Abstract
There is growing appreciation of the functional relevance of unfolded proteins in biology. However, unfolded states of proteins have proven inaccessible to the usual techniques for high-resolution structural and energetic characterization. Unfolded states are still generally conceived of as statistical coils, based on the pioneering work of Flory [(1969) Statistical Mechanics of Chain Molecules (Wiley, New York)] and Tanford [(1968) Adv. Protein Chem. 23, 121–282]. Recently, several lines of independent evidence have raised doubts about the random coil model and offer support for alternative views. Here, we show that polyproline II conformation is dominant in a host–guest peptide model AcGGXGGNH2 (X ≠ glycine), in equilibrium predominantly with β-structure. This result is inconsistent with a random coil model and the general view that these peptides are unstructured. By calculating a set of apparent ΔG values from the measured coupling constants of the backbone amides, we can construct a polyproline II scale that correlates negatively with β-sheet scales.
The process by which a protein acquires its native structure is among the most complex reactions known, and challenges remain in defining the nature of the transition state(s), the structure and role of intermediates, and the properties of the starting ensemble of states (1–4). According to Flory (5) and Tanford (6), unfolded proteins can be represented as statistical random coils, in which a given residue has no strong preference for any specific conformation. Confirming earlier conclusions by Tiffany and Krimm (7–9), recent evidence from a variety of spectroscopic probes (10–22), theoretical studies (23–34), and coil library surveys (35–43) consistently point to a major role for the polyproline II (PPII, Φ = –75°, ψ = +145°) conformation in oligo-Ala (for review, see ref. 3 and related articles in the same volume), oligo-Lys, and oligo-Glu peptides (44). We have reported that in a seven-Ala peptide model PPII converts to a β-like structure with increasing temperature (13). These findings raise several important questions regarding the structure of unfolded proteins: Although alanine is arguably a reasonable model for the unperturbed peptide backbone, is PPII also present in unfolded peptide chains composed of nonalanine nonproline residues? Is there an intrinsic PPII propensity for each individual side chain? If PPII is in equilibrium with β-structure, is there a correlation between scales of PPII propensity and analogous β-sheet scales? To what extent is PPII sequence and context dependent?
Here, we address these questions by analyzing a series of end-blocked host pentapeptides AcGGXGGNH2, where X denotes 19 natural amino acids except glycine. Members of the series are found to differ in their extent of PPII conformation as determined by NMR and CD spectroscopy. Our results lead to the following conclusions: PPII is present as a dominant conformation in the majority of AcGGXGGNH2 peptides. Different side chains show distinct propensities to adopt PPII in these unfolded molecules. Importantly, we find an inverse correlation between the determined PPII scale and the β-sheet-forming propensities derived from a zinc-finger model system (45) when 18 aa (except Gly and Pro) are divided into two groups: one, the nonpolar β-branched and bulky aromatic residues (VIWFY) and the other all of the remaining side chains. Finally, we find a correlation between our PPII scale in AcGGXGGNH2 and a PPII scale derived from alternative model peptides such as AcPPPXPPPGYNH2 (46). Still there are indications that the PPII scale is likely to be sequence and context dependent (47).
Materials and Methods
Peptides Synthesis and Purification. Peptides were assembled on Rink Amide resin (Advanced ChemTech) with a Rainin Instruments PS3 solid-phase synthesizer by using fluorenylmethoxycarbonyl (Fmoc) chemistry. Fmoc-amino acids, 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate, 1-hydroxybenzotriazole, and N,N′-diisopropylcarbodiimide were purchased from Nova Biochem. Acetic anhydride was used to cap the N termini of peptides after assembly on the solid matrix. Trifluoroacetic acid was used to cleave peptides from the matrix in the presence of the scavengers anisole, thioanisole, and water. Crude peptides were purified on a reverse-phase HPLC C-18 column (Vydac, 2.2 × 25 cm, 300 Å) with water and acetonitrile as eluents. The fractions containing product were collected and lyophilized. The molecular weight of each peptide was confirmed by MALDI-TOF MS (Bruker, Billerica, MA) by using α–cyano-4-hydroxycinnamic acid as matrix.
Sedimentation. The peptide GGWGG was studied for aggregation by using analytical ultra-centrifugation (Beckman Optima XL-A). The results show this peptide remains monomeric up to a concentration of 5 mM in water, above the experimental concentrations used in CD and NMR.
