Abstract
The T-box transcription factor TBX5 plays essential roles in cardiac and limb development. Various mutations in the TBX5 gene have been identified in patients with Holt–Oram syndrome, which is characterized by congenital defects in the heart and upper extremities. In this study, we identified a WW-domain-containing transcriptional regulator TAZ as a potent TBX5 coactivator. TAZ directly associates with TBX5 and markedly stimulates TBX5-dependent promoters by interacting with the histone acetyltransferases p300 and PCAF. YAP, a TAZ-related protein with conserved functional domains, also stimulates TBX5-dependent transcription, possibly by forming a heterodimer with TAZ. TBX5 lacks a PY motif, which mediates the association of other proteins with TAZ, and interacts with TAZ through multiple domains including its carboxyl-terminal structure. Truncation mutants of TBX5 identified in patients with Holt–Oram syndrome were markedly impaired in their ability to associate with and be stimulated by TAZ. These findings reveal key roles for TAZ and YAP in the control of TBX5-dependent transcription and suggest the involvement of these coactivators in cardiac and limb development.
Keywords: transcriptional coactivator, organ development, hereditary birth defects
The activation of specific programs of gene expression during cell differentiation and organ development depends on combinatorial interactions among DNA-binding transcription factors, transcriptional cofactors, and histone-modifying enzymes (1, 2). Additional specificity and fine-tuning of gene expression is achieved by signal-dependent modulation of the expression and activity of the components of such multiprotein transcriptional complexes.
Members of the T-box family of transcription factors regulate a variety of developmental processes in vertebrates and invertebrates, including specification of mesoderm, development of the heart, vasculature, and limbs and tumorigenesis (3–5). The T-box, which encodes a conserved 180-amino acid DNA-binding domain, has been identified in at least 18 mammalian T-box genes. In many cases, haploinsufficiency of T-box genes results in dramatic morphological abnormalities, emphasizing the importance of specific thresholds of transcriptional activity of T-box factors for developmental decisions (4). For example, heterozygous mutations in TBX1 have been implicated in 22q11 deletion (DiGeorge) syndrome, characterized by abnormalities in the aortic arch arteries due to defects in neural crest cell migration (4, 6), and mutations in TBX5 cause Holt–Oram syndrome (HOS), which manifests as a variety of cardiac and upper limb abnormalities (7–11).
TBX5 is expressed in the embryonic heart and forelimbs and regulates transcription of downstream genes, such as those encoding atrial natriuretic factor (ANF) and fibroblast growth factor 10 (Fgf10) by binding to TBX-binding DNA elements (TBEs) (12–17). Targeted deletion of the Tbx5 gene in mice results in embryonic lethality with severe cardiac defects (13), and mice with limb-specific Tbx5 deletion show no forelimb formation (15). Despite extensive analyses of the developmental functions of TBX5, relatively little is known of mechanisms of transcriptional activation by TBX5. Given the remarkable sensitivity of diverse developmental processes to precise levels of T-box protein expression (3–5), transcriptional coactivators, corepressors, and signaling molecules are likely to have profound effects on TBX5-dependent gene expression.
Here, we show that the WW-domain-containing transcriptional regulator TAZ (18) acts as a potent TBX5 coactivator. TAZ physically associates with TBX5 and histone acetyltransferase (HAT) proteins and mediates TBX5-dependent gene activation. These findings provide insights into the mechanism of action of TBX5 and suggest that TAZ plays important roles in the control of TBX5-dependent genes during cardiac and limb development.
Materials and Methods
Plasmids. Mammalian expression constructs of human TBX5, mouse TAZ, YAP, and PGC1 were prepared by inserting PCR fragments into the pcDNA3.1 vector (Invitrogen) with a Flag or Myc tag or into the pM Gal4-DBD vector. Point mutations were introduced by using the QuickChange XL site-directed mutagenesis kit (Stratagene). Luciferase reporters containing two copies of a TBE or its mutant (TBE, 5′-TCACACCTTTGAAGTG-3′; mutant, 5′-TCAGACCTTTGAAGTG-3′) (19) were prepared by using the pLUC-MCS plasmid (Stratagene). The same TBE mutations were also introduced into ANF-luciferase. Plasmids encoding p300, Tip60, Grip1, PCAF, human ANF-luciferase, Fgf10-luciferase, and control β-galactosidase were described in ref. 15 and refs. 20–26), and a UAS-luciferase plasmid, pFR-Luc, was purchased from Stratagene.
Luciferase Reporter Assays. Plasmid transfection into COS-1, 293T, and NIH 3T3 cells was carried out by using FuGENE6 reagent (Roche). Primary neonatal rat cardiac myocytes were prepared as described in ref. 26 and were transfected by using Lipofectamine Plus reagent (Invitrogen). Luciferase reporter activity was examined 48 h after transfection and was normalized to β-galactosidase activity, as described in ref. 26. The results of luciferase assays were reproduced in at least three independent experiments performed in triplicate, and the representative data are shown in the figures.
