Abstract
The diurnal cycle strongly influences many plant metabolic and physiological processes. Arabidopsis thaliana rosettes were harvested six times during 12-h-light/12-h-dark treatments to investigate changes in gene expression using ATH1 arrays. Diagnostic gene sets were identified from published or in-house expression profiles of the response to light, sugar, nitrogen, and water deficit in seedlings and 4 h of darkness or illumination at ambient or compensation point [CO2]. Many sugar-responsive genes showed large diurnal expression changes, whose timing matched that of the diurnal changes of sugars. A set of circadian-regulated genes also showed large diurnal changes in expression. Comparison of published results from a free-running cycle with the diurnal changes in Columbia-0 (Col-0) and the starchless phosphoglucomutase (pgm) mutant indicated that sugars modify the expression of up to half of the clock-regulated genes. Principle component analysis identified genes that make large contributions to diurnal changes and confirmed that sugar and circadian regulation are the major inputs in Col-0 but that sugars dominate the response in pgm. Most of the changes in pgm are triggered by low sugar levels during the night rather than high levels in the light, highlighting the importance of responses to low sugar in diurnal gene regulation. We identified a set of candidate regulatory genes that show robust responses to alterations in sugar levels and change markedly during the diurnal cycle.
INTRODUCTION
Plants are exposed to changing environmental conditions, one of the most dramatic being the daily alternation between light and darkness. These changes of irradiance act via multiple red and blue light receptors to regulate gene expression (Schaffer et al., 2001; Tepperman et al., 2004) and have wide-reaching indirect consequences for many metabolic and physiological processes.
In the light, photosynthetic carbon fixation in leaves supports the synthesis and export of sucrose to the remainder of the plant to support metabolism, storage, and growth. At night, the plant becomes a net consumer of carbon. It is important to buffer against these diurnal fluctuations of the carbon balance. Even a few hours of carbon depletion lead to an inhibition of growth, which is only slowly reversed when carbon becomes available again (Gibon et al., 2004a). Part of the photosynthate is accumulated in the leaves in the light, usually as starch, and is remobilized to support sucrose synthesis and export at night (Stitt et al., 1987; Geiger and Servaites, 1994; Stitt, 1996; Geiger et al., 2000). Sugar levels nevertheless undergo marked diurnal changes in leaves (Geiger and Servaites, 1994; Scheible et al., 1997a, 1997b; Matt et al., 2001a, 2001b) and nonphotosynthetic sink tissues, such as stems (Kehr et al., 1998), roots (Scheible et al., 1997a; Gibon et al., 2004a), and tubers (Geigenberger and Stitt, 2000).
Fixed carbon and reducing equivalents are required to convert inorganic nitrogen and sulfate into amino acids, nucleotides, and cofactors. Nitrate assimilation is stimulated in the light, resulting in a decrease of nitrate and accumulation of amino acids (Stitt and Krapp, 1999; Matt et al., 2001a, 2001b). These changes are amplified by photorespiration because Gly decarboxylation releases ammonium that must be reassimilated via the GOGAT pathway (Matt et al., 2001b).
In the light, stomata open to facilitate CO2 uptake for photosynthesis (Dietrich et al., 2001). The accompanying increase in transpiration leads to a decrease of the leaf water potential (McDonald and Davies, 1996). There is a gradual inhibition of leaf expansion as the light period progresses (Schmundt et al., 1998), which is accentuated at suboptimal water supplies (Kitano, 1993; Salah and Tardieu, 1997; Schurr, 1998). Changes of the leaf water potential are transmitted by the xylem to the rest of the plant. Potato (Solanum tuberosum) tubers decrease in volume after illumination and recover after darkening (Kehr et al., 1998).
All eukaryotes and some bacteria possess sophisticated biological clocks, which share common organizational features but differ in their molecular components (Harmer et al., 2001; Schaffer et al., 2001; Schultz and Kay, 2003; Webb, 2003). The clock plays a major role in the regulation of processes that are linked to daylength, such as floral induction (Hayama and Coupland, 2003). It also regulates the expression of genes that are involved in metabolism and growth (Harmer et al., 2000; Schaffer et al., 2001). Transcripts for different sets of genes peak at different times in a free-running 24-h cycle (Harmer et al., 2000, 2001), leading to the proposal that circadian regulation anticipates diurnal changes. Growth rates are decreased in clock mutants in short days (Green et al., 2002) or when the lengths of the circadian and diurnal cycle differ (Dodd et al., 2005).
The question arises how irradiance, carbon and nitrogen metabolism, water relations, and the circadian clock interact to regulate metabolism, cellular function, and growth during the diurnal cycle. At this time, global expression profiling provides the most complete readout of the output of signaling pathways. Light (Tepperman et al., 2004; Thum et al., 2004), sugars (Koch, 1996; Price et al., 2004; Thimm et al., 2004; Thum et al., 2004), nutrients (Wang et al., 2003; Scheible et al., 2004), water deficit (Seki et al., 2002; Kawaguchi et al., 2004), and circadian regulation (Harmer et al., 2000; Schaffer et al., 2001) trigger large changes in global gene expression. Using a 7.8K array, Schaffer et al. (2001) estimated that 11% of genes undergo diurnal changes of expression. The following article uses triplicated sampling and analysis with ATH1 arrays to show that >30% of the genes expressed in Arabidopsis thaliana rosettes undergo diurnal changes of expression, identifies which genes and functional areas are most strongly affected, and explores strategies to identify and visualize the contributions of different inputs to this complex multifactorial response.
RESULTS
Characterization of Metabolism during the Diurnal Cycle in Wild-Type Arabidopsis in a 12-h-Light/12-h-Dark Cycle
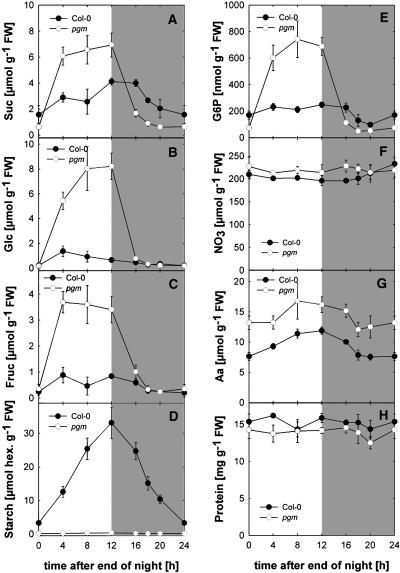
Rosettes were harvested from 5-week-old Columbia-0 (Col-0) growing in a 12-h-light/12-h-dark cycle after 4, 8, and 12 h of illumination and 4, 8, and 12 h of darkness. Figure 1 shows typical responses of metabolites (data for phosphoglucomutase [pgm] are considered later). Sucrose (Figure 1A) and reducing sugars (Figures 1B and 1C) rise in the light and fall during the night. Starch (Figure 1D) accumulates in the light and is almost fully remobilized by the end of the night, as is typical in rapidly growing plants (Fondy and Geiger, 1985; Stitt et al., 1987; Geiger and Servaites, 1994; Matt et al., 1998; Zeeman et al., 1998). The slight decline of glucose-6-phosphate (Figure 1E) at the end of the night is consistent with an impending carbohydrate shortage. This metabolite drops strongly within 2 h when the night is extended (data not shown). Nitrate was high and stable in these growth conditions (Figure 1F), amino acids (Figure 1G) rise during the day and decrease at night, and protein (Figure 1H) did not change.
Figure 1.
Changes of Metabolites during the Diurnal Cycle in Wild-Type and pgm Rosettes.
Wild-type Col-0 and pgm were grown in a 12-h-light/12-h-dark cycle as described in Methods. The whole rosette was harvested at the end of the night, 4, 8, and 12 h into the light period, and 4 and 8 h into the dark period. The results are given as the mean ± sd (n = 5 independent samples, each consisting of three rosettes). The results of one typical experiment from three biological replicates conducted at 2-month intervals are shown. FW, fresh weight.
(A) Sucrose.
(B) Glucose.
(C) Fructose.
(D) Starch.
(E) Glucose-6-phosphate.
(F) Nitrate.
(G) Total amino acids.
(H) Protein.
Global Changes of Gene Expression during the Diurnal Cycle
Three independent experiments performed over a period of 12 months were analyzed with ATH1 arrays. The raw data (CEL files) were processed with Robust Multiarray Analysis (RMA) software (Bolstad et al., 2003). The data are provided in Supplemental Table 1 online. Depending on the experiment, 33 to 38% of the genes were called “absent” at all six time points by Affymetrix microarray suite software (MAS version 5.0) software (see Supplemental Tables 1 and 2 online). Reproducibility between the three biological replicates was assessed by comparing signals for the remaining 13,690 genes at each of the six time points in three pair-wise plots. The correlation coefficients of these 18 plots were between 0.976 and 0.992 (see Supplemental Table 3 online). For each gene, the average signal at each time point was estimated and used to calculate the amplitude of the diurnal change (defined as the log2-scaled ratio of the maximum to the minimum signal; see Supplemental Table 1 online). The amplitude exceeded 0.25, 0.5, 0.8, 1, and 2 for 9255, 6755, 3590, 2464, and 542 genes. This is equivalent to 68, 49, 26, 18, and 4% of the expressed genes or 41, 30, 16, 11, and 2.4% of the genes on the array (Table 1). Application of three statistical methods (Table 1, see Methods for details; see Supplemental Table 1 online for individual genes) revealed that most are bona fide diurnal changes, especially when the amplitude is >0.8. At least 30% and possibly up to half of the genes expressed in wild-type rosettes undergo diurnal changes.
Table 1.
Statistical Analysis of the Global Diurnal Changes of Transcripts in Col-0
| Genes Grouped According to the Amplitude of Their Apparent Diurnal Change [log2(maximum/minimum expression)] |
|||||||
|---|---|---|---|---|---|---|---|
| Total | 0–0.25 | 0.25–0.5 | 0.5–0.8 | 0.8–1 | 1–2 | >2 | |
| No. of Genes | 13,690 | 720 | 5,479 | 3,802 | 1,220 | 1,921 | 548 |
| t Test P-value <0.05 | 8,680 | 191 | 2,372 | 2,703 | 1,056 | 1,812 | 546 |
| Correlation cc >0.5 | 9,402 | 147 | 2,500 | 3,165 | 1,168 | 1,876 | 546 |
| GeneTS P-value <0.1 | 6,104 | 209 | 1,448 | 1,624 | 773 | 1,536 | 512 |
| t Test + cc | 7,142 | 50 | 1,323 | 2,394 | 1,028 | 1,802 | 545 |
| GeneTS + cc | 5,249 | 53 | 872 | 1,527 | 769 | 1,517 | 511 |
| GeneTS + t test | 4,486 | 23 | 627 | 1,431 | 750 | 1,504 | 510 |
| GeneTS + t test + cc | 4,423 | 18 | 575 | 1,408 | 748 | 1,502 | 510 |
The data for three biological replicates were analyzed. All 13,690 genes were included that were called present at one or more of the six time points in every biological replicate. The genes are grouped horizontally according to the amplitude of the apparent diurnal change. Three different statistical tests were applied: (1) a t test on the triplicate values at the time points where the average signals were highest and lowest using log-normalized values and a false discovery rate (FDR)-controlled P-value cutoff of >0.05, (2) the average correlation coefficients of three pair-wise plots of the signal at all six sampling times with a cutoff for the average correlation coefficient (cc) of <0.5, and (3) the occurrence of a reproducible sinusoidal curve detected using GeneTS with an FDR-controlled P-value cutoff of >0.1. The number of genes identified as showing significant diurnal changes by at least two tests and all three tests is also listed.
Classes of Genes Showing Diurnal Rhythms
Table 2 shows the proportion of genes in different functional classes that exhibit diurnal changes with a maximum/minimum log ratio >0.8, >1, or >2. Functional classes where the frequency of diurnal changes was more than twofold higher or lower than the average response are indicated by bold and italic type, respectively. The classification uses groups defined in MapMan (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/). Two files (see Supplemental Tables 4 and 5 online) were also generated to display the amplitude of the diurnal change and the timing of the maximum on a gene-for-gene basis in the visualization tool MapMan (Thimm et al., 2004; Usadel et al., 2005; for web-based and downloadable versions, see http://gabi.rzpd.de/projects/MapMan/). Figure 2 shows one example, and more screenshots are available in Supplemental Figure 2 online.
Table 2.
