Abstract
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a cytokine with potential therapeutic value against cancers because of its selective cytotoxicity to many transformed, but not normal, cells. The “decoy receptors” TRAIL-R3 (TR3) and TRAIL-R4 (TR4) were believed to negatively regulate TRAIL-induced cytotoxicity by competing for ligand binding with TRAIL-R1 (TR1) and TRAIL-R2 (TR2). Here, we show that inhibition of TRAIL-induced apoptosis by TR4 critically depends on its association with TR2 via the NH2-terminal preligand assembly domain overlapping the first partial cysteine-rich domain of both receptors. By contrast, ligand binding by TR4 is dispensable for its apoptosis inhibitory function, thereby excluding the possibility that TR4 was a “decoy” to inhibit apoptosis by binding up TRAIL. In primary CD8+ T cells, which express only TR2 and TR4 and are resistant to TRAIL-induced apoptosis, stimulation with phorbol myristate acetate abrogated the ligand-independent interaction between TR2 and TR4 and enhanced their sensitivity to TRAIL-induced apoptosis. Hence, whereas most TNF receptors normally form only homotrimeric complexes, the preligand assembly domains in TR2 and TR4 permit mixed complex formation as a means to regulate apoptosis induction. We propose that TR4 is a “regulatory” rather than “decoy” receptor that inhibits apoptosis signaling by TRAIL through this previously uncharacterized ligand-independent mechanism.
Keywords: decoy receptors
The regulation of cell death by members of the TNF family plays a critical role in immune function and homeostasis (1). TRAIL is a TNF-like cytokine that selectively induces apoptosis in many tumor cells, but not in normal cells. Administration of recombinant TRAIL or antibodies against TR2 in several experimental tumor models exhibited potent antitumor activity with minimal hepatic toxicity (2-6). Moreover, recombinant TRAIL or agonist TRAIL receptor antibody often synergizes with chemotherapy or radiation to induce tumor-cell apoptosis (7-10). This unique property of TRAIL has prompted many to vaunt it as a potential therapeutic agent against malignant diseases. Despite its potency against tumor cells, the physiological function of TRAIL is largely unknown, although some reports have implicated TRAIL to be involved in tumor surveillance (11), target cell killing by various immune effector cells (12, 13), and the regulation of innate immune responses (14).
TRAIL binds to five distinct TNF receptor (TNFR)-like receptors, TR1 (TRAIL-R1/DR4), TR2 (TRAIL-R2/DR5/Killer/Trick), TR3 (TRAIL-R3/DcR1/LIT/TRID), TR4 (TRAIL-R4/DcR2/TRUNDD), and the soluble receptor osteoprotegerin (OPG). OPG is a soluble receptor that also binds another TNF-like cytokine called TRANCE/RANK-L and may have a more prominent role in bone and myeloid cell development than in regulating TRAIL-induced apoptosis. The four membrane-anchored TRAIL receptors contain two complete cysteine-rich domains (CRDs) for ligand binding that are preceded at the NH2 termini by a highly conserved partial CRD with unknown function (15, 16). TR1 and TR2 signal for apoptosis through their cytoplasmic death domains (DDs). Similar to Fas/CD95/APO-1, stimulation of TR1 or TR2 results in the recruitment of FADD and subsequently the initiator caspases caspase-8 and caspase-10 (17). The recruited caspase-8 and caspase-10 undergo autocatalytic cleavage and activation to trigger the caspase cascade that ultimately leads to the apoptotic death of the cell.
Unlike TR1 and TR2, neither TR3 nor TR4 contain intact cytoplasmic DDs that signal for apoptosis. TR4 possesses a partially truncated DD, whereas TR3 is anchored on the membrane via glycosyl-phosphatidylinositol (GPI) linkage. Because they can bind TRAIL but do not signal for apoptosis, TR3 and TR4 have been proposed to serve as “decoys” that inhibit apoptosis by sequestering TRAIL from the death-inducing TRAIL receptors. Alternatively, the decoy receptors may inhibit apoptosis by complexing with TR1 and/or TR2 upon binding to the trimeric ligand (18). Both of these ligand-dependent models of inhibition of apoptosis by the decoy receptors require the decoy receptors to bind TRAIL with relatively high affinities. Moreover, they predict that high expression of the decoy receptors will correlate with resistance to TRAIL-induced apoptosis.
Here, we report that inhibition of apoptosis by the decoy receptor TR4 does not depend on ligand binding of the receptor. Rather, inhibition of apoptosis critically depends on the formation of ligand-independent complexes between TR2 and TR4. We found that the NH2 termini overlapping the first partial CRD of TR2 and TR4, termed the preligand assembly domains (PLADs), are essential for the formation of this ligand-independent, death-inhibitory complex. The formation of this death-inhibitory complex regulates sensitivity of primary human CD8+ T cells to TRAIL-induced apoptosis. Thus, PLAD-mediated preligand receptor assembly is a previously uncharacterized mechanism by which TR4 regulates cellular sensitivity to TRAIL-induced apoptosis.