Coimmunoprecipitation and Oligonucleotide Pull-Down Assays. Proteins were expressed by plasmid transfection of 293T cells or as GST-fusion proteins in Escherichia coli. Immunoprecipitation was performed by using anti-Flag (Sigma) or anti-HA antibody (Santa Cruz Biotechnology), and oligonucleotide pull-down assays were performed by using biotin-labeled DNA fragments containing the TBEs (underlined) (5′-AATATCACACCTGTACAATATCACACCTGTACAATATCACACCTGTAC-3′), as described in ref. 27.
RNA Interference Experiment. Small interfering (si)RNAs against mouse TAZ and YAP were chemically synthesized (TAZ-site #1, 5′-AAUCACCACAUGGCAAGACUU-3′ and 5′-GUCUUGCCAUGUGGUGAUUUU-3′; TAZ-site #2, 5′-AGAGAUACUUCCUUAAUCAUU-3′ and 5′-UGAUUAAGGAAGUAUCUCUUU-3′; YAP-site #1, 5′-GCCAUGACUCAGGAUGGAGUU-3′ and 5′-CUCCAUCCUGAGUCAUGGCUU-3′; YAP-site #2, 5′-AGAAAGCUUUCUCACGUGGUU-3′ and 5′-CCACGUGAGAAAGCUUUCUUU-3′). The effects of siRNAs were assessed by cotransfection with TAZ and YAP expression plasmids. TAZ and YAP siRNAs significantly decreased the expression of Flag-TAZ and -YAP, respectively (data not shown). Primary neonatal mouse cardiac myocytes were prepared by the method used for rat myocyte culture (26). siRNA against TAZ or YAP or control siRNA (Qiagen) was cotransfected with ANF-luciferase plasmid into mouse myocytes by using Lipofectamine Plus reagent, and luciferase assays were performed 48 h after transfection.
Electrophoretic Mobility Shift Assay. Electrophoretic mobility shift assays were performed by using glutathione-S-transferase-fused TBX5 proteins and 32P-labeled TBE oligonucleotide probe as described in refs. 19 and 27.
in Vitro HAT Assay. Myc-TAZ, Myc-YAP, Myc-TBX5, and HA-p300 were expressed in 293T cells and immunoprecipitated with anti-Myc or anti-HA antibody. In vitro HAT assays were performed by using p300/CBP IP-HAT assay kit (Upstate Biotechnology, Lake Placid, NY). Protein expression was confirmed by Western blot analysis.
Results
Identification of TAZ as a Potent Coactivator for TBX5. To identify transcriptional coactivators for TBX5, we performed a candidate-based search using a luciferase reporter controlled by promoter of the ANF gene, a cardiac-specific target of TBX5 (13, 19). Various transcriptional cofactors were coexpressed with TBX5 in COS-1 cells, and the expression of ANF-luciferase was examined as an indicator of TBX5-dependent transcriptional activity. In this screen, we found that a WW-domain-containing protein TAZ (18), also called WWTR1, dramatically activated TBX5-dependent ANF-luciferase expression in a dose-dependent manner, whereas transcriptional coactivators, such as p300 (21), Grip1 (22), Tip60 (23), PGC1 (24), and PCAF (25), did not strongly stimulate ANF-luciferase expression (Fig. 1 A and B). TAZ also stimulated TBX5-dependent ANF-luciferase activity in other cells, such as 293T, NIH 3T3, and neonatal rat cardiac myocytes (data not shown).
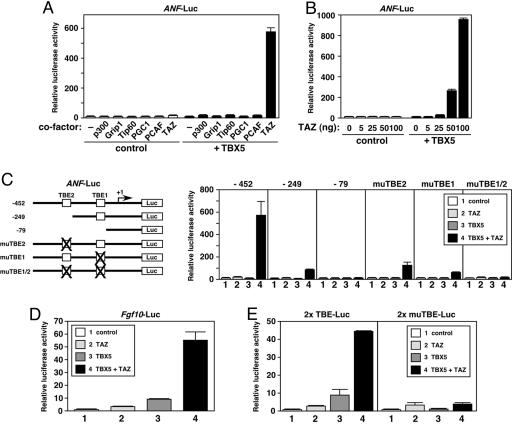
Fig. 1.
