Abstract
Winter-annual accessions of Arabidopsis thaliana are often characterized by a requirement for exposure to the cold of winter to initiate flowering in the spring. The block to flowering prior to cold exposure is due to high levels of the flowering repressor FLOWERING LOCUS C (FLC). Exposure to cold promotes flowering through a process known as vernalization that epigenetically represses FLC expression. Rapid-cycling accessions typically have low levels of FLC expression and therefore do not require vernalization. A screen for mutants in which a winter-annual Arabidopsis is converted to a rapid-cycling type has identified a putative histone H3 methyl transferase that is required for FLC expression. Lesions in this methyl transferase, EARLY FLOWERING IN SHORT DAYS (EFS), result in reduced levels of histone H3 Lys 4 trimethylation in FLC chromatin. EFS is also required for expression of other genes in the FLC clade, such as MADS AFFECTING FLOWERING2 and FLOWERING LOCUS M. The requirement for EFS to permit expression of several FLC clade genes accounts for the ability of efs lesions to suppress delayed flowering due to the presence of FRIGIDA, autonomous pathway mutations, or growth in noninductive photoperiods. efs mutants exhibit pleiotropic phenotypes, indicating that the role of EFS is not limited to the regulation of flowering time.
INTRODUCTION
In plants, successful reproduction is dependent on flowering at the correct time. Plants monitor both environmental and internal signals in order to ensure that reproduction occurs at the appropriate time of year and stage of development. Many species have evolved pathways that sense environmental cues, such as daylength and temperature, and endogenous signals, such as plant age, to regulate the timing of the floral transition. In Arabidopsis thaliana, a facultative long-day plant, flowering is accelerated by environmental factors such as long days (LD) and prolonged exposure to cold (the process by which exposure to cold promotes flowering is known as vernalization). In addition, the autonomous floral promotion pathway and the plant hormone gibberellin promote flowering largely in response to developmental signals (reviewed in Boss et al., 2004; Putterill et al., 2004; He and Amasino, 2005).
Much natural variation in flowering habit exists among Arabidopsis accessions. One component of this variation is the degree to which vernalization promotes flowering. Many winter-annual accessions are late flowering unless vernalized, whereas rapid-cycling accessions flower rapidly in the absence of cold treatment. The genetic difference between these vernalization responses is often due to allelic variation at FRIGIDA (FRI) and/or FLOWERING LOCUS C (FLC) (Burn et al., 1993; Lee et al., 1993, 1994; Clarke and Dean, 1994; Koornneef et al., 1994). Both FRI and FLC activity are required for late flowering. FLC, a MADS box transcription factor, is a floral repressor (Michaels and Amasino, 1999; Sheldon et al., 1999), and FRI upregulates FLC expression to a level that inhibits flowering (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). Most winter-annual accessions have dominant alleles of FRI and FLC, whereas most rapid-cycling accessions that have been examined contain mutations that eliminate FRI or FLC activity (Johanson et al., 2000; Gazzani et al., 2003; Michaels et al., 2003).
In addition to the positive regulation of FLC by FRI, a group of seven genes known collectively as the autonomous pathway act to repress FLC expression. In rapid-cycling accessions that lack FRI activity, the autonomous pathway genes promote flowering by suppressing FLC expression. Thus, in rapid-cycling backgrounds, mutations in autonomous pathway genes lead to elevated FLC levels and a late-flowering phenotype. In winter-annual accessions, the repression of FLC by the autonomous pathway is overridden by FRI (i.e., FRI is epistatic to the autonomous pathway). Vernalization results in a permanent epigenetic repression of FLC expression despite the presence of autonomous pathway mutations or FRI; therefore, winter annuals or rapid-cycling accessions containing autonomous pathway mutations flower rapidly after vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999). Hence, the regulation of FLC expression is the convergence point of FRI, the autonomous pathway, and vernalization.
Recent studies have begun to reveal the molecular mechanisms that control FLC expression. Genetic and molecular studies have shown that FRI, the autonomous pathway, and vernalization all influence the state of FLC chromatin (reviewed in He and Amasino, 2005). In FRI-containing winter annuals, the level of trimethylation of histone H3 at Lys 4 (H3-K4) of FLC chromatin is elevated (He et al., 2004). The autonomous pathway repressors FLD and FVE are involved in deacetylating FLC chromatin (He et al., 2003; Ausin et al., 2004; Kim et al., 2004). Vernalization leads to repressive histone modifications, such as dimethylation of histone H3 at Lys 9 and Lys 27 of FLC chromatin (Bastow et al., 2004; Sung and Amasino, 2004). Thus, chromatin modification is emerging as a major regulator of FLC expression.
Genetic analyses of mutations that render a FRI-containing winter-annual line early flowering have led to the identification of several loci that are required for FLC expression: PHOTOPERIOD INDEPENDENT EARLY FLOWERING1 (PIE1) (Noh and Amasino, 2003), VERNALIZATION INDEPENDENCE4 (VIP4) (Zhang and van Nocker, 2002), VIP3 (Zhang et al., 2003), EARLY FLOWERING7 (ELF7) (He et al., 2004), ELF8/VIP6 (He et al., 2004; Oh et al., 2004), and VIP5 (Oh et al., 2004). ELF7, ELF8, VIP4, and VIP5 are likely to form an RNA Polymerase II Associated Factor 1 (PAF1)-like complex that promotes FLC expression (He et al., 2004; Oh et al., 2004). In yeast, the PAF1 complex promotes gene expression in part by recruiting a histone H3-K4 methyl transferase–containing complex to target gene chromatin (Krogan et al., 2003a; Ng et al., 2003). Increased levels of histone H3-K4 trimethylation is often associated with actively transcribed genes (Krogan et al., 2003a; Ng et al., 2003). Similar to the yeast PAF1 complex, the PAF1-like complex in Arabidopsis may also recruit an H3-K4 methyl transferase to FLC to regulate its expression.