Materials and Methods
Reagents. CD8+ T cells, Jurkat cells, and BW5147 thymoma were cultured in RPMI medium 1640. 293T cells were cultured in DMEM. All media were supplemented with 10% FCS, 100 units/ml penicillin, 100 μg/ml streptomycin, 30 μg/ml l-glutamine, and 60 μM 2-mercaptoethanol. Recombinant TRAIL, antibodies against TRAIL receptors, and TRAIL receptor fusion proteins were obtained from R & D Systems and Axxora Biochemicals.
TRAIL-Induced Cell Death in CD8+ T Cells. CD8+ T cells were purified from peripheral blood mononuclear cells by using the CD8 T cell purification system (R & D Systems). FACS staining of TRAIL receptor expression was performed by using specific antibodies from Axxora Biochemicals. Cell death was induced with recombinant TRAIL upon treatment with 20 ng/ml phorbol myristate acetate (PMA) for 30-240 min. Immunoprecipitation (IP) in CD8+ T cells were performed by using an antibody specific for TR2 (R & D Systems). Western blots were performed by using TR2 and TR4 antibodies. Small interference RNA was introduced into primary CD8+ T cells by using the Amaxa nucleofection method. RNA interference (RNAi) sequences against TR4 were: si506, r(GGAAGCUUCCAGGAUAAAA)dTdT; and si1142, r(GGACAUGCAAAGGAAACAA)dTdT. Transfection efficiency was typically >90% as determined by transfection with a FITC-conjugated oligonucleotide duplex. Maximal silencing of TRAIL-R4 expression was normally achieved 4 days posttransfection.
293T Coimmunoprecipitation. 293T cells were transfected by using FuGENE 6 (Roche). Twenty-four to 48 h posttransfection, cells were harvested and lysed in IP buffer (150 mM NaCl/20 mM Tris·Cl, pH 7.5/1% Nonidet P-40) supplemented with Complete protease inhibitors (Roche). After the lysates were cleared by centrifugation, IP was performed by using monoclonal antibodies against GFP (Roche), which also reacts with yellow fluorescent protein (YFP), for 2 h at 4°C with protein G coupled to agarose beads. The beads were washed twice with IP buffer, twice with IP buffer supplemented with 500 mM NaCl, and once with IP buffer. Immunoprecipitates and whole-cell lysates were resolved on 10% Bis/Tris NuPAGE gels (Invitrogen).
TRAIL-Binding Assay. Murine BW5147 thymoma was transfected with the respective TRAIL receptor expression plasmids encoding TRAIL receptor fusions to the YFP by electroporation. Twenty-four hours later, cells were stained with recombinant FLAG-tagged TRAIL, followed by anti-FLAG tag antibody (M2, Sigma) and goat anti-mouse IgG antibody conjugated to phycoerythrin.
Functional Dominant Interference Assay. Jurkat cells were transfected by electroporation with the BTX Electro Cell Manipulator 600 (Genetronics). Transfected cells were treated with TRAIL for 6 h at 37°C. In some experiments, TR fusion proteins were added to the cells 15 min before stimulation with TRAIL. Apoptosis of the transfected cells was analyzed by using phycoerythrin-conjugated Annexin V (BD Pharmingen) and YFP fluorescence. The percentage of apoptotic cells in the transfected population was determined by the percentage of YFP-positive cells that also exhibited positive Annexin V staining. Percentage of cell loss was determined as described in ref. 19.
Results
Successful expansion of memory CD8+ T cells during recall responses requires CD4+ T cell help and priming when the CD8+ T cells are challenged during the primary response. The lack of CD4+ T cell help during primary antigenic challenge led to abortive memory CD8+ T cell expansion during recall response due to TRAIL-induced apoptosis (20). This sensitization to TRAIL-induced cell death could be attributed to the partial activation signals the cells received during the initial antigen priming stage in the absence of CD4+ T cell help. We sought to understand the mechanism that underlies the sensitization of human primary CD8+ T cells to TRAIL-induced apoptosis by using PMA to mimic partial activation of CD8+ T cells. We found that human peripheral CD8+ T cells or human CD8+ T cell clones that recognize the influenza M1 peptide were normally resistant to TRAIL-induced apoptosis. However, pretreatment with PMA for as little as 30 min sensitized the cells to TRAIL-induced apoptosis (Fig. 1A). Because CD8+ T cells express predominantly TR2 and TR4 on the cell surface (Fig. 1B), we speculated that the PMA-induced sensitization to TRAIL-induced apoptosis might be caused by changes in cell-surface expression of the inhibitory receptor TR4. Such a scenario would be consistent with the previously reported sequestration of decoy TRAIL receptors in an intracellular compartment as a means to modulate TRAIL responses (21). Indeed, PMA stimulation did modestly reduce TR4 expression on CD8+ T cells (Fig. 1C, mean fluorescence intensity from 146 to 110). The expression of TR2 was also reduced slightly in response to PMA (Fig. 1C, mean fluorescence intensity from 5.99 to 3.82). Hence, changes in cell-surface expression of TR2 and TR4 may account for the sensitization of CD8+ T cells to TRAIL-induced apoptosis.
Fig. 1.