TAZ potently activates TBX5-dependent transcription. (A) TAZ potently activates ANF-luciferase expression in the presence of TBX5. COS-1 cells were transiently transfected with plasmids for ANF-luciferase (ANF-Luc, 100 ng) and control β-galactosidase (10 ng), TBX5 expression plasmid (10 ng), and the expression plasmids of various transcriptional coregulators (100 ng). Basal luciferase activity without expression of TBX5 and coregulators was given a value of 1. (B) TAZ activates TBX5-dependent ANF-luciferase expression in a dose-dependent manner. TBX5, 10 ng; TAZ, 5–100 ng. (C) TBEs are essential for TAZ-mediated activation of ANF-luciferase expression. (Right) Luciferase assays with the ANF-luciferase constructs (–452, –249, and –79 bp) and its mutants (muTBE). TBX5, 10 ng; TAZ, 100 ng. (Left) Structures of the ANF-luciferase constructs (–452, –249, and –79 bp) and its mutants are shown. The transcriptional initiation site is shown as +1. TBX5, 10 ng; TAZ, 100 ng. (D) TAZ activates TBX5-dependent Fgf10-luciferase (Fgf10-Luc) expression. TBX5, 50 ng; TAZ, 100 ng. (E) TAZ stimulates the activity of a luciferase reporter that contains tandem TBEs (2× TBE-Luc), but not the reporter with mutated TBEs (2× muTBE-Luc). TBX5, 10 ng; TAZ, 100 ng. In B–E, COS-1 cells were transfected with the expression plasmids of TBX5 and TAZ together with luciferase and β-galactosidase plasmids. Basal activity of each luciferase reporter without TBX5 and TAZ expression was given a value of 1.
DNA Binding of TBX5 Is Essential for TAZ-Mediated Transcriptional Activation. The –452-bp human ANF promoter contains two TBEs, TBE1 and TBE2 (Fig. 1C) (13, 20). Analysis of promoter deletion mutants suggested that the action of TAZ was mediated through these TBEs (Fig. 1C). In fact, point mutations in either TBE significantly decreased TAZ-dependent transcriptional activation, and mutations of both TBEs completely abolished it (Fig. 1C). As shown in Fig. 1D, TAZ also activated the Fgf10 promoter, a known TBX5 target gene (13–15). TAZ also stimulated the activity of a luciferase reporter that contained tandem TBEs and a minimal promoter, whereas it failed to activate the reporter with mutated TBEs (Fig. 1E). These results clearly indicated that TAZ-mediated transcriptional activation relied on the binding of TBX5 to the TBEs.
TAZ and Its Related Protein YAP Are Important for Endogenous ANF Promoter Activity. TAZ shows significant structural similarity to another WW-domain-containing protein YAP (Fig. 2A) (18, 28). TAZ and YAP share conserved domains, such as WW domain(s), a coiled-coil region, and a PDZ-binding motif. As shown in Fig. 2B, YAP also activated TBX5-dependent transcription in a dose-dependent manner, although the effect of YAP was significantly weaker than that of TAZ. During mouse embryonic development, Taz and Yap are broadly expressed, including in the heart and forelimbs (M.M., James A. Richardson, E.N.O., and O.N., unpublished results). To assess the importance of endogenous TAZ and YAP for the ANF promoter activity in cardiac myocytes, we examined the effect of siRNA against TAZ (TAZ-site #1 and -site #2) or YAP (YAP-site #1 and -site #2) (Fig. 2C). Either TAZ or YAP siRNAs significantly decreased ANF-luciferase reporter expression in mouse cardiac myocytes, suggesting that endogenous TAZ and YAP play important roles in cardiac ANF expression.
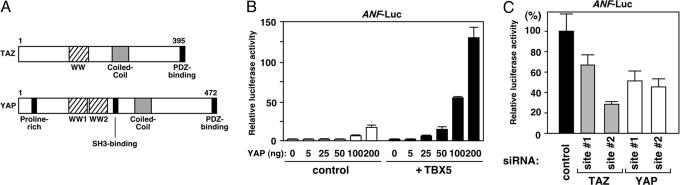
Fig. 2.
TAZ and YAP play important roles in cardiac ANF expression. (A) Structure of mouse TAZ and YAP proteins. WW, WW1, and WW2, WW domain. TAZ and YAP share WW domain(s), a coiled-coil motif, and a PDZ-binding motif. YAP contains a proline-rich domain and an SH3-binding motif. (B) YAP activates TBX5-dependent ANF-luciferase expression in a dose-dependent manner. TBX5, 10 ng; YAP, 5–200 ng. Basal luciferase activity without TBX5 and YAP expression was given a value of 1. (C) siRNA against TAZ or YAP reduced ANF-luciferase activity in cardiac myocytes. Primary neonatal mouse cardiac myocytes were cotransfected with siRNA and ANF-luciferase plasmid. siRNA, control (siRNA without known homology with mammalian sequences) (Qiagen); TAZ-site #1 and -site #2, against mouse TAZ; YAP-site #1 and -site #2, against mouse YAP. Luciferase activity with control siRNA cotransfection was given a value of 100%.
