Abstract
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants and procarcinogens that require activation by host metabolism. Metabolic activation of PAHs by Aldo-Keto-Reductases (AKRs) leads to formation of reactive and redox active o-quinones, which may cause oxidatively generated DNA damage. Spectrophotometric assays showed that NADPH caused PAH o-quinones to enter futile redox-cycles, which result in the depletion of excess cofactor. Copper (II) amplified NADPH-dependent redox-cycling of the o-quinones. Concurrent with NADPH oxidation, molecular oxygen was consumed, indicating the production of ROS. To determine whether PAH o-quinones can cause 8-oxo-dGuo formation in salmon testis DNA, three pre-requisite experimental conditions were satisfied. Quantitative complete enzymatic hydrolysis of DNA was achieved, adventitious oxidation of dGuo was eliminated by the use of chelex and desferal, and basal levels of less than 2.0 8-oxo-dGuo/105 dGuo were obtained. The HPLC-ECD analytical method was validated by spiking the DNA with standard 8-oxo-dGuo and demonstrating quantitative recovery. HPLC-ECD analysis revealed that in the presence of NADPH and Cu(II), submicromolar concentrations of PAH o-quinones generated > 60.0 8-oxo-dGuo adducts/105 dGuo. The rank order of 8-oxo-dGuo generated in isolated DNA was NP-1,2-dione > BA-3,4-dione > 7,12-DMBA-3,4-dione > BP-7,8-dione. The formation of 8-oxo-dGuo by PAH o-quinones was concentration-dependent. It was completely or partially inhibited when catalase, tiron or a Cu(I) specific chelator, bathocuproine were added, indicating the requirement for H2O2, O2− and Cu(I), respectively. Methional which is a copper-hydroperoxo complex (Cu(I)OOH) scavenger also suppressed 8-oxo-dGuo formation. By contrast, mannitol, sodium benzoate and sodium formate, which act as hydroxyl radical scavengers, did not block its formation. Sodium azide which can act as both a hydroxyl radical and 1O2 scavenger abolished the formation of 8-oxo-dGuo. These data showed that the production of 8-oxo-dGuo was dependent on Cu(II) /Cu(I) catalyzed redox cycling of PAH o-quinones to produce ROS and that the immediate oxidant was not hydroxyl radical or Cu(I)OOH and that it is more likely 1O2 which can produce a 4,8-endoperoxide-dGuo intermediate.
Keywords: Polycyclic aromatic hydrocarbons, o-quinones, 8-oxo-dGuo, ROS, singlet oxygen
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants and procarcinogens (1,2). They are present in tobacco smoke and are candidate human lung carcinogens (3). PAHs require activation by host xenobiotic metabolism to exert their harmful effects. They are activated to their corresponding final reactive electrophiles via three proposed enzymatic pathways.
The first pathway involves the formation of PAH-radical cations catalyzed by P450 peroxidases (4). The second pathway involves initial epoxidation by P450 1A1/P450 1B1 to form an arene oxide and subsequent hydrolysis by epoxide hydrolase yields the corresponding trans-dihydrodiol, a proximate carcinogen (5). Once formed, the trans-dihydrodiol undergoes secondary monooxygenation to yield a diol-epoxide by the same P450 isoforms. The third pathway involves oxidation of PAH trans-dihydrodiols by aldo-keto reductases (AKRs) to produce reactive and redox-active o-quinones (Scheme 1) (6-8). In this pathway, AKRs catalyze the NADP+-dependent oxidation of a PAH trans-dihydrodiol to form a ketol that spontaneously re-arranges to a catechol. The catechol is extraordinarily air-sensitive and undergoes two sequential one-electron oxidations to yield an o-semiquinone radical anion and subsequently the fully oxidized o-quinone.
Scheme 1.
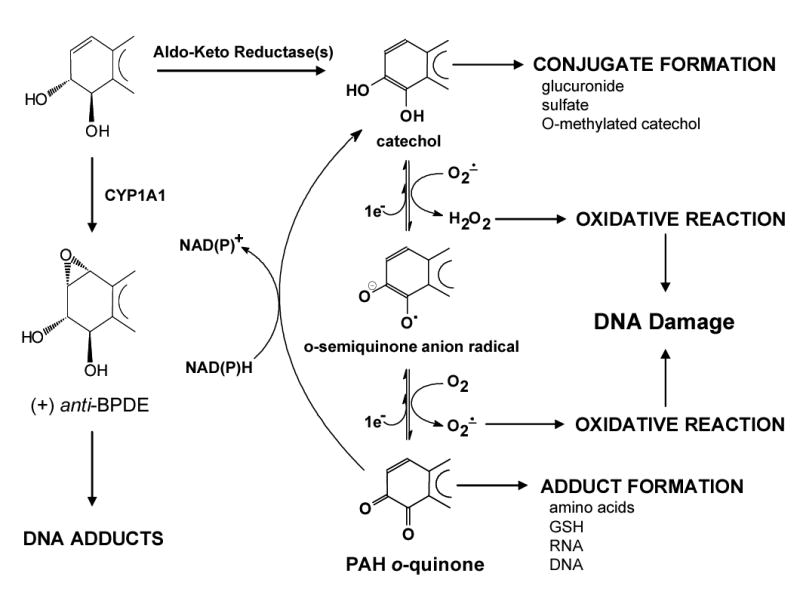
Metabolic activation of PAHs by Aldo-Keto Reductases and ROS formation
The three classes of reactive PAH metabolites (radical cations, diol-epoxides and o-quinones) can cause DNA adducts that may lead to mutation. For example, the BP radical cation can yield N7 and C8 depurinating adducts with guanine and adenine that have resulted in H-ras mutation in mouse skin (9,10). (±)-Anti-BP-7,8-diol-9,10-epoxide a representative diol-epoxide can form stable N2-adducts with 2′-deoxyguanosine in vivo and in vitro (11–14) and leads to the mutation of p53 and H-ras (3,15). BP-7,8-dione can produce a stable N2-adduct with 2′-deoxyguanosine by either a 1,4-Michael addition to form a hydrated adduct or by a 1,6-Michael addition to yield a hydrated cyclized adduct (16,17). BP-7,8-dione also forms depurinating N7 adducts with guanine (18) but which adduct dominates in DNA in vitro and in vivo remains to be determined. BP-7,8-dione was also found to be a potent mutagen of p53 in vitro but only under redox cycling conditions (19), leading to G to T transversions and a mutational pattern reminiscent of that seen in lung cancer patients (20,21). The mutations were abolished by ROS scavengers, suggesting that they were ROS-mediated.
Although formation of covalent PAH-DNA adducts is a widely accepted mechanism for the initiation of PAH carcinogenesis, PAH-mediated oxidative DNA damage has been also linked to carcinogenesis (22). Parent PAH and PAH trans-dihydrodiols have been shown to cause oxidative DNA damage in vitro and in vivo (23–28). The AKR pathway of PAH activation provides a route to ROS generation and ROS-mediated DNA lesions seen with PAH (Scheme 1).
PAH o-quinones produced from the AKR pathway can enter into futile redox cycles in the presence of cellular reducing equivalents (e.g. NADPH and NADH) and amplify ROS e.g. hydroxyl radical (•OH), superoxide anion (O2•−) and hydrogen peroxide (H2O2) until the reducing cofactor is depleted. Once formed, ROS can generate excess oxidative stress and overwhelm the cellular defense mechanisms. While ROS are short-lived species, PAH o-quinones can be delivered to the nucleus since they are ligands for the aryl-hydrocarbon receptor (29). This pathway could provide the explanation for oxidative DNA damage in PAH exposed sites and cells.
In the present investigation, four PAH o-quinones including NP-1,2-dione, BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione have been examined for their ability to undergo redox cycling in the presence of NADPH and to form 8-oxo-dGuo in salmon testis DNA in vitro. It was found that sub-micromolar concentrations of PAH o-quinones produced significant amounts of 8-oxo-dGuo provided NADPH and CuCl2 were both present. 8-Oxo-dGuo formation was abolished by ROS scavengers and by methional which scavenges Cu(I)OOH. However, 8-oxo-dGuo formation was not abolished by •OH scavengers but was eliminated by sodium azide which is both a •OH radical and 1O2 scavenger. Our data are consistent with a Cu(I)OOH acting as a source of 1O2 which would produce 8-oxo-dGuo via a 4,8-endoperoxide-dGuo intermediate.
Experimental Procedures
Caution: All PAH o-quinones are potentially hazardous and should be handled in accordance with “NIH Guidelines for the Laboratory Use of Chemical Carcinogens”.
Chemicals and Reagents.
NP-1,2-dione and BP-7,8-dione, BA-3,4-dione and 7,12-DMBA-3,4-dione were synthesized according to published methods (30–32). All compounds were analyzed for purity and identity by LC/MS before use. Standard 8-oxo-dGuo was purchased from Cayman Chemical Co. (Ann Arbor, MI). 2′-deoxyguanosine, 2′-deoxyadenosine, 2′-deoxycytidine and thymidine were obtained from ICN Biomedical Inc, (Irvine, CA). Salmon testis DNA, DNase I (type II), alkaline phosphatase (type III from Escherichia coli), superoxide dismutase (4,980 units/mg from bovine erythrocytes), catalase (17,600 units/mg from bovine liver), deferoxamine mesylate (Desferal), 4,5-dihydroxy-1,3-benzene disulfonic acid (Tiron), Bathocuproine disulfonic acid, 3-methylthiopropionaldehyde (Methional), sodium azide, cupric chloride, cuprous chloride and hydrogen peroxide were acquired from Sigma-Aldrich Chemical Co. (St. Louis, MO). Phosphodiesterase I (type II) from Crotalus adamanteus venom was obtained from Worthington Biochemical Corp. (Lakewood, NJ). Mannitol, sodium formate and sodium benzoate were purchased from Fischer Chemicals (Fairlawn, NJ). β-Nicotinamide adenine dinucleotide phosphate, reduced form (NADPH) was obtained from Boehringer Manheim Biochemicals (Indianapolis, IN). Chelex-100 resin was purchased from Bio-Rad Laboratories (Hercules, CA). All other chemicals were of the highest grade available and all solvents were HPLC grade.