In this report, we present the identification and characterization of a putative histone H3-K4 methyl transferase involved in modulating FLC expression: EARLY FLOWERING IN SHORT DAYS (EFS), a relative of the Drosophila melanogaster H3-K4 methyl transferase ABSENT SMALL HOMEOTIC DISCS1 (ASH1). efs mutations suppress FLC expression in FRI-containing or autonomous pathway mutant backgrounds. Lesions in EFS also reduce the level of histone H3-K4 trimethylation in FLC chromatin.
RESULTS
Identification of efs Alleles as Suppressors of FRI
Extensive genetic screens have been effective in identifying a large number of genes that regulate flowering in rapid-cycling Arabidopsis accessions. Less is known regarding the genes that are responsible for the creation of the winter-annual habit because the rapid-cycling accessions do not exhibit elevated FLC expression; thus, mutations that prevent FRI from elevating FLC expression are difficult to identify in screens of such accessions. To identify genes required for the late-flowering habit of winter annuals, a line containing FRI introgressed into the Columbia background (FRI-Col) was mutagenized by fast-neutron radiation and random T-DNA insertions (Michaels and Amasino, 1999). The resulting M2 generations were screened for early-flowering mutants.
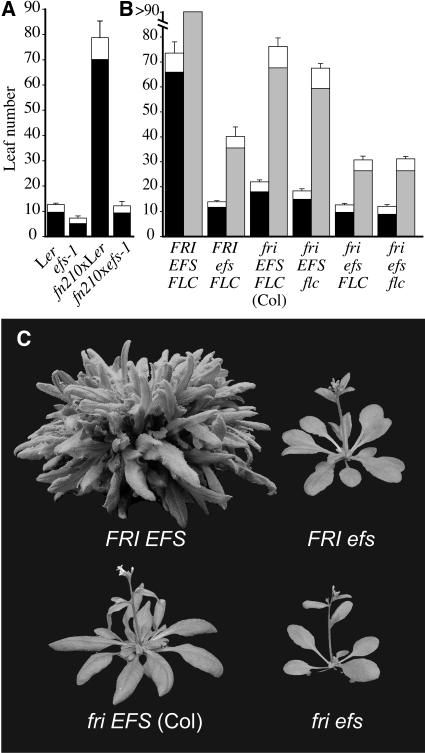
One group of six mutants identified in this screen strongly suppressed the late-flowering phenotype of the FRI-Col line (see below). In addition, these mutants also showed a number of other distinctive phenotypes, such as reduced plant size (the rosette diameter of the mutants was ∼80% of the wild type), leaves that are rounder and slightly paler than the wild type, and reduced fertility (∼80% of wild-type seed set when self-pollinated). Pairwise crosses between these mutants produced early-flowering F1 and F2 plants, indicating that these mutations were allelic. Because the phenotypes of these mutants are similar to that of the previously described efs mutant (Soppe et al., 1999), allelism tests were performed between a fast-neutron allele of this complementation group (fn210) and efs-1, which is in the Landsberg erecta (Ler) genetic background. The F1 plants resulting from the fn210 × efs cross were early flowering, indicating allelism between this complementation group and efs (Figure 1A). Allelism was also confirmed in the F2 generation (all plants were early flowering; data not shown). Thus, these mutants are alleles of efs and were designed as efs-3 through efs-8. efs-3 (fn210) was used in all subsequent experiments.
Figure 1.
efs Mutations Suppress the Late-Flowering Phenotype of FRI.
(A) Allelism tests between efs-1 in the Ler background and fn210 in the FRI-Col background. The closed portion of the bars indicates the number of rosette leaves formed by the primary shoot apical meristem prior to flowering. The open portion of the bars indicates the number of cauline leaves. Error bars indicate 1 sd.
(B) Number of leaves formed prior to flowering for the indicated genotypes. Black and gray bars represent plants grown in LD and SD, respectively. Error bars indicate 1 sd.
(C) The effect of efs mutations on flowering time in FRI-Col background (top) or Col background (bottom). Plants were grown in LD.
EFS Is Required for FLC Expression in Backgrounds Containing FRI or Autonomous Pathway Mutations
efs mutations result in a large reduction in the number of leaves formed prior to flowering in the FRI-Col background (Figures 1B and 1C). Because FRI delays flowering in winter-annual accessions by increasing FLC expression, a possible explanation for the early-flowering phenotype of efs in the FRI-Col background is that EFS is required for elevated FLC expression. To test this hypothesis, FLC mRNA levels were determined in wild-type FRI-Col and efs mutant seedlings. In FRI-Col, as expected, FLC is highly expressed (Figure 2A). In the efs mutant, however, FLC expression is greatly reduced. A previously described flowering-time gene, PIE1, has been shown to be required for the expression of FLC in shoots only (i.e., in pie1 mutants, FLC expression is reduced in the shoot apex, but not in the root apex). To determine if EFS is required for FLC expression in both the shoot and root, FLC:β-glucuronidase (GUS) expression was examined in FRI-Col and efs mutant backgrounds. In the efs mutant, GUS staining is reduced in both the shoot and root apex (Figures 3A and 3B). Thus, unlike PIE1, EFS expression is required for elevated FLC expression throughout the plant. Consistent with EFS acting as a positive regulator of FLC, expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), a promoter of flowering that is negatively regulated by FLC, is also affected by efs mutations. In a FRI-containing background, SOC1 expression is suppressed by high levels of FLC (Figure 3C); however, in a FRI efs background, SOC1 is highly expressed.