Assignment of Diurnally Regulated Genes to Different Functional Categories
| Amplitude Limit |
|||||||
|---|---|---|---|---|---|---|---|
| No. of Genes |
Percentage of Genes |
||||||
| Functional Category | Total | >0.8 | >1 | >2 | >0.8 | >1 | >2 |
| Polyamine metabolism | 9 | 4 | 4 | 1 | 41.2 | 41.2 | 11.8 |
| N-metabolism | 23 | 9 | 6 | 2 | 39.1 | 26.1 | 8.7 |
| Major CHO metabolism | 100 | 39 | 32 | 16 | 38.7 | 32.2 | 16.1 |
| S-assimilation | 13 | 5 | 4 | 1 | 38.5 | 30.8 | 7.7 |
| Metal handling | 69 | 25 | 16 | 3 | 35.3 | 23.4 | 4.0 |
| Fermentation | 12 | 4 | 2 | 1 | 33.3 | 16.7 | 8.3 |
| Amino acid metabolism | 225 | 68 | 48 | 11 | 30.1 | 21.5 | 4.7 |
| Redox regulation | 178 | 50 | 38 | 10 | 27.9 | 21.4 | 5.6 |
| Hormone metabolism | 474 | 113 | 91 | 31 | 23.9 | 19.3 | 6.5 |
| Cofactor and vitamins | 36 | 9 | 6 | 2 | 23.6 | 16.7 | 4.2 |
| Photosynthesis | 153 | 35 | 19 | 6 | 22.7 | 12.6 | 3.9 |
| Transport | 849 | 191 | 136 | 30 | 22.5 | 16.0 | 3.5 |
| Lipid metabolism | 347 | 78 | 51 | 7 | 22.5 | 14.7 | 2.0 |
| Tetrapyrrole synthesis | 33 | 7 | 6 | 5 | 22.4 | 19.3 | 15.3 |
| Nucleotide metabolism | 127 | 28 | 13 | 2 | 22.1 | 9.9 | 1.6 |
| Secondary metabolism | 358 | 79 | 58 | 10 | 21.9 | 16.1 | 2.9 |
| C1 metabolism | 24 | 5 | 3 | 0 | 21.9 | 13.7 | 0.0 |
| Minor CHO metabolism | 105 | 23 | 17 | 9 | 21.5 | 15.8 | 8.6 |
| Gluconeogenesis/glyoxylate | 9 | 2 | 1 | 0 | 21.4 | 10.7 | 0.0 |
| Stress | 764 | 158 | 114 | 39 | 20.7 | 14.9 | 5.2 |
| Development | 402 | 81 | 56 | 19 | 20.1 | 13.9 | 4.7 |
| Miscellaneous | 1110 | 210 | 160 | 38 | 18.9 | 14.4 | 3.4 |
| Protein postmodification | 948 | 175 | 112 | 28 | 18.5 | 11.8 | 2.9 |
| Signaling | 760 | 137 | 85 | 19 | 18.0 | 11.1 | 2.4 |
| RNA regulation | 2274 | 406 | 280 | 73 | 17.9 | 12.3 | 3.2 |
| Glycolysis | 61 | 11 | 10 | 2 | 17.4 | 15.7 | 3.3 |
| Cell wall | 463 | 80 | 61 | 12 | 17.3 | 13.2 | 2.5 |
| Oxidative pentose phosphate | 28 | 5 | 3 | 0 | 16.4 | 10.9 | 0.0 |
| Cell | 557 | 84 | 48 | 8 | 15.0 | 8.7 | 1.4 |
| TCA/organic acid metabolism | 71 | 10 | 7 | 1 | 14.2 | 9.2 | 1.4 |
| Not assigned | 8653 | 1149 | 765 | 139 | 13.3 | 8.8 | 1.6 |
| RNA transcription | 62 | 8 | 4 | 1 | 12.9 | 6.5 | 1.6 |
| Protein degradation | 1058 | 122 | 70 | 16 | 11.5 | 6.6 | 1.5 |
| Mitochondrial electron transport | 112 | 11 | 6 | 1 | 10.1 | 5.6 | 0.9 |
| Protein targeting | 173 | 16 | 10 | 0 | 9.0 | 5.8 | 0.0 |
| RNA processing | 201 | 17 | 8 | 0 | 8.2 | 4.0 | 0.0 |
| DNA | 977 | 63 | 29 | 3 | 6.4 | 3.0 | 0.3 |
| Protein synthesis | 474.2 | 30 | 16 | 1 | 6.4 | 3.4 | 0.2 |
Subsets of genes that show increasingly large diurnal changes in transcript levels were identified by filtering out all genes called “absent” at all six time points and all genes where the average correlation coefficient between three pair-wise comparisons of the biological triplicates was <0.5 and then applying three increasingly stringent filters of a diurnal change with a maximum:minimum ratio of >0.8, >1, or >2 on a log2 scale. The genes were grouped using the functional categories defined in MapMan (Thimm et al., 2004; version 1.4.3, http://gabi.rzpd.de/projects/MapMan/). The functional categories are ordered according to their frequency in the gene set with the smallest diurnal cycle. The average response for an amplitude >0.8, >1, or >2 was 16, 11, and 2.4%, respectively. Classes of genes that show a twofold higher, a higher, and a lower frequency of diurnal changes than the average are shown in bold, normal, and italic type, respectively, in the columns giving the percentage of response. CHO, carbohydrate; TCA, tricarboxylic acid.
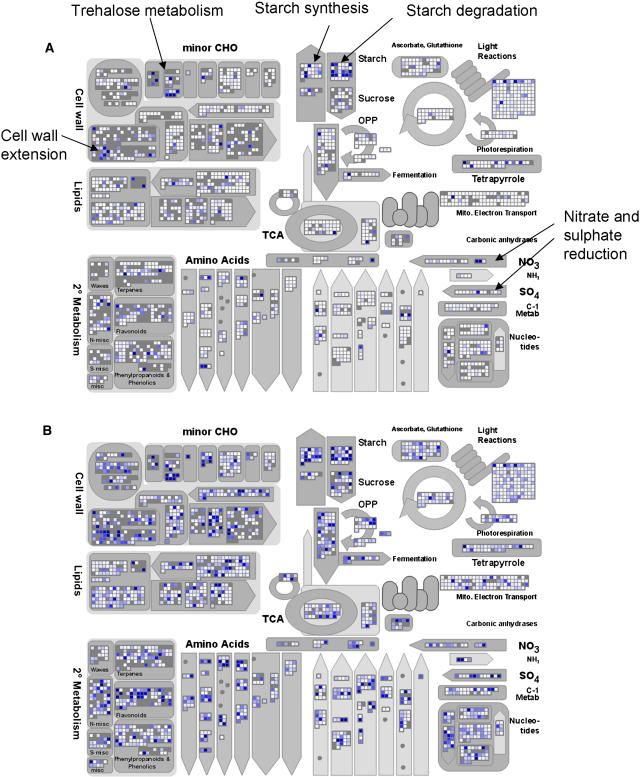
Figure 2.
Amplitude of the Diurnal Changes in the Expression of Genes Assigned to Central Metabolism Displayed in MapMan.
Genes with no significant change in the amplitude are depicted in white, and genes with an increasingly large amplitude are shown as an increasingly intense blue, with the scale saturating at an amplitude [log2(maximum/minimum)] of 3. TCA, tricarboxylic acid; CHO, carbohydrate; OPP, oxidative pentose phosphate.
(A) Wild-type Col-0. The average amplitude is shown for three experiments, including 13,690 probe sets that were detected at least once throughout a diurnal cycle.
(B) pgm mutant. The amplitude for 16,559 probe sets that were detected at least once throughout a diurnal cycle.
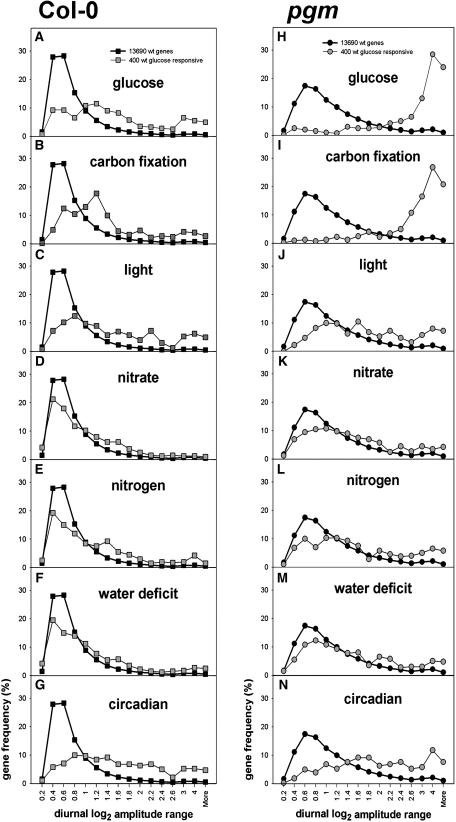
Diurnal changes are especially frequent for genes assigned to starch and sucrose metabolism, trehalose metabolism, nutrient uptake, and assimilation and redox regulation and affect other sectors of metabolism, including glycolysis, gluconeogenesis, the tricarboxylic acid cycle, amino acid, lipid, nucleotide, secondary, and polyamine metabolism, photosynthesis, and tetrapyrrole synthesis (Table 2, Figure 2A). Although a lower proportion of genes assigned to regulation and development was affected, large changes were found for individual genes assigned to cell wall modification, abscisic acid (ABA) and ethylene metabolism and sensing, specific protein kinases, phosphatases, and proteases, a very large proportion of the CONSTANS, MYB-related, and pseudoresponse regulator (APRR) transcription factor families and, to a lesser extent, the AP2-EREBP, MYB, bZIP, and AUX-IAA transcription factor families (see supplemental data online). When only large diurnal changes are considered, the bias to starch and sucrose metabolism becomes even more marked and the frequency remains high for nitrogen and sulfate metabolism and rises for tetrapyrrole synthesis, minor carbohydrate metabolism, gluconeogenesis, redox regulation, and hormone synthesis/sensing (Table 2).
Temporal Distribution of the Changes in Gene Expression
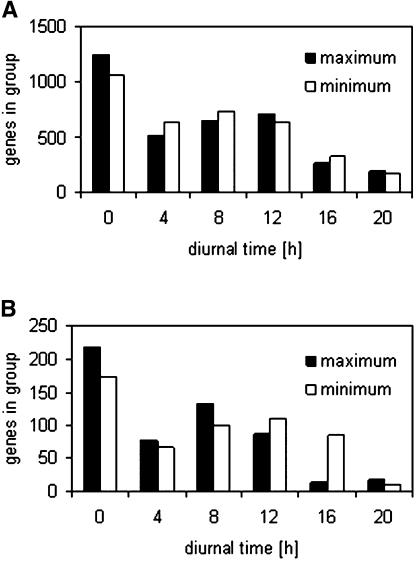
Figure 3 groups genes according to the timing of their diurnal changes. Many genes peaked at the end of the night and were lowest at the end of the light period or early in the night. Another large group peaked during or at the end of the light period and was lowest at the end of the night. This differs from the distribution during a free-running circadian cycle (Harmer et al., 2000; see below for further discussion).
Figure 3.
Frequency of the Timing of the Maxima and Minima for Genes That Show a Diurnal Change.
The results are averaged for three biological replicates. The plots show the results for 3590 genes that have a maximum:minimum ratio >0.8 (A) and 456 genes that have a maximum:minimum ratio >2 (B) on a log2 scale. All genes for which the average correlation coefficient for three pair-wise comparisons of the three biological replicates was <0.5 were omitted.
Transcripts for low-sugar-induced genes such as ASN1 and GDH1 (Koch, 1996; Thimm et al., 2004) peak at the end of the night, indicating that the rosette is becoming carbon depleted. Transcripts for several genes assigned to trehalose metabolism peak at the end of the night. This may also represent an early response to sugar depletion, as these transcripts rise steeply when the night is extended (Thimm et al., 2004) and decrease after sugar addition (Price et al., 2004; see also below). Genes required for photosynthesis peak preferentially at the end of the night or the start of the light period, for nitrate and sulfate uptake and nitrate assimilation at the end of the night (see also Stitt and Krapp, 1999), for sucrose synthesis during the light period, and for organic acid synthesis (these are needed for the use of fixed nitrogen; see Scheible et al., 2000) during the light period. As already reported (Smith et al., 2004), many genes involved in starch degradation peak at the end of the day, but others peak at the end of the night. Coordinated changes of genes assigned to other areas of metabolism, including mitochondrial electron transport and lipid and secondary metabolism, are noted in the supplemental data online. A subset of genes that might be involved in extension growth (cell wall modifying and degrading enzymes, two tonoplast intrinsic proteins, and many of the genes annotated as plasmalemma intrinsic proteins) peak toward the end of the night, whereas a subset of the genes involved in DNA synthesis (in particular histones), cell division, and cell cycle regulation peak during or at the end of the light period (see below for more data). Several cytokinin response regulators and a large set of auxin-regulated genes peak toward the end of the night, while several genes involved in ABA synthesis and sensing peak in the light period.
To identify the inputs driving these complex diurnal changes, published and in-house expression profiles were inspected to identify subsets of diagnostic genes whose expression is particularly responsive to sugars, light, nitrogen, water status, or the circadian clock. For each subset, we asked whether the diurnal changes of transcripts match those predicted from the diurnal changes of the input.
Comparison of the Diurnal Expression Pattern of Sets of Sugar-Responsive Genes with the Changes of Sugars
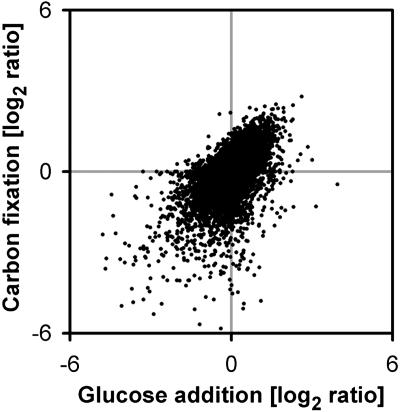
Two approaches were used to identify sugar-responsive genes. One investigated the response after adding sugars to carbon-starved seedlings. Seedlings were grown in liquid culture in full nutrient medium with sucrose and weak continuous light for 7 d (see Scheible et al., 2004), transferred to zero sugar for 2 d to deplete carbohydrates, and then resupplied with exogenous glucose for 3 h. The 200 genes that showed the largest induction and the 200 genes that showed the largest repression (see Supplemental Table 6 online) were termed glucose-responsive genes. An analogous experiment was analyzed to compile a set of sucrose-responsive genes (see Supplemental Table 6 online). The second approach involved a study of the response to endogenous changes of sugars. Five-week-old wild-type Col-0 was illuminated for 4 h in the presence of ambient or <50 ppm [CO2]. The latter is close to compensation point, so there is no net photosynthesis, and sugar levels resemble those in the dark (see legend to Figure 4). The results of two biological replicates are available in Supplemental Table 7 online. There is relatively good agreement between the global responses to carbon fixation in 5-week-old rosettes and glucose addition in seedlings (Figure 4). The 200 genes showing the largest induction and the 200 genes showing the largest repression in ambient compared with low [CO2] were termed carbon fixation-responsive genes (see Supplemental Table 6 online). Comparison with the glucose-responsive set revealed that 45 and 34% of the repressed and induced genes were present in both sets, a further 29 and 23% of the repressed and induced genes showed qualitatively similar responses (i.e., changed by >2 in both treatments but were in the 200 most strongly affected genes in only one treatment), and only 2.8% showed opposite responses (see Supplemental Table 8 online).