Determination of Redox Cycling of PAH o-Quinones.
The assay was conducted on a Beckman DU-7 UV/VIS spectrophotometer at 26 °C. The mixture contained 10 mM potassium phosphate (pH 6.0), 180 μM NADPH, increasing concentrations (0.16 ~ 20 μM) of PAH o-quinones in 8% DMSO, 10 μM CuCl2 in a total volume of 1.0 mL. The reaction was initiated by the addition of NADPH and monitored at 340 nm. The rate of NADPH oxidation was calculated using 6,270 M−1cm−1 as the molar extinction coefficient.
Oxygen Consumption during Redox Cycling of PAH o-Quinones.
The consumption of molecular oxygen was measured using a computerized Instech oxygen electrode under atmospheric pressure. The reaction mixture contained 10 mM potassium phosphate (pH 6.0), 180 μM NADPH, 20 μM of PAH o-quinone, 8% DMSO, 10 μM CuCl2 in a total volume of 600 μL. The output of the electrode was connected through an amplifier (Instech) that allowed the simultaneous display of oxygen concentration and the rate of oxygen consumption as a function of time. The data acquisition software allowed the recording of 1 to 1000 data points/min. Oxygen consumption by redox cycling of PAH o-quinones was monitored over 100 min and data points were taken every 6 s. All oxygen consumption measurements were performed at 26 °C, assuming that the amounts of oxygen in the reaction chamber are 0.126 nmole at 26 °C at 760 mmHg.
Cu(I)-bathocuproine Complex Formation during Redox Cycling of Cu(II) to Cu(I).
The formation of Cu(I)-bathocuproine complex in the presence of CuCl2 and NADPH, CuCl2 or CuCl alone was monitored on DU-7 UV/VIS spectrophotometer at 26 °C. The buffer contained 10 mM potassium phosphate (pH 6.5) and 200 μM bathocuproine plus 8% DMSO as co-solvent. The poor solubility of CuCl was overcome by using DMSO as solvent. The reaction was initiated by the addition of either 180 μM NADPH and 20 μM CuCl2, 20 μM CuCl2 or 20 μM CuCl alone. Complex formation was measured immediately and again at 5 and 60 min intervals using a molar extinction coefficient of ɛ = 12,900 M−1cm−1.
8-Oxo-dGuo Formation in Salmon Testis DNA in the Presence of PAH o-Quinones.
8-Oxo-dGuo formation in salmon testis DNA following PAH o-quinone treatment was measured as follows. Briefly, a dried salmon testis DNA pellet was dissolved in 10 mM potassium phosphate buffer (pH 6.5) containing 0.1 mM Desferal® overnight at 4 °C. The concentration of the hydrated DNA was determined by measuring the absorbance at 260 nm and the DNA solution was divided into 80 μg aliquots. The DNA was precipitated by adding 5 M NaCl to yield a final concentration of 1 M and two volumes of ice-cold 100% ethanol. The samples were kept at –20 °C until use.
The precipitated DNA was recovered by centrifugation at 13,000 rpm for 15 min and rinsed with 70% ethanol to remove excess salts. The pellets were dissolved in 250 μL of 10 mM potassium phosphate buffer (pH 6.5) containing 0.16 ~ 10 μM PAH o-quinone, 180 μM NADPH plus 8% DMSO as co-solvent. Other incubations contained 0 ~ 10 μM CuCl2. Samples were incubated for 3 h at 37 °C. As negative controls, incubations were also performed with DNA containing NADPH and CuCl2, or NADPH and CuCl2 alone. The effects of various scavenging agents and transition metal chelators on the formation of 8-oxo-dGuo in PAH o-quinone-treated salmon testis DNA were also measured. After incubation, the samples were extracted twice with equal volumes of chloroform and isopropyl alcohol (24:1) to remove PAH o-quinone. The resulting clear supernatant was collected by centrifugation at 13,000 for 5 min. Then, 62.5 μL of 5 M NaCl and two volumes of ice-cold ethanol were added again to completely precipitate DNA overnight at −20 °C. The samples were centrifuged at 13,000 rpm for 15 min and the pellets were rinsed with 70% ethanol. The resulting DNA pellets were re-dissolved in 200 μL of 10 mM Tris-HCl buffer (pH 7.4) containing 100 mM MgCl2 and 0.1 mM Desferal® for enzymatic digestion.
Digestion of Salmon Testis DNA.
Three enzymes were used to quantitatively digest the salmon testis DNA. The DNA pellet was dissolved in 200 μL of 10 mM Tris-HCl buffer (pH 7.4) containing 100 mM MgCl2 and 0.1 mM Desferal® and digested with 1 unit DNase I for 1.5 h at 37 °C. The pH was adjusted to 9.0 with 0.2 M glycine-acetate buffer (pH 10.0) and the mixture was digested with 0.025 units phosphodiesterase I for 1.5 h at 37 °C. Finally, the pH was adjusted to pH 8.6 with 50 mM Tris-HCl buffer (pH 7.4) containing 50 mM MgCl2 and digested with 0.1 unit alkaline phosphatase for 1.5 h at 37 °C. The digest was adjusted again to pH 5.5 with 0.5 M acetic acid and stored −20 °C until for HPLC analysis.
Elimination of Adventitious Oxidation of DNA.
To prevent the adventitious formation of 8-oxo-dGuo during the DNA work-up, all buffers were treated with Chelex resin and contained Desferal (final concentration 0.1 mM). Optimal suppression of the background level of 8-oxo-dGuo was achieved when the treated buffers were used in the initial DNA solubilization step and during the DNA-digestion procedure.
RP-HPLC analysis of DNA Digest.
To confirm that complete digestion of the DNA sample was achieved, an aliquot of the digest was analyzed on a Beckman UV-HPLC system. The HPLC column used was a Zorbax-ODS (5 μm; 4.6 x 250 mm) and normal 2′-deoxyribonucleosides were resolved using a flow rate of 0.5 mL/min with 50 mM ammonium acetate buffer (pH 4.5) with a 8 to 30% MeOH gradient and detected at 254 nm. In this system, the resulting retention times were observed at 9.9 min dCyt, 23.1 min dGuo, 27.1 min Thd and 38.4 min dAdo. Digestion was considered complete when the ratios of dGuo to dCyt and Thd to dAdo were 1:1, respectively. Because dGuo in salmon testis DNA contains 22% dGuo (33), the amounts of dGuo and dCyt were less than the observed with Thd and dAdo. All samples analyzed for 8-oxo-dGuo were checked for complete DNA digestion and recovery of the four 2′-deoxyribonucleosides. 5.3 μg of DNA routinely gave 3.3, 3.6, 4.6 and 5.1 nmol of dCyt, dGuo, Thd and dAdo, respectively (Fig. 1).
Figure 1.
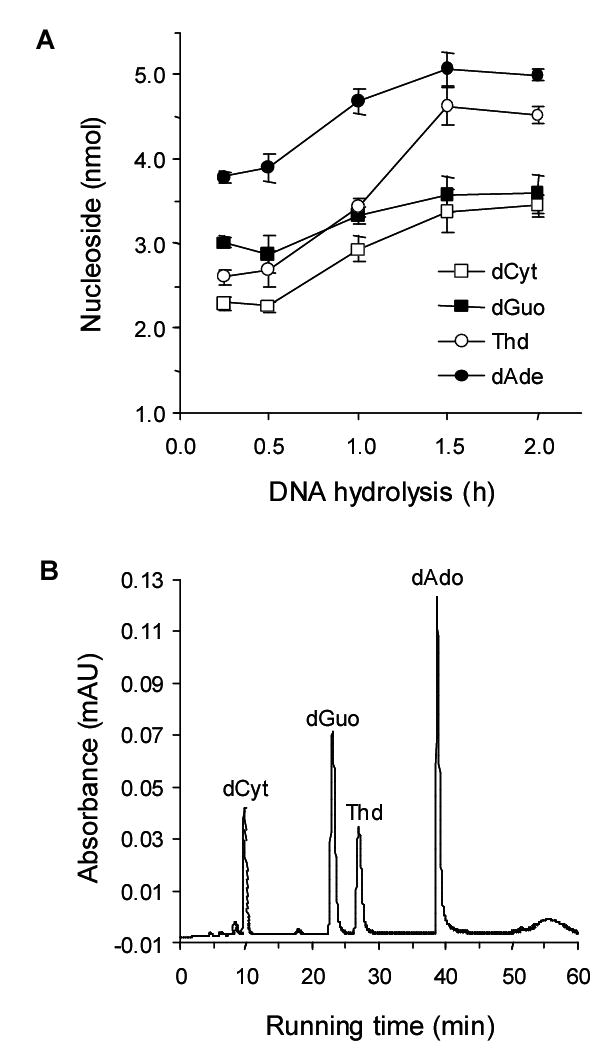
Quantitative enzymatic hydrolysis of 5.3 μg salmon testis DNA (A) and HPLC-UV chromatogram of completely digested DNA (B).
8-Oxo-dGuo Detection by HPLC-ECD.
8-Oxo-dGuo in DNA digests was measured by HPLC-ECD using a Coularray detector (ESA Inc., Chelmsford, MA). The four channel electrochemical detector had potential settings of 200 ~ 300 to detect 8-oxo-dGuo, and 700 ~ 800 for the detection of dGuo. A mobile phase of 100 mM sodium acetate pH 5.2 containing 5% methanol (v/v) was used to develop a YMC basic S column (3 μm; 4.6 x 150 mm). In this system, dGuo and 8-oxo-dGuo had retention times of 7.9 and 11.0 min, respectively. Since 8-oxo-dGuo is a trace analyte, the sensitivity of detection was optimized to be linear in the fmol range. Also, this method reports 8-oxo-dGuo as 8-oxo-dGuo per 105 dGuo, therefore a standard curve was also developed for the internal std dGuo. The dGuo standard curve was non-linear above the range of 1.0 nmol providing an estimate of upper limit of the dynamic range that can be used, if 8-oxo-dGuo is to be quantified relative to dGuo. Before each analysis, fresh standard curves for 8-oxo-dGuo and dGuo were constructed to ensure that the detection limits were reproducible. In addition, the analytical method was checked to ensure that the retention time of the analyte showed no difference and that the potentials of the electrode were stable. Representative standard curves and HPLC-ECD chromatograms are shown in Fig. 2.