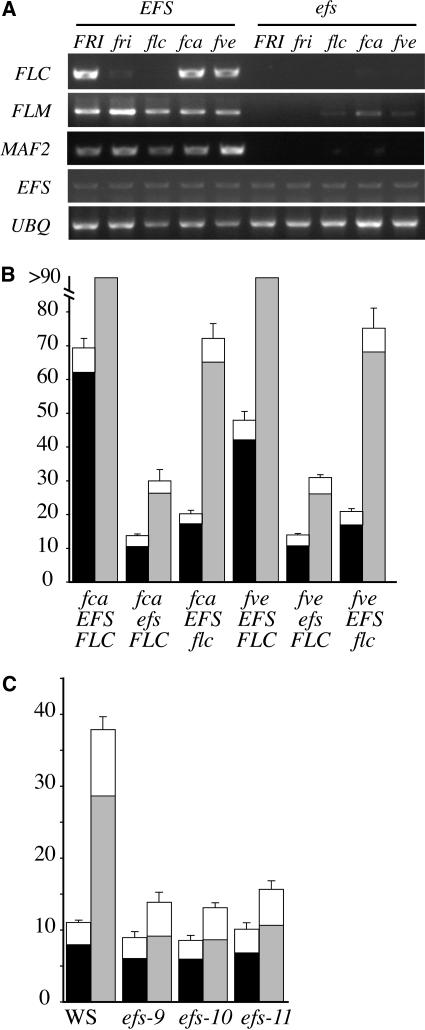
Figure 2.
Effect of EFS on Gene Expression and Flowering Time of Autonomous Pathway Mutants.
(A) RT-PCR analysis of flowering time gene expression in wild-type and efs mutant backgrounds. RNA was isolated from 14-d-old seedlings grown in LD. Tissue was harvested 4 h after lights on. UBIQUITIN (UBQ) was used as a control for loading.
(B) and (C) Effect of efs on the flowering time of fca and fve mutants (B) and the effect of efs alleles in the Ws genetic background (C). The closed portion of the bar indicates the number of rosette leaves formed by the primary shoot apical meristem prior to flowering, while the open portion indicates the number of cauline leaves. Black and gray bars represent plants grown in LD and SD, respectively. Error bars indicate 1 sd.
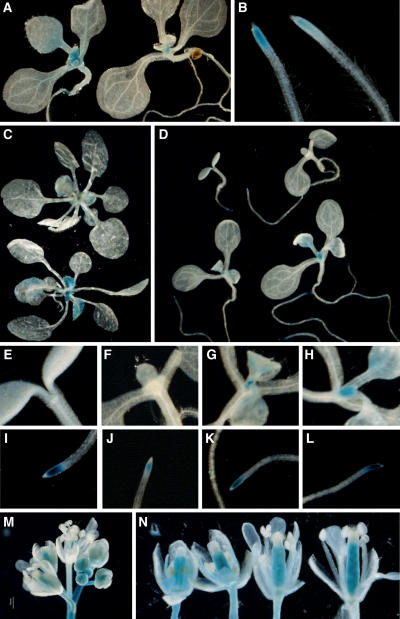
Figure 3.
Histochemical Analysis of Gene Expression.
To minimize variation in GUS staining, plants that appear in the same panel were stained in parallel. Unless otherwise mentioned, all plants were grown in LD and are in the FRI-Col background.
(A) and (B) FLC:GUS expression in shoots and roots in the wild-type FRI-Col (left) or efs mutant background (right).
(C) SOC1:GUS expression in wild-type FRI-Col (top) or efs mutant background (bottom).
(D) EFS:GUS expression during vegetative development. Plants were harvested 2 (top left), 4 (top right), 6 (bottom left), and 8 DAG.
(E) to (L) Higher magnification images of the shoot apex and root apex of the plants shown in (D). 2 DAG ([E] and [I]), 4 DAG ([F] and [J]), 6 DAG ([G] and [K]), and 8 DAG ([H] and [L]).
(M) EFS:GUS expression in the inflorescence.
(N) EFS:GUS expression in flowers.
Like FRI-containing winter annuals, rapid-cycling accessions that contain loss-of-function mutations in autonomous pathway genes are also late flowering due to elevated levels of FLC expression. Previous work has shown that efs mutations effectively suppress the late-flowering phenotype of the autonomous pathway mutations fca and fve in the Ler genetic background (Soppe et al., 1999). Because Ler contains an atypical weak allele of FLC (Michaels et al., 2003), we investigated the ability of efs mutations to suppress the late-flowering phenotype of fca and fve in the Col background, which contains a strong FLC allele. efs was crossed to fve-4 and fca-9, and in the resulting F2 populations, efs fca and efs fve double mutants were isolated that were homozygous for the Col allele of fri (genotypes were verified using molecular markers). In the double mutants, efs strongly suppressed the late-flowering phenotype of fca and fve (Figure 2B). To determine if, as in the FRI-containing background, the efs lesion blocks the expression of FLC, we examined FLC mRNA expression in the efs fca and efs fve double mutants (Figure 2A). In the fca and fve single mutants, FLC mRNA is expressed at high levels (similar to that seen with FRI). In the efs fca and efs fve double mutants, however, FLC expression is strongly suppressed (Figure 2A). Thus, EFS is required for high levels of FLC expression in both FRI-containing lines and autonomous pathway mutants. We also found that efs could not suppress FLC expression from the constitutive cauliflower mosaic virus 35S promoter (data not shown; the construct tested was the complete 35S promoter and 5′ untranslated region joined to start codon of an FLC genomic clone).