Figure 4.
Comparison of the Response of Genes to Addition of 100 mM Glucose to Carbon-Starved 7-d-Old Seedlings and the Response of Genes for 4 h in the Light at Ambient (350 ppm) [CO2] or Compensation Point (<50 ppm) [CO2].
A further treatment (data not shown) involved a 4-h extension of the night. The raw data are provided in the supplemental data online. The sugar levels in the material were as follows: 1.4 μmol·g FW sucrose, 0.7 μmol·g FW glucose, 0.8 μmol·g FW fructose after 4 h in the dark at compensation point [CO2], 1.3 μmol·g FW sucrose, 0.6 μmol·g FW glucose, 1.1 μmol·g FW fructose after 4 h in the light at compensation point [CO2] and 3.5 μmol·g FW sucrose, and 1.3 μmol·g FW glucose and 0.8 μmol·g FW fructose after 4 h light under ambient [CO2].
Sugars are high in the light and fall at night (Figure 1). We reasoned that if these changes contribute to the diurnal regulation of expression, genes that are induced by sugars should peak in the light and genes that are repressed should peak at night. The diurnal responses of the 200 glucose-repressed (Figure 5A) and 200 glucose-induced genes (Figure 5B) were k-means clustered into seven groups and visually inspected. Many showed the predicted response. The data were condensed by subjectively grouping the clusters into four classes (Table 3) corresponding to (1) genes that show large or (2) small changes that are consistent with the expected response, (3) genes that do not show a detectable diurnal change, and (4) genes that show an inconsistent response. For example, for glucose-repressed genes, these classes correspond to clusters where genes are (1) strongly or (2) weakly repressed during the night and induced during the day, (3) do not show diurnal changes, or (4) are induced during the night and repressed during the day. Two-thirds of the glucose-responsive genes show a diurnal response consistent with sugars contributing to the diurnal regulation of gene expression, 30% did not show a diurnal change (see below for further discussion), and 4% showed a contrary response.
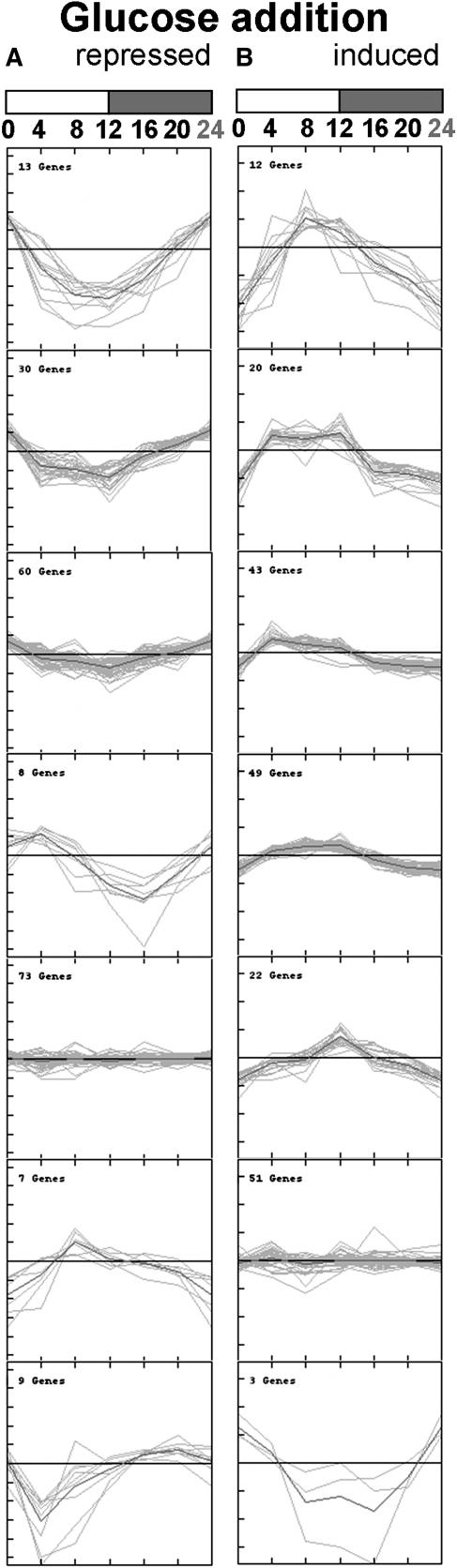
Figure 5.
Clustering of 400 Glucose-Responsive Genes According to Their Response in the Diurnal Cycle in Wild-Type Rosettes.
(A) The 200 glucose-repressed genes.
(B) The 200 glucose-induced genes.
The glucose-responsive genes were identified from an experiment in which 7-d-old Arabidopsis seedlings growing in liquid culture in full nutrient medium under weak continuous light were subjected to carbon starvation for 2 d, before adding 100 mM glucose for 3 h, and correspond to the 200 genes that show the largest repression and the 200 genes that show the largest induction. The genes are listed in Supplemental Table 6 online. The genes in the two sets were k-means clustered based on their diurnal response. For each gene, clustering was performed on the log2-transformed average values at each of the six times during the diurnal, normalized to the average level in the diurnal cycle. The 24-h point of time was copied from time 0. The number of genes in each cluster is given in each panel. The day/night cycle is indicated at the top of the first panel by a white (day) and a gray (night) bar.
Table 3.
Qualitative Comparison of the Diurnal Changes of Sets of Diagnostic Genes That Respond to Glucose, Sucrose, Light, Nitrogen Metabolite, and Water Deficit and the Diurnal Changes of Sugars, Light, Nitrogen Metabolites, and Water Deficit
| Wild-Type Col-0 |
pgm |
|||||||
|---|---|---|---|---|---|---|---|---|
| (%) | (%) | |||||||
| Treatment | Good Fit | Weak Fit | No Change | Negative | Good Fit | Weak Fit | No Change | Negative |
| Glucose | 63 | 4 | 31 | 4 | 88 | 5 | 9 | 1 |
| Sucrose | 35 | 9 | 53 | 4 | 62 | 0 | 23 | 10 |
| Carbon fixation | 48 | 16 | 31 | 3 | 88 | 12 | 0 | 0 |
| Photomorphogenesis | 3 | 24 | 49 | 24 | 23 | 0 | 55 | 21 |
| Light | 23 | 20 | 40 | 17 | 38 | 16 | 28 | 17 |
| N 30 min | 7 | 19 | 45 | 27 | 26 | 8 | 47 | 19 |
| N 3 h | 22 | 17 | 48 | 11 | 15 | 28 | 65 | 15 |
| Water deficit | 26 | 27 | 41 | 5 | 22 | 31 | 42 | 5 |
Sets of 400 genes corresponding to the 200 most strongly induced and the 200 most strongly repressed genes were identified in published or in-house profiles showing the global changes of expression 3 h after adding 15 mM glucose or 15 mM sucrose to 9-d-old seedlings in liquid culture that had been starved of carbon for 2 d (D. Osuna, R. Morcuende, W.-R. Scheible, Y. Gibon, O. Bläsing, O. Thimm, M. Höhne, M. Günther, B. Usadel, M. Udvardi, B. Kamlage, R. Trethewey, and M. Stitt, unpublished data), from 5-week-old plants illuminated for 4 h under ambient (450 ppm) or low (<50 ppm) [CO2], from dark-grown seedlings 4 h after exposure to weak white light (AtGenExpress), from 5-week-old plants exposed to low (<50 ppm) [CO2] under 4 h of illumination or 4 h of prolonged darkness, 30 min and 3 h after adding 3 mM nitrate to 9-d-old seedlings that had been depleted of nitrogen for the previous 2 d (Scheible et al., 2004), and 3 h after adding 100 mM mannitol to 9-d-old seedlings (W.-R. Scheible, unpublished data). The lists of genes and responses are provided in Supplemental Table 6 online. The diurnal changes of the eight sets of 200 induced genes and the eight sets of 200 repressed genes were subjected to k-means clustering (see Figures 5A and 5B for the glucose-responsive genes and Supplemental Figure 3 online for the genes responsive to sucrose, carbon fixation, photomorphogenesis, light, nitrogen, and mannitol). They were compared with the diurnal changes of glucose and sucrose (Figure 1), light, nitrate, and amino acids (Figure 1), and water deficit (it was assumed that the water deficit increases in the light) to identify clusters where the diurnal change was in good or weak agreement with the physiological input contributing to the diurnal change in expression, clusters where there was no diurnal change, and clusters where the diurnal changes were opposed to those expected if that particular physiological input were contributing to the diurnal changes in gene expression. The average value for the signal at each time point was used to generate the clusters (Figures 5 and 6; see supplemental data online), which are the basis of the calculations for this table.
An analogous procedure was applied to a set of glucose-responsive genes extracted from Price et al. (2004), the sucrose-responsive genes, and the carbon dioxide–responsive genes (the clusters are shown in Supplemental Figure 3 online). For the sucrose-responsive genes, >40% gave a response consistent with sugars contributing to diurnal regulation, half showed no marked diurnal changes, and 4% gave a contrary response (Table 3). For the carbon fixation-responsive genes, two-thirds gave a response consistent with sugars contributing to diurnal regulation, 31% showed no marked diurnal changes, and 3% gave a contrary response (Table 3).
Diurnal Changes of Transcripts for Sugar-Regulated Genes Are Accentuated in the Starchless pgm Mutant
To provide genetic evidence that sugars contribute to the diurnal regulation of gene expression, the studies were extended to the starchless pgm mutant (Caspar et al., 1985). In this mutant, sugars are very high during the day and very low at night (Figure 1; see also Caspar et al., 1985; Gibon et al., 2004b), whereas the irradiance regime and the diurnal changes of nitrate and amino acids are unaltered (Figure 1). Genes whose diurnal changes of expression are driven by sugars should show an accentuated response in pgm.
Expression profiling was performed with two biological replicates at the end of the light period and the night (correlation coefficients were 0.989 and 0.985, respectively) and without replication at four other times. The original data are provided in Supplemental Table 1 online. In total, >4000 genes showed a more than twofold larger diurnal change in pgm than in Col-0. Increased amplitudes were found in almost all sectors of metabolism (cf. Figures 2A and 2B), cell cycle, cell division, RNA synthesis and processing, amino acid activation, and protein processing (see Supplemental Figure 2 online for further screen shots). Comparison with the response to glucose addition or carbon fixation confirmed that the modified diurnal pattern of gene expression in the pgm mutant is mainly due to changes of sugar levels (see below and Supplemental Figure 4 online).
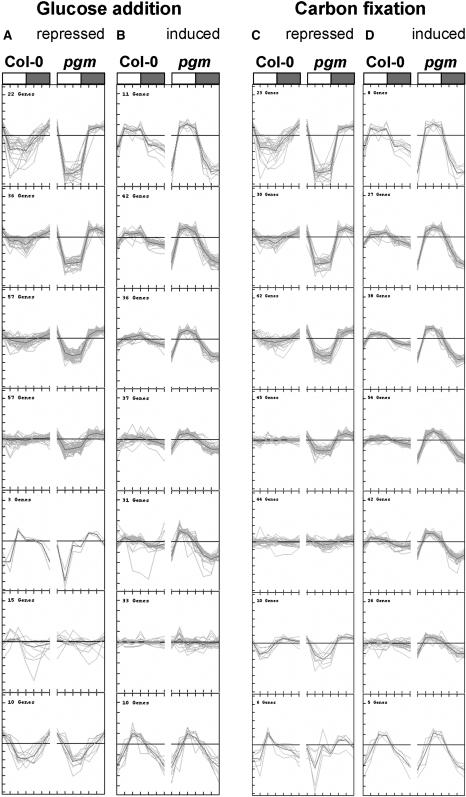
The data sets for Col-0 and pgm were combined and clusters generated for glucose-repressed and -induced genes (Figures 6A and 6B) and carbon fixation–induced and –repressed (Figures 6C and 6D) genes. More than 70% (285) of the glucose-responsive genes showed a response consistent with sugars contributing to their diurnal regulation in Col-0 (Figures 6A and 6B, Table 3), slightly more than when Col-0 data are clustered on their own (see Figures 5A and 5B) because additional genes are recruited due to the larger diurnal changes of sugars in pgm. Almost all (265) of these 285 genes showed a larger diurnal change in pgm than in Col-0. Another 57 genes did not change in Col-0 but changed in pgm in a manner matching the response expected from the larger diurnal changes of sugars. Only a few genes did not show a diurnal change in pgm (33) or showed a diurnal change that was inconsistent (25) with it being due to changes in the sugar levels. Analysis of the set of sucrose-responsive genes (Table 3) confirmed that the contribution of sugars to the diurnal changes of gene expression increases in pgm. An even more striking result was obtained for carbon fixation–responsive genes (see Figures 6C and 6D for the clusters), where every gene responded in pgm in a manner consistent with them being regulated by sugar (Table 3).
