Abstract
A short exposure to light in the middle of the night causes inhibition of flowering in short-day plants. This phenomenon is called night break (NB) and has been used extensively as a tool to study the photoperiodic control of flowering for many years. However, at the molecular level, very little is known about this phenomenon. In rice (Oryza sativa), 10 min of light exposure in the middle of a 14-h night caused a clear delay in flowering. A single NB strongly suppressed the mRNA of Hd3a, a homolog of Arabidopsis thaliana FLOWERING LOCUS T (FT), whereas the mRNAs of OsGI and Hd1 were not affected. The NB effect on Hd3a mRNA was maximal in the middle of the 14-h night. The phyB mutation abolished the NB effect on flowering and Hd3a mRNA, indicating that the NB effect was mediated by phytochrome B. Because expression of the other FT-like genes was very low and not appreciably affected by NB, our results strongly suggest that the suppression of Hd3a mRNA is the principal cause of the NB effect on flowering in rice.
INTRODUCTION
Transition from the vegetative phase to the reproductive phase is one of the most important developmental switches in plants. The floral induction is controlled by genetic as well as environmental factors, such as light and temperature (Koornneef et al., 1998; Araki, 2001; Boss et al., 2004). Among them, the photoperiod, or daylength, is one of the most important regulators of flowering for successful reproduction. Plants adjust their flowering time by measuring the daylength, which changes depending on the time of the year at a particular location, to survive most successfully (Yanovsky and Kay, 2003; Jack, 2004; Searle and Coupland, 2004). Flowering plants can be classified into three groups depending on their responses to the photoperiod: long-day plants (LDPs), which promote flowering under long-day (LD) conditions; short-day plants (SDPs), which flower faster under short-day (SD) conditions; and day-neutral plants, which flower independently of the photoperiod (Thomas and Vince-Prue, 1997).
Extensive molecular–genetic studies on the photoperiodic control of flowering in Arabidopsis thaliana, an LDP, have revealed a number of genes that play important roles in the measurement of daylength (Koornneef et al., 1998; Yanovsky and Kay, 2003; Searle and Coupland, 2004; Corbesier and Coupland, 2005). They are categorized into three groups based on their general functions. The first group includes genes involved in the regulation of circadian clocks. It became clear that circadian clocks play an important role in the determination of flowering because many mutations that have defects in the circadian clock function show altered control of flowering (Yanovsky and Kay, 2003). The second group of genes that have important functions in the photoperiodic control of flowering includes those encoding photoreceptors (Lin, 2000). They receive light signals and transduce them to the genes that have more direct effects on the regulation of flowering. The third group includes those whose functions are more or less specific to the regulation of daylength-controlled flowering. They include GIGANTEA (GI) (Fowler et al., 1999; Park et al., 1999), CONSTANS (CO) (Putterill et al., 1995; Suarez-Lopez et al., 2001), and FLOWERING LOCUS T (FT) (Kardailsky et al., 1999; Kobayashi et al., 1999), and their mRNA levels show diurnal changes over a 24-h period. The role of GI in the control of the circadian clock has also been demonstrated (Park et al., 1999; Mizoguchi et al., 2005). Genes in the third group integrate signals from the circadian clock and photoreceptors and promote or inhibit flowering under appropriate environmental conditions. Regulation of the CO protein, a key regulator of flowering in Arabidopsis, can be achieved at the transcriptional as well as the posttranscriptional level to ensure the precise regulation of flowering time (Imaizumi et al., 2003, 2005; Valverde et al., 2004).