FLC-Independent Effects of efs on Flowering Time
To determine if the early-flowering phenotype of efs is solely due to the suppression of FLC expression, the phenotypes of FRI and autonomous pathway mutations in efs and flc mutant backgrounds were compared. If the effect of efs on flowering time is entirely due to the suppression of FLC expression, the flowering time of FRI and autonomous pathway mutations should be similar in the efs and flc mutant backgrounds. In the FRI-Col background, however, efs mutants flower significantly earlier than either a fri null or a fri flc double mutant (Figure 1B). Similarly, efs mutations in the fca and fve mutant backgrounds flower earlier than fca flc and fve flc double mutants (Figure 2B). Thus, the early-flowering phenotype of the efs mutation in FRI and autonomous pathway mutant backgrounds cannot be fully explained by the suppression of FLC expression. This FLC-independent acceleration of flowering is also observed in the absence of FRI or autonomous pathway mutations. Plants containing efs in the Col background (which lacks FRI activity) or in an fl-3 mutant background (which lacks both FRI and FLC activity) flower earlier than wild-type Col or an flc mutant (Figure 1B).
In addition to the efs alleles that were isolated as suppressors of the late-flowering phenotype conferred by FRI (described above), we also isolated three alleles of efs by screening a T-DNA–mutagenized population in the Wassilewskija (Ws) background for early flowering in short days (SD). Like the original efs alleles that were isolated from a similar screen performed in the Ler genetic background, these lines have a strong early-flowering phenotype in SD (Figure 2C). flc-null mutants have not been reported in the Ler or Ws background; therefore, the contribution of the loss of FLC expression to the early flowering in SD cannot be determined in these genetic backgrounds. flc-null mutants in the Col background, however, have only a modest early-flowering phenotype in SD (Michaels and Amasino, 2001), suggesting that the strong early-flowering phenotype of efs in SD cannot be entirely explained by the suppression of FLC. To investigate this possibility, we compared the effects of efs and flc mutations on flowering in SD in the same genetic background. In the Col background, the flc mutant flowered with approximately eight fewer leaves than the wild type, whereas the efs mutant flowered much earlier, forming approximately 40 fewer leaves than the wild type (Figure 1B). Indeed, in all genetic backgrounds tested (Col, FRI-Col, fve, and fca), mutations in efs caused earlier flowering than mutations in flc (Figures 1B and 2B) in SD. Thus, the suppression of FLC by efs accounts for only part of the early-flowering phenotype of the efs mutant in SD.
EFS Is Required for the Expression of Additional MADS Box Transcription Factors in the FLC Clade
Recent work has shown that the genes ELF7 and ELF8/VIP6 (He et al., 2004; Oh et al., 2004) are required for the late-flowering phenotype of winter-annual Arabidopsis, and like efs, elf7/8 mutants are also early flowering in SD (He et al., 2004). elf7/8 mutations lead to decreased H3-K4 trimethylation of FLC chromatin and reduced FLC mRNA levels (He et al., 2004). Interestingly, in elf7/8 mutants, the expression of two other MADS box transcription factors from the FLC clade are also suppressed (He et al., 2004). Loss-of-function mutations in these genes, FLM/MADS AFFECTING FLOWERING1 (MAF1) and MAF2, are early flowering in SD (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001). Thus, it appears that the early-flowering phenotype of elf7/8 mutants in SD may be due to the cumulative effect of suppression of FLC, FLM, and MAF2 (He et al., 2004). To investigate whether EFS might also be required for the expression of FLM and MAF2, mRNA levels were determined in the presence or absence of EFS activity. In all genetic backgrounds tested (FRI-Col, Col, flc, fca, and fve), the efs mutation suppressed the expression of both FLM and MAF2 in addition to FLC (Figure 2A). Thus, like ELF7/8, EFS is required for the expression of a group of related flowering repressors in the FLC clade.
EFS Encodes a SET Domain–Containing Transcription Factor
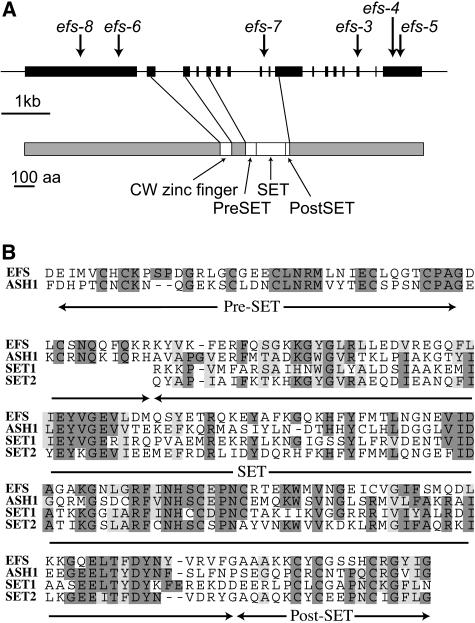
Because the efs-4 through efs-8 mutations were created by T-DNA mutagenesis, these lines were used to identify the EFS gene by isolating genomic DNA flanking the sites of T-DNA integration. Flanking DNA from the efs T-DNA alleles mapped to the predicted coding region of At1g77300 (Figure 4A). At1g77300 is located on the bottom of chromosome I, and its position is consistent with the previously published mapping data for EFS (Soppe et al., 1999). Analysis of the efs-3 allele (generated by fast-neutron radiation) identified a 23-bp deletion (TATTAGAGTATCTTGCCACAAGG) in At1g77300, which creates a frame shift (Figure 4A). As additional proof that lesions in At1g77300 are responsible for the mutant phenotype of efs, efs-3 was transformed with a 12.2-kb genomic DNA fragment that is not predicted to contain any genes other than At1g77300. The resulting T1 plants were late flowering and did not display any of the pleiotropic phenotypes associated with the efs mutation. Thus, At1g77300 encodes EFS. It should also be noted that this gene has also been referred to as SDG8 (Springer et al., 2003).