Figure 2.
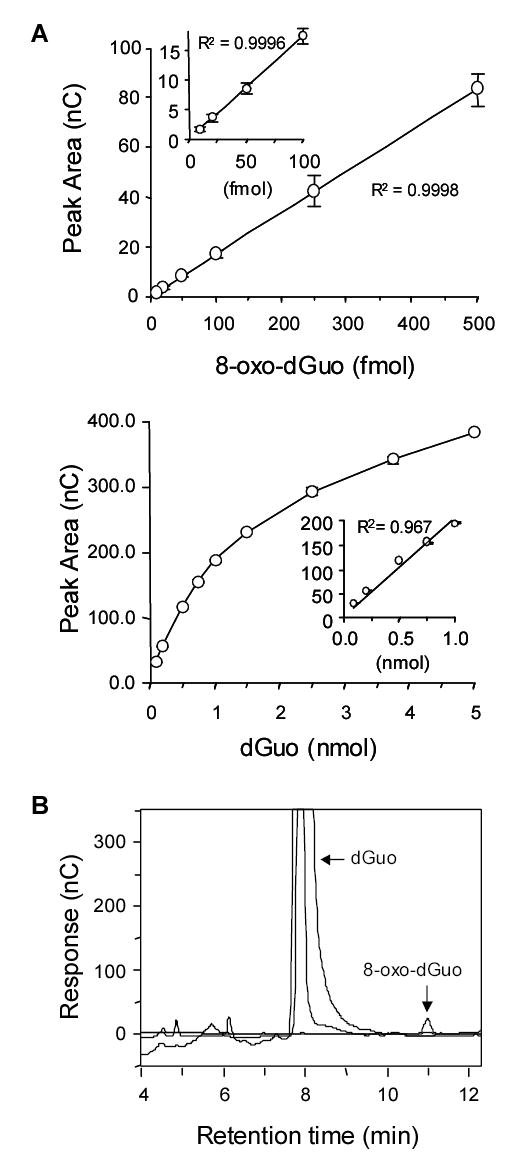
Sensitivity and linearity of 8-oxo-dGuo and dGuo detection by HPLC-ECD analysis. Insets show the working range of the respective std curves to permit the detection of 8-oxo-dGuo relative to the internal standard dGuo. A) Standard curves of 8-oxo-dGuo and dGuo. B) Typical HPLC-ECD chromatogram of completely digested DNA sample (equivalent to 1.8 μg DNA).
Statistical Analysis of the Data.
Each experimental condition was replicated between two and 18 times, and the results were tabulated with means and standard deviations. Statistical hypothesis testing was conducted via regression analysis, using a two-sided significance level of 0.05. Each experimental condition was assessed by comparing the condition with no PAH o-quinone added to the same condition in which varying amounts of PAH o-quinone were added. The effect of the amount of PAH o-quinone was assessed both as a linear function and using categorical indicator variables. To make comparisons among all four PAHs used, F-tests with three degrees of freedom were employed. We used a standard ANOVA F-test to compare the production of 8-oxo-dGuo with and without various scavenging agents. We assessed the robustness of the F-test results by applying the non-parametric Kruskal-Wallis test, which uses the ranks or ordering of the data rather than the actual values, and confirmed the results in all or most cases. The F-test p-values are reported. All statistical analyses were conducted using the SAS System, Version 8 (SAS, Inc., Cary, NC).
Results
Non-Enzymatic Redox Cycling of PAH o-quinones.
Our goal was to compare the redox-cycling capabilities of PAH o-quinones with their ability to form oxidatively generated DNA damage. For our studies we chose four structurally different PAH o-quinones: NP-1,2-dione (which is the simplest PAH o-quinone), BP-7,8-dione (a bay region o-quinone), and BA-3,4-dione and 7,12-DMBA-3,4-dione (non-methylated and methylated bay region quinones from the same structural series). Previously, we showed that in the presence of a 9-fold molar excess of NADPH each of these o-quinones underwent non-enzymatic redox-cycling (34). In each case the number of moles of NADPH oxidized to NADP+ exceeded the total number of moles of quinone present. Furthermore, the reactions proceeded until NADPH was depleted. Results reported here support these findings. Based on velocity data, 20 nmoles of the four o-quinones tested would deplete a 9-fold molar excess of NADPH within 40–100 min, but were not consumed themselves (Fig. 3A and Table 1). Assuming that 1 mole of NADPH is oxidized per mole of o-quinone, it would be anticipated that the quinones would be depleted within 5–20 mins but this was not observed. The rank order of non-enzymatic redox-cycling was NP-1,2-dione > 7,12-DMBA-3,4-dione > BA-3,4-dione > BP-7,8-dione.
Figure 3.
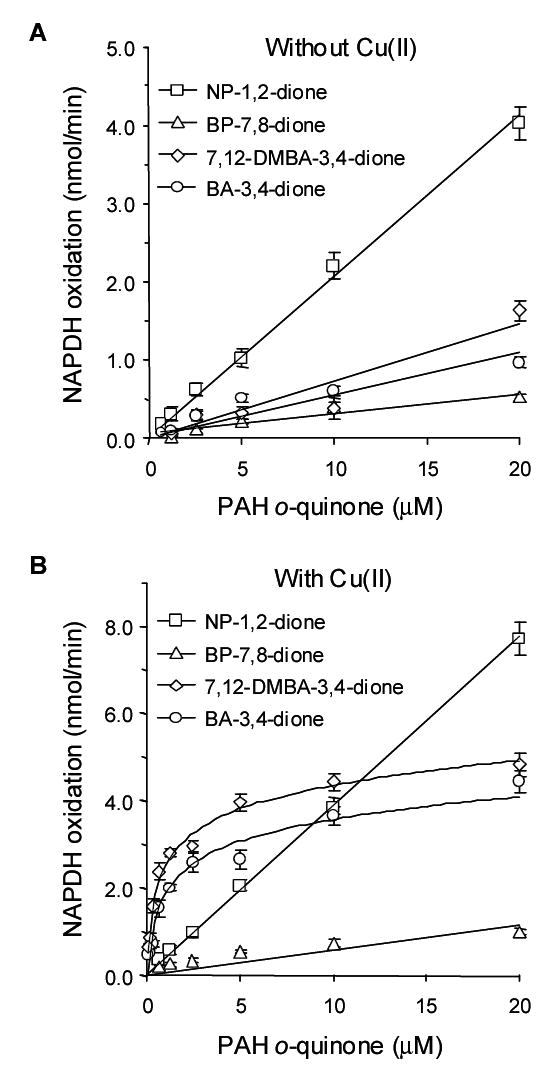
Non-enzymatic redox cycling of PAH o-quinones with and without copper chloride. See the experimental procedures for details.
Table 1.
NADPH reduction and oxygen consumption during the redox cycling of PAH o-quinone with and without copper chloride
| Oxidized NADPH (nmol/min)
|
Consumed oxygen (nmol/mim)
|
|||
|---|---|---|---|---|
| PAH o-quinones (20 nmole/mL) | − Cu(II) | + Cu(II) | − Cu(II) | + Cu(II) |
| NP-1,2-dione | 4.03 ± 0.21 | 7.53 ± 0.37 | 4.00 ± 0.43 | 5.67 ± 0.54 |
| BP-7,8-dione | 0.52 ± 0.05 | 1.00 ± 0.06 | 0.49 ± 0.09 | 0.45 ± 0.05 |
| 7,12-DMBA-3,4-dione | 1.63 ± 0.13 | 3.83 ± 0.24 | 0.98 ± 0.07 | 2.23 ± 0.28 |
| BA-3,4-dione | 0.98 ± 0.07 | 4.72 ± 0.28 | 0.67 ± 0.05 | 2.83 ± 0.32 |
Previously, we showed that the presence of Cu(II) increased PAH o-quinone mediated DNA strand scission and p53 mutation (19,35,36). We now show that Cu(II) increased the non-enzymatic rate of o-quinone redox-cycling. Using a range of PAH o-quinone concentrations, we found that the rates of redox-cycling observed with BA-3,4-dione and 7,12-DMBA-3,4-dione were now curvilinear (Fig. 3B)
Reactions were also replicated so that the rate of oxygen consumption that occurred during the redox-cycling of the PAH o-quinone could be monitored. Substantial rates of oxygen consumption were observed provided the o-quinone, NADPH and Cu(II) were all present (Fig. 4). The moles of oxygen consumed per min were compared with the moles of NADPH consumed per min and are shown in Table 1. We found that in the absence of Cu(II) there was almost a stoichiometric relationship between the rate of NADPH consumed and the rate of molecular oxygen consumed consistent with;
Figure 4.