Because Arabidopsis is an LDP, similar molecular–genetic studies on flowering time on an SDP are required to understand the differences between LDPs and SDPs and to study the evolution of the photoperiodic regulation of flowering in plants. Rice (Oryza sativa), a second higher plant whose nearly complete genome information is available, is an SDP and has been used to study the regulation of flowering in SDPs (Yano et al., 2001; Izawa et al., 2003; Hayama and Coupland, 2004). The general conclusions from such studies are that major genes, which play important roles in photoperiodic flowering, are highly conserved in Arabidopsis and rice; the orthologs of Arabidopsis GI, CO, and FT play key roles in flower induction, although their regulation is altered to adjust to flowering under SD conditions (Simpson, 2003; Hayama and Coupland, 2004).
The night break (NB) effect on flowering has been known for a long time, being discovered in 1938 (Hamner and Bonner, 1938). The NB effect on flowering is most evident with SDPs, in which it inhibits flowering by a very short exposure of light during the night (Thomas and Vince-Prue, 1997). The NB effect on LDPs, which is shown as the promotion of flowering, is clearly observed in only a limited number of species and generally requires longer times of light exposure (Thomas and Vince-Prue, 1997). For instance, the NB effect has been examined in various photomorphogenic mutants of Arabidopsis, and clear promotion of flowering was observed only with a 1-h exposure of light given every day in the middle of the night until flowering (Goto et al., 1991). On the other hand, because SDPs are generally sensitive to short NB treatment, various SDPs have been used extensively for NB experiments. Early studies on the light quality required for NB indicated that phytochromes are major photoreceptors used for NB in SDPs (Borthwick et al., 1952). An early indication of the involvement of the circadian clock in the photoperiodic regulation of flowering came from NB studies in various SDPs (Carr, 1952; Coulter and Hamner, 1964; Takimoto and Hamner, 1964). The observation that light has a direct effect on flowering in addition to its effect through the circadian clock came from an NB study using morning glory (Pharbitis nil) (Lumsden and Furuya, 1986). The results of that study supported the original hypothesis proposed by Erwin Bünning (Bünning, 1936), namely, that the time measurement in seasonal responses is operated by the circadian clock. Later, this model was refined to become a hypothesis called an external coincidence model, which essentially proposes that the interaction of the external light signal and a circadian rhythm is the basis of daylength measurements (Pittendrigh and Minis, 1964).
Although numerous physiological studies of flowering time determination have been performed using NB as an experimental tool (Thomas and Vince-Prue, 1997), virtually nothing is known about the molecular basis of the NB effect. The lack of molecular study on the NB has been attributable primarily to the lack of information on genes involved in the daylength control of flowering in SDPs, because they show clear effects on flowering. Although some early studies in morning glory identified genes whose expression patterns were affected by the NB treatment (O'Neill et al., 1994; O'Neill and Zheng, 1998; Sage-Ono et al., 1998b) and orthologs of genes that play important roles in the control of flowering time in Arabidopsis (Liu et al., 2001), molecular–genetic studies of the NB effect on flowering have not yet been reported.
In this study, we used rice, an SDP, to study the NB effect on flowering, because several genes that play important roles in the photoperiodic regulation of flowering in rice were cloned recently and their functions in flowering are becoming clear. We initially established the conditions required for an efficient NB effect on flowering in rice. Studies on the expression of key flowering-time genes after the NB treatment and studies using rice mutants with altered flowering time and for individual phytochrome genes clearly showed that the suppression of Hd3a mRNA is the principal cause of the NB effect on flowering in rice and that phytochrome B transduces the NB signal to inhibit flowering.