Figure 4.
The EFS Gene and Predicted Protein.
(A) Schematic representation of the EFS gene (top; black boxes indicate exons) and protein (bottom). Positions of lesions in efs mutants are indicated.
(B) Clustal alignment of the SET, pre-SET, and post-SET domains of EFS, ASH1, SET1, and SET2. Identical residues are shaded in dark gray and similar residues in light gray.
The Arabidopsis Genome Initiative (AGI) annotation predicts that the EFS gene contains 17 exons with 5280 bp of coding sequence (www.arabidopsis.org). To verify the predicted cDNA sequence, the EFS cDNA was amplified by RT-PCR and sequenced. Due to the large size of the transcript, we were unable to amplify the entire transcript in one reaction. Therefore, the EFS cDNA was amplified as a series of overlapping RT-PCR products. The empirically determined cDNA sequence was identical to that predicted by AGI.
The predicted EFS protein is 1759 amino acids in length. BLAST and InterProScan searches identified several domains indicating that EFS is likely to play a role in regulating gene activity by modifying chromatin structure. EFS contains a SET [for Su(var)3-9, Enhancer-of-zeste, Trithorax] domain and two other domains often found with SET domains, a Cys-rich post-SET domain and a pre-SET domain (Figure 4A) (Trievel et al., 2002; Wilson et al., 2002). Many SET domain proteins have been shown to act as histone methyltransferases (e.g., Rea et al., 2000; Nakayama et al., 2001), and the SET domain itself appears to comprise the catalytic site (Xiao et al., 2003). The set domain of EFS is similar to ASH1 in Drosophila and SET1 and SET2 in Saccharomyces cerevisiae (Figure 4B; data not shown). ASH1 and SET1 have been biochemically shown to methylate Lys 4 of histone H3 (Briggs et al., 2001; Roguev et al., 2001; Beisel et al., 2002; Byrd and Shearn, 2003), and SET2 has been shown to methylate Lys 36 of histone H3 (Krogan et al., 2003b). EFS also contains a CW domain, which is predicted to be a four-Cys zinc-finger motif (Perry and Zhao, 2003). CW domains are found in a number of proteins that contain other domains that are involved in binding DNA or chromatin (Perry and Zhao, 2003). These domains suggest a role for EFS in the regulation of gene expression through changes in chromatin structure via histone modifications (see below).
EFS Is Not Regulated by FRI, by Autonomous Pathway Genes, or by Vernalization
Flowering inputs from FRI, the autonomous pathway, and vernalization converge at the level of FLC regulation. FLC is positively regulated by FRI and repressed by autonomous pathway genes and vernalization. Because EFS acts as a positive regulator of FLC, it is possible that one or more of these flowering inputs could regulate FLC levels by modulating EFS expression. To test this model, we examined the EFS mRNA levels in various genetic backgrounds and in response to vernalization. EFS steady state mRNA levels were not affected by FRI, by the autonomous pathway, or by vernalization (Figures 2A and 5A). We also examined the effect of EFS overexpression by placing the EFS gene under control of the strong 35S cauliflower mosaic virus promoter. When placed into efs mutant plants in the FRI background, the 35S:EFS construct restored a late-flowering phenotype, demonstrating that the construct is functional. When placed into the Col background, however, 35S:EFS construct did not cause late flowering (data not shown). Thus, it seems that EFS expression alone is insufficient to activate FLC expression and delay flowering.
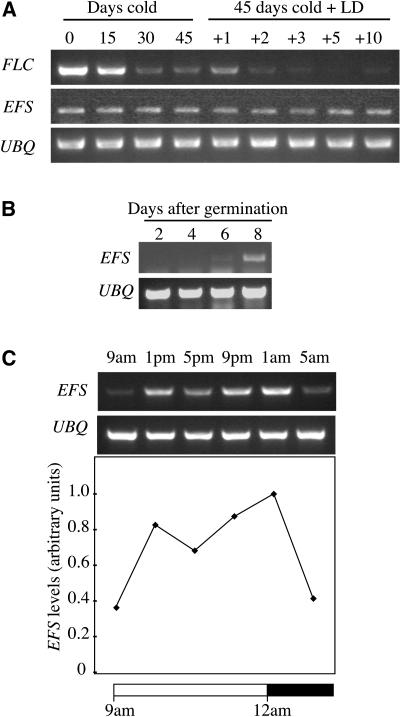
Figure 5.
RT-PCR Analysis of EFS Expression.
(A) EFS and FLC expression during vernalization. Eight-day-old seedlings were exposed to cold for 0, 15, 30, or 45 d and harvested for RNA isolation. Also, seedlings exposed to cold for 45 d were grown at 22°C for 1, 2, 3, 5, or 10 d prior to harvest.
(B) EFS expression during vegetative development. Shoots were harvested at 2, 4, 6, and 8 DAG.
(C) Daily fluctuations in EFS expression. Eight-day-old seedlings were harvested at the indicated times. UBQ was used as a control for loading in all experiments.
EFS Is Developmentally Regulated and Is Preferentially Expressed in the Dividing Cells of Apical Regions
Whereas FLC levels are relatively constant throughout vegetative development (see Supplemental Figure 1 online; Michaels and Amasino, 1999; Sheldon et al., 1999), EFS mRNA levels were found to be developmentally regulated. EFS mRNA levels were low immediately following germination and increased over the first 8 d after germination (DAG) (Figure 5B). EFS mRNA levels were also found to fluctuate over the course of a day, being lower before dawn (Figure 5C).