TAZ Physically Associates with TBX5. TAZ significantly activated transcription of a UAS-luciferase reporter by the Gal4 DNA-binding domain fused to TBX5 (Fig. 3A), suggesting that TAZ is recruited to DNA by direct interaction with TBX5. Consistent with this hypothesis, TAZ showed physical association with TBX5 in coimmunoprecipitation assays (Fig. 3B), and TAZ was pulled down with a TBE oligonucleotide in the presence of TBX5 (Fig. 3C).
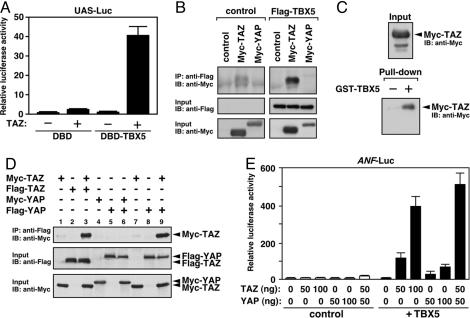
Fig. 3.
Physical interactions among TBX5, TAZ, and YAP. (A) TAZ activates UAS-luciferase reporter expression by Gal4 DNA-binding domain fused to TBX5 (DBD-TBX5). DBD or DBD-TBX5, 10 ng; TAZ, 100 ng. (B) TAZ, but not YAP, was coimmunoprecipitated with TBX5. Flag-TBX5, Myc-TAZ, and Myc-YAP were expressed in 293T cells and coimmunoprecipitation was performed by using anti-Flag antibody. Western blot analysis (IB) was performed on immunoprecipitates (IP) and total cell lysates (Input) by using anti-Myc or -Flag antibody. (C) TAZ associates with TBE DNA fragments through TBX5. Biotin-labeled TBE oligonucleotide fragments were bound to streptoavidin beads and incubated with Myc-TAZ in the presence of GST or GST-TBX5. Recovery of Myc-TAZ with streptoavidin beads was examined by Western blotting using anti-Myc antibody. (D) TAZ forms a homodimer and a heterodimer with YAP. Flag-TAZ or -YAP was immunoprecipitated by using anti-Flag antibody, and associated proteins were detected by Western blotting using anti-Myc antibody (Top). Expression of TAZ and YAP was confirmed by using anti-Myc or -Flag antibody (Middle and Bottom). (E) Coexpression of TAZ potently enhances YAP-mediated activation of TBX5-dependent transcription. COS-1 cells were cotransfected with TBX5 expression plasmid (10 ng) and different amounts of TAZ and YAP plasmids. Basal luciferase activity was given a value of 1.
The WW domain of TAZ is known to mediate interactions with transcription factors that contain PY-motifs (18, 29). Interestingly, however, TBX5 lacks a PY-motif, and a mutation of the WW domain in TAZ did not affect association with TBX5 (data not shown). Instead, deletion mutant analysis using coimmunoprecipitation assays suggested that TBX5 interacts with TAZ through multiple domains, including its carboxyl-terminal sequences (data not shown).
YAP May Activate TBX5-Dependent Transcription Through Complex Formation with TAZ. Despite the structural similarity between TAZ and YAP, YAP did not show detectable physical association with TBX5 (Fig. 3B), even though YAP significantly activated TBX5-dependent transcription in COS-1 cells (Fig. 2B). Because TAZ and YAP contain coiled-coil domains and are endogenously expressed in COS-1 cells at low levels (data not shown), we hypothesized that YAP might be recruited to TBX5 by association with TAZ. Indeed, we found that YAP forms a stable heterodimer with TAZ (Fig. 3D, lane 9). TAZ, but not YAP, could also form a homodimer (Fig. 3D, lanes 3 and 6), and the coiled-coil domain was important for homodimer formation (see Fig. 6, which is published as supporting information on the PNAS web site). Consistently, the effects of YAP on TBX5-dependent transcription were strongly augmented by coexpression of TAZ, although the stimulatory activity of YAP on TBX5 was significantly weaker than that of TAZ (Fig. 3E). These results suggest that YAP may activate TBX5-dependent transcription through association with TAZ and that the complex formation of TAZ/YAP proteins may be necessary for their transcriptional activity.
TAZ Associates with HAT Proteins to Activate TBX5-Dependent Transcription. TAZ functions as a transcriptional coactivator for DNA-binding proteins, such as Runx2, TTF-1, and TEF-1 (30–32). However, the precise mechanisms by which TAZ stimulates transcription remain unclear. We therefore sought to define the mechanisms of TAZ-dependent activation of TBX5. TAZ did not show intrinsic HAT activity against histone H4 peptides (Fig. 4A). However, TAZ physically associated with HAT proteins, such as p300 and PCAF (Fig. 4B). Additionally, p300 or PCAF potently enhanced TBX5-dependent ANF-luciferase activity in the presence of TAZ, in contrast to their weak activity without TAZ coexpression (Fig. 4C). These results suggested that TAZ is essential for the activation of TBX5-dependent transcription by these HAT proteins.