CD Measurements. Far-UV CD spectra were recorded on an Aviv Associates (Lakewood, NJ) 202 CD spectrometer with a 0.1-cm pathlength cuvette (Hellma, Forest Hills, NJ). CD measurements were carried out with 1–2 mM peptide in 20 mM phosphate with pH adjusted to 3–5. Wavelength scans were performed from 250 to 190 nm at temperatures of 4°C, 45°C, and 90°C.
NMR Spectroscopy. NMR samples were made up with 1–4 mM peptide in the same buffer as used in CD experiments with the addition of 10% D2O and 0.02% NaN3. Small volumes of 0.2 M HCl were added to adjust the pH to 3.0–4.0. The peptides GGDGG and GGEGG were adjusted to pH 5 to deprotonate the side chains. 3J(Hα-HN)(3JαN) coupling constants were measured on a Varian INOVA 600 spectrometer over a temperature range from 0°C to 60°C in 10°C increments. NMR spectra were collected by using 64,000 real data points and 32 scans averaged over a spectral width of 6,400 Hz by using a 1D Watergate or presaturation pulse sequence. Coupling constants were either read directly from the peak splitting of amide protons or α protons with simultaneous decoupling of β protons. No window function except line broadening (lb = 0.1–0.5) was applied to the original free induction decay before Fourier transform. The coupling constants were derived directly from the deconvolution function provided in the vnmr 6.3 (Varian) program. Data collection and processing were identical for each peptide sample. 2D rotating-frame Overhauser effect spectroscopy/NOESY measurements were carried out on selected peptides at 20°C by using a mixing time of 200 or 400 ms. The resulting 2D data sets were processed in vnmr 6.3.
Results and Discussion
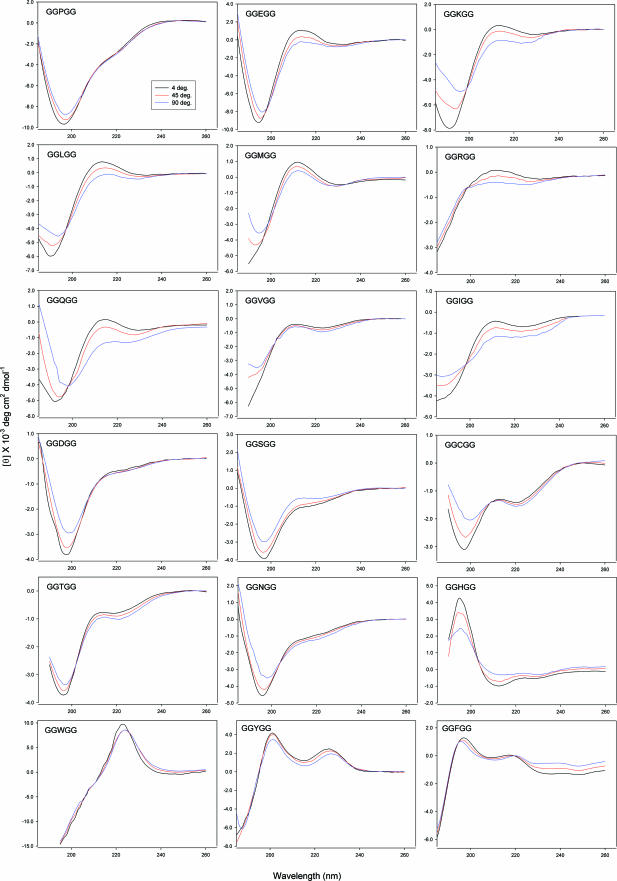
Model Peptides AcGGXGGNH2 and Their CD Spectroscopy. We recently reported that the peptide AcGGAGGNH2 has a high degree of PPII structure in water at room temperature and undergoes a transition to β-structure with increasing temperature (20), consistent with previous findings from a soluble alanine heptamer (13). Peptides with flanking Gly residues have been used to model unstructured states and extensively studied for many years in efforts to calibrate the NMR chemical shifts and coupling constants of the amino acids in unfolded proteins (48). In principle, flanking glycines minimize the steric interactions that can influence the backbone conformation of the central residue. To avoid charge effects, the peptides of this study have both ends blocked. CD spectra for most AcGGXGGNH2 except those with ring side chains (H, W, Y, F) show the characteristic far-UV CD signature of PPII conformation, with a strong negative band at ≈195 nm and a weak positive band at ≈215 nm (Fig. 1 and ref. 3). We believe that the positive band in AcGGPGGNH2 is weak because of the presence of a small population of turn structure and the cis Pro conformation. The CD signals for all AcGGXGGNH2 except those obscured by aromatic side-chain effects show a decrease in intensity of both the 190- to 200-nm and 210- to 220-nm bands with increasing temperature (Fig. 1). This trend is consistent with the temperature-dependent CD signal observed in the model alanine peptides [AcX2A7O2NH2 (13) and AcGGAGGNH2 (20)] that we interpret as a transition from PPII to β-structure. The