Figure 6.
Comparison of the Changes of Expression of Sugar-Responsive Genes during the Diurnal Cycle in Wild-Type Col-0 and in the Starchless pgm Mutant.
Clustering of the diurnal responses of 200 glucose-repressed genes (A), 200 glucose-induced genes (B), 200 carbon fixation–repressed genes (C), and 200 carbon fixation–induced genes (D) in the diurnal cycle in wild-type Col-0 (left part of each panel) and pgm rosettes (right part of each panel). Glucose-responsive genes were identified, and the corresponding transcript data for a combined data set, including all diurnal time points for wild-type Col-0 and pgm, were subjected to k-clustering as in Figure 9. The day/night cycle is exemplarily indicated at the top of the first panel by a white (day) and a gray (night) bar.
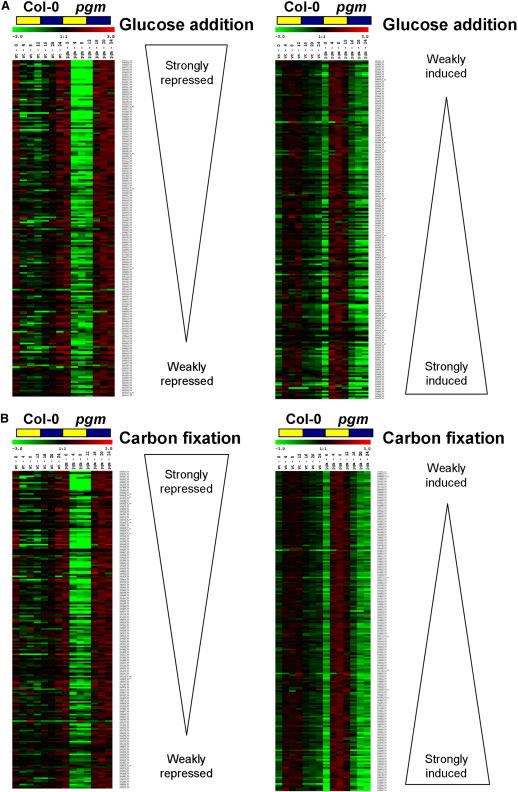
The 400 glucose-responsive genes were ranked according to the extent of the change after adding glucose to seedlings. With few exceptions, genes that show the largest changes in expression after adding glucose to seedlings show the largest diurnal changes in pgm (Figure 7A). A similar result was obtained for the carbon fixation–responsive genes (Figure 7B). This trend was apparent although less clear-cut in Col-0.
Figure 7.
Quantitative Comparison of the Changes of Expression of Sugar-Responsive Genes during the Diurnal Cycle in Wild-Type Col-0 and in the Starchless pgm Mutant.
The 200 genes that were most strongly repressed (left side) or induced (right side) after adding 100 mM glucose to carbon-starved seedlings were ranked according to the size of the change in expression (A). The same approach was used for carbon fixation–responsive genes (B). The changes in expression at six time points during the diurnal cycle in wild-type Col-0 (left points) and pgm (right points) are shown on a false color scale. For Col-0, each data point is the average of three biological replicates, whereas for pgm, the data points are the average of duplicates at the end of the night and the end of the day and single determinations at the other four time points. These signals were normalized to the average level in the diurnal cycle in the respective genotype.
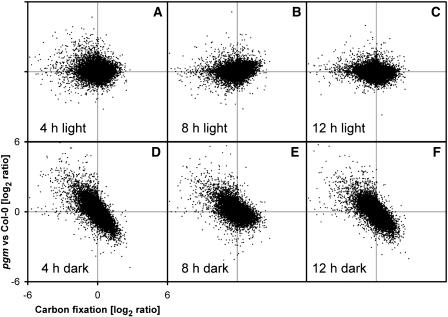
The Accentuated Diurnal Changes in pgm Are Mainly Due to the Lower Sugar Levels at Night
Figure 8 shows whether the exaggerated diurnal changes of transcripts in pgm are due to the higher levels of sugars in the light or the lower levels of sugars at night. Each subpanel shows a different time during the diurnal cycle, the y axis shows transcript levels for 22,746 genes in pgm relative to Col-0, and the x axis shows how expression of each gene responds to sugar. This plot uses the difference between 4-h illumination in the presence of ambient and 50 ppm [CO2] (similar results were obtained using the response to glucose addition; data not shown). In the dark (Figures 8D to 8F), many genes show large differences in their transcript levels between pgm and Col-0, and the direction and extent of the change correlates with the response of the gene to carbon fixation. In the light (Figures 8A to 8C), there is no consistent change of transcript levels in pgm, even though sugars are 10-fold higher than in Col-0. These results indicate that the large diurnal changes of expression in pgm are mainly triggered by the low levels of sugar at night. A similar conclusion was reached after calculating the average levels of transcripts across the entire diurnal cycle: these fell for genes that were induced by sugars and rose for genes that were repressed by sugars, as expected if the changed gene expression in pgm is driven by the low sugar levels in the night (see Supplemental Figure 4 online).
Figure 8.
The Relation between the Changed Levels of Transcripts for Individual Genes in pgm Compared with Col-0 and the Response of These Genes to Glucose.
For each of the 22,746 genes on the ATH1 array, the files (CEL) for Col-0 and pgm were combined and processed with RMA, and the level of each gene in pgm relative to Col-0 was calculated for each of the six harvest times and plotted on the y axis. For each gene, the response to endogenous changes of sugars was extracted from the experiment in which rosettes were illuminated for 4 h at ambient or compensation point [CO2] and plotted on the x axis.
Further support for this conclusion is provided by inspection of Figures 6A to 6D. Almost all genes in the subsets “induced by glucose addition” and “induced by carbon fixation” are more strongly repressed at night in pgm than Col-0, and many genes in the subsets “repressed by glucose addition” and “repressed by carbon fixation” are more strongly or more rapidly induced in the night in pgm than Col-0. Several clusters from the subsets “repressed by glucose addition” and “repressed by carbon fixation” show decreased expression in the light in pgm compared with Col-0, revealing that some highly sugar-responsive genes are repressed by the high sugars in the light.
Investigation of the Diurnal Expression Pattern of Light-Responsive Genes
Two sets of genes were used to investigate whether changes in irradiance contribute directly to the diurnal regulation of gene expression. The public resource AtGenExpress (http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenextable3.htm, treatments 2 and 16) was used to identify the 200 most strongly induced and 200 most strongly repressed genes (see Supplemental Table 6 online) after illuminating dark-grown seedlings with weak white light for 4 h. These genes (termed photomorphogenesis responsive) were clustered (see Supplemental Figure 3 online) and grouped, assuming that transcripts of light-induced genes should increase in the light and decrease at night and light-repressed genes should show the opposite response (Table 3). Half did not show a marked diurnal change, and the remainder fell into two roughly equal groups whose responses were weakly consistent with or inconsistent with that expected if signaling pathways that play a predominant role in seedling photomorphogenesis contribute to diurnal changes of gene expression in mature plants.
Light signaling in light-grown plants may differ from that during photomorphogenesis. A second set of light-responsive genes was identified by comparing rosettes that were illuminated for 4 h at <50 ppm [CO2] (this is compensation point and prevents the light-dependent increase of sugars; see above) with rosettes after a 4 h extension of the night. The experiment was repeated twice. The data are available in Supplemental Table 7 online. The 200 genes that showed the largest induction and the 200 genes that showed the largest repression after illumination at compensation point [CO2] (termed light-responsive genes) are listed in the Supplemental Tables 7 and 8 online. In Col-0, 40% of the light-responsive genes did not show a marked diurnal change, and 43 and 17% showed a change consistent and inconsistent with light acting as an input. In pgm, a higher proportion (54%) of the genes showed changes consistent with light being an input. As the light regime was unaltered, this is presumably because these genes respond to sugars.
Comparison of the Diurnal Expression Pattern of Nitrogen-Responsive Genes with the Changes in Nitrogen Metabolites
Two sets of nitrogen-responsive genes were identified from experiments in which 3 mM nitrate was provided to 9-d-old seedlings that had been depleted of nitrogen for 2 d (Scheible et al., 2004; see Supplemental Table 6 online for lists of the genes). The first set contains 200 genes that are strongly induced and 200 genes that are strongly repressed 30 min after adding nitrate. As there are negligible changes of amino acids at this time, many will be responding to nitrate (Scheible et al., 2004). These genes were clustered (see Supplemental Figure 3 online) and grouped (Table 3), assuming that physiologically available nitrate is highest at the end of the night (see Stitt and Krapp, 1999). The second set contained 400 genes that show large changes 3 h after adding nitrate, by which time there are large changes of amino acids (Scheible et al., 2004). These genes were clustered and grouped based on the measurements showing that amino acids peak at the end of the light period (Figure 1). The diurnal response did not show a bias toward the expected response for either set of genes (see Supplemental Figures 3 and 5 online for the clusters and Table 3 for a summary of the responses). These results do not exclude the possibility of a contribution of nitrogen to the diurnal regulation of a small number of genes.
Investigation of the Diurnal Expression Pattern of Water Deficit–Responsive Genes with the Changes in Sugars
A set of mild water stress–responsive genes was identified by treating 9-d-old seedlings with 100 mM mannitol for 3 h. A list of the 200 induced and 200 repressed genes is given in Supplemental Table 6 online. Mannitol induced one isoform of sucrose phosphate synthase and a set of genes involved in ABA sensing and repressed a large set of xyloglucan endotransglycosylases, other cell wall modifying enzymes, and cell wall degrading enzymes. The diurnal responses were clustered (see Supplemental Figures 3 and 5 online) and grouped assuming that leaf water deficit increases in the light (see Introduction). Of the genes that were induced by mannitol, >50% peaked and only 5% showed a minimum during the light period. Of the genes that were repressed by mannitol, >50% were repressed and only 5% were induced during the light period (Table 3). These results indicate that changes of water deficit contribute to the global changes of gene expression during the diurnal cycle.
Investigation of the Diurnal Expression Pattern of Circadian-Regulated Genes
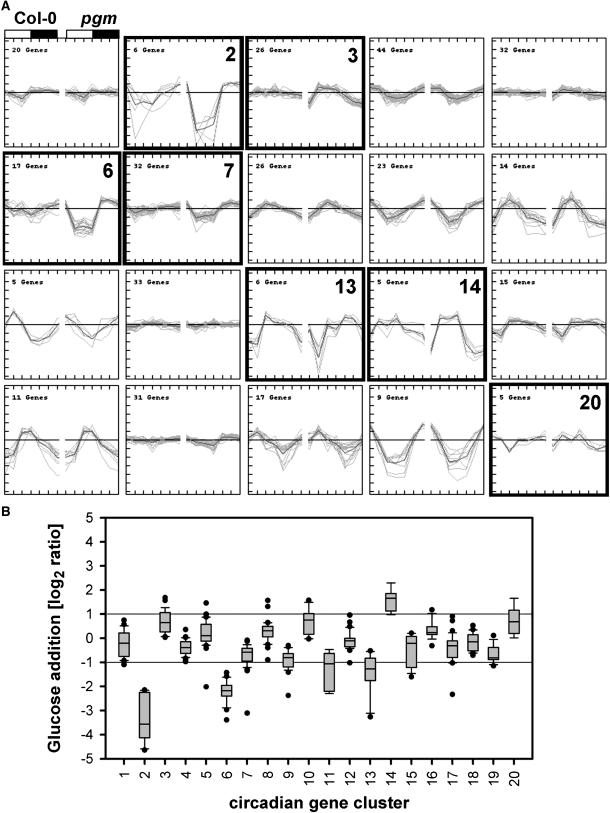
Harmer et al. (2000) found that 6% of the genes on the 7.8K Affymetrix array are subject to circadian regulation when plants entrained to a 12-h-light/dark cycle are transferred to a free-running cycle. The 22K ATH1 array contains 373 unique probe sets for these genes (see Supplemental Table 11 online). As this set of genes is larger than the others, 20 k-means clusters were generated from their diurnal changes in a combined data set for Col-0 and pgm (Figure 9A). There was a clear bias toward large diurnal changes of transcripts (see also Figure 11G). In Col-0, 26% showed a marked, 40% showed a small, and 32% a weak or negligible diurnal change (Figure 9A). A similar proportion of the circadian-regulated genes showed diurnal changes in pgm. Some circadian-regulated genes showed modified responses in pgm (Figure 9A; see below for discussion). We also observed a clear shift of the peaking time for circadian genes in the diurnal rhythm when compared with the free-running cycle (Figure 10; see below for discussion).
Figure 9.
Responses of a Subset of Circadian-Regulated Genes during the Diurnal Cycle in Wild-Type Plants and pgm.
(A) K-means clustering of the responses of 373 circadian genes (taken from Harmer et al., 2000) during a diurnal cycle in wild-type Col-0 (left part of the panels) and pgm (right parts of the panels). The day/night cycle is exemplarily indicated at the top of the first panel by a white (day) and a gray (night) bar. Black boxed panels (clusters 2, 3, 6, 7, 12, 13, and 20) contain circadian genes that behave differently in wild-type and pgm mutant plants. When compared with the results of the glucose starvation–readdition experiment, genes from panels 2, 6, and 7 are induced under low sugar and repressed under sugar starvation, whereas genes from panels 3 and 14 are induced by high sugar and repressed under low sugar.