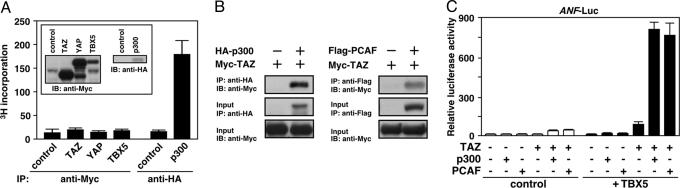
Fig. 4.
TAZ associates with HAT proteins to activate transcription. (A) TAZ and YAP do not show HAT activity. In vitro HAT assays were performed by using the proteins that were expressed in 293T cells and purified by immunoprecipitation (IP). (Inset) Protein expression was confirmed by Western blot analysis. (B) TAZ physically associates with p300 and PCAF. Coimmunoprecipitation was performed as described above. (C) p300 and PCAF enhance TBX5-dependent transcription in the presence of TAZ. COS-1 cells were transfected with the expression plasmids (TBX5, 10 ng; TAZ, 50 ng; p300 or PCAF, 100 ng) and ANF-luciferase plasmid.
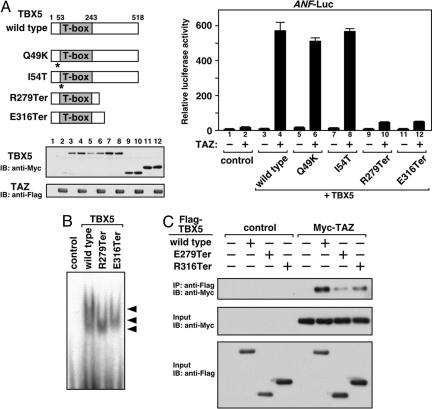
Functional Interaction of TBX5 and TAZ in HOS Patients. The apparent dependence of TBX5 on TAZ for efficient transcriptional activation prompted us to examine whether any TBX5 mutants identified in HOS patients might be compromised in their ability to recruit TAZ. We were especially curious about a class of TBX5 mutants that retains the ability to bind DNA but, for unknown reasons, is unable to activate transcription. Two such mutants, TBX5-R279Ter (9) and -E316Ter (10), which contain intact T-box DNA-binding domains but are truncated prematurely, were unable to cooperate with TAZ to activate ANF-luciferase expression (Fig. 5A). Activation of Fgf10-luciferase by TAZ was also impaired when TBX5-R279Ter or -E316Ter was coexpressed (data not shown). In contrast, the point mutants TBX5-Q49K, -I54T (Fig. 5A), -G169R, and -S252I (data not shown) were unimpaired with respect to their responsiveness to TAZ.
Fig. 5.
Effects of TAZ on various TBX5 mutants identified in HOS patients. (A) Premature-termination mutants identified in HOS patients, TBX5-R279Ter and -E316Ter, show marked reduction of TAZ-mediated transactivation. (Left) Structures of TBX5 mutants are shown. Numbers indicate the positions of the T-box and amino-/carboxyl-termini of full-length protein in the amino acid sequence. Asterisks indicate the positions of point mutations. COS-1 cells were cotransfected with expression plasmids encoding TBX5 or HOS mutants (10 ng) and/or TAZ plasmid (100 ng). Expression of TBX5 proteins and TAZ was confirmed by Western blotting. Lane numbers on the gel correspond to transfections (Right). (B) DNA-binding activity of TBX5-R279Ter and -E316Ter mutants is not affected. Electrophoretic mobility shift assay is shown. Arrowheads indicate shifted bands. (C) TBX5-R279Ter and -E316Ter show inefficient interaction with TAZ. Coimmunoprecipitation was performed as described in Fig. 3.
As expected, TBX5-R279Ter and -E316Ter showed normal binding to the TBE (Fig. 5B). The T-box domain of TBX5 is known to mediate association with Nkx2.5 (19), and TBX5-R279Ter and -E316Ter physically interacted with Nkx2.5 in coimmunoprecipitation assays (data not shown). However, association of these two TBX5 mutants with TAZ was significantly reduced (Fig. 5C), suggesting that abnormalities of TBX5–TAZ complex formation might contribute, at least in part, to the defects of TBX5-dependent transcriptional regulation in HOS.
Discussion
The results of this study demonstrate that TAZ acts as a powerful coactivator for TBX5, a transcription factor implicated in HOS. TAZ directly associates with TBX5 and the HAT proteins p300 and PCAF, thereby acting as a central component in TBX5-dependent transcriptional complexes. The TAZ-related protein YAP also stimulates TBX5 activity, and its influence on TBX5 is potentiated by TAZ, with which it forms a heterodimer.