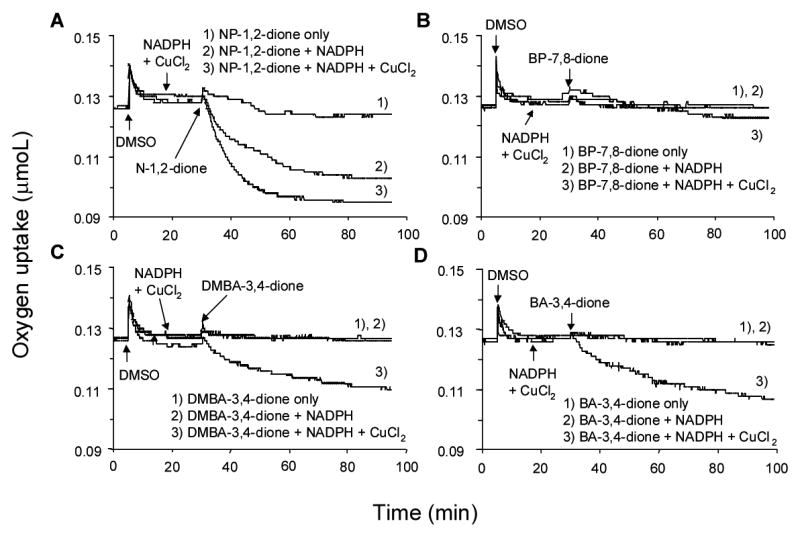
Consumption of molecular oxygen during the redox-cycling of PAH o-quinones in the presence of NADPH and CuCl2. A) NP-1,2-dione. B) BP-7,8-dione. C) 7,12-DMBA-3,4-dione. D) BA-3,4-dione treatment. See the experimental procedures for details.
| (Eq. 1) |
| (Eq. 2) |
In the presence of Cu(II) the amount of NADPH consumed doubled while the amount of oxygen consumed was one half of this value. This is consistent with:
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
| (Eq. 4) |
| (Eq. 5) |
| (Eq. 6) |
Precautions in Measuring 8-oxo-dGuo as a Surrogate Marker of Oxidative DNA Damage.
To detect oxidatively generated DNA damage under conditions in which PAH o-quinones redox-cycle, posses a challenge since the surrogate marker 8-oxo-dGuo is a notoriously difficult analyte to measure accurately. Three major problems that exist with the detection of this analyte are (i) variability in the digestion of the DNA sample; (ii) adventitious oxidation of dGuo to 8-oxo-dGuo that can occur in sample work-up; and (iii) lack of validation of the analytical HPLC-ECD method. In the experimental procedures section we demonstrate that our DNA-digestion protocol is quantitative between samples.
Adventitious oxidation of dGuo could occur at three steps during our DNA handling procedure: (i) DNA solubilization; (ii) PAH o-quinone treatment; and (iii) DNA digestion. We used Chelex-treated buffers and desferal containing buffers to remove endogenous transition metals (37) to suppress the adventitious formation of 8-oxo-dGuo in salmon testis DNA.
Without desferal present, the basal level of 8-oxo-dGuo was 3.17 ± 0.42 8-oxo-dGuo per 105 dGuo. This level was reduced to 2.14 ± 0.06 when desferal was included throughout the work-up (Table 2). When desferal was used throughout the work-up except for the quinone treatment step, the level of 8-oxo-dGuo was elevated to 4.00 ± 0.51 8-oxo-dGuo per 105 dGuo (Table 3). In subsequent experiments, desferal was not added to the PAH o-quinone treatment step because of its ROS scavenging capability (37).
Table 2.
Effects of Desferal on the reduction of the adventitious formation of 8-oxo-dGuo in untreated salmon testis DNA using Chelex-treated buffer
| Treatment | Analyzed DNA digest (μg) | Detected 8-oxo-dGuo (fmol) | Detected dGuo (nmol) | 8-oxo-dGuo adduct/105dGuo |
|---|---|---|---|---|
| No Desferal treatment | 0.86 | 21.1 ± 2.4 | 0.66 ± 0.42 | 3.17 ± 0.42 |
| Desferal treatment | 0.86 | 13.0 ± 1.0 | 0.60 ± 0.03 | 2.14 ± 0.06* |
50 μg of salmon testis DNA was used for this experiment and 0.86 μg of digest DNA was analyzed.
p<0.05.
Table 3.
Effects of Desferal on DNA treated with NP-1,2-dione using Chelex-treated buffer
| Treatment | Analyzed DNA digest (μg) | Detected 8-oxo-dGuo (fmol) | Detected dGuo (nmol) | 8-oxo-dGuo adduct/105dGuo |
|---|---|---|---|---|
| Desferal treatment - except for PAH treatment | ||||
| Control | 0.86 | 10.4 ± 0.3 | 0.44 ± 0.01 | 2.35 ± 0.02 |
| NP-1,2-dione + NADPHa | 0.86 | 20.2 ± 3.5 | 0.50 ± 0.05 | 4.00 ± 0.51 |
| Desferal treatment - all incubations | ||||
| Control | 0.86 | 12.9 ± 1.7 | 0.52 ± 0.03 | 2.47 ± 0.20 |
| NP-1,2-dione + NADPH | 0.86 | 14.8 ± 0.8 | 0.52 ± 0.01 | 2.83 ± 0.15 |
0.625 μM of NP-1,2-dione and 180 μM of NADPH were used.
50 μg of salmon testis DNA was used for this experiment and 0.86 μg of digest DNA was analyzed.
In the experimental procedure we describe the sensitivity of the HPLC-ECD method. Since we are reporting 8-oxo-dGuo as a ratio of 8-oxo-dGuo to dGuo the linearity of the standard curves for the two analytes was also established so that for the unknowns the ratio would be reported using the linear range of both standard curves.
To validate the HPLC-ECD method we spiked three different amounts of the 8-oxo-dGuo analyte into DNA-samples at the re-solubilization step and then used the analytical method to determine whether we could quantitatively detect the spiked amount. Over a dynamic range of 33.3 fmoles to 666.7 fmoles, we were able to recover > 96.8% of the spiked sample in the 8-oxo-dGuo peak (Table 4). This shows that our protocol does not result in a loss of the analyte or an overestimation of 8-oxo-dGuo present.
Table 4.
Validation of the 8-oxo-dGuo detection method by spiking standard 8-oxo-dGuo into the DNA sample
| Treatment /Analyzed amount | Detecteda 8-oxo-dGuo (fmol) | Retention time (min) | Recovery (%) |
|---|---|---|---|
| Standards, 8-oxo-dGuo (n=1, 3 times inj.) | |||
| 33.3 (fmol) | 32.6 ± 0.5 | 9.57 | |
| 66.7 (fmol) | 70.7 ± 2.0 | 9.61 | |
| 666.7 (fmol) | 744.5 ± 0.1 | 9.58 | |
| Spiked 8-oxo-dGuo (n=3, 2 times inj.) | |||
| 1.8 μg of DNA digest only | 50.9 ± 2.2 | 9.55 ± 0.01 | - |
| 1.8 μg of DNA digest + 33.3 (fmol) | 85.2 ± 3.4 | 9.53 ± 0.01 | 105.1 ± 10.3 |
| 1.8 μg of DNA digest + 66.7 (fmol) | 119.2 ± 1.8 | 9.52 ± 0.01 | 96.6 ± 2.6 |
| 1.8 μg of DNA digest + 666.7 (fmol) | 771.3 ± 4.2 | 9.54 ± 0.01 | 96.8 ± 0.6 |
The mobile phase used for this experiment was 100 mM sodium acetate pH 5.2 with 6 % methanol.
Detection of 8-oxo-dGuo in Salmon Testis DNA treated with PAH o-Quinones.
Our redox-cycling experiments established the conditions under which 8-oxo-dGuo should be detected in salmon testis DNA, the results of these experiments are shown in Fig. 5. The basal level of 8-oxo-dGuo in untreated DNA was found to be 1.6 adducts per 105 dGuo. Other treatment conditions slightly elevated the amount of 8-oxo-dGuo detected from 1.7 to 3.0 adducts per 105 dGuo. However, two treatment groups substantially increased the amount of 8-oxo-dGuo detected. One group was the DNA, NADPH and Cu(II) treatment which reached a value of 36.0 adducts per 105 dGuo, and the other group was DNA, NADPH, Cu(II) plus an o-quinone, where the amount of 8-oxo-dGuo detected ranged from 66.0 to 133.0 adducts per 105 dGuo. These amounts were observed in the presence of 160 nM o-quinone but were not observed when Cu(II) was absent from the incubation. Concentration dependence of formation of 8-oxo-dGuo was observed with each of the PAH o-quinones, as shown in Fig. 6. At the highest concentrations of PAH o-quinones (2.5 ~ 10 μM) used the amount of 8-oxo-dGuo increased to 500 ~ 1,000 adducts per 105 dGuo. The rank order of 8-oxo-dGuo observed was NP-1,2-dione > BA-3,4-dione > 7,12-DMBA-3,4-dione > BP-7,8-dione. This rank order is reminisecent of that observed for the oxygen uptake measurements described earlier.
Figure 5.
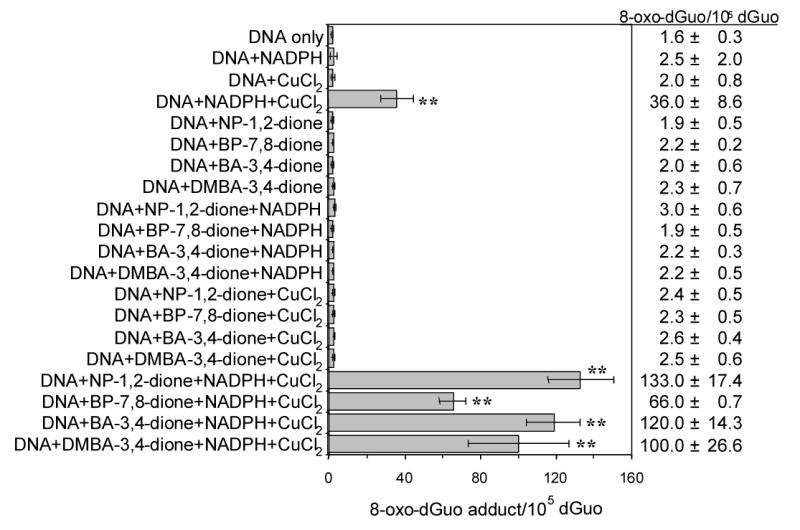
8-Oxo-dGuo formation in salmon testis DNA treated with PAH o-quinones. Of the conditions used, only those containing DNA + NADPH + CuCl2 (p < .0001) and those containing DNA + quinone + NADPH + CuCl2 (p < .0001 ) were significantly different. Irrespective of the quinone the p value was the same. In addition, these two conditions were significantly different from each other (p < .0001).
