. & Schmittgen, T. D. (2001) Methods 25, 402-408. [DOI] [PubMed] [Google Scholar]
- 26.Tamborini, E., Bonadiman, L., Greco, A., Gronchi, A., Riva, C., Bertulli, R., Casali, P. G., Pierotti, M. A. & Pilotti, S. (2004) Clin. Cancer Res. 10, 938-943. [DOI] [PubMed] [Google Scholar]
- 27.Bonci, D., Cittadini, A., Latronico, M. V. G., Borello, U., Aycock, J. K., Drusco, A., Innocenzi, A., Follenzi, A., Lavitrano, M., Monti, M. G., et al. (2003) Gene Ther. 10, 630-636. [DOI] [PubMed] [Google Scholar]
- 28.Ricci-Vitiani, L., Pedini, F., Collinari, C., Condorelli, G., Bonci, D., Bez, A., Colombo, A., Parati, E., Peschle, C. & De Maria, R. (2004) J. Exp. Med. 200, 1275-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziegler, B., Valtieri, M., Almeida Porada, G., De Maria, R., Müller, R., Masella, B., Gabbianelli, M., Casella, I., Pelosi, E., Bock, T., et al. (1999) Science 285, 1553-1558. [DOI] [PubMed] [Google Scholar]
- 30.Vitelli, L., Condorelli, G. L., Lulli, V., Hoang, T., Luchetti, L., Croce, C. M. & Peschle, C. (2000) Mol. Cell. Biol. 20, 5330-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munugalavadla, V., Dore, L. C., Tan, B. L., Hong, L., Vishnu, M., Weiss, M. J. & Kapur, R. (2005) Mol. Cell. Biol. 25, 6747-6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, E., Nocka, K., Beier, D. R., Chu, T. Y., Buck, J., Lahm, H. W., Wellner, D., Leder, P. & Besmer, P. (1990) Cell 63, 225-233. [DOI] [PubMed] [Google Scholar]
- 33.Duffaud, F. & Blay, J. Y. (2003) Oncology 3, 187-197. [DOI] [PubMed] [Google Scholar]
- 34.Gilliland, D. G. (2002) Semin. Hematol. 4, 6-11. [DOI] [PubMed] [Google Scholar]
- 35.Kitamura, Y. & Hirotab, S. (2004) Cell Mol. Life Sci. 23, 2924-2931. [DOI] [PubMed] [Google Scholar]
- 36.Ida, J. A., Dubois-Dalcq, M. & McKinnon, R. D. (1993) J. Neurosci. Res. 5, 596-606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.