isodichroic points ≈205 nm observed in the temperature spectra profiles for most AcGGXGGNH2 peptides are consistent with apparent two-state transition behavior. The NMR spectra show strong dαN(i, i + 1) NOE cross peaks in each case, whereas dNN(i, i + 1) NOEs are not measurable or extremely weak, and no medium-range NOEs are detected (data not shown). Together with the CD data, these results indicate that these peptides are present predominantly in the extended β or PPII basins (in the cases of Pro, Asp, and Asn, a minor population (≈ 10%) of turn structures are expected to be present), in agreement with previous findings by Merutka et al. (48). Following our earlier analysis of the data (13, 20), we conclude that each peptide samples predominantly the extended PPII or β-structure but not the α basin.
Fig. 1.
CD spectra as a function of temperature for the GGXGG series.
PPII Contents and the Correlation of PPII Propensities with β Scales. A more quantitative account of the sampled conformations can be derived from the 3JαN values of each residue X in AcGGXGGNH2. 3JαN is directly related to the backbone Φ angle by a Karplus equation (49). 3JαN values at 20°C for AcGGXGGNH2 span the range from 5.7 to 7.8 Hz, indicating that the population of PPII varies among the peptide series (Table 1). Our previous work demonstrated that the transition from PPII to β-like structure is noncooperative in oligo–alanines (50) and not a simple two-state equilibrium. Given each peptide presents in solution predominantly in the extended PPII or β conformation, we can calculate the PPII population (P%) of each peptide with the equation:
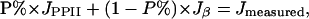 |
[1] |
where JPPII and Jβ are the reference 3JαN values for PPII and β conformations of each amino acid, respectively. In our earlier work, we assigned standard 3JαN values for the Ala in PPII/β to be 5.45/9.81 Hz (13). However, differences in the size and chemical nature of the side chains in principle should result in differences in the respective PPII and β conformations and correspondingly distinct 3JαN values. Indeed, Swindells et al. (36) have found that each amino acid has its own intrinsic Φ/ψ propensities as derived from the coil regions of known structures. Recently, Avbelj and Baldwin (39) surveyed a large body of dihedral angles from the coil library of the Protein Data Bank and estimated the occurrence frequency as a function of Φ, g(Φ), for each residue. They found that g(Φ) can be fitted to a sum of two Gaussian functions that describe the Φ angle distribution between PPII and β basins. The derived Φ angle distributions are those between β and PPII only as the coil library we used excludes the αL and αR conformations (see the Table 1 legend for details). It is obvious that they are substantially different among the amino acids. Taken together, these results imply that different side chains adopt distinctive residue-specific PPII and β conformations, as has been proposed for tripeptides in the series AXA (19). In our analysis this assumption defines a set of unique residue-specific 3JαN values for the PPII and β conformations, allowing us to estimate a set of apparent ΔG corresponding to PPII ↔ β in each AcGGXGGNH2 peptide from the 3JαN measurements (Table 1).