Figure 6.
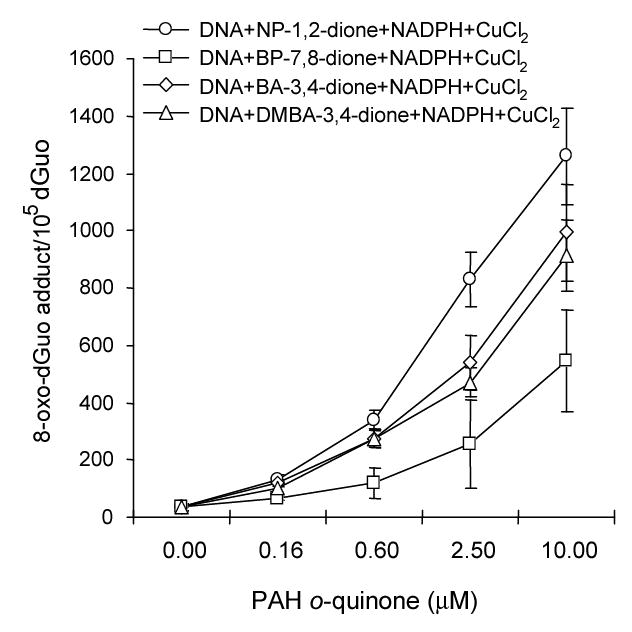
The formation of 8-oxo-dGuo in salmon testis DNA by PAH o-quinones is concentration-dependent. It was found that there was a strong positive effect of dose on 8-oxo-dGuo formation for each quinone tested where p < .0001 in each instance. The linear trend with dose was also assessed for the four agents. To determine whether there were significant differences in the linear trend for the four quinones as a whole a F-test with three degrees of freedom was performed. BP-7,8-dione was set as the reference agent since it had the lowest production of 8-oxo-dGuo. It was found that NP-1,2-dione was significantly different from BP-7,8-dione after adjusting for dose (p < .0001). By contrast BA-3,4-dione and DMBA-3,4-dione did not appear to be significantly different from BP-7,8-dione in their linear trends. An F-test was also performed to determine whether the four agents produced the same or different amounts of 8-oxo-dGuo after adjusting for dose. A p-value of < .005 was obtained. Thus among the group of four agents there were significant differences in the level of 8-oxo-dGuo production after adjusting for dose.
ROS Scavengers and 8-oxo-dGuo Formation.
To identify the ROS (superoxide anion, hydrogen peroxide, hydroxyl radical or 1O2) responsible for 8-oxo-dGuo formation in salmon testis DNA treated with PAH o-quinones a number of scavenging agents were used. Irrespective of the PAH o-quinone used, the pattern of results observed was the same (Fig. 7). Superoxide dismutase did not inhibit 8-oxo-dGuo formation, whereas catalase did, showing the dependence on hydrogen peroxide. Tiron a superoxide anion scavenger also attenuated 8-oxo-dGuo formation. By contrast hydroxyl radical scavengers including 1.0 mM sodium benzoate, 1.0 mM sodium formate and 1.0 mM mannitol failed to block 8-oxo-dGuo formation. The Cu(I) chelator bathocuproine (200 μM) also abolished 8-oxo-dGuo formation under all conditions. When 0.2 mM methional a copper-hydroperoxo complex (Cu(I)OOH) scavenger was used 8-oxo-dGuo formation was not suppressed. When these experiments were replicated using 0.1 M of each scavenging reagent identical results were obtained except now methional abolished 8-oxo-dGuo formation. We also used 0.1M sodium azide a known hydroxyl radical and 1O2 scavenger and found that 8-oxo-dGuo formation was now eliminated. The ability of this scavenger to work when other hydroxyl radical scavengers failed suggests that 1O2 is involved.
Figure 7.
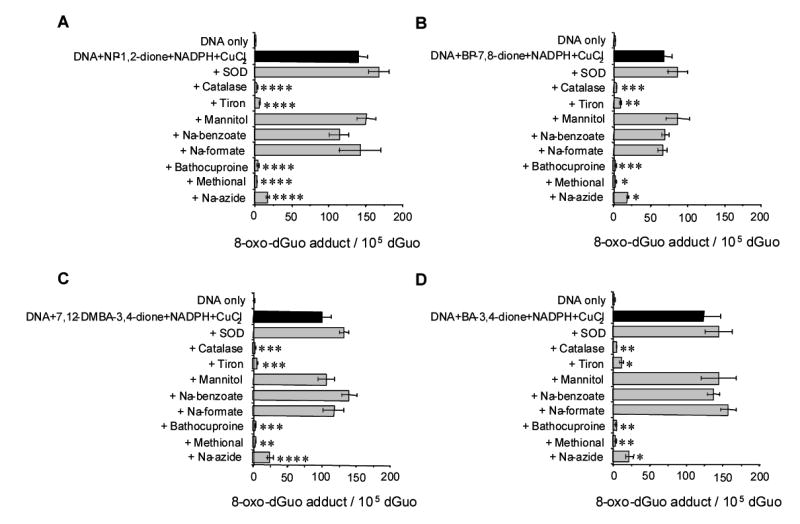
The effects of ROS scavenging agents on the formation of 8-oxo-dGuo in PAH o-quinones treated salmon testis DNA. A) NP-1,2-dione. B) BP-7,8-dione. C) 7,12-DMBA-3,4-dione. D) BA-3,4-dione. The concentration of each scavenger or chelators was as follows. SOD and catalase were added as 200 U/mL and 800 U/mL, respectively. Tiron, mannitol, sodium benzoate, sodium formate, methional and sodium azide were added to achieve a final concentration of 0.1 M. Bathocuproine was added to achieve a final concentration of 200 μM. Identical results were obtained when the concentration of ROS scavengers were reduced from 0.1 M to 1.0 mM. However, in those experiments methional was ineffective at concentrations of 0.2 mM. In the data presented the effect of each scavenger on the level of 8-oxo-dGuo was compared with the level of 8-oxo-dGuo in the control group (complete system). Several treatments significantly reduced the level of 8-oxo-dGuo across group and the significance was scored using a standard ANOVA F-test as follows: p = < 0.05*, p = < 0.001**, p= < 0.0005*** and p = < 0.0001****.
The Role of Hydrogen Peroxide, Cu(II) and Cu(I) on 8-oxo-dGuo Formation.
To determine the role of hydrogen peroxide, copper(I) and copper(II) on 8-oxo-dGuo formation additional observations were made. First, hydrogen peroxide did not cause 8-oxo-dGuo formation even at 500 μM H2O2 (Fig. 8A), showing that in the absence of any added transition metal 8-oxo-dGuo formation could not occur. Cu(II) also failed to generate 8-oxo-dGuo formation, irrespective of the concentration used (Fig. 8B). Whereas, when Cu(I) was used the level of 8-oxo-dGuo formation increased in a concentration-dependent manner (Fig. 8C). The addition of hydrogen peroxide stimulated the ability of Cu(II) to produce significant amounts of 8-oxo-dGuo (Fig. 8D). Since the amount of 8-oxo-dGuo formed with H2O2 and Cu(II) was much greater than seen with Cu(I), Cu(I) mediated Fenton chemistry was not occurring.
Figure 8.
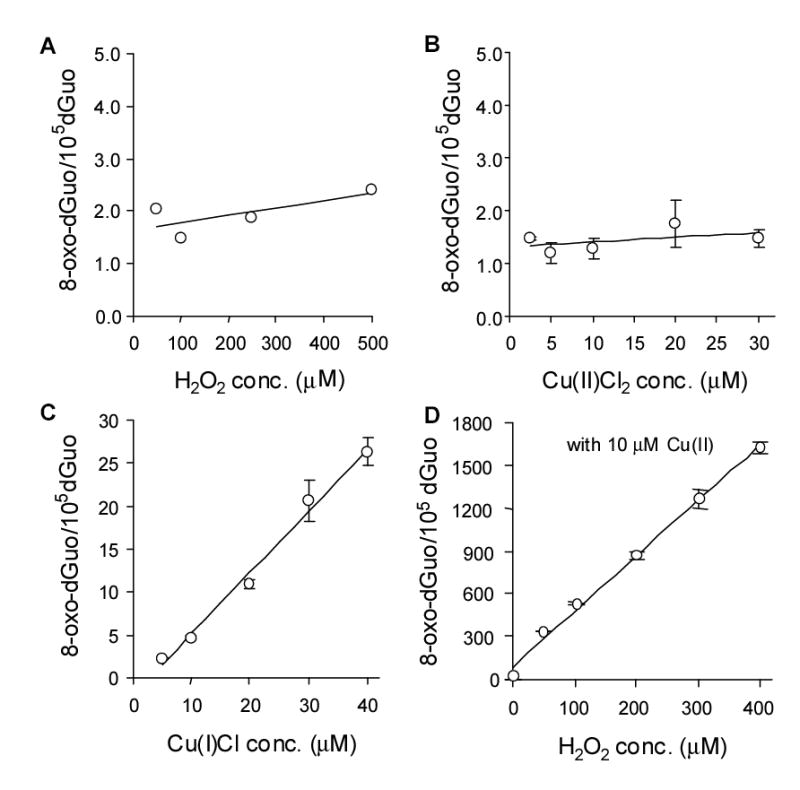
8-oxo-dGuo formation in salmon testis DNA by hydrogen peroxide (A), Cu(II) (B), and Cu(I) (C), and Cu(II) plus hydrogen peroxide (D).
Formation of the Cu(I)-bathocuproine complex at λmax 475 nm offered additional clues to the redox events occurring. Cu(II) failed to form a measurable bathocuproine complex as expected (Fig. 9A). Surprisingly, Cu(I) produced a minimal complex with bathocuproine since it was readily oxidized to Cu(II) with the formation of O2•− (Fig. 9B). When Cu(II) and NADPH were used a large complex was formed indicating reduction back to Cu(I) (Fig. 9C). These findings support the redox-cycling of Cu(I) and Cu(II) during the incubations employed and the formation of NADP•.