(B) Response of the genes in each of the individual clusters to the addition of 15 mM glucose to carbon-starved seedlings.
Figure 11.
Comparison of the Amplitudes of the Diurnal Changes of Genes Responsive to Sugar, Light, Nitrogen, Water Stress, and Circadian Clock Regulation Compared with the Diurnal Changes of All Detectable Genes on the ATH1 Array.
The results are taken from Figure 6 for glucose- and carbon fixation–responsive genes, from the corresponding data for sucrose, light, photomorphogenetic light, nitrogen, and water deficit in the supplemental data online, and from Figure 9 for circadian-regulated genes.
Figure 10.
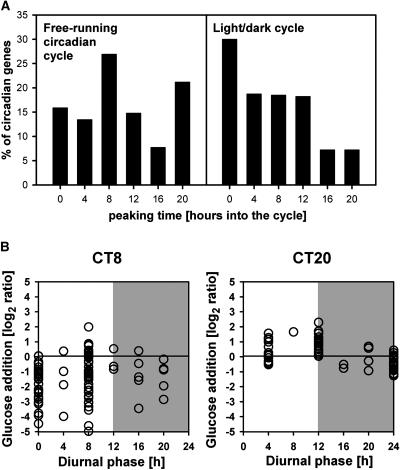
Responses of a Subset of Circadian-Regulated Genes during the Diurnal Cycle in Wild-Type Plants.
(A) Peaking times of 373 circadian clock regulated genes in a free-running circadian cycle (Harmer et al., 2000) and in the diurnal cycle in wild-type Col-0 extracted from the mean of the three biological replicates.
(B) Phase shift of the circadian peaking time (CT) into the diurnal time (x axis) in wild-type Col-0 and the corresponding sugar response (y axis) in a seedling culture after adding 15 mM glucose to carbon-starved seedlings. The phase shift is shown for CT8 and CT20 and for the remaining times in Supplemental Figure 6 online.
Quantitative Comparison of the Amplitudes of the Diurnal Changes of Sugar-, Nitrogen-, Light-, Water Deficit–, and Circadian-Regulated Genes
In Figure 11, the amplitudes of the diurnal changes of the sets of glucose- (Figure 11A), carbon fixation– (Figure 11B), light- (Figure 11C), nitrogen status– (Figures 11D and 11E), water stress– (Figure 11F), and circadian-responsive (Figure 11G) genes are compared with those of the 13,690 genes that are called present at one or more time points in Col-0. Genes with a diurnal response that was inconsistent with a contribution of the proposed input (see Table 3) are excluded from these plots.
Glucose-responsive and carbon dioxide fixation–responsive genes show a clear bias to large diurnal changes. Similar results (see Supplemental Figure 7 online) were obtained for 400 sucrose-responsive genes. Circadian-regulated genes also show a clear bias to large diurnal changes. The responses of light-responsive and especially the nitrogen- and water deficit–responsive genes show a smaller shift. This plot may still overestimate the contribution from these inputs. Many genes in these sets have already been removed because they responded in a manner inconsistent with the input contributing to their diurnal response (see Table 2), and some of the genes in the diagnostic subsets also respond to sugars or circadian regulation (see Supplemental Tables 8 and 12 online). The response in pgm is shown in Figures 11H to 11N. Almost all glucose- and carbon fixation–responsive genes show strongly accentuated diurnal changes, and many circadian-regulated genes still show large changes, whereas there is only a small shift for the nitrogen-, light-, and water deficit–responsive genes. These results show that sugars and circadian inputs make major contributions to the diurnal regulation of expression in Col-0 and that this contribution increases in pgm.
Principle Component Analysis Confirms a Major Role for Sugars in the Diurnal Regulation of Gene Expression
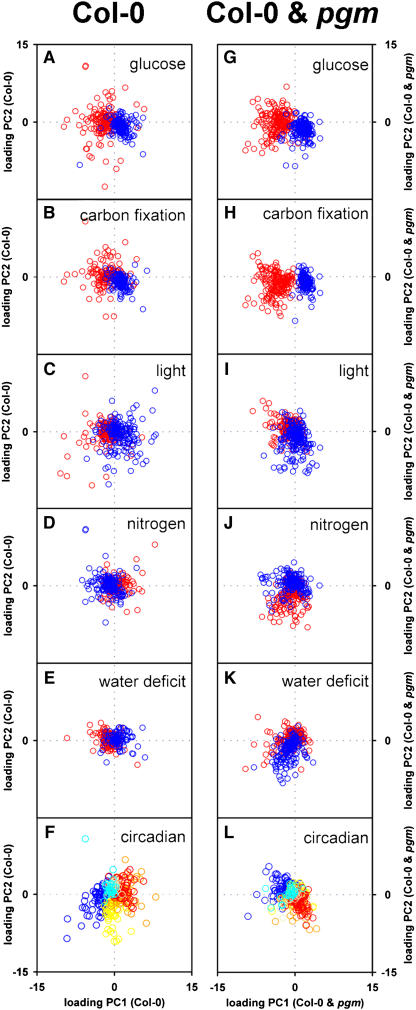
The data analysis in Table 3 and Figures 5 to 11 requires assumptions about the timing of the physiological and environmental inputs. An alternative unbiased approach is taken in Figures 12 and 13. Principle component analysis of the 18 ATH1 arrays separated them into six groups, which correspond to the six harvest times during the diurnal cycle (Figures 12A and 12B). The first two components captured >60% of the total variation (see below). The weightings (eigenvectors) of individual genes in the principle components reflect their contribution to the total variation during the diurnal cycle. The different panels of Figure 13 show the weightings of members of the sets of glucose-, carbon fixation–, light-, nitrogen-, water deficit–, and circadian-responsive genes in the first (x axis) and second (y axis) components. Genes that are induced and repressed by a given input are colored blue and red, respectively. If an input makes a large contribution to a component, the genes it affects will have high and opposite weightings for the genes that it induces and represses (i.e., they will be separated in opposite directions along the axis on which the component is displayed). In the case of circadian regulation, genes that peak at different times during the free-running cycle are distinguished by six different colors.
Figure 12.
Principle Component Analysis of Samples Collected at Different Times during the Diurnal Cycle from Wild-Type Col-0 and pgm.
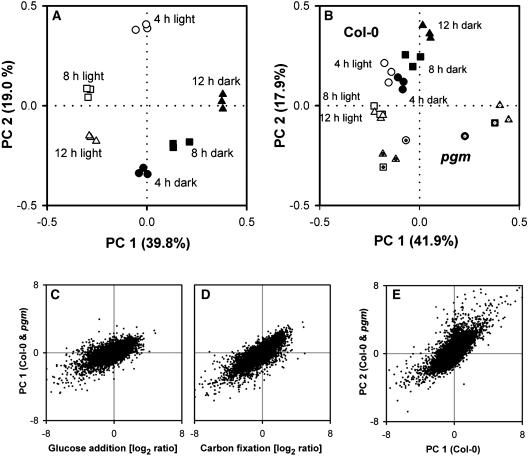
(A) Separation of Col-0 samples by the first and second components. These are shown on the x and y axes and account for 40 and 19% of the total variation, respectively. Each biological replicate is shown separately. The data set contains triplicate samples for all six time points in the Col-0 diurnal cycle and includes 13,690 genes that were called present for at least one time point in each biological replicate. Circles, boxes, and triangles indicate time points at 4, 8, and 12 h into the light (open items) and dark (closed items) periods, respectively.
(B) Separation of a combined data set for wild-type Col-0 and pgm by the first and second components. These are shown on the x and y axes and account for 42 and 18% of the total variation, respectively. The data set contains triplicate samples for all six time points in the Col-0 diurnal cycle, duplicate samples for the end of the night and end of the dark period in pgm, and single samples for other four time points in pgm and includes 16,559 genes that were called present for at least one time point in one genotype. Circles, boxes, and triangles indicate time points at 4, 8, and 12 h into the light (open items) and dark (closed items) periods, respectively. Symbols for pgm contain a dot.
(C) and (D) Correlation between the response of gene expression 3 h after addition of glucose to carbon-starved seedlings (C) or the response of gene expression to carbon fixation and the weightings of genes in the first principle component ([D]; see [B]) separating diurnal samples from Col-0 and pgm during the diurnal cycle.
(E) Comparison of the first component separating diurnal Col-0 samples ([A], x axis) and the second component separating diurnal samples from Col-0 and pgm ([B], y axis).
Figure 13.
Weightings in the First and Second Principle Components of the Subsets of Genes That Are Responsive to Glucose, Carbon Fixation, Light, Nitrogen, Water Stress, and Circadian Regulation.
The genes in the subsets of 200 glucose-, carbon fixation–, light-, nitrogen- (3 h), and water deficit–induced and 200 glucose-, carbon fixation–, light-, nitrogen- (3 h), and water deficit–repressed genes (see Supplemental Table 6 online) and the 373 circadian-regulated genes (see Supplemental Table 11 online) were compared with the weightings of all genes in the first and second components of Figures 12A and 12B (see Supplemental Table 1 online for the individual values) to identify the weightings for these selected subsets of genes. The weightings (loadings) in the first two components that separate Col-0 samples ([A] to [F]) and the first two components that separate the combined set of Col-0 and pgm samples ([F] to [L]) were plotted for all members of the gene subsets responsive to glucose ([A] and [G]), carbon fixation ([B] and [H]), light ([C] and [I]), nitrogen ([D] and [J]), water deficit ([E] and [K]), and circadian regulation ([F] and [L]). Each gene is shown as a point, and its weightings in the first and second components by its position along the x axis and y axis, respectively. As a weighting can be positive or negative, the axes intersect in the center of the plot. For genes responsive to glucose, carbon fixation, light, nitrogen, and water deficit, different colors were used to distinguish between induced (blue) and repressed (red) genes. Induced genes are plotted over the repressed genes, with the result that the data for repressed genes is obscured when they are located in an area that has a high density of induced genes. Similar results were obtained by plotting all genes that showed a more than twofold change to each of these inputs (see Supplemental Figure 9 online). Circadian-regulated genes are shown in color according to the phase when they peak in the diurnal cycle. Gray, magenta, and dark blue indicate peak phases of 4, 8, and 12 h in the dark, while yellow, orange, and red indicate the peaking in 4, 8, and 12 h of light. The average value for the signal at each time point was used for the clusters shown in Figures 5 and 7 and the supplemental data online, which are the basis of the calculations for this figure.
In Col-0, the first component accounted for 40% of the variation. It separated samples harvested toward the end of the night from samples harvested at the end of light period (Figure 12A, x axis), when there are differences in the levels of sugars, amino acids (Figure 1), and water deficit (see Introduction). It did not separate samples harvested 4 h into the light from samples harvested 4 h into the night. The weightings of genes in this component are shown on the x axis in Figures 13A to 13E. Most glucose-induced genes had large and opposite weightings to glucose-repressed genes (Figure 13A). A similar picture is found for carbon fixation–induced and –repressed genes (Figure 13B). There was very poor separation of genes that are induced and repressed by light (Figure 13C), nitrogen (Figure 13D), or water deficit (Figure 13E). Many circadian-regulated genes show a high weighting, with a good separation of genes that peak at the end of the subjective light and dark periods in a free-running circadian cycle (Figure 13F).
The second component (Figure 12A, y axis) accounted for 19% of the variation. It separated samples harvested 4 h into the light from samples harvested 4 h into the dark period but did not separate samples collected at the end of the day from samples collected at the end of the night. The weightings of genes are shown on the y axis in Figures 13A to 13F. Circadian-regulated genes show a different pattern to the first component, with a particularly clear separation of genes that peak in the first part of the subjective day and the night in a free-running circadian cycle (Figure 13F). A small proportion of the light-induced and -repressed genes was separated (Figure 13C), but there was no separation of genes that are induced and repressed by glucose, carbon fixation, nitrogen, or water deficit (Figures 13A, 13B, 13D, and 13E).
The same procedure was taken for a data set that combines Col-0 and pgm. Samples from pgm group close to Col-0 at the end of the light period but are clearly separated at the end of the night (Figure 12B) when (see Gibon et al., 2004a) they resemble Col-0 after an extension of the dark period. The first component (x axis, 42% of the variation) separated samples in the following order: Col-0 at the end of the light period and pgm in or at the end of the day, Col-0 during the day or night, and Col-0 at the end of the night and pgm in the second part of the night. This mirrors their sugar content but not the amino acid content or water deficit (see Figure 1 and Introduction). Glucose-induced and -repressed genes and carbon fixation–induced and –repressed are separated in the first component even better than for Col-0 on its own (cf. Figures 13A, 13B, 13G, and 13H). Genes that are induced or repressed by light, nitrogen, or water stress are not separated (Figures 13I to 13K), and the weightings of circadian-regulated genes are decreased (Figure 13L). There is a striking correlation between the weighting of genes in the first component for this combined data set and the changes of gene expression after adding glucose to carbon-starved seedlings or increasing [CO2] from 50 ppm to ambient levels (Figures 12C and 12D). The second component of the combined data set (accounting for 18% of the variation) separated some genes that are induced or repressed by light (Figure 13I), nitrogen (Figure 13J), and water deficit (Figure 13K) and has a high weighting for many circadian-regulated genes (Figure 13L). A regression plot reveals a correlation between the second component of the combined data set and the first component when Col-0 is analyzed on its own (Figure 12E).