Protection against TRAIL-induced apoptosis in CD8+ T cells requires TR4. (A) PMA- or DMSO-treated CD8+ T cells were stimulated with recombinant TRAIL for 4 h before cell death determination by Annexin V staining. (B) Peripheral blood mononuclear cells were stained with antibodies specific for CD8 and TRAIL receptors as indicated. TRAIL receptor expression in CD8+ T cells was analyzed by histogram analysis. The shaded curves and the solid lines represent staining with a control IgG and TRAIL receptor antibodies, respectively. (C) PMA stimulation led to decreased surface expression of TR4. The thin lines and the heavy lines represent DMSO- and PMA-treated cells, respectively. (D) CD8+ T cells were transfected with control RNAi or TR4-specific RNAi and analyzed for cell-surface expression of TR2 or TR4. The shaded curves represent staining with control IgG. (E) CD8+ T cells transfected with control RNAi or TR4-specific RNAi (si506 and si1142) were treated with TRAIL, TNF, or FasL as indicated and analyzed for cell death by flow cytometry. (F) TR2 was immunoprecipitated from untreated or PMA-stimulated CD8+ T cells. The presence of TR4 and the two isoforms of TR2 (TR2L and TR2s) in the immune complex (IP) and whole-cell extract (WCE) was detected by Western blot (WB) as indicated. Results are representative of at least three experiments.
Although PMA treatment sensitized cells to TRAIL-induced apoptosis with a concomitant reduction in TR4 surface expression, it is possible that PMA might trigger other molecular events that contribute to the enhanced sensitivity to TRAIL. However, we found no change in the expression of FADD, caspase-8, and cFLIPL in the PMA-treated cells (data not shown). To determine whether TR4 expression alone was sufficient to confer protection against TRAIL in CD8+ T cells, we transiently knocked down TR4 expression using RNAi. We found that RNAi against TR4 specifically reduced expression of TR4, but not TR2 expression, in CD8+ T cells (Fig. 1D). Moreover, RNAi-mediated silencing of TR4 resulted in an enhanced sensitivity of CD8+ T cells to TRAIL-induced apoptosis (Fig. 1E). The effect of the TR4 RNAi was specific to TRAIL because CD8+ T cells were not sensitized to TNF-induced apoptosis (Fig. 1E). Moreover, RNAi against TR4 did not affect Fas ligand-induced apoptosis in CD8+ T cells (Fig. 1E). Hence, TR4 plays a critical role in the protection of CD8+ T cells against TRAIL-induced apoptosis.
Ligand-independent formation of receptor complexes plays a crucial role in the function and signaling of TNF and Fas receptors (22, 23). Although TNF and Fas receptors appear to form only homotypic receptor complexes via the extracellular PLAD, we hypothesized that the TRAIL receptors might form mixed receptor complexes because of the high degree of homology in their extracellular CRDs. Indeed, we found that TR2 and TR4 associated with each other in CD8+ T cells (Fig. 1F). Strikingly, treatment with PMA abolished this interaction (Fig. 1F). Because these interactions occurred in the absence of exogenously added TRAIL, our results strongly suggest that TR4 may inhibit apoptosis by complexing with TR2 before binding exogenous TRAIL.
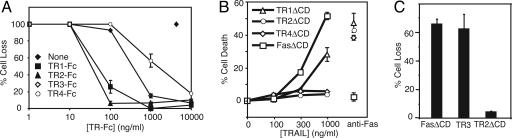
The inhibitory receptors TR3 and TR4 were thought to protect cells against TRAIL-induced apoptosis by competing with the death-inducing receptors for ligand binding. Alternatively, the trimeric ligand TRAIL may drive the formation of an abortive receptor complex containing TR1 or TR2 and the decoy receptors (18). However, our data in CD8+ T cells implied that sensitivity to TRAIL might be regulated through ligand-independent TR2-TR4 interaction. We therefore sought to determine the precise role of ligand binding by TR4 in the protection against TRAIL-induced cell death. We reasoned that any ligand-dependent mechanisms of inhibition by TR3 or TR4 would require them to bind TRAIL with relatively high affinities. Thus, we first evaluated the ligand-binding affinities of the different TRAIL receptors using a cellular assay. Jurkat cells express only TR2 and are highly sensitive to TRAIL-induced apoptosis (24). We therefore compared the efficacy of soluble TR-Fc fusion proteins [extracellular domain (ECD) of TR fused to the constant region of Ig] to inhibit TRAIL-induced apoptosis in Jurkat T cells as an indirect measurement of the ligand-binding affinity of TRAIL receptors. We found that the half maximal inhibitory dose (ID50) for TR3-Fc and TR4-Fc were 10- to 100-fold greater than that of TR1-Fc and TR2-Fc (Fig. 2A), indicating that TR3 and TR4 bind TRAIL with lower affinities than TR1 and TR2. The weak inhibition by TR3-Fc and TR4-Fc was not due to differences in protein stability or misfolding (Fig. 7, which is published as supporting information on the PNAS web site). Our results are therefore consistent with that of Truneh et al. (25), who showed that TR3 (Kd = 200 nM) and OPG (Kd = 400 nM) bind TRAIL with much lower affinities than that of TR1 (Kd = 70 nM) and TR2 (Kd = 2 nM). Hence, TR3 and TR4 bind TRAIL with far lower affinities than TR1 and TR2, which does not support the model that they are effective decoys for TRAIL.