An EFS:GUS fusion was constructed to investigate the developmental and spatial expression pattern of EFS. Similar to FLC:GUS, EFS:GUS activity was strongest in the shoot and root apex (Figures 3D to 3H) (this expression pattern was verified by RT-PCR; data not shown). Consistent with the RT-PCR data (Figure 5B), EFS:GUS activity was lower in the shoot apex of seedlings 2 and 4 DAG (Figures 3E and 3F) but was detected 6 and 8 DAG (Figures 3G and 3H). Interestingly, however, EFS:GUS activity was detected in the root at all stages tested (Figures 3D and 3I to 3L). EFS:GUS activity was also detected in the inflorescence and in the carpels (Figures 3M and 3N).
EFS Is Required for Elevated Trimethylation of H3-K4 in FLC Chromatin
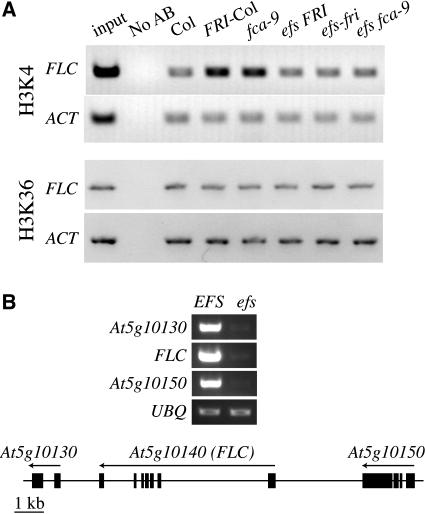
The SET domain of EFS shows amino acid similarity with both SET2, which acts as an H3-K36 methyltransferase, and with ASH1 and SET1, which act as H3-K4 methyltransferases. This suggests that the suppression of FLC expression in efs mutants might be due to a reduction in H3-K4 or H3-K36 methylation. In support of this model, we have recently shown that H3-K4 hypertrimethylation is associated with actively transcribed FLC chromatin (He et al., 2004). To determine if EFS has an effect on histone methylation at the FLC locus, we examined the H3-K4 trimethylation and H3-K36 dimethylation status of FLC chromatin in wild-type and efs mutants. Compared with Col, the trimethylated H3-K4 levels were elevated in a FRI-containing line (FRI-Col) as reported previously (He et al., 2004) and in the fca mutant in which FLC is actively transcribed (Figure 6A); introduction of efs into FRI-Col and fca eliminated the H3-K4 trimethylation increase in FLC chromatin associated with FRI and fca (Figure 6A). These data indicate that EFS is necessary for elevated FLC expression and elevated H3-K4 trimethylation levels. By contrast, no significant changes were observed in H3-K36 dimethylation status in any of the genotypes tested (Figure 6A). Thus, EFS is required for the hypertrimethylation of H3-K4 in FLC chromatin.
Figure 6.
efs Mutations Affect Histone H3-K4 Trimethylation and Gene Expression around the FLC Locus.
(A) Chromatin immunoprecipitation analysis of histone H3-K4 trimethylation and H3-K36 dimethylation state of FLC chromatin in efs and related lines. The input is Col chromatin before immunoprecipitation. “No AB” refers to the control sample lacking the antitrimethyl H3-K4 or antidimethyl H3-K36 antibody. ACTIN (ACT) served as an internal control.
(B) RT-PCR analysis of genes flanking FLC in wild-type FRI-Col and efs mutant backgrounds. UBQ was used as a control for loading.
efs Mutations Suppress the Expression of Genes Flanking FLC
Recent studies have shown that vernalization lead to repressive histone modifications in FLC chromatin (deacetylation of core histones and methylation of H3 at Lys 9 and Lys 27), resulting in epigenetic suppression of FLC (Bastow et al., 2004; Sung and Amasino, 2004). The recent observation that the expression of several genes around the FLC locus is suppressed by vernalization suggests that the vernalization-induced histone modifications may suppress gene expression in a domain surrounding FLC (Finnegan et al., 2004). This model is supported by the fact that the expression of foreign genes inserted into the FLC locus is suppressed by vernalization (Finnegan et al., 2004). We investigated whether an efs mutation would suppress the expression of other genes around the FLC locus. The mRNA levels of At5g10130, FLC (At5g10140), and At5g10150 were determined in wild-type FRI-Col and efs mutant seedlings by RT-PCR. The expression of all three genes is suppressed by the lack of EFS activity. Thus, EFS is also required for the expression of genes flanking FLC.
DISCUSSION
In this study, we have identified and characterized a putative histone H3-K4 methyl transferase that is required for elevated FLC expression in FRI-containing lines and in autonomous pathway mutants. Thus, EFS is required for the vernalization-responsive delayed flowering characteristic of the winter-annual habit. Lesions in EFS suppress the H3-K4 hypertrimethylation of FLC chromatin and prevent FLC expression. In addition to the strong suppression of FLC expression, efs mutants also display FLC-independent effects on flowering time. Under LD or SD, efs mutants flower earlier than flc null mutants; thus, efs mutations must promote flowering through other mechanisms in addition to FLC suppression. In Arabidopsis, there are five FLC relatives, FLM/MAF1, MAF2, MAF3, MAF4, and MAF5 (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001), and two of these, FLM and MAF2, have also been shown to play a role in the regulation of flowering time. Like FLC, FLM and MAF2 act as inhibitors of flowering; mutations in FLM and MAF2 lead to early flowering in both LD and SD (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001; Y. He and R. Amasino, unpublished data). We found that EFS is also required for expression of FLM and MAF2. Thus, it seems likely that EFS may also affect H3-K4 methylation in FLM and MAF2 chromatin to activate expression of these FLC-related genes.