Table 1. Reference 3JαN values for PPII/β of each amino acid and the experimentally determined 3JαN (293K) and derived apparent ΔG and PPII contents at 293K.
| Amino acids | 3JαN (PPII), Hz* | 3JαN (β), Hz* | 3JαN (293K), Hz† | PPII%, 293K‡ | Apparent ΔG, kcal/mol‡ |
|---|---|---|---|---|---|
| Ala | 4.81 | 9.87 | 5.73 | 81.8 | -0.88 |
| Ser | 5.52 | 8.97 | 6.30 | 77.4 | -0.72 |
| Trp | 5.46 | 9.86 | 6.50 | 76.4 | -0.68 |
| Val | 6.09 | 9.82 | 7.05 | 74.3 | -0.62 |
| Glu | 5.42 | 9.72 | 6.78 | 68.4 | -0.45 |
| Asn | 6.63 | 9.45 | 7.57 | 66.7 | -0.40 |
| Gln | 5.95 | 9.85 | 7.30 | 65.4 | -0.37 |
| Phe | 5.34 | 9.86 | 6.97 | 63.9 | -0.33 |
| Arg | 5.89 | 9.51 | 7.20 | 63.8 | -0.33 |
| Tyr | 5.19 | 9.84 | 6.91 | 63.0 | -0.31 |
| Lys | 5.16 | 9.79 | 7.10 | 58.1 | -0.19 |
| Leu | 5.60 | 9.24 | 7.15 | 57.4 | -0.17 |
| Cys | 5.93 | 9.70 | 7.60 | 55.7 | -0.13 |
| Thr | 6.06 | 9.62 | 7.65 | 55.3 | -0.12 |
| Asp | 5.72 | 9.69 | 7.50 | 55.2 | -0.12 |
| Ile | 5.54 | 9.66 | 7.52 | 51.9 | -0.04 |
| Met | 5.69 | 9.69 | 7.70 | 49.8 | 0.005 |
| His | 5.65 | 9.41 | 7.80 | 42.8 | 0.17 |
The 3JαN values are calculated by using the equation by Vuister and Bax (49) for PPII/β having Φ values corresponding to the fitted Gaussian maxima from the coil library of Avbelj and Baldwin (52), the coil library used includes only the PPII and β backbone conformations as the restrictions (90 < ψ < 180 and - 180 < Φ <0) are applied (39).
The coupling constants were measured from either the amide- or α-proton with simultaneous decoupling of β protons. Doublets were fit to Lorentz functions. The fitted 3JαN values can be reproduced within 0.05 Hz.
PPII contents were calculated with Eq. 1 and the apparent ΔG values were calculated with the equation: ΔG = -RTIn(P%/(1 - P%).
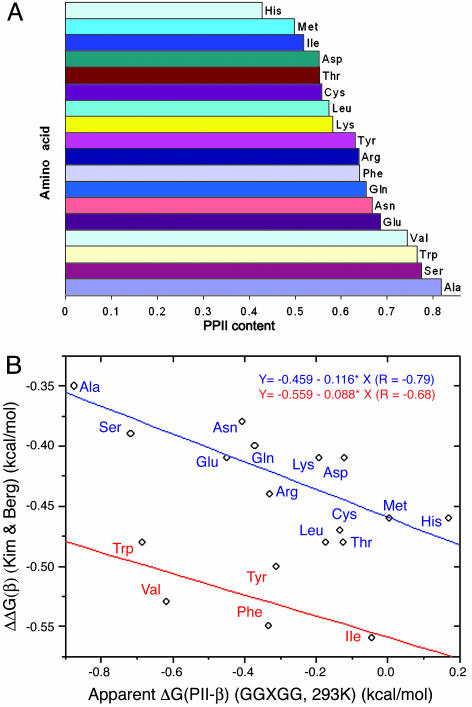
The PPII conformation dominates to differing extent in this series of peptides; the propensities are in the order given in Fig. 2A. If unfolded proteins in general exist predominantly as an equilibrium mixture of PPII and β, we would anticipate an anticorrelation between our PPII scale and existing scales for β-structure formation (45, 51). To our gratification, by separating the amino acids into two groups (VIWFY form one group, and the remainder form the second group) and linearly fitting the data independently, we find a reasonably strong negative correlation between our PPII scale and the β-sheet scale based on a zinc-finger model (45) with two lines parallel to each other (Fig. 2B). The line corresponding to the nonpolar β-branched and bulky aromatic residues is shifted by ≈1.0 kcal/mol in our scale (see discussion below).
Fig. 2.
PPII propensity scale from GGXGG and comparison with a scale of β-sheet propensity. (A) The derived PPII content scale of GGXGG at 293K. PPII contents were derived from the measured 3JαN values of GGXGG. (B) The correlation of our derived apparent ΔG (293K) for each residue and the β-sheet scale by Kim and Berg (45). The ΔΔG values for β-sheet are taken from ref. 45.