Fig. 2.
The decoy receptor TR4 is a low-affinity receptor for TRAIL but effectively inhibits apoptosis signaling by TR2 in Jurkat cells. (A) Jurkat 4E3 cells were treated with TRAIL in the presence of the indicated amount of soluble TRAIL receptor Fc fusion proteins. Cell death was determined by flow cytometry. (B) The indicated plasmids were transfected into Jurkat 4E3 cells and stimulated with the indicated doses of TRAIL or anti-Fas antibody. Apoptosis was monitored by Annexin V staining. (C) Jurkat 4E3 cells were transfected with the indicated plasmids, stimulated with TRAIL, and analyzed for cell death. Results are representative of three independent experiments.
Although soluble TR4-Fc did not bind TRAIL with high affinity, transfection of full-length TR4 or TR4 with a cytoplasmic domain (CD) substitution with the YFP conferred strong protection against TRAIL-induced apoptosis in Jurkat cells (Fig. 2B and data not shown). The inhibition conferred by TR4 was as strong as that conferred by a similarly built dominant negative TR2 and stronger than that of a dominant negative TR1 mutant (Fig. 2B). By contrast, a dominant negative version of Fas did not interfere with TRAIL-induced apoptosis, although it potently suppressed Fas-induced apoptosis. None of the dominant negative TRAIL receptors had any effects on Fas-induced apoptosis, indicating that the inhibitory effect was specific against TRAIL (Fig. 2B). Flow cytometric measurement of YFP fluorescence also revealed comparable expression between the different transfected samples (Fig. 8, which is published as supporting information on the PNAS web site). Surprisingly, transient or stable transfection of TR3 did not protect against TRAIL-induced death in Jurkat cells (Fig. 2C and data not shown). Collectively, these data indicate that TR4 could potently interfere with TR2-mediated apoptosis, but ligand-dependent mechanisms are unlikely to account for its inhibitory effect.
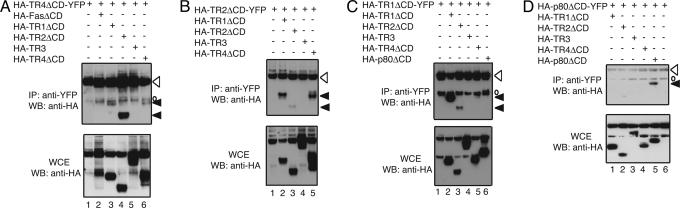
Because ligand-independent receptor complex formation is crucial for the function and signaling of TNF and Fas receptors (22, 23, 26), we postulated that it might similarly control apoptosis induction of TRAIL. Specifically, we reasoned that the highly conserved extracellular CRDs between TR2 and TR4 might permit ligand-independent formation of mixed, abortive receptor complexes. Such an event would produce heterotrimeric complexes between TR2 and TR4 that would lack three functional intracytoplasmic DDs and therefore would interfere with apoptosis induction (27). We tested this proposition by examining the interaction between different TRAIL receptors lacking the cytoplasmic DDs. Because previous work indicates that ligand-independent interactions occur in the ECD of the TNF and Fas receptors and that the cytoplasmic DDs have a strong tendency to aggregate with one another (27), we removed the cytoplasmic DDs from the TRAIL receptors before testing them for ligand-independent associations. We found that the ECDs of TR4 and TR2 interacted strongly with each other (Fig. 3 A and B, lanes 4 and 5, respectively). Remarkably, the interaction between TR2 and TR4 appeared to be stronger than that for homo-specific interaction [Fig. 3 A (compare lanes 4 and 6) and B (compare lanes 3 and 5)]. In addition, we also observed mixed receptor interactions between the ECDs of TR1 and TR4 (Fig. 3A, lane 3), and TR1 and TR2 (Fig. 3 B and C, lanes 2 and 3, respectively), although the interaction between TR4 and TR1 seemed to be much weaker than that between TR2 and TR4. Similar experiments with TR1 ECD as bait confirmed our previous observation that TR1 can form strong homotypic complexes with itself (Fig. 3C, lane 2) (22). Surprisingly, we did not observe any interactions between TR3 and other TRAIL receptors (Fig. 3 A-C, lanes 5, 4, and 4, respectively). Control experiments with TNFR-2 p80 as bait revealed interaction with the ECD of p80 but not with any other TRAIL receptor ECDs, demonstrating the specificity of interactions among the TRAIL receptors (Fig. 3D, lane 5).
Fig. 3.
The ECDs of TRAIL receptors mediate ligand-independent interactions. HEK 293T cells were transfected with the indicated plasmids. Cells were harvested for IP 24 h later by using an antibody against GFP, which also reacts with YFP. Western blots (WB) were performed by using HRP-conjugated HA-specific antibody to detect expression of the receptors in IPs and whole-cell extracts (WCE), respectively. Open arrowheads, baits; filled arrowhead, preys; open circles, nonspecific signals produced by truncation of the bait YFP proteins. Results are representative of three experiments.