How do TAZ and YAP stimulate the transcriptional activity of TBX5? Our results suggest that TBX5 recruits TAZ/YAP to downstream target genes, resulting in remarkable augmentation of transcription. The physical association of TAZ with the HAT proteins p300 and PCAF stimulates TBX5-dependent transcription, presumably by promoting acetylation of histones associated with TBX5 target genes. It is also conceivable that TAZ and YAP act through additional mechanisms, for example by recruiting other coactivators, displacing repressors, or stabilizing interactions between TBX5 and other components of the transcriptional machinery.
TBX5 Mutations Associated with HOS. Numerous different mutations in the TBX5 gene have been reported in patients with HOS, varying from single amino acid substitutions to large deletions or truncations (7–11, 19). Proteins truncated by nonsense mutations are often produced at reduced levels because of nonsense-mediated mRNA degradation (33), although the transcripts for a zebrafish Tbx5 mutant, heartstrings, which is similar to the human TBX5 truncation mutants TBX5-R279Ter and -E316Ter, are expressed at wild-type levels (34). Our results suggest that such truncated TBX5 mutants do not efficiently activate downstream gene expression, at least in part, because of defects in association with TAZ. HOS patients display a variety of clinical symptoms, but clear genotype–phenotype relationships have not been found among different TBX5 mutations (7–11, 19). We speculate that differences in transcriptional-complex formation by mutant TBX5 proteins may contribute to the diversity of clinical symptoms in HOS.
It will be of interest to investigate the potential influence of TAZ and YAP on other T-box proteins, many of which have been implicated in human disease (4). In this regard, there is evidence for the existence of modifier genes that influence the activity of T-box factors and the developmental programs they regulate (35, 36). TAZ and YAP display the properties of modifiers, raising the possibility that mutations in these factors could affect T-box-dependent processes in humans.
Diverse Functions of TAZ and YAP. TAZ and YAP interact with numerous transcription factors and have been implicated in a variety of biological processes (18, 28, 30–32, 37–40). For example, TAZ induces osteoblast differentiation of mesenchymal stem cells by coactivating Runx2 and repressing peroxisome proliferator-activated receptor γ (38), whereas a Drosophila orthologue of YAP, Yorkie, has been identified as a critical target of the Wts/Lats protein kinase, which regulates cell proliferation and apoptosis (39). TAZ and YAP are broadly coexpressed in embryonic tissues, and each has distinct repertoires of DNA-binding proteins as direct association partners, suggesting that TAZ and YAP may fulfill distinct but partially overlapping roles in various organs in vivo. Notably, TAZ and YAP also act as transcriptional repressors in some settings (38, 40); thus, their effects on specific genes likely depend on their association with DNA-binding proteins, coactivators, and corepressors.
Implications. The ANF promoter serves as a target of cardiac-specific and stress-dependent gene regulation. The remarkable sensitivity of this promoter to TBX5 and TAZ/YAP points to a key role of this protein partnership in these transcriptional programs in vivo. Phosphorylation of TAZ and YAP controls their subcellular localization and transcriptional activity (18, 37), suggesting that TAZ and YAP may provide TBX5 with responsiveness to regulatory cues. It will be intriguing to examine how these factors are involved in the marked induction of ANF gene expression in cardiac hypertrophy and failure (41, 42).
In parallel with the candidate-based search described in this article, we have performed an unbiased expression screen of the embryonic heart cDNA library using the ANF-luciferase reporter system and identified several cofactors and signaling molecules that modulate the activity of this promoter (data not shown). Understanding the combinatorial interactions and signal-dependent regulation of TAZ and YAP with other transcriptional regulators will provide insights into the molecular mechanisms of cardiac development and disease.
Supplementary Material
Acknowledgments
We thank Drs. D. Garry and R. Bassel-Duby for valuable comments on the manuscript; Dr. H. Kurihara for sharing unpublished data; Drs. Y. Nakatani (Dana–Farber Cancer Institute and Harvard Medical School, Boston), M. Stallcup (University of Southern California, Los Angeles), R. Eckner (University of Zurich, Zurich), B. Bruneau (Hospital for Sick Children and University of Toronto, Toronto), Y. Saito (Kyoto University, Kyoto, Japan), and K. Kuwahara (Kyoto University, Kyoto, Japan) for plasmids; Dr. T. Okamoto for information on mouse myocyte culture; Dr. James Richardson and J. Shelton for histological analysis; and J. Bartos, J. McAnally, and J. Page for technical and secretarial assistance. E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, and the Robert A. Welch Foundation. O.N. was supported by grants from the American Heart Association Texas Affiliate and the Muscular Dystrophy Association. M.M. was supported by a fellowship from the Japan Heart Foundation and by Kumamoto University Institute of Molecular Embryology and Genetics.