Fig. 2B indicates that VIWFY as a group has significantly higher PPII content than estimated from the zinc-finger model. This observation might reflect effects of the flanking Gly residues or perhaps solubility issues with these peptides. It appears that small flexible Gly pairs can relieve a great deal of the adverse effect caused by nonpolar branched and bulky aromatic side chains as predicted by Avbelj and Baldwin (52). They point out that bulky side chains such as those of Val and Ile exert a larger shielding effect on electrostatic interactions between solvent and backbone and therefore have a lower propensity for PPII (52). We have observed such an effect directly (47). Flanking Gly pairs may restore these solvation interactions and thereby stabilize PPII relative to β. In addition, a minor population of non-PPII/non-β conformations might play a role in unfolded states. As a group, VIWFY has substantially lower α-helix propensity than the remaining side chains (53). However, our calculations indicate that this effect ranges from 0.1 to 0.3 kcal/mol, and opposite to the direction observed (i.e., it increases the shift to ≈1.1–1.3 from ≈1.0 kcal/mol). To assess the role of solubility, we measured the sedimentation equilibrium distribution of GGWGG and found that it is monomeric at and above the concentrations used in the CD and NMR experiments. In addition, we added urea to show that the coupling constants we measured are independent of low levels of urea that should affect aggregation. Thus we believe our data correspond to the monomeric species in each case.
It is further encouraging that our data are qualitatively consistent with the scale of Rucker et al. (46) derived from a Pro-rich peptide background. The two scales are based on completely different host models and use independent measures (NMR vs. CD). The lack of quantitative agreement suggests that the PPII scale is likely to be sequence and context dependent. The correlation between our scale and the alternative β-sheet scale of Smith et al. (51) is slightly improved if two outliers (Asp and Ser) are excluded, although the overall correlation is still weaker than that with the scale of Kim and Berg (45). In addition, we can establish a reasonable correlation between the PPII scale of Rucker et al. (46) by using derived apparent ΔGs for PPII ↔ β (from the PPII contents determined by Rucker et al.) and either β-sheet scale (45, 51).
Consistent with the findings from other studies, this study finds that extended PPII and β conformations dominate in the AcGGXGGNH2 peptide series. Anticorrelation between the PPII and β scales provides indirect support for the contention that the structure of unfolded proteins consists mainly of extended PPII and β conformations (3, 48) interspersed by minor populations of turns and a variety of other local structures (54). Strictly speaking, the α and other conformations are small but not negligible; in the case of α, the corresponding Φ angles are similar to those of PPII, and therefore the observed negative correlation could be interpreted simply as that the preference for β conformation in AcGGXGGNH2 correlates with existing β-sheet scales. Nevertheless, our finding demonstrates that the backbone of a single residue blocked by glycine pairs at both ends samples mainly extended PPII or β conformation, in contrast to the general view that such peptides are unstructured. Together with recent lines of evidence that reveal an important role of PPII structure in a variety of small-model unfolded peptides, this conclusion provides a bench mark for understanding the conformation in unfolded proteins (55, 56) and thereby the unfolded state in protein folding.
Acknowledgments
We thank Franc Avbelj and Buzz Baldwin for sharing their coil library data and Min Lu for the sedimentation experiment. This work was supported by the Office of Naval Research.
Author contributions: K.C. and N.R.K. designed research; Z.S., K.C., Z.L., A.N., and W.C.B. performed research; Z.L. contributed new reagents/analytic tools; Z.S. and K.C. analyzed data; and Z.S., K.C., and N.R.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PPII, polyproline II; 3JαN, 3J(Hα-HN).
References
- 1.Rose, G. D. (2002) Adv. Protein Chem. 62, xv–xxi. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, R. L. & Zimm, B. H. (2000) Proc. Natl. Acad. Sci. USA 97, 12391–12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi, Z., Woody, R. W. & Kallenbach, N. R. (2002) Adv. Protein Chem. 62, 163–240. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, R. L. (2002) Adv. Protein Chem. 62, 361–367. [DOI] [PubMed] [Google Scholar]
- 5.Flory, P. J. (1969) Statistical Mechanics of Chain Molecules (Wiley, New York).