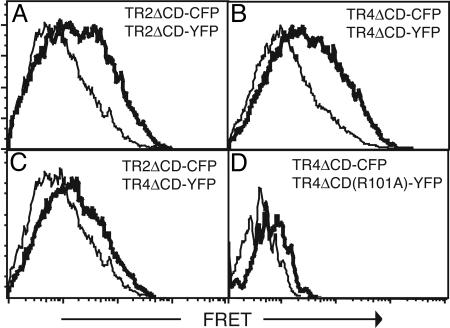
The interaction between the ECD of TR2 and TR4 was confirmed by FRET analysis with cyan fluorescent protein (CFP)- and YFP-tagged receptors. An increase in FRET signal was observed in cells cotransfected with TR2-CFP/TR2-YFP, TR4-CFP/TR4-YFP, and TR2-CFP/TR4-YFP receptor pairs (Fig. 4 A-C, bold lines) compared with the noninteracting receptor pairs TR2-CFP/Fas-YFP (Fig. 4 A-C, thin lines). These interactions did not require an intact ligand-binding site, because the non-ligand-binding mutant TR4(R101A)-YFP could still interact with wild-type TR2-CFP and TR4-CFP (Fig. 4D and data not shown). Hence, the ECDs of TR2 and TR4 interact with each other strongly in the absence of ligand.
Fig. 4.
Flow cytometric analysis of FRET revealed homotypic and heterotypic interactions of TR2 and TR4. HEK 293T cells were transfected with the indicated FRET pairs and analyzed for FRET by flow cytometry (heavy lines). The thin normal lines represent baseline FRET signals between the noninteracting receptor pairs TR2ΔCD-CFP and FasΔCD-YFP. Results are representative of three independent experiments.
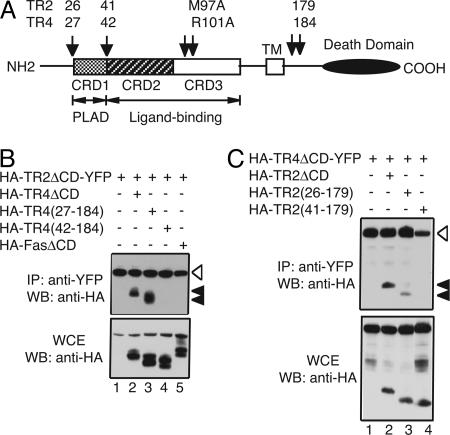
We next sought to further delineate the domain within the ECDs of TR2 and TR4 that mediate this ligand-independent interaction. TR2 binds TRAIL through two patches of residues within the two complete CRDs of the receptor while the NH2-terminal partial CRD contributes little to ligand binding (Fig. 5A, CRD2 and CRD3) (15, 16, 28). Because the PLADs in TNFR-1, TNFR-2, and Fas are located within the membrane-distal first CRD and are physically distinct from the ligand-binding domain (22, 23), we hypothesized that the PLADs located within the NH2-terminal partial CRD of TR2 and TR4 mediate formation of ligand-independent homo- and heterotypic receptor complexes (Fig. 5A). Indeed, deletion of the NH2-terminal partial CRD in TR4 [HA-TR4(42-184)], but not the non-CRD sequences preceding it [HA-TR4(27-184)], abolished the association of TR4 with the ECD of TR2 (Fig. 5B, compare lanes 2-4). Analysis of the whole-cell extracts showed that all proteins were expressed at similar levels. Similar deletions in TR2 revealed that the partial CRD at the NH2 terminus was required for the interaction with TR4 (Fig. 5C, compare lanes 2-4). Again, the interaction measured in this assay is specific, because the Fas receptor was not precipitated (Fig. 5B, lane 5). Thus, PLADs exist for TR2 and TR4 within the partial CRD at the NH2 termini of the receptors. This arrangement is similar to the PLADs in TNFR-1, TNFR-2, and Fas (22, 23).
Fig. 5.
The PLADs of TR2 and TR4 are located within the partial CRD at the NH2 termini. (A) Schematic diagram of the domain structure of TR2 and TR4. The numbers above the arrows represent the boundaries of the deletions and the positions of the alanine substitutions in the ligand-binding pocket of TR2 and TR4. The indicated deletion mutants of TR4 (B) and TR2 (C) were tested for association with the ECDs of TR2 or TR4 in 293T cells. Results are representative of three experiments.