Figure 9.
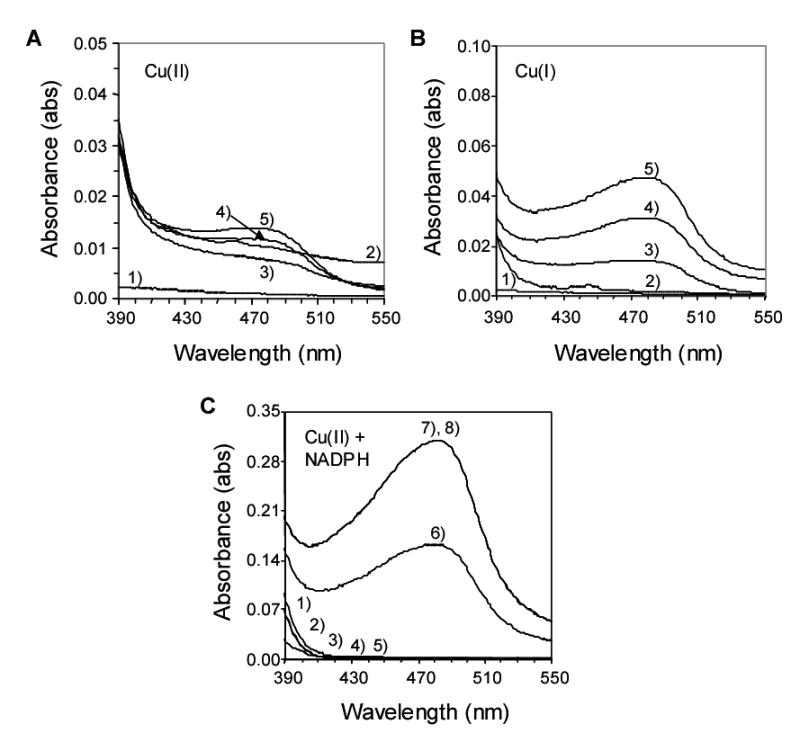
Formation of the Cu(I)-bathocuproine complex and evidence for the redox-cycling of Cu(II) to Cu(I). A) The effect of Cu(II) on the complex formation. The samples were treated as follows: 1, CuCl2 only; 2, Bathocuproine only; 3, 9 s after mixing; 4, 5 min after mixing and 5, 60 min after mixing. B) The effect Cu(I) on the complex formation. 1, CuCl only; 2, bathocuproine only; 3, 9 s after CuCl and bathocuproine were mixed; 4, 5 min after mixing and 5, 60 min after mixing. C) The effect of Cu(II) and NADPH on the complex formation. 1, NADPH only; 2, CuCl2 only; 3, bathocuproine only; 4, NADPH and CuCl2; 5, CuCl2 and bathocuproine; 6, 8 s after CuCl2, NADPH and bathocuproine were mixed; 7, 5 min after mixing and 8, 60 min after mixing.
Together these data would be consistent with the following:
| (Eq. 7) |
| (Eq. 8) |
| (Eq. 9) |
| (Eq. 10) |
Discussion
Polycyclic aromatic hydrocarbons (PAH)s are known to cause oxidative DNA damage in vitro and in vivo. The types of DNA damage detected upon PAH exposure were 8-oxo-dGuo, thymine glycol and etheno adducts (23–26). Irrespective of the final lesion the mechanism by which PAH cause this oxidative damage is uncertain. We now show that PAH o-quinones derived from AKR pathway of activation may account for the oxidatively generated DNA damage caused by the parent PAHs. Furthermore, our recent findings have shown that BP-7,8-dione mutated p53 cDNA, causing G to T transversions and that its mutagenicity was completely abrogated by the combined use of SOD and catalase, suggesting a direct relation between quinone mediated oxidatively generated DNA damage and these mutations (19).
The non-K region trans-dihydrodiol (BA-3,4-dihydrodiol) has been shown to mediate 8-oxo-dGuo formation via BA-3,4-dihydro-2,3,4-triol. The amount of 8-oxo-dGuo formed was dependent upon Cu(II) and NADPH and was greater when the dihydrodiol was used instead of the o-quinone (BA-3,4-dione) (28). As BA-3,4-dihydrodiol is converted to the BA-3,4-dione by the AKR pathway this would imply that the triol pathway is superior to the AKR pathway in producing 8-oxo-dGuo. In these earlier studies either 50 μM trans-dihydrodiol or o-quinone were used, yet we observe substantial 8-oxo-dGuo formation in the presence of 160 nM BA-3,4-dione. The reason for this difference is not yet clear, but it may be related to the quality of the synthetic PAH-metabolites used in the respective studies.
Subsequently, 8-oxo-dGuo formation was detected in DNA when 10 μM BA-1,2-trans-dihydrodiol was incubated with murine AKR1C22 (mouse dihydrodiol dehydrogenase) implicating AKR mediated quinone formation in the generation of this lesion (38). Even though the more relevant reaction is the AKR mediated oxidation of the non-K region BA-3,4-diol to BA-3,4-dione (7) and not the oxidation of BA-1,2-diol this observation supports the hypothesis presented here. Unlike these earlier studies with trans-dihydrodiols of BA, we conclude that Cu(I)OOH is not the oxidant responsible for 8-oxo-dGuo formation, and find that it was likely caused by its decomposition to produce 1O2. This ROS has been hypothesized to form a 4,8-endoperoxide-dGuo which acts as a precursor of 8-oxo-dGuo (39).
Redox-cycling of the PAH o-quinones in the presence of NADPH and CuCl2 was required (Fig. 5) to observe significant amounts of 8-oxo-dGuo. Under these conditions, 160 nM PAH o-quinones was sufficient to induce 8-oxo-dGuo formation (p < 0.05) and the levels increased in a concentration dependent manner (Fig. 6). The rank order was NP-1,2-dione > BA-3,4-dione > 7,12-DMBA-3,4-dione > BP-7,8-dione. The difference in the formation of 8-oxo-dGuo among the PAH o-quinones was positively correlated to the consumption of both NADPH and molecular oxygen during the redox cycling (Fig. 5 and Table 1). Thus, a high level of NADPH and oxygen consumption led to the high level of 8-oxo-dGuo formation and vice versa.
8-oxo-dGuo formation by the o-quinones had a strict requirement for NADPH and Cu(II). These reagents alone caused a significant amount of 8-oxo-dGuo formation but this value was significantly lower than when o-quinones were added to the system. The detection of Cu(I)-bathocuproine complex formation (Fig. 9) and the induction of 8-oxo-dGuo formation by Cu(I) but not Cu(II) (Fig. 8), indicated that the redox-cycling of Cu(II) to Cu(I) played a critical role in PAH o-quinone-mediated oxidative DNA damage.
PAH o-quinone-mediated 8-oxo-dGuo formation was completely inhibited by catalase and bathocuproine (Fig. 7), confirming that H2O2 and Cu(I) were indispensable for the observed 8-oxo-dGuo formation (p < 0.0001 – < 0.001). Tiron, a O2•− scavenger, also attenuated 8-oxo-dGuo formation (p < 0.0001 – < 0.001), whereas superoxide dismutase slightly stimulated 8-oxo-dGuo formation. The effect of SOD was expected since this will catalytically accelerate the dismutation of O2•− forming H2O2. By contrast, typical •OH scavengers such as mannitol, sodium benzoate and sodium formate did not show any inhibitory effects on the quinone-mediated 8-oxo-dGuo formation even at concentrations as high as 0.1M. H2O2 did not induce any 8-oxo-dGuo formation until CuCl2 was added, showing a strict dependence on H2O2 and Cu(II) that was independent of •OH formation (Fig. 8). Methional, which is known as a copper-hydroperoxo complex (Cu(I)OOH) chelator also blocked the quinone-mediated 8-oxo-dGuo formation (p < 0.0001 – < 0.0015). Sodium azide, a known hydroxyl and 1O2 scavenger (40) abolished 8-oxo-dGuo formation (p < 0.0001 – < 0.002). The ability of this scavenger to be effective when other hydroxyl radical scavengers failed suggests that 1O2 is the reactive oxygen species responsible for 8-oxo-dGuo formation as suggested by Cadet (39). In this mechanism 8-oxo-dGuo is derived from an unstable 4,8-endoperoxide intermediate.
Frelon et al (46) have shown that the dominant oxidant produced by Cu2+ and H2O2 is likely singlet oxygen which almost exclusively forms 8-oxo-dGuo while the level of other hydroxyl radical lesions are suppressed. This would explain our previous findings that DNA strand scission can be observed with PAH o-quinones, CuCl2 and NADPH provided micromolar concentrations of quinone are used (36). Under these conditions sufficient hydroxyl radical is likely produced to attack 2′-deoxyribose (36).
Although oxidatively generated DNA damage from hydrogen peroxide with metals and redox active organic compounds has been extensively reviewed, the exact identity of the DNA damaging species has been disputed (41–44). Cu(I)OOH, singlet oxygen 1O2 and Cu(III) have been proposed as the responsible agents (28,44–47). The Cu(I)OOH mechanism is based on a site-specific DNA-Cu(I)-H2O2 complex that is believed to release •OH in close proximity to sDNA and cause an oxidative lesion thus preventing intervention by hydroxyl radical scavengers. This may account for why hydroxyl radical scavengers in our system did not have effect on 8-oxo-dGuo formation. The inhibitory effect of methional suggests that Cu(I)OOH is involved. However, the ability of sodium azide to abolish 8-oxo-dGuo in the absence of effects from other hydroxyl radicals implicates 1O2. Unlike •OH radical which attacks nucleobases indiscriminately 1O2 is specific for dGuo modification (39) and it has been proposed that it is formed from the complexation of Cu(II) with DNA resulting in a Cu(I)OOH complex (44–46).