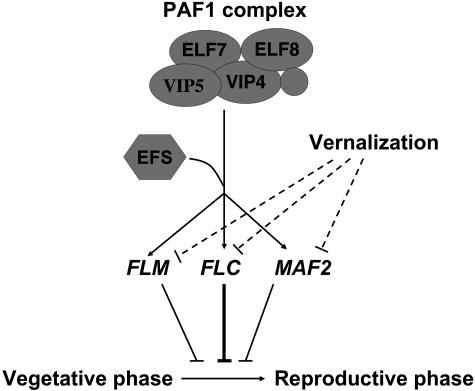
Previously, we have shown that genes encoding components of the PAF1-like complex are also required for the expression and elevated histone H3-K4 trimethylation of FLC clade genes (He et al., 2004). Lesions in ELF7 (encoding the relative of yeast PAF1) and ELF8 (encoding the relative of yeast CTR9) suppress H3-K4 hypertrimethylation in FLC and FLM chromatin and prevent their expression (He et al., 2004). The yeast PAF1 complex is a five-member complex consisting of PAF1, CTR9, LEO1, RTF1, and CDC73 (Krogan et al., 2002; Squazzo et al., 2002). Arabidopsis relatives of yeast LEO1 and RTF1 also have been identified and characterized (Zhang and van Nocker, 2002; Oh et al., 2004). Loss of function of VIP5 (encoding the relative of yeast RTF1) also prevents expression of the FLC clade genes (Oh et al., 2004), and lesions in VIP4 (encoding the relative of yeast LEO1) were shown to prevent FLC expression (Zhang and van Nocker, 2002). Furthermore, it has been shown that VIP4 protein physically interacts with VIP6/ELF8 protein in vivo (Oh et al., 2004). In yeast, the PAF1 complex is required to recruit the SET1 complex (SET1 is a H3-K4 methyltransferase; Santos-Rosa et al., 2002) and SET2 (an H3-K36 methyltransferase; Strahl et al., 2002) to target gene chromatin, resulting in coordinated methylation of H3-K4 and H3-K36 and the promotion of target gene expression (Krogan et al., 2003b; Ng et al., 2003). Thus, it is possible that EFS is a histone methyltransferase recruited by a PAF1 complex to chromatin of FLC clade genes (Figure 7).
Figure 7.
Model of Activation of the FLC Clade Genes by EFS.
EFS protein is recruited to the FLC clade loci by the PAF-like complex and presumably methylates H3-K4, resulting in activating expression of these genes. Vernalization represses expression of the FLC clade genes to accelerate floral transition. Lines with arrows indicate upregulation/activation of gene expression, and lines with bars indicate repression.
Over the entire length of the protein, the closest known relative of EFS in other organisms is Drosophila ASH1, a protein that can methylate H3-K4 (Beisel et al., 2002; Byrd and Shearn, 2003). We have shown that loss of EFS activity leads to reduced levels of H3-K4 trimethylation at FLC. Thus, EFS may act as an H3-K4 methyltransferase at FLC. We cannot rule out the possibility, however, that the primary target(s) of EFS is located elsewhere in the genome and the reduction in H3-K4 methylation seen at the FLC locus in efs mutants might be an indirect effect. Alternatively, the biochemical activity of EFS may be to methylate histones at a position other than H3K4 (ASH1 has also been shown to catalyze H4K20 and H3K9 methylation as well as H3K4), and the changes in H3K4 methylation observed in efs mutants may be a result of coordinated changes in chromatin modifications.
Interestingly, we observed that EFS expression appears to be developmentally regulated. EFS mRNA levels were low immediately following germination and increased over the first 8 DAG, whereas FLC mRNA levels are relatively constant throughout vegetative development. One possibility to account for the different developmental profiles of FLC and EFS is that although EFS expression was lower during early-stage development of seedlings, there still is sufficient EFS protein for full FLC expression. Another possibility is gene redundancy; Arabidopsis has at least nine putative histone H3-K4 methyl transferases (Baumbusch et al., 2001), and additional methyl transferases may be involved in methylating FLC chromatin during early stages of seedling development. Finally, FLC expression during early development may be independent of H3-K4 methyl transferase activity.
METHODS
Plant Materials and Growth Conditions
FRI (Lee et al., 1994), flc-3 (Michaels and Amasino, 1999), fca-9 (Bezerra et al., 2004), and fve-4 (Michaels and Amasino, 2001) are in the Col background and have been described previously. The T-DNA insertional and fast-neutron mutagenized populations used to identify efs alleles have also been described previously (Michaels and Amasino, 1999). Plants were grown under cool-white fluorescent light (∼100 μmol m−2 s−1. LD consisted of 16 h light followed by 8 h darkness, and SD consisted of 8 h light followed by 16 h darkness. A minimum of eight plants per genotype was under each experimental condition.
Gene Expression Analysis
For RT-PCR analysis, RNA isolation, reverse transcription, and PCR were preformed as described previously (Michaels et al., 2004). Primers used for the detection of FLC (Michaels et al., 2004), FLM (Scortecci et al., 2003), MAF2 (He et al., 2004), and UBQ (Michaels et al., 2004) have been described previously. For EFS (5′-CATCAAGTGAAAGTGCCGTGG-3′ and 5′-AGAGGATTTTCTCAGATGGCGAG-3′), At5g10130 (5′-CGTTGGCGTAGTGACGAGTA-3′ and 5′-CGCTTTGGATTCGAGACAAT-3′), and At5g10150 (5′-CCCATTTTCCGAAGAGTCCAAG-3′ and 5′-CGTCGTGGAAGATGTGTAACTC-3′), the indicated primers were used. For all experiments, the data shown are representative of at least three independent experiments.