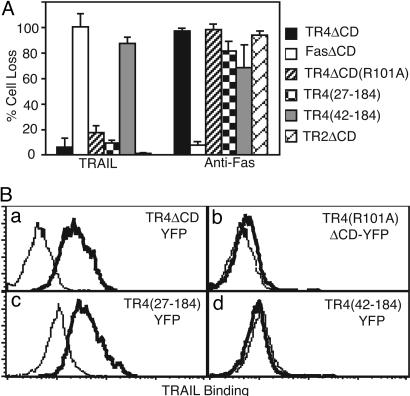
Our results thus far strongly implicate that ligand-independent mechanisms are involved in the inhibition of apoptosis by TR4. However, it is possible that both ligand-dependent and -independent mechanisms contribute to the inhibition of apoptosis by TR4. To more definitively evaluate the contributions of ligand binding and PLAD-mediated association to the inhibitory effect of TR4 on TRAIL-induced apoptosis, we tested the PLAD-deleted and non-ligand-binding TR4 mutants for protection against TRAIL-induced apoptosis in Jurkat cells. Strikingly, deletion of the PLAD completely eliminated the protective effect of TR4 [Fig. 6A, TR4(42-184)-YFP]. As expected, the DD-deleted TR2 and TR4 conferred strong protection (>95%) against TRAIL-induced death but not Fas-induced death (Fig. 6A). Moreover, the removal of the NH2-terminal non-CRD sequences that were not essential for PLAD-mediated association did not perturb the protective effect of TR4 [Fig. 6A, TR4(27-184)-YFP]. Similar deletion mutants of TR2 revealed that the NH2-terminal partial CRD was also required for dominant interference of apoptosis by a tailless TR2 (Fig. 9, which is published as supporting information on the PNAS web site). A tailless, dominant-interfering Fas receptor had no effect on TRAIL killing but strongly suppressed death induced by an anti-Fas agonistic antibody (Fig. 6A). Strikingly, we found that deletion of the PLAD, but not the NH2-terminal non-PLAD sequences, abolished TRAIL binding by TR4 (Fig. 6B c and d). The loss of TRAIL binding is unlikely to be caused by misfolding of the receptor because the PLAD-deleted TR4 was expressed on the cell surface and could be recognized by antibody specific for TR4 (Fig. 10, which is published as supporting information on the PNAS web site). Hence, the PLAD in TR4 is crucial for receptor association and efficient ligand binding, similar to TNFR-1 and TNFR-2 (22).
Fig. 6.
Ligand-independent inhibition of TR2-induced apoptosis by TR4 requires an intact PLAD domain but not the ligand-binding domain. (A) The indicated YFP-tagged chimeric receptors were transfected into Jurkat 4E3 cells, stimulated with TRAIL or anti-Fas antibody, and analyzed for cell death by flow cytometry. (B) The indicated TR4ΔCD-YFP receptors were transfected into murine BW5147 cells, and TRAIL binding was determined by flow cytometry. The histograms show TRAIL binding by the control FasΔCD-YFP receptor (thin lines) and the different TR4 receptors (solid lines). Results are representative of five experiments.
Although our results indicate that PLAD-mediated, ligand-independent interaction between TR2 and TR4 impairs receptor signaling of cell death, it is possible that deletion of the PLAD resulted in severe disruption of the ligand-binding domain and rendered the truncated receptor nonfunctional. To conclusively show that ligand was not required for PLAD-mediated inhibition of TR2, we tested the non-ligand-binding TR4 mutant R101A for inhibition against TRAIL-induced apoptosis in Jurkat cells. The R101A mutation abolished ligand binding but did not interfere with PLAD-mediated receptor interaction [Figs. 4D and 6B (compare a and b)]. Significantly, the R101A mutant strongly protected Jurkat cells against TRAIL-induced apoptosis at a level comparable to that conferred by wild-type TR4 ECD (Fig. 6A). Similarly, an analogous mutation in TR2 (M97A) that abolished ligand binding had no effect on the ability of a tailless, dominant interfering TR2 to inhibit apoptosis (Fig. 9). Taken together, our data clearly demonstrate that TR4 associates with TR2 via the PLAD and that this ligand-independent association between the two receptors is critically important for the inhibitory effect of TR4 on TR2-induced apoptosis.
Discussion
TR3 and TR4 were originally proposed to regulate TRAIL-induced cytotoxicity through ligand competition or ligand-driven formation of mixed trimer complexes with TR1 or TR2 (18). We demonstrated here that TR4 inhibits TR2-mediated cytotoxicity through ligand-independent association with TR2 via the PLAD. Contrary to previous reports, we found no evidence that TR4 acts as a “decoy” that competes for ligand binding. In fact, our evidence rules out that possibility. Because our data conclusively overrule the “decoy” model for TR4 inhibition of TRAIL signaling, we propose the term “regulatory receptor” for TR4. Formation of TR2-TR4 heterotrimeric complexes explains how the low-affinity receptor TR4 could effectively inhibit apoptosis signaling by the high-affinity receptor TR2. In this case, as in the other dominant interfering TNFR superfamily receptors, a deficiency of the DD potently reduces the transmission of the death signal (23, 27). Evidently, three intact DDs must be present for signaling to occur normally. The PLADs in both receptors allow both homotypic as well as heterotypic associations, unlike the homo-specific PLADs found in TNFR-1, TNFR-2, and Fas. This property is likely attributable to the strong sequence conservation within the PLADs of the TRAIL receptors. It is noteworthy that the other decoy receptor TR3 did not protect against TRAIL-induced apoptosis in Jurkat cells or interact with other TRAIL receptor ECDs. It is possible that TR3 may only interact with other TRAIL receptors in a complex containing three distinct receptors. Further experiments will be needed to evaluate the mechanism by which TR3 regulates TRAIL-induced apoptosis.
Many cytokine receptors signal as multisubunit entities. The association of the α, β, and γ subunits of the IL-2 receptor increases the affinity of the receptor. Swapping the α subunit of the IL-2 receptor with that from the IL-15 receptor changes the ligand specificity of the receptor. By contrast, the TR2-TR4 hetero-complexes did not alter the ligand specificity of the receptor. Rather, TR2-TR4 hetero-complexes dramatically affect the biological outcome of TRAIL stimulation through inhibition of apoptosis. Although the TR2-TR4 complexes are deficient for apoptosis induction, it is unclear whether they are also defective in NF-κB or MAP kinase signaling. Nonetheless, the PLAD-mediated mixed complexes have a dramatic regulatory effect on the cellular response to TRAIL.