This unbiased analysis confirms that sugars play a major role in the diurnal regulation of gene expression in Col-0, which is balanced and integrated with other inputs, including circadian regulation, light, water deficit, and nitrogen. In pgm, sugars take over the dominant role in the global expression of gene expression, displacing the other inputs into the second component. Identical results were obtained when all genes that changed by more than twofold in response to a particular input were visualized (see Supplemental Figure 9 online). Information about the weightings of individual genes is available in Supplemental Table 1 and Supplemental Figure 8 online.
Interaction between Sugars and the Circadian Clock
We next investigated the interaction between the two major inputs, sugars and the circadian clock. Figure 9 reveals how the diurnal responses of circadian genes are affected by accentuated changes of sugars. Approximately half of the clock-responsive genes show similar responses in Col-0 and pgm (Figure 9A). Another 97 genes (26% of the total) showed a similar phase but larger amplitude in pgm (Figure 9A). Of these, 55 genes (clusters 2, 6, and 7) show an accentuated decrease in the light period and faster recovery in the night, and 31 (clusters 3 and 14) show a slightly accentuated increase in the light period and faster decrease in the night. A possible explanation is that clusters 2, 6, and 7 are sugar induced and clusters 3 and 14 are sugar repressed and that in pgm the amplified response to the large diurnal changes of sugars is superimposed on the circadian rhythm. To test this, we inspected how these genes respond after adding glucose to carbon-starved seedlings (see Supplemental Table 8 online for the original data). Genes in clusters 2, 6, 7, 11, and 13 are strongly, moderately, and weakly repressed by glucose, and genes in clusters 14 and 3 are strongly and weakly induced by glucose (see box plot in Figure 9B). These results indicate that for approximately one-quarter of circadian-regulated genes, the diurnal changes of sugars in a light/dark cycle reinforce the circadian regulation.
A few genes showed a phase shift of 4 (six genes in cluster 13) or 12 h (five genes in cluster 20) in pgm (Figure 9A). Figure 10 provides further evidence that sugars modify the phase of a subset of the clock-regulated genes. Figure 10A compares the time at which 373 circadian-regulated genes peak in a free-running circadian cycle (Harmer et al., 2000) and a diurnal light/dark cycle in Col-0. In a free-running cycle, the highest frequency is after 8 and 20 h (i.e., 8 h into the subjective light period and 8 h into the subjective dark period). In a light/dark cycle, the highest frequency is at the end of the night (see also Figure 3A, which shows a similar response for all 13,690 genes). Figure 10B shows how the phase of the genes that peak at 8 and 20 h in a free-running cycle is modified in a light/dark cycle (most of the genes that peak at the other four time points in a light/dark cycle peak at the same time or with only a 4-h shift in a free-running circadian cycle; see Supplemental Figure 6 online). The y axis in these plots shows the response to glucose addition. Many genes that peak at 8 h in a free-running cycle are repressed by glucose and peak toward the end of the night in a light/dark cycle. Many of the genes that peak at 20 h in a free-running cycle are induced by glucose and peak in or at the end of the light period in a light/dark cycle. In total, 24% of the clock-regulated genes show a phase shift of 8 to 12 h between a free-running and a light/dark cycle, which is probably due to diurnal changes of sugar counteracting the input from the circadian clock.
Identification of Genes Potentially Involved in Sugar-Dependent Changes of Gene Expression in the Diurnal Cycle
The data sets generated in this article were mined to identify candidate genes that may participate in signaling processes triggered by physiological changes of sugars under nonstressful growth conditions. Genes were short listed whose transcript levels show (1) more than twofold changes after glucose addition, (2) a similar more than twofold change in response to carbon fixation, (3) a large diurnal change in Col-0 that is accentuated in pgm, and (4) a qualitatively consistent response in an extended night (Thimm et al., 2004). These were further inspected to exclude genes where timing of the diurnal changes is inconsistent with that predicted from the diurnal changes of sugars and the impact of sugars on expression. Genes that fulfilled these criteria and whose TIGR5 annotations and MapMan functional categories indicate a role in signaling are listed in Supplemental Table 13 online.
This analysis identified 29 known or putative transcription factors from a wide range of families and several protein kinases, including receptor kinases, a mitogen-activated protein kinase, and a putative AMP-regulated protein kinase (AKINβ1). Some were induced and others were repressed by sugars. One of the sugar-repressed transcription factors (ZAT10) has been implicated as a functional homolog of yeast (Saccharomyces cerevisiae) SNF4 (Kleinow et al., 2000), which lies downstream of SNF1 in yeast. The receptor-like protein kinase ATR1/MYB34, which is sugar induced and peaks during the day, has been implicated as a key homeostatic regulator of Trp and indolic glucosinolate metabolism (Celenza et al., 2005). AKINβ1 belongs to the SNF1-related protein kinases, which are implicated in sugar signaling in yeast and in plants phosphorylate enzymes including HMG-CoA reductase, nitrate reductase, and sucrose phosphate synthase (Sugden et al., 1999).
A set of sugar-repressed genes involved in ubiquitin-dependent protein degradation rose toward the end of the night, including ubiquitin-conjugating enzyme UBC17, five F-box proteins that are components of E3 ligases, and two BTB/POZ proteins that assemble with cullins to form E3 ligases (Gingerich et al., 2005). These results indicate that incipient sugar depletion triggers a reprogramming of targeted protein degradation. Another sugar-regulated gene whose expression rose toward the end of the night was autophagy protein 8e, a member of a gene family that plays a key role in the assembly of autophagosomes for nontargeted protein degradation (Thompson and Vierstra, 2005). APG8 is activated by the ATP-conjugating enzyme AGP7. It has previously been shown that APG7 and APG8 transcripts rise during senescence and that apg7 mutants show early senescence (Doelling et al., 2002).
A set of sugar-induced genes involved or implicated in DNA, RNA, and protein synthesis are induced during the day, indicating that falling sugars repress these processes during the night. Candidates include two DNA licensing factors, two histone deacetylases, nucleolus organizers (two transducins and NOP56; Calikowski et al., 2003), a DNA-dependent RNA polymerase I, genes involved in RNA processing, including FIB2 (Saez-Vasquez et al., 2004), ribosome organizers (two brix domain-containing proteins; Eisenhaber et al., 2001), elongation factor 1B α-subunit 1, CNX1, and several chaperonins.
DISCUSSION
A Large Proportion of the Genes That Are Expressed in Rosettes Show Diurnal Changes of Their Transcript Levels
Between 30 and 50% of the genes expressed in rosettes undergo reproducible diurnal changes of their transcript levels in Arabidopsis wild-type plants growing in a 12-h-light/12-h-dark cycle with an ample supply of nutrients and water. The amplitude varied from small to >100-fold, with ∼44, 18, and 4% of expressed genes having a maximum >50, >100, and >300% above the minimum. In a study with a 7.8K array, Schaffer et al. (2001) reported that 11% of genes show diurnal changes in expression. The larger number found in our experiments may reflect the higher quality of the ATH1 array and the use of more time points and biological triplicates.
Diurnal changes are especially frequent and large for genes assigned to sucrose and starch metabolism, nutrient acquisition and assimilation, and redox regulation. However, almost all functional areas contain genes with marked diurnal changes of expression. The most frequent time for the maximum is the end of the night, followed by the end of the day. Relatively few genes peak during the day and even less during the night. This differs from a free-running circadian cycle, where most genes peak during the subjective day or night (Harmer et al., 2000; see below for further discussion).
Many genes that are known to be induced by low sugar peak at the end of the night, indicating that carbohydrate levels decline to critical levels by the end of the night. Genes involved in photosynthesis tend to peak at the end of the night or start of the day, nitrate and sulfate assimilation at the end of the night, and genes involved in the synthesis of sucrose and organic acids toward the end of the day. However, genes assigned to a particular metabolic function often show diverging diurnal responses (see also Smith et al., 2004). This may be because annotations based on sequence indicate a general function rather than the precise role. Interestingly, transcripts for genes required for cell division, chromosome replication, and DNA, RNA, and protein synthesis peak toward the end of the day, whereas a set of genes that may be involved in expansion growth peak at the end of the night. This is accompanied by reciprocal changes of genes assigned to auxin and cytokinin sensing, which peak at the end of the night, and ABA synthesis and sensing, which peak during the day. This might speculatively be linked to changes in water deficit (see Introduction). It is also striking that a set of sugar-regulated genes involved in targeted and general protein degradation are induced toward the end of the night.
The Major Inputs of the Diurnal Regulation of Gene Expression Are from Sugars and the Circadian Clock
As outlined in the Introduction, changes of irradiance drive complex diurnal changes of sugars, nitrogen metabolites, and leaf water status. Published and in-house expression profiles were inspected to identify sets of diagnostic genes whose expression is markedly altered by each of these inputs. Comparison of the diurnal changes of the transcripts and the corresponding input indicated that sugars make a major contribution to the diurnal changes of gene expression, while light, nitrogen metabolites, and water deficit may make smaller contributions. For sugars, similar results were obtained when diagnostic genes were identified from experiments in which sugars were added to seedlings, and physiological treatments were used to generate changes of endogenous sugar levels in rosettes. This underscores the robustness of the approach. To assess the contribution of circadian regulation, a subset of 373 genes was identified from a 7.8K expression profile (Harmer et al., 2000). Most underwent marked diurnal changes in Col-0, identifying circadian regulation as a second major input to the diurnal regulation of expression in a light/dark cycle.
The Contribution of Different Inputs in Multifactorial Interactions Can Be Visualized by Displaying Weightings of Genes in Principle Components
The same data set was analyzed using a second approach, which provides a compact visualization of the relative importance of different inputs in a multifactorial situation without prior assumptions about the timing of the inputs. Principle component analysis was used to separate samples harvested at different times of the diurnal cycle. The weightings of genes in the principle components were then inspected to discover if genes that are induced and repressed by a given input have high and opposite weightings in one of the principle components. By inspecting the principle component plot, it is also possible to check if this component separates samples according to the expected intensity of the input.
This approach confirmed that the global changes of expression in Col-0 reflect a balance between sugars and circadian regulation, while light, nitrogen, and water deficit make a much smaller contribution. Most glucose-induced and -repressed genes had high and opposite weightings in the first component, as did most carbon fixation–induced and –repressed genes, but only a small proportion of genes that are induced or repressed by light, nitrogen, or water deficit. Many circadian-regulated genes also separated in accordance with their responses in a free-running cycle. The separation of samples in the first component was in good agreement with the levels of sugars at the time of harvest. Many circadian-regulated genes and some light-induced and -repressed genes had high and opposite weightings in the second component.
Genetic Evidence for a Major Role for Sugars in the Diurnal Regulation of Gene Expression
The starchless pgm mutant lacks the buffering effect of diurnal starch turnover (see Introduction) and experiences much larger diurnal changes of sugars than Col-0. These are accompanied by amplified diurnal changes of transcripts for thousands of genes. Almost every diagnostic gene identified by correlative evidence as responding to changes of sugars during the diurnal cycle in Col-0 showed a qualitatively similar but quantitatively amplified diurnal change in pgm. This provides strong evidence that expression of these genes is responding to diurnal changes of sugars. A further set of genes that did not show diurnal changes in Col-0 showed diurnal changes in pgm. This is consistent with them responding when sugars move outside the range found during the diurnal cycle in wild-type plants.
When an expanded data set including samples from pgm was subjected to principle component analysis, the weightings of the diagnostic sugar-induced and -repressed genes separated even more clearly in the first component. Genes that are induced and repressed by light, nitrogen metabolites, and water deficits were not separated or were only separated in the second component. This analysis confirms that diurnal changes of sugars play the dominant role when the buffering effect of transitory starch accumulation is absent, displacing other inputs into a secondary role.
Expression Responds More Sensitively to Low Than to High Sugars during the Diurnal Cycle
Our analysis of diurnal changes of gene expression underscores the importance of sugar regulation for the daily orchestration of metabolism and physiology. Responses to sugars have been divided into feast and famine programs, reflecting responses to excessive sugar and to sugar starvation (Koch, 1996). There has been considerable interest in the genetic and molecular analysis of responses to high sugar (Gibson, 2005; Rolland and Sheen, 2005). Comparison of the diurnal changes in Col-0 and pgm allows us to assess the global impact on expression when endogenous sugar levels temporarily rise above or fall below the range found in a normal diurnal cycle.
At night, when pgm has lower levels of sugars than Col-0, there are large changes of transcript levels for thousands of genes. At a gene-to-gene level, the changes match those predicted from the response to low sugar. In the light, when pgm has much higher levels of sugars than wild-type plants, the global changes of transcript levels are rather small and do not match those predicted from the sugar responsiveness of individual genes. The average transcript level across the entire diurnal cycle also matched the response predicted from the response of the gene to low, not high, sugar. These results show that global gene expression is rapidly and strongly modified when endogenous sugar levels fall to low levels but is not widely affected when sugars exceed the levels found in a wild-type diurnal cycle. A small group of genes, which are highly sensitive to repression by sugars, was more strongly repressed in pgm than Col-0 in the light.