- 6.Tanford, C. (1968) Adv. Protein Chem. 23, 121–282. [DOI] [PubMed] [Google Scholar]
- 7.Krimm, S. & Tiffany, M. L. (1974) Israel J. Chem. 12, 189–200. [Google Scholar]
- 8.Tiffany, M. L. & Krimm, S. (1968) Biopolymers 6, 1379–1382. [DOI] [PubMed] [Google Scholar]
- 9.Tiffany, M. L. & Krimm, S. (1968) Biopolymers 6, 1767–1770. [DOI] [PubMed] [Google Scholar]
- 10.Poon, C. D., Samulski, E. T., Weise, C. F. & Weisshaar, J. C. (2000) J. Am. Chem. Soc. 122, 5642–5643. [Google Scholar]
- 11.Woutersen, S. & Hamm, P. (2000) J. Phys. Chem. B 104, 11316–11320. [Google Scholar]
- 12.Schweitzer-Stenner, R., Eker, F., Huang, Q. & Griebenow, K. (2001) J. Am. Chem. Soc. 123, 9628–9633. [DOI] [PubMed] [Google Scholar]
- 13.Shi, Z. S., Olson, C. A., Rose, G. D., Baldwin, R. L. & Kallenbach, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asher, S. A., Mikhonin, A. V. & Bykov, S. (2004) J. Am. Chem. Soc. 126, 8433–8440. [DOI] [PubMed] [Google Scholar]
- 15.Woutersen, S. & Hamm, P. (2001) J. Chem. Phys. 114, 2727–2737. [Google Scholar]
- 16.Woutersen, S., Pfister, R., Hamm, P., Mu, Y. G., Kosov, D. S. & Stock, G. (2002) J. Chem. Phys. 117, 6833–6840. [Google Scholar]
- 17.Blanch, E. W., Morozova-Roche, L. A., Cochran, D. A., Doig, A. J., Hecht, L. & Barron, L. D. (2000) J. Mol. Biol. 301, 553–563. [DOI] [PubMed] [Google Scholar]
- 18.Schweitzer-Stenner, R., Eker, F., Griebenow, K., Cao, X. & Nafie, L. A. (2004) J. Am. Chem. Soc. 126, 2768–2776. [DOI] [PubMed] [Google Scholar]
- 19.Eker, F., Griebenow, K., Cao, X., Nafie, L. A. & Schweitzer-Stenner, R. (2004) Proc. Natl. Acad. Sci. USA 101, 10054–10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, L., Chen, K., Santini, P. A., Shi, Z. & Kallenbach, N. R. (2003) J. Am. Chem. Soc. 125, 8092–8093. [DOI] [PubMed] [Google Scholar]
- 21.McColl, I. H., Blanch, E. W., Hecht, L., Kallenbach, N. R. & Barron, L. D. (2004) J. Am. Chem. Soc. 126, 5076–5077. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. S., Wang, J. P. & Hochstrasser, R. M. (2005) J. Phys. Chem. B 109, 7511–7521. [DOI] [PubMed] [Google Scholar]
- 23.Han, W. G., Jalkanen, K. J., Elstner, M. & Suhai, S. (1998) J. Phys. Chem. B 102, 2587–2602. [Google Scholar]
- 24.Pappu, R. V., Srinivasan, R. & Rose, G. D. (2000) Proc. Natl. Acad. Sci. USA 97, 12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mu, Y. G., Kosov, D. S. & Stock, G. (2003) J. Phys. Chem. B 107, 5064–5073. [Google Scholar]
- 26.Sreerama, N. & Woody, R. W. (1999) Proteins 36, 400–406. [PubMed] [Google Scholar]
- 27.Mezei, M., Fleming, P. J., Srinivasan, R. & Rose, G. D. (2004) Proteins 55, 502–507. [DOI] [PubMed] [Google Scholar]
- 28.Kentsis, A., Mezei, M., Gindin, T. & Osman, R. (2004) Proteins 55, 493–501. [DOI] [PubMed] [Google Scholar]
- 29.Hu, H., Elstner, M. & Hermans, J. (2003) Proteins 50, 451–463. [DOI] [PubMed] [Google Scholar]
- 30.Zaman, M. H., Shen, M. Y., Berry, R. S., Freed, K. F. & Sosnick, T. R. (2003) J. Mol. Biol. 331, 693–711. [DOI] [PubMed] [Google Scholar]