Ligand-independent association between TR2 and TR4 may explain the resistance of CD8+ T cells and other untransformed cells to TRAIL. The lack of CD4+ T cell help during priming of CD8+ T cells causes TRAIL-induced apoptosis of these cells, which limits the expansion of the memory CD8+ T cells during secondary recall responses (20). Using PMA as a mimic for partial activation without CD4+ T cell help, we found that primary CD8+ T cells were rendered sensitive to TRAIL-induced apoptosis. Strikingly, this change in cellular sensitivity to TRAIL accompanied the abolition of ligand-independent interaction between TR2 and TR4. The heightened response to TRAIL in PMA-treated CD8+ T cells does not involve changes in expression of apoptosis inhibitors such as cFLIP (data not shown). Rather, RNAi-mediated knock down of TR4 expression was sufficient to sensitize cells to TRAIL. These data strongly implicate that ligand-independent interaction between TR2 and TR4 is a regulatory mechanism that controls cellular sensitivity to TRAIL. Similar regulation on the assembly of ligand-independent Fas complexes and sensitivity to Fas-induced apoptosis in CD4+ T cells has recently been reported, suggesting that ligand-independent receptor assembly is a general mechanism that regulates cellular sensitivity to death receptor stimulation (29). Finally, mutations in the PLAD or ligand-binding domain of TRAIL receptors have been identified in certain lung cancers (30, 31). These mutations may affect ligand-independent receptor assembly and cellular responses to TRAIL-induced apoptosis. Modulation of ligand-independent interactions between TRAIL signaling and regulatory receptors may be a useful strategy in cancer therapies.
Supplementary Material
Acknowledgments
We thank R. Welsh and J. Kang for critical reading of the manuscript. This work was supported by National Institutes of Health Grant AI065877 and the departmental startup fund (to F.K.-M.C.). F.K.-M.C. is a recipient of the Smith Family New Investigator Award and the Cancer Research Institute Investigator Award. Core resources supported by Diabetes Endocrinology Research Center Grant DK32520 were also used.
Author contributions: M.J.L. and F.K.-M.C. designed research; L.C., K.M., K.A., M.W., J.M., and F.K.-M.C. performed research; J.M. and G.S. contributed new reagents/analytic tools; M.J.L. and F.K.-M.C. analyzed data; and F.K.-M.C. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFP, cyan fluorescent protein; CRD, cysteine-rich domain; DD, death domain; ECD, extracellular domain; IP, immunoprecipitation; PLAD, preligand assembly domain; PMA, phorbol myristate acetate; RNAi, RNA interference; YFP, yellow fluorescent protein.
References
- 1.Chan, F. K., Siegel, M. R. & Lenardo, J. M. (2000) Immunity 13, 419-422. [DOI] [PubMed] [Google Scholar]
- 2.Ichikawa, K., Liu, W., Zhao, L., Wang, Z., Liu, D., Ohtsuka, T., Zhang, H., Mountz, J. D., Koopman, W. J., Kimberly, R. P. & Zhou, T. (2001) Nat. Med. 7, 954-960. [DOI] [PubMed] [Google Scholar]
- 3.Chuntharapai, A., Dodge, K., Grimmer, K., Schroeder, K., Marsters, S. A., Koeppen, H., Ashkenazi, A. & Kim, K. J. (2001) J. Immunol. 166, 4891-4898. [DOI] [PubMed] [Google Scholar]
- 4.Ashkenazi, A., Pai, R. C., Fong, S., Leung, S., Lawrence, D. A., Marsters, S. A., Blackie, C., Chang, L., McMurtrey, A. E., Hebert, A., et al. (1999) J. Clin. Invest. 104, 155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., et al. (1999) Nat. Med. 5, 157-163. [DOI] [PubMed] [Google Scholar]
- 6.Takeda, K., Yamaguchi, N., Akiba, H., Kojima, Y., Hayakawa, Y., Tanner, J. E., Sayers, T. J., Seki, N., Okumura, K., Yagita, H. & Smyth, M. J. (2004) J. Exp. Med. 199, 437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gliniak, B. & Le, T. (1999) Cancer Res. 59, 6153-6158. [PubMed] [Google Scholar]