A short period of sugar starvation triggers a rapid inhibition of growth, which is not immediately reverted when carbon becomes available again (Gibon et al., 2004a). Within hours, carbon depletion triggers a widespread reprogramming of expression affecting hundreds of genes involved in carbohydrate production and utilization (Contento et al., 2004; Thimm et al., 2004). The results in this article show that many sugar-regulated genes start to respond to falling sugars toward the end of the night in wild-type plants, even in an undisturbed diurnal cycle. This response anticipates the massive changes in the starchless pgm mutant (see above) or after a short extension of the night (Thimm et al., 2004). There is an interesting contrast between the sensitivity, speed, and breadth of the response of gene expression to low sugar and the more limited response to high sugar. A period of high sugar is probably much less stressful for a plant than a period of low sugar. Similarly, a period of darkness is not a stress, providing the carbohydrate supply is maintained until the plant is reilluminated. In agreement, light per se makes a relatively small contribution to the changes of expression during a diurnal cycle.
Interactions between Sugar and Circadian Regulation
The data were analyzed to reveal how the two major inputs, sugars and circadian regulation, interact during the diurnal cycle. Approximately half of the circadian-regulated genes showed a very similar response in Col-0 and pgm. This shows that the clock is unaffected by a major disruption of carbon metabolism, in which sugars are exhausted 6 to 8 h earlier in the 24-h cycle. An analogous shortening of the light regime would have led to a marked shortening of the clock period (Harmer et al., 2001).
Sugars do, however, interact with clock output pathways. Approximately one-quarter of the circadian-regulated genes show quantitative changes of their diurnal response (i.e., increased amplitude without a change of phase), which are consistent with a quasi-additive interaction between sugars and circadian regulation. Another one-quarter of the circadian-regulated genes show a qualitatively different response in a light/dark cycle compared with published data for their response in a free-running cycle (viz a marked shift of 8 to 12 h in the timing of the maximum). Intriguingly, expression of many of these genes responds to sugar, and the phase shift matches that expected if the diurnal changes of sugar in a 12-h-light/12-h-dark cycle override circadian regulation. More research is needed to understand the biological significance of this interaction. Two possibilities are that daylength modulates the response to changes in sugar levels or that this interaction allows a more sensitive response when changes in the growth conditions lead to a change in the diurnal responses of sugars.
Biological Significance of Diurnal Changes of Transcripts
Diurnal changes of transcripts provide information about the changing output of networks that regulate gene transcription. If the transcription of a gene is regulated by a particular input, and this input changes during the diurnal cycle, the corresponding transcript will exhibit diurnal changes. However, these changes do not necessarily have a direct impact on biological processes. This will require changes in the level of the encoded protein and will depend on the rate of protein synthesis and turnover.
Diurnal changes of transcripts are followed by changes in the levels of many proteins that are closely related to clock function, such as CCA1, LHY, TOC1, and ELF3 (Wang and Tobin, 1998; Kim et al., 2003; Mas et al., 2003), proteins that mediate clock-dependent responses to the photoperiod, such as CCR2/ATGRP7 (Heintzen et al., 1997) or CONSTANS (Hayama and Coupland, 2003), and some genes in metabolism, for example, NIA (Scheible et al., 1997b; Gibon et al., 2004b). Interestingly, however, transformants that constitutively overexpress NIA show similar changes of NIA protein (Vincentz and Caboche, 1991; Ferrario et al., 1995), emphasizing the critical contribution of posttranscriptional and transcriptional regulation.
In other cases, diurnal changes of transcripts do not lead to diurnal changes of the encoded protein. A set of 23 enzymes in central carbon and nitrogen metabolism showed marked changes of transcripts but smaller and slower changes of activity, both in the diurnal cycle and after transfer to continuous darkness (Gibon et al., 2004b). Large diurnal changes of transcripts encoding enzymes involved in starch breakdown are not accompanied by marked changes of the encoded proteins (Smith et al., 2004; Lu et al., 2005). For such genes, diurnal changes of transcription will not modulate fluxes or other biological processes within the same 24-h cycle. This is not necessary if the encoded protein or other proteins in the pathway are allosterically or posttranslationally regulated. Changes of protein might even be counterproductive: they would waste energy, and the protein level would be inappropriately adjusted to the previous rather than the momentary situation. Rather, diurnal changes in the level of thousands of transcripts may allow various external and internal inputs to be sensed in a recurrent and integrative manner, resulting in gradual changes in the levels of the slowly turning-over proteins if a modified response is sustained over several days (Gibon et al., 2004b).
The results in this article highlight the importance of the response to falling sugars at the end of the night. Functional analysis of genes and proteins that show an early and strong response to falling sugars could reveal how carbohydrate formation and utilization is balanced. By combining the large and independent data sets generated in this study, it was possible to identify a robust set of candidate regulatory genes, which undergo large diurnal changes of expression that were very likely to be triggered by endogenous changes of sugars. These included many putative transcription factors, protein kinases, genes involved in the regulation of DNA, RNA, and protein synthesis, and components of the ubiquitin-dependent pathway that will facilitate the targeted degradation of specific proteins as well as components for nontargeted protein degradation.
METHODS
Plant Growth
Arabidopis thaliana ecotype Col-0 and pgm (Col-0 background) were grown on soil in 12 h light/12 h dark at a light intensity of 130 μmol/m2 s (Thimm et al., 2004) for 5 weeks, at which time flowering had not commenced. Plants were harvested by transferring rosettes into liquid nitrogen under ambient irradiance. Typically, five replicate samples, each containing three rosettes, were collected 4, 8, and 12 h into the light period and 4, 8, and 12 h into the night. Three independent biological experiments were performed at 2-month intervals for wild-type Col-0. For pgm, biological replicates were obtained for the end of the day and end of the night and single samples after 4 and 8 h of light and 4 and 8 h of dark. For the low [CO2] treatment, four replicates with three plants were harvested after 4 h of illumination under ambient [CO2] and under low [CO2] and after 4 h of prolonged darkness under low [CO2]. The CO2 concentration was adjusted to <50 ppm 30 min before the start of the light period. The experiment was repeated twice at an interval of 2 months.
Arabidopsis seedlings were grown in liquid culture as described by Scheible et al. (2004). Treatments to alter the nitrogen supply were exactly as described by Scheible et al. (2004). Treatments to alter sugars were performed in a similar manner, expect that in this case the seedlings were transferred on day 7 to medium from which sucrose was omitted, and on day 9 the seedlings were provided with 15 mM glucose or 15 mM sucrose and harvested 3 h later. By day 9, sugar and starch levels had fallen to low levels (data not shown). To identify water deficit–responsive genes, seedlings were provided with 100 mM mannitol for 3 h. This concentration does not plasmolyze the seedlings (data not shown).
Extraction and Assay of Metabolites
Sucrose, glucose, and fructose were determined in ethanol extracts as described by Geigenberger et al. (1996), starch as described by Hendriks et al. (2003), glucose-6-phosphate as described by Gibon et al. (2002), and nitrate according to the nitrate determination kit of Roche Diagnostics (number 1746081). Assays were prepared in 96-well microplates using a Multiprobe II pipetting robot (Perkin-Elmer). The absorbance was read at 340 nm or at 570 nm in a Synergy or an ELX-800-UV microplate reader (Bio-Tek Instruments).
RNA Isolation and Expression Analysis with 22K Affymetrix Arrays
Isolation of total RNA, cDNA synthesis, cRNA labeling, and the hybridization on the GeneChip Arabidopsis ATH1 genome array was done as described by Scheible et al. (2004) and Thimm et al. (2004) and recommended by the manufacturer (part number 900385; Affymetrix UK). The microarray suite software package (MAS 5.0; Affymetrix) was used to evaluate probe set signals of the array. The generated data files (CEL) were the input for the software package RMAExpress to normalize and estimate signal intensities (Bolstad et al., 2003). This data set was reduced from 22,746 to 13,690 probe sets for the wild-type diurnal cycle, which were called present at least once in each of the three biological replicated sets of six time points. For the pgm data set, 16,552 probe sets passed this filter.
Functional Categories and Gene Classification
The display and gene classifications are based on MapMan (Thimm et al., 2004; http://gabi.rzpd.de/projects/MapMan/, version 1.4.3, mapping file version 5.01).
Statistical Analysis
Standard operations, including the calculation of amplitudes, regression plots, and correlation coefficients, were performed in Microsoft Excel. Tables for assigning Affymetrix probe set identifiers to Arabidopsis Genome Initiative codes were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org). Table associations were done with Microsoft Access.
Three biological replicates were performed for all time points in the diurnal cycle in Col-0, and the data were statistically analyzed using three different approaches. The first compared the individual values at the times when transcripts for a given gene were at their maximum and minimum, using a t test of log2-scaled values under FDR control (Benjamini and Hochberg, 1995). Of the 13,690 genes called present, 8680, 3308, and 1619 genes showed significant changes at FDR-controlled P-values of <0.05, <0.01, and <0.005 (see Supplemental Table 1 online). This approach uses the two most divergent of a large number of pair-wise comparisons. The second approach used experiment-wise correlation of the time series to assess reproducibility across all six time points. For each gene, Pearson correlation coefficients were calculated from three pair-wise plots of the six signals and then averaged (see Supplemental Table 1 online). The correlation coefficient was high for almost every gene that had a high maximum/minimum log ratio. The coefficient varied from −0.5 to +0.9 for genes with a low maximum/minimum log ratio (see Supplemental Figure 1 online). As random noise should generate a similar distribution of negative and positive correlation coefficients, >+0.5 was taken as a cutoff. This threshold was exceeded by 6755 of 7591 genes whose maximum/minimum log ratio was >0.5 and by 2647 of the 6199 genes that had a smaller maximum/minimum log ratio. The third approach used an average periodogram and FDR-controlled P-values for detecting significant sinusoidal gene activity calculated using the software package GeneTS (Wichert et al., 2004) implemented in the R software environment for statistical computing (R version 1.9; http://www.r-project.org/) running on a Microsoft Windows 2000 computer system. The average periodogram and P-values for periodic gene expression in wild-type Arabidopsis are listed in Supplemental Figure 1 and Supplemental Table 1 online. With FDR-controlled P-values of 0.05 or 0.1 as filters, 4596 and 6104 genes were classified as showing a diurnal change. This may be an underestimate because GeneTS will not recognize nonsinusoidal changes and noise will mask small changes. When genes identified with GeneTS were compared with those that passed the primary filter (correlation coefficient >0.5), GeneTS identified 84 to 94% of the genes where the secondary filter for the amplitude [log2 (maximum/minimum)] was >1 but less if the amplitude was smaller.
Two biological replicates were performed for the end of the night and end of the day for pgm, which correspond to the times when the vast majority of genes in this mutant show their diurnal maximum or minimum. The evaluation of which genes show an amplitude of the diurnal change was based on replicated results in both experiments. All experiments to define sets of the 400 genes that respond most strongly to different challenges were also performed in duplicate, and all selected genes responded in both replicates.
K-means clustering with euclidean distance and principle component analysis of variance were performed with the software tool TIGR MeV (Saeed et al., 2003; http://www.tigr.org/software/tm4/mev.html) after mean scaling and log2 transformation. For small gene sets, the figure of merit was visually inspected to obtain the point where more clusters do not significantly contribute to gene separation and to avoid the use of too many clusters. For each gene, clustering was performed on the average values (for Col-0, the average of three biological replicates, for pgm, the average of duplicates at the end of the night and the end of the day, and single determinations at the other four time points) at each of the six times during the diurnal cycle, normalized to the average level in the diurnal cycle.
Accession Numbers
The raw data of the transcript profiles described in Methods were deposited in the public Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) under the accession codes GSE3416 (Col-0 wild-type diurnal rhythm), GSE3424 (Col-0 pgm mutant diurnal rhythm), and GSE3423 (Col-0 carbon fixation and light regulation experiment).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Expression Profiling Data for Wild-Type Col-0 and pgm.
Supplemental Table 2. MapMan Files for Amplitudes of Diurnal Gene Expression in Wild-Type Col-0 and pgm.
Supplemental Table 3. MapMan Files for Timing of Diurnal Gene Expression in Wild-type Col-0 and pgm.
Supplemental Table 4. Correlations between Three Independent Wild-Type Col-0 Experiments.
Supplemental Table 5. MAS 5 Call Statistics for the Col-0 Wild-Type Experiment.
Supplemental Table 6. Lists of Top 200 Most Strongly Induced and 200 Most Strongly Repressed Genes of Various Treatments (Diagnostic Sets).
Supplemental Table 7. Expression Profiling Data in Darkness and after 4 h Illumination at Ambient or Compensation Point [CO2] in Wild-Type Col-0.
Supplemental Table 8. Shared Responses of Top 200 Carbon Fixation– and Glucose-Responsive Genes.
Supplemental Table 9. Genes with Significantly Shifted Average Expression in pgm when Compared with Wild-Type Col-0.
Supplemental Table 10. Genes with Significantly Shifted Phase between Wild-Type Col-0 and pgm.
Supplemental Table 11. Response of Circadian Genes in the Diurnal Cycle of Wild-Type Col-0 and pgm.
Supplemental Table 12. Shared Genes in Diagnostic Sets.
Supplemental Table 13. Specific Sugar-Regulated Genes Involved in Signaling.
Supplemental Figure 1. Supporting Material to the Statistical Analysis in Table 1.
Supplemental Figure 2. MapMan Screenshots of the Diurnal Rhythm.
Supplemental Figure 3. Clustering of Sets of Diagnostic Genes Based on Their Diurnal Response in Wild-Type Col-0.
Supplemental Figure 4. Comparison of the Average Transcript Level across the Entire Diurnal Cycle in Wild-Type Col-0 and pgm.