RESULTS
Effects of NB on Flowering in Rice
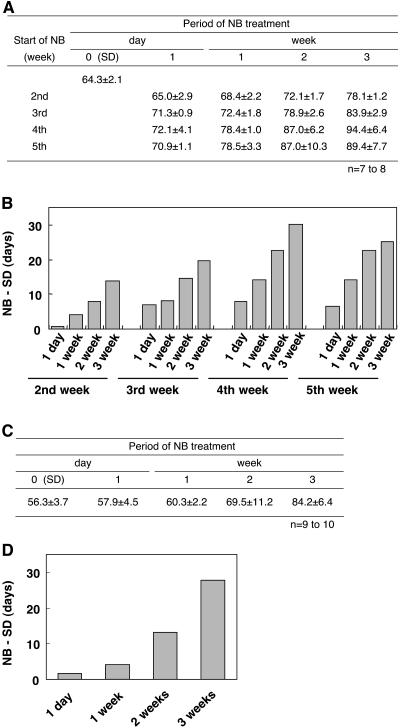
To initiate a molecular–genetic study of an NB response in rice, we first determined which developmental stage was the most sensitive to NB. We germinated rice seeds and grew plants under SD conditions for 2 to 5 weeks before starting the NB treatment. Each NB was scheduled to last 2 h and to begin at 6 h into a 14-h night. We applied NB treatments for various lengths of time, which included 1 d, 1 week, 2 weeks, and 3 weeks. Plants were returned to SD conditions, and the number of days required for flowering was measured. The results (Figures 1A and 1B) showed that the NB effect on the delay in flowering was detectable even at the earliest stage examined; however, the most sensitive developmental stage was found to be 4 to 5 weeks after sowing. The results also showed that even a single 2-h NB treatment could cause a measurable delay in flowering and that the degree of delay in flowering was proportional to the number of nights with an NB.
Figure 1.
Effect of NB on Flowering in Rice.
(A) The flowering times of rice plants treated with a 2-h NB at different developmental stages (2, 3, 4, and 5 weeks after sowing) for different periods (1 d, 1 week, 2 weeks, and 3 weeks) were measured. The flowering times of plants without NB treatment, 0 d (SD), were also measured as a control. Data are means ± sd of seven to eight plants.
(B) Degrees of delay in flowering as expressed by the number of days. The difference was calculated by subtracting values for control plants (SD) from those of NB-treated plants, as shown in (A).
(C) and (D) The flowering times of plants treated with a 10-min NB for various periods are indicated. Data are means ± se of 9 to 10 plants.
Because we found that the 2-h NB had a clear inhibitory effect on flowering in rice, we next tested the effects of a 10-min NB on the inhibition of flowering. The results showed that the 10-min NB had clear effects on flowering when applied for various numbers of days (Figures 1C and 1D). Therefore, although we did not observe complete suppression of flowering in rice by the NB treatment, the results indicate that rice can be used for the molecular–genetic analysis of the NB effect on flowering.
Suppression of Hd3a mRNA but Not of OsGI or Hd1 mRNAs by the NB Treatment
To examine the NB effect on gene expression, we chose three genes that are known to play important roles in the flowering of rice: OsGI (Hayama et al., 2002), Hd1 (Yano et al., 2000), and Hd3a (Kojima et al., 2002). OsGI, Hd1, and Hd3a are orthologs of Arabidopsis GI (Fowler et al., 1999; Park et al., 1999), CO (Putterill et al., 1995; Suarez-Lopez et al., 2001), and FT (Kardailsky et al., 1999; Kobayashi et al., 1999), respectively, and Hd3a has been shown to be a floral activator in rice (Kojima et al., 2002). It was recently shown that, under inductive SD conditions, OsGI activates Hd1 expression and Hd1 activates Hd3a expression (Hayama et al., 2003). Under noninductive LD conditions, Hd1 suppresses Hd3a expression (Hayama et al., 2003). Therefore, this reversal of Hd3a regulation by Hd1 in rice relative to Arabidopsis under LD conditions has been proposed to be the molecular basis of the difference between rice, an SDP, and Arabidopsis, an LDP (Hayama et al., 2003; Simpson, 2003; Hayama and Coupland, 2004).