- 8.Chinnaiyan, A. M., Prasad, U., Shankar, S., Hamstra, D. A., Shanaiah, M., Chenevert, T. L., Ross, B. D. & Rehemtulla, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankar, S. & Srivastava, R. K. (2004) Drug Resist. Updat. 7, 139-156. [DOI] [PubMed] [Google Scholar]
- 10.Wajant, H., Pfizenmaier, K. & Scheurich, P. (2002) Apoptosis 7, 449-459. [DOI] [PubMed] [Google Scholar]
- 11.Schmaltz, C., Alpdogan, O., Kappel, B. J., Muriglan, S. J., Rotolo, J. A., Ongchin, J., Willis, L. M., Greenberg, A. S., Eng, J. M., Crawford, J. M., et al. (2002) Nat. Med. 8, 1433-1437. [DOI] [PubMed] [Google Scholar]
- 12.Kayagaki, N., Yamaguchi, N., Nakayama, M., Kawasaki, A., Akiba, H., Okumura, K. & Yagita, H. (1999) J. Immunol. 162, 2639-2647. [PubMed] [Google Scholar]
- 13.Fanger, N. A., Maliszewski, C. R., Schooley, K. & Griffith, T. S. (1999) J. Exp. Med. 190, 1155-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diehl, G. E., Yue, H. H., Hsieh, K., Kuang, A. A., Ho, M., Morici, L. A., Lenz, L. L., Cado, D., Riley, L. W. & Winoto, A. (2004) Immunity 21, 877-889. [DOI] [PubMed] [Google Scholar]
- 15.Hymowitz, S. G., Christinger, H. W., Fuh, G., Ultsch, M., O'Connell, M., Kelley, R. F., Ashkenazi, A. & de Vos, A. M. (1999) Mol. Cell 4, 563-571. [DOI] [PubMed] [Google Scholar]
- 16.Mongkolsapaya, J., Grimes, J., Chen, N., Xu, X., Stuart, D., Jones, E. & Screaton, G. (1999) Nat. Struct. Biol. 6, 1048-1053. [DOI] [PubMed] [Google Scholar]
- 17.Kischkel, F. C., Lawrence, D. A., Tinel, A., LeBlanc, H., Virmani, A., Schow, P., Gazdar, A., Blenis, J., Arnott, D. & Ashkenazi, A. (2001) J. Biol. Chem. 276, 46639-46646. [DOI] [PubMed] [Google Scholar]
- 18.Ashkenazi, A. & Dixit, V. M. (1999) Curr. Opin. Cell Biol. 11, 255-260. [DOI] [PubMed] [Google Scholar]
- 19.Chan, F. K. M. & Lenardo, M. J. (2000) Eur. J. Immunol. 30, 652-660. [DOI] [PubMed] [Google Scholar]
- 20.Janssen, E. M., Droin, N. M., Lemmens, E. E., Pinkoski, M. J., Bensinger, S. J., Ehst, B., Griffith, T. S., Green, D. R. & Schoenberger, S. P. (2005) Nature 434, 88-93. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, X. D., Franco, A. V., Nguyen, T., Gray, C. P. & Hersey, P. (2000) J. Immunol. 164, 3961-3970. [DOI] [PubMed] [Google Scholar]
- 22.Chan, F. K., Chun, H. J., Zheng, L., Siegel, R. M., Bui, K. L. & Lenardo, M. J. (2000) Science 288, 2351-2354. [DOI] [PubMed] [Google Scholar]
- 23.Siegel, R. M., Frederiksen, J. K., Zacharias, D. A., Chan, F. K., Johnson, M., Lynch, D., Tsien, R. Y. & Lenardo, M. J. (2000) Science 288, 2354-2357. [DOI] [PubMed] [Google Scholar]
- 24.Sprick, M. R., Weigand, M. A., Rieser, E., Rauch, C. T., Juo, P., Blenis, J., Krammer, P. H. & Walczak, H. (2000) Immunity 12, 599-609. [DOI] [PubMed] [Google Scholar]
- 25.Truneh, A., Sharma, S., Silverman, C., Khandekar, S., Reddy, M. P., Deen, K. C., McLaughlin, M. M., Srinivasula, S. M., Livi, G. P., Marshall, L. A., et al. (2000) J. Biol. Chem. 275, 23319-23325. [DOI] [PubMed] [Google Scholar]
- 26.Chan, F. K. M., Siegel, R. M., Zacharias, D., Swofford, R., Holmes, K. L., Tsien, R. Y. & Lenardo, M. J. (2001) Cytometry 44, 361-368. [DOI] [PubMed] [Google Scholar]
- 27.Boldin, M. P., Mett, I. L., Varfolomeev, E. E., Chumakov, I., Shemer-Avni, Y., Camonis, J. H. & Wallach, D. (1995) J. Biol. Chem. 270, 387-391. [DOI] [PubMed] [Google Scholar]
- 28.Cha, S. S., Sung, B. J., Kim, Y. A., Song, Y. L., Kim, H. J., Kim, S., Lee, M. S. & Oh, B. H. (2000) J. Biol. Chem. 275, 31171-31177. [DOI] [PubMed] [Google Scholar]
- 29.Muppidi, J. R. & Siegel, R. M. (2004) Nat. Immunol. 5, 182-189. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., Shin, M. S., Kim, H. S., Lee, H. K., Park, W. S., Kim, S. Y., Lee, J. H., Han, S. Y., Park, J. Y., Oh, R. R., et al. (1999) Cancer Res. 59, 5683-5686. [PubMed] [Google Scholar]
- 31.Fisher, M. J., Virmani, A. K., Wu, L., Aplenc, R., Harper, J. C., Powell, S. M., Rebbeck, T. R., Sidransky, D., Gazdar, A. F. & El-Deiry, W. S. (2001) Clin. Cancer Res. 7, 1688-1697. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.