Singlet oxygen is known to cause specific mutations in DNA. G to T transversion mutations (50.8% of 61 base substitutions) predominated in a 1O2 treated-shuttle vector that was allowed to replicate in COS7 cells (48). The same mutational pattern (G to T transversions, 63.0 % of 65 base substitutions) was obtained after transfection of 1O2-treated M13lacZ bacteriophage DNA in Escherichia coli (49). Also, 8-oxo-dGuo is a frequent lesion detected in DNA exposed to 1O2 (39,50–52). These findings suggest that 1O2 causes 8-oxo-dGuo formation and this accounts for the G to T transversions. This would explain the mutational pattern observed with BP-7,8-dione in p53 cDNA using yeast reporter system (19). G to T transversions (46.0 % of 63 base substitutions) predominated and these mutations were blocked by ROS scavengers.
Overall the mechanism of PAH o-quinone-mediated 8-oxo-dGuo formation in the presence of NADPH and CuCl2 can be explained by Scheme 2 which accounts for all the observations: (a) Cu2+ /Cu+ stimulated redox-cycling of o-quinones; (b) failure to block 8-oxo-dGuo formation with hydroxyl radical scavengers; (c) the ability of methional to attenuate 8-oxo-dGuo formation and (d) the ability to form 8-oxo-dGuo in the presence of Cu(II) and H2O2.
Scheme 2.
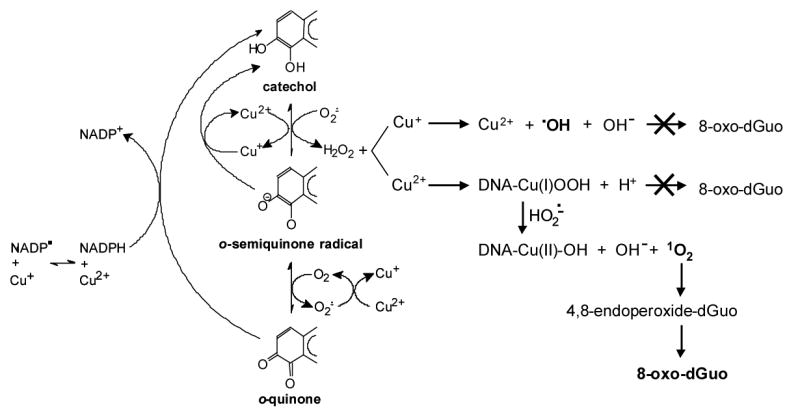
Generation of reactive species during redox cycling of PAH o -quinones in the presence of NADPH and copper(II) chloride and the formation of 8-oxo-dGuo
If Cu(II) is required for 8-oxo-dGuo formation by PAH o-quinones, is Cu(II) present in the cellular environment? Cu(II) is protein bound and is present in higher concentrations in the nucleus than in the cytosol (53). In the nucleus, copper exists in a nuclear histone protein complex located at the base of DNA loops, where it maintains the nuclear matrix and DNA folding (54,55). In DNA, it binds mostly to N7 position of purines, especially guanine (56). The high prevalence of copper in the nucleus and DNA suggests that it plays a role in oxidative damage in DNA mediated by redox-active compounds.
In conclusion, we report that PAH o-quinones in the presence of NADPH and Cu(II) produced significant levels of 8-oxo-dGuo, which leads to G to T transversion mutations. The 8-oxo-dGuo formation was promoted by reactive species such as O2•−, H2O2 and Cu(I). The most plausible mechanism involves the formation of Cu(I)-hydroperoxy complex with DNA and the resultant production of 1O2. This study suggests that PAH o-quinones derived by AKR enzymes may contribute significantly to PAH-associated carcinogenesis and mutagenesis.
Acknowledgments
This work was supported by grants from NIH (P01-CA-092537) and (R01-CA-39504) awarded to TMP. We thank Dr. Ian A. Blair for his advice and for reading this manuscript.
Footnotes
Abbreviations: AKR, aldoketo-reductase; (±)-anti-BP-7,8-diol-9,10-epoxide, (±)-anti-7,8-dihydroxy-9α,10α–epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene; bathocuproine, bathocuproinedisulfonic acid; BP-7,8-dione, benzo[a]pyrene-7,8-dione; BA-3,4-dione, benz[a]anthracene-3,4-dione; P450, cytochrome P450; Desferal®, deferoxamine mesylate; dGuo, 2′-deoxyguanosine; 7,12-DMBA-3,4-dione, 7,12-dimethylbenz[a]anthracene-3,4-dione; methional, 3-(methylthio)propionaldehyde; NADPH, β-nicotinamide adenine dinucleotide phosphate (reduced); NP-1,2-dione, naphthalene-1,2-dione; 8-oxo-dGuo, 8-oxo-7,8-dihydro-2′-deoxyguanosine; PAH, polycyclic aromatic hydrocarbon; PDE I, phosphodiesterase I; ROS, reactive oxygen species; SOD, superoxide dismutase; Tiron, 4,5-dihydroxy-1,3-benzene disulfonic acid.
References
- 1.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 2.Dipple, A. (1985) Polyaromatic hydrocarbon carcinogenesis: An introduction. In Polycyclic hydrocarbon and carcinogenesis (Harvey, R. G., Ed.) ACS Symposium Series 283, pp 1–17, American Chemical Society, Washington, DC.
- 3.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 4.Cavalieri EL, Rogan EG. Central role of radical cations in metabolic activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 5.Gelboin HV. Benzo[a]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 6.Smithgall TE, Harvey RG, Penning TM. Regio- and stereospecificity of homogeneous 3α-hydroxysteroid-dihydrodiol dehydrogenase for trans-dihydrodiol metabolites of polycyclic aromatic hydrocarbons. J Biol Chem. 1986;261:6184–6191. [PubMed] [Google Scholar]
- 7.Smithgall TE, Harvey RG, Penning TM. Oxidation of the trans-3,4-dihydrodiol metabolites of the potent carcinogen 7,12-dimethylbenz[a]anthracene and other benz[a]anthracene derivatives by 3α-hydroxysteroid-dihydrodiol dehydrogenase: effects of methyl substitution on velocity and stereochemical course of trans-dihydrodiol oxidation. Cancer Res. 1988;48:1227–1232. [PubMed] [Google Scholar]
- 8.Smithgall TE, Harvey RG, Penning TM. Spectroscopic identification of ortho-quinones as the products of polycyclic aromatic trans-dihydrodiol oxidation catalyzed by dihydrodiol dehydrogenase. A potential route of proximate carcinogen metabolism. J Biol Chem. 1988;263:1814–1820. [PubMed] [Google Scholar]
- 9.Chakravarti D, Pelling JC, Cavalieri EL, Rogan EG. Relating aromatic hydrocarbon-induced DNA adducts and c-H-ras mutations in mouse skin papillomas: the role of apurinic sites. Proc Natl Acad Sci U S A. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todorovic R, Ariese F, Devanesan P, Jankowiak R, Small GJ, Rogan E, Cavalieri EL. Determination of benzo[a]pyrene- and 7,12-dimethylbenz[a]anthracene-DNA adducts formed in rat mammary glands. Chem Res Toxicol. 1997;10:941–947. doi: 10.1021/tx970003y. [DOI] [PubMed] [Google Scholar]
- 11.Jennete KW, Jeffrey AM, Blobstein SH, Beland FA, Harvey RG, Weinstein IB. Nucleoside adducts from the in vitro reaction of benzo[a]pyrene-7,8-dihydrodiol 9,10-oxide or benzo[a]pyrene 4,5-oxide with nucleic acids. Biochemistry. 1977;16:932–938. doi: 10.1021/bi00624a019. [DOI] [PubMed] [Google Scholar]
- 12.Osborne MR, Beland FA, Harvey RG, Brookes P. The reaction of (+/−)-7α, 8β-dihydroxy-9β, 10β-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene with DNA. Int J Cancer. 1976;18:362–368. doi: 10.1002/ijc.2910180315. [DOI] [PubMed] [Google Scholar]
- 13.Jeffrey AM, Jennette KW, Blobstein SH, Weinstein IB, Beland FA, Harvey RG, Kasai H, Miura I, Nakanishi K. Benzo[a]pyrene-nucleic acid derivative found in vivo: structure of a benzo[a]pyrenetetrahydrodiol epoxide-guanosine adduct. J Am Chem Soc. 1976;98:5714–5715. doi: 10.1021/ja00434a060. [DOI] [PubMed] [Google Scholar]
- 14.Koreeda M, Moore PD, Wiskocki PG, Levin W, Yagi H, Jerina DM. Binding of benzo[a]pyrene 7,8-diol-9,10-epoxides to DNA, RNA, and protein of mouse skin occurs with high stereoselectivity. Science. 1978;199:778–781. doi: 10.1126/science.622566. [DOI] [PubMed] [Google Scholar]
- 15.Marshall CJ, Vousden KH, Phillips DH. Activation of c-Ha-ras-1 proto-oncogene by in vitro modification with a chemical carcinogen, benzo[a]pyrene diolepoxide. Nature. 1984;310:586–589. doi: 10.1038/310586a0. [DOI] [PubMed] [Google Scholar]
- 16.Balu N, Padgett WT, Lambert GR, Swank AE, Richard AM, Nesnow S. Identification and characterization of novel stable deoxyguanosine and deoxyadenosine adducts of benzo[a]pyrene-7,8-quinone from reactions at physiological pH. Chem Res Toxicol. 2004;17:827–838. doi: 10.1021/tx034207s. [DOI] [PubMed] [Google Scholar]
- 17.Shou M, Harvey RG, Penning TM. Contribution of dihydrodiol dehydrogenase to the metabolism of (+/−)-trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene in fortified rat liver subcellular fractions. Carcinogenesis. 1992;13:1575–1582. doi: 10.1093/carcin/13.9.1575. [DOI] [PubMed] [Google Scholar]
- 18.McCoull KD, Rindgen D, Blair IA, Penning TM. Synthesis and characterization of polycyclic aromatic hydrocarbon o-quinone depurinating N7-guanine adducts. Chem Res Toxicol. 1999;12:237–246. doi: 10.1021/tx980182z. [DOI] [PubMed] [Google Scholar]
- 19.Yu D, Berlin JA, Penning TM, Field J. Reactive oxygen species generated by PAH o-quinones cause change-in-function mutations in p53. Chem Res Toxicol. 2002;15:832–842. doi: 10.1021/tx010177m. [DOI] [PubMed] [Google Scholar]