- 31.Gnanakaran, S. & Garcia, A. E. (2003) J. Phys. Chem. B 107, 12555–12557. [Google Scholar]
- 32.Garcia, A. E. (2004) Polymer 45, 669–676. [Google Scholar]
- 33.Drozdov, A. N., Grossfield, A. & Pappu, R. V. (2004) J. Am. Chem. Soc. 126, 2574–2581. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan, V., Ranbhor, R. & Durani, S. (2004) J. Am. Chem. Soc. 126, 16332–16333. [DOI] [PubMed] [Google Scholar]
- 35.Adzhubei, A. A. & Sternberg, M. J. (1993) J. Mol. Biol. 229, 472–493. [DOI] [PubMed] [Google Scholar]
- 36.Swindells, M. B., MacArthur, M. W. & Thornton, J. M. (1995) Nat. Struct. Biol. 2, 596–603. [DOI] [PubMed] [Google Scholar]
- 37.O'Connell, T. M., Wang, L., Tropsha, A. & Hermans, J. (1999) Proteins 36, 407–418. [PubMed] [Google Scholar]
- 38.Hovmoller, S., Zhou, T. & Ohlson, T. (2002) Acta Crystallogr. D 58, 768–776. [DOI] [PubMed] [Google Scholar]
- 39.Avbelj, F. & Baldwin, R. L. (2003) Proc. Natl. Acad. Sci. USA 100, 5742–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adzhubei, A. A. & Sternberg, M. J. (1994) Protein Sci. 3, 2395–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreerama, N. & Woody, R. W. (1994) Biochemistry 33, 10022–10025. [DOI] [PubMed] [Google Scholar]
- 42.Stapley, B. J. & Creamer, T. P. (1999) Protein Sci. 8, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cubellis, M. V., Caillez, F., Blundell, T. L. & Lovell, S. C. (2005) Proteins 58, 880–892. [DOI] [PubMed] [Google Scholar]
- 44.Mikhonin, A. V., Myshakina, N. S., Bykov, S. V. & Asher, S. A. (2005) J. Am. Chem. Soc. 127, 7712–7720. [DOI] [PubMed] [Google Scholar]
- 45.Kim, C. A. & Berg, J. M. (1993) Nature 362, 267–270. [DOI] [PubMed] [Google Scholar]
- 46.Rucker, A. L., Pager, C. T., Campbell, M. N., Qualls, J. E. & Creamer, T. P. (2003) Proteins 53, 68–75. [DOI] [PubMed] [Google Scholar]
- 47.Chen, K., Liu, Z., Zhou, C., Shi, Z. & Kallenbach, N. R. (2005) J. Am. Chem. Soc. 127, 10146–10147. [DOI] [PubMed] [Google Scholar]
- 48.Merutka, G., Dyson, H. J. & Wright, P. E. (1995) J. Biomol. NMR 5, 14–24. [DOI] [PubMed] [Google Scholar]
- 49.Vuister, G. W. & Bax, A. (1993) J. Am. Chem. Soc. 115, 7772–7777. [Google Scholar]
- 50.Chen, K., Liu, Z. & Kallenbach, N. R. (2004) Proc. Natl. Acad. Sci. USA 101, 15352–15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, C. K., Withka, J. M. & Regan, L. (1994) Biochemistry 33, 5510–5517. [DOI] [PubMed] [Google Scholar]
- 52.Avbelj, F. & Baldwin, R. L. (2004) Proc. Natl. Acad. Sci. USA 101, 10967–10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakrabartty, A., Kortemme, T. & Baldwin, R. L. (1994) Protein Sci. 3, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shortle, D. & Ackerman, M. S. (2001) Science 293, 487–489. [DOI] [PubMed] [Google Scholar]
- 55.Fitzkee, N. C. & Rose, G. D. (2004) Proc. Natl. Acad. Sci. USA 101, 12497–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fitzkee, N. C., Fleming, P. J., Gong, H., Panasik, N., Jr., Street, T. O. & Rose, G. D. (2005) Trends Biochem. Sci. 30, 73–80. [DOI] [PubMed] [Google Scholar]