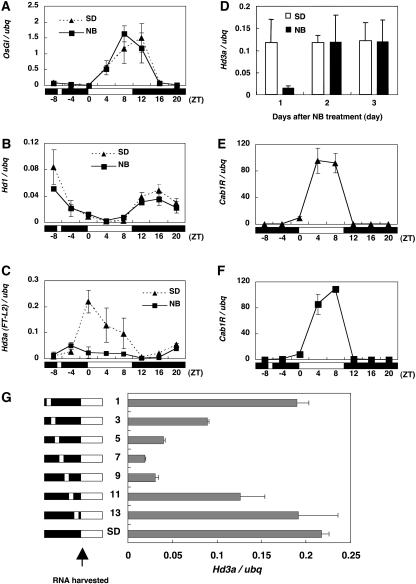
We measured the mRNA levels of the three genes by real-time PCR over a 24-h period under SD conditions in the presence or absence of NB. For the analysis of mRNA expression, we applied 10 min of light at 7 h into the 14-h night. The results showed that a single 10-min NB in the middle of a 14-h night strongly suppressed Hd3a mRNA (Figure 2C). Under normal SD conditions, Hd3a mRNA became visible 4 h before dawn, peaked at dawn, and remained high during the day (Figure 2C). In contrast with Hd3a mRNA, no clear NB effect was found on Hd1 mRNA (Figure 2B). For OsGI mRNA, no suppression was detected, but a shift of the peak toward morning was observed with the NB treatment (Figure 2A). Probably, this was a phase shift caused by the light pulse during the night, because GI has been shown to be controlled by the circadian clock (Park et al., 1999; Mizoguchi et al., 2005) and the phase shift of mRNA by a short light pulse has been shown in rice (Sugiyama et al., 2001). Thus, the NB effect on the suppression of Hd3a mRNA was specific. To determine how long the effect of an NB on Hd3a mRNA suppression continued after a single NB treatment, we examined Hd3a mRNA at dawn at 3 d after the NB treatment. The results showed that the suppression of Hd3a mRNA completely disappeared on the second morning (Figure 2D), suggesting that the Hd3a suppression was reversible and that the Hd3a mRNA level resumed the next day. These results are consistent with those on the NB effect on flowering (Figure 1), showing that the NB effect on flowering was reversible and that the degree of delay in flowering is proportional to the number of nights with an NB.
Figure 2.
Effect of a Single Short (10-min) NB Treatment on Gene Expression, and Determination of the Most Effective Time for Hd3a mRNA Suppression during the Night.
(A) to (C) A single short NB causes the suppression of Hd3a mRNA but has no effects on OsGI or Hd1 mRNA. Quantification of OsGI (A), Hd1 (B), and Hd3a (C) mRNA under SD and NB conditions. The white and black bars at bottom represent the light and dark periods, respectively.
(D) Hd3a mRNA is suppressed by a single 10-min NB but recovers the next day with SD treatment. Leaves of four plants were bulked and harvested at zeitgeber time-0 (lights on) for RNA isolation. Hd3a mRNA was quantified with ubq mRNA. Data are means ± sd of three separate RNA extractions.
(E) and (F) A single short NB causes no effect on Cab1R mRNA under SD (E) and NB (F) conditions. Each mRNA was quantified relative to the ubq mRNA level, and data were standardized by three separate RNA extractions (means ± sd).
(G) The middle of the night is the most sensitive time for NB with regard to Hd3a mRNA suppression. Quantification of Hd3a mRNA after a single 10-min NB at various times during the dark period. Leaves of four plants were harvested and bulked at zeitgeber time-0, and RNA was isolated. Hd3a mRNA was quantified with ubq mRNA. The white and black bars at left represent the light and dark periods, respectively, and the small white bars indicate NB treatments. The numbers shown in the middle of the figure indicate the length of the dark periods in hours before the NB was given. The total length of the night period was 14 h. Data are means ± sd of two separate RNA extractions.