Supplemental Figure 5. Clustering of Sets of Diagnostic Genes Based on Their Diurnal Responses in a Combined Data Set for Wild-Type Col-0 and pgm
Supplemental Figure 6. Phase Shift between a Free-Running and a Diurnal Cycle for Genes That Peak at Six Different Times during the Diurnal Cycle and Relation to the Sugar Sensitivity of the Genes.
Supplemental Figure 7. Comparison of the Amplitudes of the Diurnal Changes of Sucrose-Responsive, Photomorphogenetic Light–Responsive, and Glucose-Responsive Genes to the Background Amplitudes of All Measured Genes on the ATH1 Array.
Supplemental Figure 8. MapMan Screenshots Displaying the First and Second Principle Components Separating Wild-Type Col-0 Samples Collected at Different Times of the Diurnal Cycle in MapMan.
Supplemental Figure 9. Weightings in the First and Second Principle Components of Genes That Are Responsive to Glucose, Carbon Fixation, Light, Nitrogen, Water Stress, and Circadian Regulation.
Supplementary Material
Acknowledgments
This research was supported by the Max Planck Society and the Bundesministerium für Bildung und Forschung–funded project Genomanalyse im Biologischen System Pflanze Verbund Arabidopsis III “Gauntlets, Carbon and Nutrient Signaling: Test Systems, and Metabolite and Transcript Profiles” (0312277A). We thank Florian Wagner at the Resource Center and Primary Database (Berlin, Germany) for carrying out the microarray hybridizations, the bioinformatics group of Joachim Selbig at the Max Planck Institute of Molecular Plant Physiology for helpful discussions, and Stacy Harmer (University of California, Davis, CA) for helpful comments on the manuscript.
The author responsible for distribution of materials integral to findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Oliver E. Bläsing (blaesing@mpimp-golm.mpg.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035261.
References
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19 185–193. [DOI] [PubMed] [Google Scholar]
- Calikowski, T.T., Meulia, T., and Meier, I. (2003). A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90 361–378. [DOI] [PubMed] [Google Scholar]
- Caspar, T., Huber, S.C., and Somerville, C.R. (1985). Alterations in growth, photosynthesis and respiration in a starch deficient mutant of Arabidopsis thaliana (L.) Heynh deficient in chloroplast phosphoglucomutase. Plant Physiol. 79 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Quiel, J.A., Smolen, G.A., Merrikh, H., Silvestro, A.R., Normanly, J., and Bender, J. (2005). The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento, A.L., Kim, S.J., and Bassham, D.C. (2004). Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd, A.N., Salathia, N., Anthony Hall, A., Réka Tóth, E.-K., Ferenc Nagy, F., Julian, M., Hibberd, J.M., Andrew, J., Millar, A.J., and Webb, A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309 630–633. [DOI] [PubMed] [Google Scholar]
- Dietrich, P., Sanders, D., and Hedrich, R. (2001). The role of ion channels in light-dependent stomatal opening. J. Exp. Bot. 52 1959–1967. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, F., Wechselberger, C., and Kreil, G. (2001). The Brix domain protein family–A key to the ribosomal biogenesis pathway? Trends Biochem. Sci. 26 345–347. [DOI] [PubMed] [Google Scholar]
- Doelling, J.H., Walker, J.M., Friedman, E.M., Thompson, A.R., and Vierstra, R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277 33105–33114. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Valadier, M.-H., Morot-Gaudry, J.-F., and Foyer, C.H. (1995). Effects of constitutive expression of nitrate reductase in transgenic Nicotiana plumbaginifolia L. in response to varying nitrogen supply. Planta 196 288–294. [Google Scholar]
- Fondy, B.R., and Geiger, D.R. (1985). Diurnal changes of allocation of newly fixed carbon in exporting sugar beet leaves. Plant Physiol. 78 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger, P., Lerchl, J., Stitt, M., and Sonnewald, U. (1996). Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ. 19 43–55. [Google Scholar]
- Geigenberger, P., and Stitt, M. (2000). Diurnal changes in sucrose, nucleotides, starch synthesis, and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 23 795–806. [DOI] [PubMed] [Google Scholar]
- Geiger, D.R., and Servaites, J.C. (1994). Diurnal regulation of photosynthetic carbon metabolism in C3 plants. Annu. Rev. Plant Biol. 45 235–256. [Google Scholar]
- Geiger, D.R., Servaites, J.C., and Fuchs, M.A. (2000). Role of starch in carbon translocation and partitioning at the plant level. Aust. J. Plant Physiol. 27 571–582. [Google Scholar]
- Gibon, Y., Bläsing, O., Palacios, N., Pankovic, D., Hendriks, J.H.M., Fisahn, J., Höhne, M., Günter, M., and Stitt, M. (2004. a). Adjustment of diurnal starch turnover to short days: A transient depletion of sugar towards the end of the night triggers a temporary inhibition of carbohydrate utilisation, leading to accumulation of sugars and post-translational activation of ADP glucose pyrophosphorylase in the following light period. Plant J. 39 847–862. [DOI] [PubMed] [Google Scholar]
- Gibon, Y., Bläsing, O.E., Hannemann, J., Carillo, P., Höhne, M., Hendriks, J.H., Palacios, N., Cross, J., Selbig, J., and Stitt, M. (2004. b). A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon, Y., Vigeolas, H., Tiessen, A., Geigenberger, P., and Stitt, M. (2002). Sensitive and high throughput metabolic assays for inorganic pyrophosphate, ADPglc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J. 30 221–235. [DOI] [PubMed] [Google Scholar]
- Gibson, S.I. (2005). Control of plant development and gene expression by sugar signaling. Curr. Opin. Plant Biol. 8 93–102. [DOI] [PubMed] [Google Scholar]
- Gingerich, D.J., Gagner, J.M., Salter, D.W., Hellmann, H., Estelle, M., Ma, L.G., and Vierstra, R.D. (2005). Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac (BTB) protein family to form essential ubiquitin-protein ligases (E3s) in Arabidopsis. J. Biol. Chem. 280 18810–18821. [DOI] [PubMed] [Google Scholar]
- Green, R.M., Tingay, S., Wang, Z.Y., and Tobin, E.M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Panda, S., and Kay, S.A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17 215–253. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2003). Shedding light on the circadian clock and the photoperiodic control of flowering. Curr. Opin. Plant Biol. 6 13–19. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., Nater, M., Apel, K., and Staiger, D. (1997). AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks, J.H.M., Kolbe, A., Gibon, Y., Stitt, M., and Geigenberger, P. (2003). ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 133 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi, R., Girke, T., Bray, E.A., and Bailey-Serres, J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38 823–839. [DOI] [PubMed] [Google Scholar]
- Kehr, J., Hustiak, F., Walz, C., Willmitzer, L., and Fisahn, J. (1998). Transgenic plants changed in carbon allocation pattern display a shift in diurnal growth pattern. Plant J. 16 497–503. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., Song, H.R., Taylor, B.L., and Carre, I.A. (2003). Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J. 22 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, M. (1993). Dynamic analyses of water relations and leaf growth in cucumber plants under midday water deficit. Biotronics 22 73–85. [Google Scholar]
- Kleinow, T., Bhalerao, R., Breuer, F., Umeda, M., Salchert, K., and Koncz, C. (2000). Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 23 115–122. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Biol. 47 509–540. [DOI] [PubMed] [Google Scholar]
- Lu, Y., Gehan, J.P., and Sharkey, T.D. (2005). Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 138 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas, P., Kim, W.Y., Somers, D.E., and Kay, S.A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426 567–570. [DOI] [PubMed] [Google Scholar]
- McDonald, A.J.S., and Davies, W.J. (1996). Keeping in touch: Responses of whole plant to deficits in water and nutrient supply. Adv. Bot. Res. 22 229–300. [Google Scholar]
- Matt, P., Geiger, M., Walch-Liu, P., Engels, C., Krapp, A., and Stitt, M. (2001. a). The diurnal changes of nitrogen metabolism in leaves of nitrate-replete tobacco are driven by an imbalance between nitrate reductase activity and the rate of nitrate uptake and ammonium and glutamine metabolism during the first part of the light period. Plant Cell Environ. 24 177–190. [Google Scholar]
- Matt, P., Geiger, M., Walch-Liu, P., Engels, C., Krapp, A., and Stitt, M. (2001. b). Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ. 24 1119–1137. [Google Scholar]
- Matt, P., Schurr, U., Krapp, A., and Stitt, M. (1998). Growth of tobacco in short day conditions leads to high starch, low sugars, altered diurnal changes of the Nia transcript and low nitrate reductase activity, and an inhibition of amino acid synthesis. Planta 207 27–41. [DOI] [PubMed] [Google Scholar]
- Price, J., Laxmi, A., St Martin, S.K., and Jang, J.C. (2004). Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, F., and Sheen, J. (2005). Sugar sensing and signaling networks in plants. Biochem. Soc. Trans. 33 269–271. [DOI] [PubMed] [Google Scholar]
- Saeed, A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Salah, H.B.H., and Tardieu, F. (1997). Control of leaf expansion rate of droughted maize plants under fluctuating evaporative demand: A superposition of hydraulic and chemical messages? Plant Physiol. 114 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez, J., Caparros-Ruiz, D., Barneche, F., and Echeverria, M. (2004). A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 24 7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Accerbi, M., Simon, V., Larson, M., and Wisman, E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Gonzalez-Fontes, A., Lauerer, M., Muller-Rober, B., Caboche, M., and Stitt, M. (1997. a). Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.-R., Gonzales-Fontes, A., Morcuende, R., Lauerer, M., Geiger, M., Glaab, J., Schulze, E.D., and Stitt, M. (1997. b). Tobacco mutants with a decreased number of functional nia-genes compensate by modifying the diurnal regulation transcription, post-translational modification and turnover of nitrate reductase. Planta 203 305–319. [DOI] [PubMed] [Google Scholar]
- Scheible, W.-R., Krapp, A., and Stitt, M. (2000). Reciprocal changes of phosphenolpyruvate carboxylase and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ. 23 1155–1167. [Google Scholar]
- Scheible, W.-R., Morcuende, R., Czechowski, T., Osuna, D., Fritz, C., Palacios-Rojas, N., Thimm, O., Udvardi, M.K., and Stitt, M. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes and signaling infrastructure by nitrogen. Plant Physiol. 136 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmundt, D., Stitt, M., Jähne, B., and Schurr, U. (1998). Quantitative analysis of the local growth rates of leaves at a high temporal and spatial resolution using image sequence analysis. Plant J. 16 505–514. [Google Scholar]
- Schultz, T.F., and Kay, S.A. (2003). Circadian clocks in daily and seasonal control of development. Science 301 326–328. [DOI] [PubMed] [Google Scholar]
- Schurr, U. (1998). Growth physiology: Approaches to a spatially and temporally varying problem. Prog. Bot. 59 355–373. [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 279–292. [DOI] [PubMed] [Google Scholar]
- Smith, S.M., Fulton, D.C., Chia, T., Thorneycroft, D., Chapple, A., Dunstan, H., Hylton, C., Zeeman, S.C., and Smith, A.M. (2004). Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and post-transcriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136 2687–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt, M. (1996). Metabolic regulation of photosynthesis. In Advances in Photosynthesis, Vol. 3, Environmental Stress and Photosynthesis, N. Baker, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 151–190.
- Stitt, M., Huber, S., and Kerr, P. (1987). Control of photosynthetic sucrose synthesis. In The Biochemistry of Plants, Vol. 10, M.D. Hatch and N.K. Boardman, eds (San Diego, CA: Academic Press), pp 327–409.
- Stitt, M., and Krapp, J.A. (1999). The molecular physiological basis for the interaction between elevated carbon dioxide and nutrients. Plant Cell Environ. 22 583–622. [Google Scholar]
- Sugden, C., Crawford, R.M., Halford, N.G., and Hardie, D.G. (1999). Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 19 433–439. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Hudson, M.E., Khanna, R., Zhu, T., Chang, S.H., Wang, X., and Quail, P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38 725–739. [DOI] [PubMed] [Google Scholar]
- Thimm, O., Blaesing, O.E., Gibon, Y., Nagel, A., Meyer, S., Krueger, P., Selbig, J., Mueller, L.A., Rhee, S.Y., and Stitt, M. (2004). Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37 914–939. [DOI] [PubMed] [Google Scholar]
- Thompson, A.R., and Vierstra, R.D. (2005). Autophagic recycling: Lessons from yeast help define processes in plants. Curr. Opin. Plant Biol. 8 165–173. [DOI] [PubMed] [Google Scholar]
- Thum, K.E., Shin, M.J., Palenchar, P.M., Kouranov, A., and Coruzzi, G.M. (2004). Genome-wide investigation of light and carbon signaling interactions in Arabidopsis. Genome Biol. 5 R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel, B., et al. (2005). Extension of the visualisation tool MapMan to allow statistical analysis of arrays, display of co-responding genes and comparison with known responses. Plant Physiol. 138 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz, M., and Caboche, M. (1991). Constitutive expression of nitrate reductase allows normal growth and development of Nicotiana plumbaginifolia plants. EMBO J. 10 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wang, R., Okamoto, M., Xing, X., and Crawford, N.M. (2003). Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 132 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, A.A.R. (2003). The physiology of circadian rhythms in plants. New Phytol. 160 281–303. [DOI] [PubMed] [Google Scholar]
- Wichert, S., Fokianos, K., and Strimmer, K. (2004). Identifying periodically expressed transcripts in microarray time series data. Bioinformatics 20 5–20. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Northrop, F., Smith, A.M., and ap Rees, T. (1998). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15 357–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.