Author contributions: E.N.O., O.N., and M.M. designed research; M.M. and M.N. performed research; M.M. and M.N. contributed new reagents/analytic tools; E.N.O., O.N., and M.M., analyzed data; and E.N.O. and O.N. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: ANF, atrial natriuretic factor; Fgf10, fibroblast growth factor 10; HAT, histone acetyltransferase; HOS, Holt–Oram syndrome; si, small interfering; TBE, TBX-binding DNA element.
References
- 1.Spiegelman, B. M. & Heinrich, R. (2004) Cell 119, 157–167. [DOI] [PubMed] [Google Scholar]
- 2.Perissi, V. & Rosenfeld, M. G. (2005) Nat. Rev. Mol. Cell Biol. 6, 542–554. [DOI] [PubMed] [Google Scholar]
- 3.Smith, J. (1999) Trends Genet. 15, 154–158. [DOI] [PubMed] [Google Scholar]
- 4.Packham, E. A. & Brook, J. D. (2003) Hum. Mol. Genet. 12, R37–R44. [DOI] [PubMed] [Google Scholar]
- 5.Plageman, T. F., Jr., & Yutzey, K. E. (2005) Dev. Dyn. 232, 11–20. [DOI] [PubMed] [Google Scholar]
- 6.Baldini, A. (2004) Curr. Opin. Cardiol. 19, 201–204. [DOI] [PubMed] [Google Scholar]
- 7.Basson, C. T., Bachinsky, D. R., Lin, R. C., Levi, T., Elkins, J. A., Soults, J., Grayzel, D., Kroumpouzou, E., Traill, T. A., Leblanc-Straceski, J., et al. (1997) Nat. Genet. 15, 30–35. [DOI] [PubMed] [Google Scholar]
- 8.Li, Q. Y., Newbury-Ecob, R. A., Terrett, J. A., Wilson, D. I., Curtis, A. R., Yi, C. H., Gebuhr, T., Bullen, P. J., Robson, S. C., Strachan, T., et al. (1997) Nat. Genet. 15, 21–29. [DOI] [PubMed] [Google Scholar]
- 9.Brassington, A. M., Sung, S. S., Toydemir, R. M., Le, T., Roeder, A. D., Rutherford, A. E., Whitby, F. G., Jorde, L. B. & Bamshad, M. J. (2003) Am. J. Hum. Genet. 73, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross, S. J., Ching, Y. H., Li, Q. Y., Armstrong-Buisseret, L., Spranger, S., Lyonnet, S., Bonnet, D., Penttinen, M., Jonveaux, P., Leheup, B., et al. (2000) J. Med. Genet. 37, 785–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, C., Liu, M. & Wang, Q. (2003) J. Biol. Chem. 278, 8780–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Esteban, C., Tsukui, T., Yonei, S., Magallon, J., Tamura, K. & Izpisua Belmonte, J. C. (1999) Nature 398, 814–818. [DOI] [PubMed] [Google Scholar]
- 13.Bruneau, B. G., Nemer, G., Schmitt, J. P., Charron, F., Robitaille, L., Caron, S., Conner, D. A., Gessler, M., Nemer, M., Seidman, C. E. & Seidman, J. G. (2001) Cell 106, 709–721. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh, T. K., Packham, E. A., Bonser, A. J., Robinson, T. E., Cross, S. J. & Brook, J. D. (2001) Hum. Mol. Genet. 10, 1983–1994. [DOI] [PubMed] [Google Scholar]
- 15.Rallis, C., Bruneau, B. G., Del Buono, J., Seidman, C. E., Seidman, J. G., Nissim, S., Tabin, C. J. & Logan, M. P. (2003) Development (Cambridge, U.K.) 130, 2741–2751. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal, P., Wylie, J. N., Galceran, J., Arkhitko, O., Li, C., Deng, C., Grosschedl, R. & Bruneau, B. G. (2003) Development (Cambridge, U.K.) 130, 623–633. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi, J. K., Koshiba-Takeuchi, K., Suzuki, T., Kamimura, M., Ogura, K. & Ogura, T. (2003) Development (Cambridge, U.K.) 130, 2729–2739. [DOI] [PubMed] [Google Scholar]
- 18.Kanai, F., Marignani, P. A., Sarbassova, D., Yagi, R., Hall, R. A., Donowitz, M., Hisaminato, A., Fujiwara, T., Ito, Y., Cantley, L. C. & Yaffe, M. B. (2000) EMBO J. 19, 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiroi, Y., Kudoh, S., Monzen, K., Ikeda, Y., Yazaki, Y., Nagai, R. & Komuro, I. (2001) Nat. Genet. 28, 276–280. [DOI] [PubMed] [Google Scholar]