Because the mRNA levels of OsGI, Hd1, and Hd3a are likely to be under the control of circadian rhythms, as shown for the orthologous genes in Arabidopsis, the NB effect on Hd3a mRNA suppression could possibly be mediated by the circadian clocks. To test this idea, we examined the NB effect on the mRNA of Cab1R, a well-characterized clock-controlled gene of rice (Sugiyama et al., 2001). The results showed that there were no differences in the Cab1R mRNA patterns between SD conditions (Figure 2E) and SD conditions in the presence of NB (Figure 2F), indicating that the suppression of Hd3a mRNA was not directly related to changes in the clock function. Therefore, it is possible that light has a direct inhibitory effect on Hd3a mRNA expression during the night.
Maximal Suppression of Hd3a mRNA Occurs in the Middle of the Night
It has been shown that the NB effect on flowering inhibition varies depending on the time during the night and that its maximum sensitivity, referred to as NB-max, occurs in the middle of a 12- to 16-h dark period in various species (Thomas and Vince-Prue, 1997). Therefore, to test whether the NB effect on the suppression of Hd3a mRNA is similarly affected by the time during the night, we applied 10-min NBs at various times in the night and examined their effects on Hd3a mRNA levels at dawn. The results clearly showed that the maximum NB effect on Hd3a suppression occurred in the middle of the night (at 7 h into the 14-h night) and that plants were not sensitive to light at the beginning and end of the night (Figure 2G). This sensitivity curve of Hd3a mRNA suppression during the night was very similar to those obtained in a number of previous experiments performed on the effects of NBs on flowering (Thomas and Vince-Prue, 1997). The clear correlation between NB-max in the suppression of Hd3a mRNA and that in flowering inhibition observed in many SDPs strongly suggests that the primary effect of NB is the suppression of Hd3a mRNA.
Expression of the Other FT-Like Genes Is Not Affected by the NB Treatment
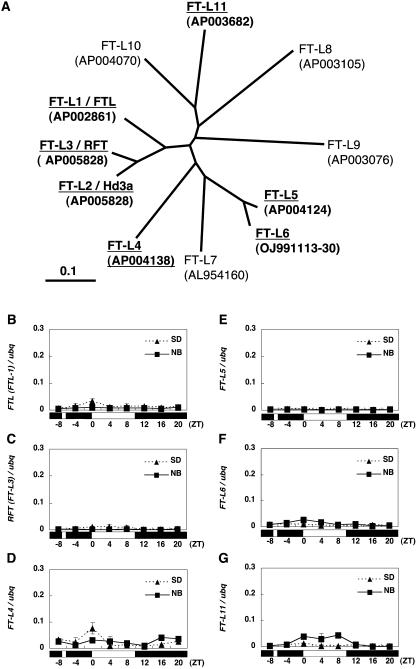
It has been shown that there are 10 FT-like (FT-L) genes in rice (Izawa et al., 2003). More recently, we identified another FT-L gene called FT-L11 (Figure 3A; see Supplemental Figure 1 online). Our RT-PCR analysis of 11 FT-L genes using the wild-type Norin 8 variety indicated that 6 of 11 FT-L genes (FTL/FT-L1, RFT/FT-L3, FT-L4, FT-L5, FT-L6, and FT-L11) showed very low levels of expression at 35 d after sowing under SD conditions, which is the most sensitive stage for NB. No evidence for expression of the four other FT-L genes (FT-L7, FT-L8, FT-L9, and FT-L10) at this developmental stage was obtained after repeated experiments (data not shown). Therefore, we decided to examine whether the expression of six FT-L genes was also suppressed by the NB treatment. As shown in Figures 3B to 3G, expression of the six FT-L genes was very low compared with that of Hd3a under SD conditions, and the NB effect was not clearly observed for their expression. These results suggest that six FT-L genes are not likely to contribute to either flowering under SD conditions or the NB effect on flowering and that Hd3a is the major activator of flowering in rice under SD conditions. Therefore, our results strongly suggest that the suppression of Hd3a mRNA is the major cause of the NB effect on flowering in rice.
Figure 3.
The Other FT-L Genes Are Not Likely to Be Involved in the NB Effect on Flowering.