- 20.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 21.Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003;21:229–239. doi: 10.1002/humu.10177. [DOI] [PubMed] [Google Scholar]
- 22.Nair J, Furstenberger G, Burger F, Marks F, Bartsch H. Promutagenic etheno-DNA adducts in multistage mouse skin carcinogenesis: correlation with lipoxygenase-catalyzed arachidonic acid metabolism. Chem Res Toxicol. 2000;13:703–709. doi: 10.1021/tx000045d. [DOI] [PubMed] [Google Scholar]
- 23.Frenkel K, Wei L, Wei H. 7,12-dimethylbenz[a]anthracene induces oxidative DNA modification in vivo. Free Radic Biol Med. 1995;19:373–380. doi: 10.1016/0891-5849(95)00046-z. [DOI] [PubMed] [Google Scholar]
- 24.Kim KB, Lee BM. Oxidative stress to DNA, protein, and antioxidant enzymes (superoxide dismutase and catalase) in rats treated with benzo[a]pyrene. Cancer Lett. 1997;113:205–212. doi: 10.1016/s0304-3835(97)04610-7. [DOI] [PubMed] [Google Scholar]
- 25.Leadon SA, Stampfer MR, Bartley J. Production of oxidative DNA damage during the metabolic activation of benzo[a]pyrene in human mammary epithelial cells correlates with cell killing. Proc Natl Acad Sci U S A 8. 1988;5:4365–4368. doi: 10.1073/pnas.85.12.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leadon SA, Sumerel J, Minton TA, Tischler A. Coal tar residues produce both DNA adducts and oxidative DNA damage in human mammary epithelial cells. Carcinogenesis. 1995;16:3021–3026. doi: 10.1093/carcin/16.12.3021. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi S, Kawanishi S. Double base lesions of DNA by a metabolite of carcinongenic benzo[a]pyrene. Biochem Biophys Res Commun. 2002;290:778–782. doi: 10.1006/bbrc.2001.6249. [DOI] [PubMed] [Google Scholar]
- 28.Seike K, Murata M, Oikawa S, Hiraku Y, Hirakawa K, Kawanishi S. Oxidative DNA damage induced by benz[a]anthracene metabolites via redox cycles of quinone and unique non-quinone. Chem Res Toxicol. 2003;16:1470–1476. doi: 10.1021/tx034103h. [DOI] [PubMed] [Google Scholar]
- 29.Burczynski ME, Penning TM. Genotoxic polycyclic aromatic hydrocarbon ortho-quinones generated by aldoketo reductases induce CYP1A1 via nuclear translocation of the aryl hydrocarbon receptor. Cancer Res. 2000;60:908–915. [PubMed] [Google Scholar]
- 30.Fieser LF. β-Naphthoquinone and α-naphthoquinone. Org Synth. 1943;2:430–435. [Google Scholar]
- 31.Sukumaran KB, Harvey RG. Synthesis of the o-quinones and dihydrodiols of polycyclic aromatic hydrocarbons from the corresponding phenols. J Org Chem. 1980;45:4407–4413. [Google Scholar]
- 32.Fu PP, Cortez C, Sukumaran KB, Harvey RG. Synthesis of isomeric phenols of benz[a]anthracene from benz[a]anthracene. J Org Chem. 1979;44:4265–4271. [Google Scholar]
- 33.Shapiro, H. S. (1975). Distribution of purines and pyrimidines in deoxyribonucleic acids. In Handbook of Biochemistry and Molecular Biology (Fasman, G. D., Ed) CRC press, pp H80-H110, Chemical Rubber Company, Cleveland.
- 34.Flowers L, Harvey RG, Penning TM. Examination of polycyclic aromatic hydrocarbon o-quinones produced by dihydrodiol dehydrogenase as substrates for redoxcycling in rat liver. Biochem J (Life Sci Adv) 1992;11:49–58. [Google Scholar]
- 35.Flowers L, Harvey RG, Penning TM. Identification of benzo[a]pyrene-7,8-dione as an authentic metabolite of (+/−)-trans-7,8-dihydroxy-7,8-dihydrobenzo[a]pyrene in isolated rat hepatocytes. Carcinogenesis. 1995;16:2707–2715. doi: 10.1093/carcin/16.11.2707. [DOI] [PubMed] [Google Scholar]
- 36.Flowers L, Ohnishi ST, Penning TM. DNA strand scission by polycyclic aromatic hydrocarbon o-quinones: role of reactive oxygen species, Cu(II)/Cu(I) redox cycling, and o-semiquinone anion radicals. Biochemistry. 1997;36:8640–8648. doi: 10.1021/bi970367p. [DOI] [PubMed] [Google Scholar]
- 37.Halliwell B. Protection against tissue damage in vivo by desferrioxamine: What is its mechanism of action? Free Rad Biol Med. 1989;7:645–651. doi: 10.1016/0891-5849(89)90145-7. [DOI] [PubMed] [Google Scholar]
- 38.Seike K, Murata M, Hirakawa K, Deyashiki Y, Kawanishi S. Oxidative DNA damage induces by benz[a]anthracene dihydrodiols in the presence of dihydrodiol dehydrogenase. Chem Res Toxicol. 2004;17:1445–1451. doi: 10.1021/tx0498814. [DOI] [PubMed] [Google Scholar]
- 39.Ravanat JL, Saint-Pierre C, Di Mascio P, Martinez GR, Medeiros MHG, Cadet J. Damage to isolated DNA mediated by singlet oxygen. Helv Chim Acta. 2001;84:3702–3709. [Google Scholar]
- 40.Halliwell, B. and Gutteridge, J. M. C. (2000) Detection of free radicals and other reactive species: trapping and fingerprinting. In Free Radicals in Biology and Medicine (Halliwell, B. and Gutteridge, J. M. C. 3rd Ed) Oxford Science Publications, pp 351–429, Oxford University Press, U.K.
- 41.Bal W, Kasprzak KS. Induction of oxidative DNA damage by carcinogenic metals. Toxicol Lett. 2002;127:55–62. doi: 10.1016/s0378-4274(01)00483-0. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez H, Druin R, Holmquist GP, O’Conner TR, Boiteux S, Laval J, Doroshow JH, Akman SA. Mapping of copper/hydrogen peroxide-induced DNA damage at nucleotide resolution in human genomic DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1995;270:17633–17640. doi: 10.1074/jbc.270.29.17633. [DOI] [PubMed] [Google Scholar]
- 43.Stohs SJ, Bachi D. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto Y, Kawanishi S. Hydroxyl free radical is not the main active species in site-specific DNA damage induced by copper (II) ion and hydrogen peroxide. J Biol Chem. 1989;264:15435–15440. [PubMed] [Google Scholar]
- 45.Schweigert N, Acero JL, von Gunten U, Canonica S, Zehnder AJB, Eggen RIL. DNA degradation by mixture of copper and catechol is caused by DNA-copper-Hydroperoxo complexes, probably DNA-Cu (I)-OOH. Environ Mol Mutagen. 2000;36:5–12. doi: 10.1002/1098-2280(2000)36:1<5::aid-em2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 46.Frelon S, Douki T, Favier A, Cadet J. Hydroxyl radical is not the main reactive oxygen species involved in the degradation of DNA bases by copper in the presence of hydrogen peroxide. Chem Res Toxicol. 2003;16:191–197. doi: 10.1021/tx025650q. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Benito JF. Reaction pathway in the decomposition of hydrogen peroxide catalyzed by copper (II) J Inorg Biochem. 2004;98:430–438. doi: 10.1016/j.jinorgbio.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 48.de Oliveira RC, Ribeiro DT, Nigro RG, Mascio PD, Menck CFM. Singlet oxygen induced mutation spectrum in mammalian cells. Nucleic Acids Res. 1992;20:4319–4323. doi: 10.1093/nar/20.16.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decuyper-Debergh D, Piette J, van de Vorst A. Singlet oxygen-induced mutation in M13 lacZ phage DNA. EMBO J. 1987;6:3155–3161. doi: 10.1002/j.1460-2075.1987.tb02626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider JE, Price S, Maidt L, Gutteridge JMC, Floyd RA. Methylene blue plus light mediates 8-hydroxy 2′-deoxyguanosine formation in DNA preferentially over strand breakage. Nucleic Acids Res. 1990;18:631–635. doi: 10.1093/nar/18.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein fprmamidopyrimidine-DNA glycosylase: excision of purine lesions in DNA produced by ionizing radiation or photosnesitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 52.Wei H, Cai Q, Rahn R, Zhang X. Singlet oxygen involvement in ultraviolet (254 nm) radiation-induced formation of 8-hydroxy-deoxyguanosine in DNA. Free Rad Biol Med. 1997;23:148–154. doi: 10.1016/s0891-5849(96)00526-6. [DOI] [PubMed] [Google Scholar]
- 53.Bryan SE, Vizard DL, Beary DA, LaBiche R, Hardy KJ. Partitioning of zinc and copper within subnuclear nucleoprotein particles. Nucleic Acids Res. 1981;9:5811–5823. doi: 10.1093/nar/9.21.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 55.George AM, Sabovljev SA, Hart LE, Cramp WA, Harris G, Hornsey S. DNA quaternary structure in the radiation sensitivity of human lymphocytes--a proposed role of copper. Br J Cancer Suppl. 1987;8:141–144. [PMC free article] [PubMed] [Google Scholar]
- 56.Gao YG, Sriram M, Wang AH. Crystallographic studies of metal ion-DNA interactions: different binding modes of cobalt(II), copper(II) and barium(II) to N7 of guanines in Z-DNA and a drug-DNA complex. Nucleic Acids Res. 1993;21:4093–4101. doi: 10.1093/nar/21.17.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]