- 20.Kuwahara, K., Saito, Y., Ogawa, E., Takahashi, N., Nakagawa, Y., Naruse, Y., Harada, M., Hamanaka, I., Izumi, T., Miyamoto, Y., et al. (2001) Mol. Cell. Biol. 21, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao, T. P., Oh, S. P., Fuchs, M., Zhou, N.-D., Ch'ng, L.-E., Newsome, D., Bronson, R. T., Li, E., Livingston, D. M. & Eckner, R. (1998) Cell 93, 361–372. [DOI] [PubMed] [Google Scholar]
- 22.Hong, H., Kohli, K., Trivedi, A., Johnson, D. L. & Stallcup, M. R. (1996) Proc. Natl. Acad. Sci. USA 93, 4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao, X. & Sudhof, T. C. (2001) Science 293, 115–120. [DOI] [PubMed] [Google Scholar]
- 24.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., et al. (2002) Nature 418, 797–801. [DOI] [PubMed] [Google Scholar]
- 25.Puri, P. L., Sartorelli, V., Yang, X. J., Hamamori, Y., Ogryzko, V. V., Howard, B. H., Kedes, L., Wang, J. Y., Graessmann, A., Nakatani, Y. & Levrero, M. (1997) Mol. Cell 1, 35–45. [DOI] [PubMed] [Google Scholar]
- 26.Kathiriya, I. S., King, I. N., Murakami, M., Nakagawa, M., Astle, J. M., Gardner, K. A., Gerard, R. D., Olson, E. N., Srivastava, D. & Nakagawa, O. (2004) J. Biol. Chem. 279, 54937–54943. [DOI] [PubMed] [Google Scholar]
- 27.Murakami, M., Kataoka, K., Fukuhara, S., Nakagawa, O. & Kurihara, H. (2004) Eur. J. Biochem. 271, 3330–3339. [DOI] [PubMed] [Google Scholar]
- 28.Yagi, R., Chen, L. F., Shigesada, K., Murakami, Y. & Ito, Y. (1999) EMBO J. 18, 2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudol, M., Chen, H. I., Bougeret, C., Einbond, A. & Bork, P. (1995) FEBS Lett. 369, 67–71. [DOI] [PubMed] [Google Scholar]
- 30.Cui, C. B., Cooper, L. F., Yang, X., Karsenty, G. & Aukhil, I. (2003) Mol. Cell. Biol. 23, 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, K. S., Whitsett, J. A., Di Palma, T., Hong, J. H., Yaffe, M. B. & Zannini, M. (2004) J. Biol. Chem. 279, 17384–17390. [DOI] [PubMed] [Google Scholar]
- 32.Mahoney, W. M., Hong, J. H., Yaffe, M. B. & Farrance, I. K. (2005) Biochem. J. 44, 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frischmeyer, P. A. & Dietz, H. C. (1999) Hum. Mol. Genet. 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 34.Garrity, D. M., Childs, S. & Fishman, M. C. (2002) Development (Cambridge, U.K.) 129, 4635–4645. [DOI] [PubMed] [Google Scholar]
- 35.Huang, T. (2002) Curr. Opin. Pediatr. 14, 691–695. [DOI] [PubMed] [Google Scholar]
- 36.Vitelli, F., Taddei, I., Morishima, M., Meyers, E. N., Lindsay, E. A. & Baldini, A. (2002) Development (Cambridge, U.K.) 129, 4605–4611. [DOI] [PubMed] [Google Scholar]
- 37.Basu, S., Totty, N. F., Irwin, M. S., Sudol, M. & Downward, J. (2003) Mol. Cell 11, 11–23. [DOI] [PubMed] [Google Scholar]
- 38.Hong, J. H., Hwang, E. S., McManus, M. T., Amsterdam, A., Tian, Y., Kalmukova, R., Mueller, E., Benjamin, T., Spiegelman, B. M., Sharp, P. A., et al. (2005) Science 309, 1074–1078. [DOI] [PubMed] [Google Scholar]
- 39.Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. (2005) Cell 122, 421–434. [DOI] [PubMed] [Google Scholar]
- 40.Zaidi, S. K., Sullivan, A. J., Medina, R., Ito, Y., van Wijnen, A. J., Stein, J. L., Lian, J. B. & Stein, G. S. (2004) EMBO J. 23, 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burnett, J. C., Jr., Kao, P. C., Hu, D. C., Heser, D. W., Heublein, D., Granger, J. P., Opgenorth, T. J. & Reeder, G. S. (1986) Science. 231, 1145–1147. [DOI] [PubMed] [Google Scholar]
- 42.Saito, Y., Nakao, K., Arai, H., Nishimura, K., Okumura, K., Obata, K., Takemura, G., Fujiwara, H., Sugawara, A., Yamada, T., et al. (1989) J. Clin. Invest. 83, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.