(A) A phylogenetic tree of rice FT-L genes. ClustalW software was used to make this tree. Seven of 11 FT-L genes (FTL/FT-L1, Hd3a/FT-L2, RFT/FT-L3, FT-L4, FT-L5, FT-L6, and FT-L11) were expressed at 35 d after sowing under SD conditions (underlined).
(B) to (G) Expression of six FT-L genes in the presence and absence of the NB treatment under SD conditions. All expression data were quantified with ubq mRNA. Data were standardized with two separate RNA extractions (means ± sd). The white and black bars at bottom represent the light and dark periods, respectively.
Hd1-Dependent and -Independent Hd3a Activation Are Both Subject to the NB Effect
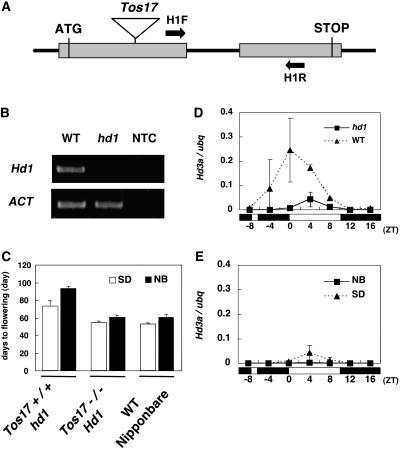
To examine whether Hd1 is required for the NB suppression of Hd3a expression, we used a Tos17-induced mutant of Hd1. This mutation was caused by the insertion of Tos17 (Hirochika, 2001) into exon 1 and was null (Figures 4A and 4B). The mutant flowered later than the wild-type plant under SD conditions (Figure 4C), as previously shown in another mutant of Hd1 (Yano et al., 2000). Interestingly, the hd1 mutant still exhibited an NB effect on flowering: it flowered later in the presence of NB treatment (Figure 4C). In this study, we used a 2-h light exposure. A single 2-h NB caused a clear delay in flowering, and the NB suppression of Hd3a mRNA was essentially similar to that with a 10-min NB (data not shown).
Figure 4.
Hd1-Dependent and -Independent Hd3a Activation Are Both Subject to the NB Effect.
(A) Diagram of the Tos17-induced hd1 mutation.
(B) RT-PCR analysis of the Tos17-induced hd1 mutant. Locations of the primers used are shown in (A).
(C) NB effect on flowering in hd1. Data are means ± sd of 9 to 10 plants.
(D) Quantification of Hd3a mRNAs under SD conditions in hd1. Results for the wild type are also shown. The white and black bars at bottom represent the light and dark periods, respectively. Each mRNA was quantified with ubq mRNA. Data were standardized with two separate RNA extractions (means ± sd).
(E) Quantification of Hd3a mRNAs with a 2-h NB in hd1. Each mRNA was quantified with ubq mRNA. Data were standardized with two separate RNA extractions (means ± sd).
Next, we examined the mRNA level of Hd3a in the hd1 mutant in the absence of NB treatment (Figure 4D). The results showed that in hd1 a low but detectable level of Hd3a expression was observed. The peak of this Hd1-independent Hd3a expression was 4 h later than the major peak in the wild type. When the hd1 mutant was treated with NB, this Hd1-independent Hd3a expression was also suppressed (Figure 4E). These results suggest that the observed NB effect on flowering in the hd1 mutant (Figure 4C) was probably caused by suppression of the Hd1-independent Hd3a expression. The relatively large effect of NB on flowering in hd1 (Figure 4C) could be partly explained by the late flowering of the mutant plants, which may increase the difference between the mutant and the wild type.