Constructs
A translational EFS:GUS fusion was created by inserting the GUS gene into the BspEI site in the 14th exon of a genomic EFS clone. The clone contained 3027 bp upstream of the predicted translational start site and 545 bp downstream of the predicted stop codon. A 35S:EFS fusion was created by fusing the 35S cauliflower mosaic virus promoter to the predicted start site of a genomic EFS clone. The construct contained the full-length EFS gene plus 545 bp downstream of the predicted stop codon.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation experiments were performed as described by Johnson et al. (2002) and He et al. (2003) using 10-d-old seedlings. The primer pair CH2 and CH12 as described by He et al. (2003) was used to amplify FLC. Antibodies were obtained from Upstate USA.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g77300 (EFS), At5g10140 (FLC), At4g00650 (FRI), At1g77080 (FLM), At5g65050 (MAF2), At4g16280 (FCA), At2g19520 (FVE), and At2g45660 (SOC1).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Expression of FLC:GUS in a FRI-Containing Background 2 and 6 DAG.
Supplementary Material
Acknowledgments
We thank Mark Doyle for providing an allele of EFS and Edward Himelblau for assistance in generating and screening our T-DNA population. Work in R.M.A.'s lab was supported by the College of Agricultural and Life Sciences, by the Graduate School of the University of Wisconsin, and by National Science Foundation Grants 0446440 and 0209786. Work in S.M.'s lab was supported by the College of Arts and Sciences of Indiana University and by National Science Foundation Grant 0447583.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard Amasino (amasino@biochem.wisc.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.034645.
References
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. [DOI] [PubMed] [Google Scholar]
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002). Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419, 857–862. [DOI] [PubMed] [Google Scholar]
- Bezerra, I.C., Michaels, S.D., Schomburg, F.M., and Amasino, R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40, 112–119. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, S.D., Bryk, M., Strahl, B.D., Cheung, W.L., Davie, J.K., Dent, S.Y., Winston, F., and Allis, C.D. (2001). Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, J.E., Smyth, D.R., Peacock, W.J., and Dennis, E.S. (1993). Genes conferring late flowering in Arabidopsis thaliana. Genetica 90, 147–155. [Google Scholar]
- Byrd, K.N., and Shearn, A. (2003). ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl. Acad. Sci. USA 100, 11535–11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.H., and Dean, C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., Sheldon, C.C., Jardinaud, F., Peacock, W.J., and Dennis, E.S. (2004). A cluster of Arabidopsis genes with a coordinate response to an environmental stimulus. Curr. Biol. 14, 911–916. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Gendall, A.R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132, 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., and Amasino, R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10, 30–35. [DOI] [PubMed] [Google Scholar]
- He, Y., Doyle, M.D., and Amasino, R.M. (2004). PAF1 complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18, 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Michaels, S.D., and Amasino, R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Hyun, Y., Park, J.Y., Park, M.J., Park, M.K., Kim, M.D., Lee, M.H., Moon, J., Lee, I., and Kim, J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36, 167–171. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W., and Peeters, T. (1994). The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild type. Plant J. 6, 911–919. [Google Scholar]
- Krogan, N.J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., Greenblatt, J.F., and Shilatifard, A. (2003. a). The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Kim, M., Ahn, S.H., Zhong, G., Kobor, M.S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J.F. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., Kim, M., Tong, A., Golshani, A., Cagney, G., Canadien, V., Richards, D.P., Beattie, B.K., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J. (2003. b). Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23, 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Bleecker, A., and Amasino, R. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Lee, I., Michaels, S.D., Masshardt, A.S., and Amasino, R.M. (1994). The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6, 903–909. [Google Scholar]
- Michaels, S., and Amasino, R. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Bezerra, I.C., and Amasino, R.M. (2004). FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., He, Y., Scortecci, K.C., and Amasino, R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. (2003). Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719. [DOI] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S., Zhang, H., Ludwig, P., and van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16, 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J., and Zhao, Y. (2003). The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Laurie, R., and Macknight, R. (2004). It's time to flower: The genetic control of flowering time. Bioessays 26, 363–373. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., and Riechmann, J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., and Jenuwein, T. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Roguev, A., Schaft, D., Shevchenko, A., Pijnappel, W.W., Wilm, M., Aasland, R., and Stewart, A.F. (2001). The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Scortecci, K., Michaels, S.D., and Amasino, R.M. (2003). Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 52, 915–922. [DOI] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., and Amasino, R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W.J., Bentsink, L., and Koornneef, M. (1999). The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126, 4763–4770. [DOI] [PubMed] [Google Scholar]
- Springer, N.M., Napoli, C.A., Selinger, D.A., Pandey, R., Cone, K.C., Chandler, V.L., Kaeppler, H.F., and Kaeppler, S.M. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132, 907–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo, S.L., Costa, P.J., Lindstrom, D.L., Kumer, K.E., Simic, R., Jennings, J.L., Link, A.J., Arndt, K.M., and Hartzog, G.A. (2002). The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., Grant, P.A., Briggs, S.D., Sun, Z.W., Bone, J.R., Caldwell, J.A., Mollah, S., Cook, R.G., Shabanowitz, J., Hunt, D.F., and Allis, C.D. (2002). Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Trievel, R.C., Beach, B.M., Dirk, L.M., Houtz, R.L., and Hurley, J.H. (2002). Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111, 91–103. [DOI] [PubMed] [Google Scholar]
- Wilson, J.R., Jing, C., Walker, P.A., Martin, S.R., Howell, S.A., Blackburn, G.M., Gamblin, S.J., and Xiao, B. (2002). Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell 111, 105–115. [DOI] [PubMed] [Google Scholar]
- Xiao, B., Wilson, J.R., and Gamblin, S.J. (2003). SET domains and histone methylation. Curr. Opin. Struct. Biol. 13, 699–705. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Ransom, C., Ludwig, P., and van Nocker, S. (2003). Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch FLOWERING LOCUS C. Genetics 164, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and van Nocker, S. (2002). The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 31, 663–673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.