Phytochrome B Is Responsible for the NB Effect on Flowering as Well as Hd3a mRNA
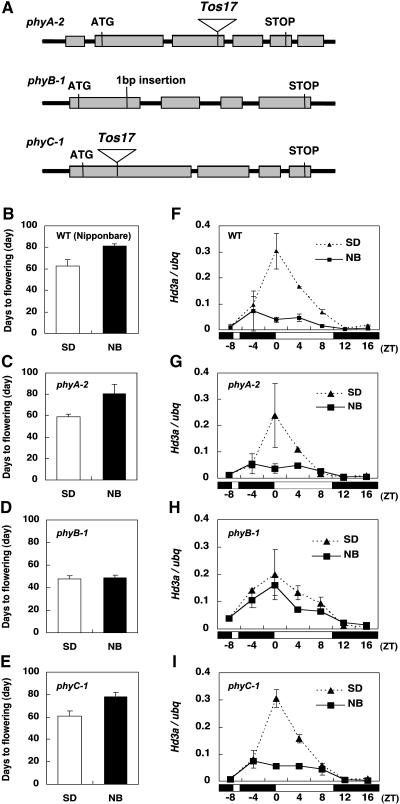
Since the first discovery of the NB effect on flowering, it has been well established that phytochrome is an important photoreceptor for the NB response (Borthwick et al., 1952; Thomas and Vince-Prue, 1997). It was shown recently that phytochromes exert an inhibitory effect on flowering in rice by suppressing Hd3a mRNA under both SD and LD conditions and that this effect is a direct effect of light and is not mediated by the circadian clock (Izawa et al., 2002). Therefore, we wanted to test whether the NB effect on Hd3a expression is mediated by phytochrome. The rice se5 mutant, which has no functional phytochromes because of the mutation in an enzyme for chromophore biosynthesis, is extremely early flowering under both SD and LD conditions (Izawa et al., 2000). In the se5 mutant, the NB effect on flowering was not observed, and no suppression of Hd3a mRNA was detected when a 2-h NB treatment was applied (see Supplemental Figure 2 online). Because mutants of the rice PHYA gene have been available (Takano et al., 2005) and more recently those of PHYB and PHYC genes were also obtained (Takano et al., 2005), we wanted to examine which of the three rice phytochromes is required for the NB effect on flowering and Hd3a suppression. The phyA and phyC mutations were caused by the insertion of Tos17, and the phyB mutation was induced by a single base insertion in exon 1 (Figure 5A). A detailed description and their effects on deetiolation and flowering under LD conditions are given by Takano et al. (2005).
Figure 5.
Phytochrome B Is Responsible for the NB Effect on Flowering as Well as Hd3a mRNA.
(A) Diagram of phy mutants used for the experiments.
(B) to (E) Flowering times of the wild type and phyA, phyB, and phyC mutants that were treated with a 30-min NB. Data are means ± sd of two to seven plants.
(F) to (I) Quantification of Hd3a mRNAs with an NB in the phy mutants and the wild type. Each mRNA was quantified with ubq mRNA. Data were standardized with two separate RNA extractions (means ± sd). The white and black bars at bottom represent the light and dark periods, respectively.
The results on the effects of individual phytochrome mutants on flowering under NB conditions clearly indicated that phyB is responsible for the NB effect on the delay of flowering (Figure 5D). The phyA and phyC mutants showed no effects on the delay of flowering by the NB treatment (Figures 5C and 5E). Similarly, the NB effect on the suppression of Hd3a mRNA was abolished in phyB (Figure 5H), whereas no clear effects were observed in phyA and phyC (Figures 5G and 5I). Some differences were detected in the patterns of OsGI (see Supplemental Figures 3A and 3C online) and Hd1 (see Supplemental Figures 3E and 3G online) mRNA between the wild type and phyB, and possible influences of these changes on flowering and Hd3a expression need to be studied in the future. Together, these results clearly demonstrated that phyB is responsible for the delayed flowering and the Hd3a suppression caused by the NB treatment in rice, supporting the hypothesis that Hd3a suppression is the main cause of the NB effect on flowering.
In addition to its effect on NB, phyB caused early flowering, whereas phyA or phyC showed virtually no effect on flowering time under SD conditions. These results indicate that phyB has an inhibitory effect on flowering under SD conditions in rice.
The NB Effect on the Suppression of Hd3a mRNA Is Possibly at the Transcriptional Level
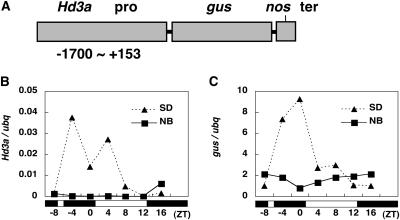
In the absence of NB, the Hd3a mRNA started to increase 4 h before dawn, reached a peak at dawn, and remained high during the day (Figure 2C). After the NB treatment, Hd3a mRNA was almost completely suppressed (Figure 2C). This rapid suppression of Hd3a mRNA could be caused by the suppression of transcription, the posttranscriptional degradation of Hd3a mRNA, or both. To gain insights into the molecular mechanism of the NB effect on Hd3a mRNA, we tried to determine whether the β-glucuronidase (gus) reporter mRNA driven by a 1.7-kb Hd3a promoter exhibited similar suppression to Hd3a mRNA by NB in transgenic rice plants. Analysis of gus mRNA in transgenic rice plants carrying a 1.7-kb Hd3a promoter-gus-nopaline synthase (Nos) terminator (Figure 6A) (Hayama et al., 2003) over a 24-h period showed that its expression pattern was essentially the same as that of Hd3a mRNA under SD conditions (Figures 6B and 6C). These results suggested that the 1.7-kb Hd3a promoter can accurately regulate gus mRNA transcription under SD conditions. Then, we analyzed gus mRNA and found that, after an NB treatment, it showed a similar expression profile to Hd3a mRNA (Figure 6C). Similar results were obtained from another transgenic line (see Supplemental Figure 4 online). These results suggest that the NB effect on Hd3a expression may be primarily at the transcriptional level and not at the posttranscriptional level, because gus mRNA having a 3′ untranslated region from the Nos terminator was also subject to suppression by an NB.
Figure 6.
The NB Effect on the Suppression of Hd3a mRNA Is Possibly at the Transcriptional Level.
(A) Scheme of the Hd3a pro-gus transgene.
(B) Hd3a mRNA levels under SD conditions and with an NB in transgenic line 5. The white and black bars at bottom represent the light and dark periods, respectively.
(C) gus mRNA levels under SD conditions and with an NB in transgenic line 5.
DISCUSSION
Our results showed that a short exposure to light in the middle of the night suppresses the subsequent accumulation of Hd3a mRNA, resulting in a delay in flowering in rice. This suppression of Hd3a mRNA accumulation disappears the next day. Analysis of the Hd3a-gus transgenic rice plants suggested that the suppression of Hd3a mRNA accumulation is primarily caused by a mechanism at the transcriptional level.
Several genes have been isolated previously from morning glory and their mRNA expression patterns examined after a brief NB treatment. For instance, the mRNA level of PnC401, which was increased during the dark period, was reduced by NB treatment (Sage-Ono et al., 1998a). However, PnC401 is a novel gene and its role in flowering remains to be studied. The HMG1 mRNA level was shown to be increased by the NB treatment, but its physiological role is not known (O'Neill and Zheng, 1998). The homolog of the Arabidopsis CO gene, PnCO, has been isolated, and expression analysis indicates that its mRNA level was not altered by the NB treatment (Liu et al., 2001). This is in agreement with our results on the NB effect on Hd1 mRNA level (Figure 2B). Therefore, although the NB effect on mRNA levels of various genes in morning glory has been examined, little is known about the molecular effects of the NB treatment.
Our results clearly demonstrate that phyB is required for the NB to suppress flowering as well as Hd3a mRNA. It has been shown that phyB has a general inhibitory effect on flowering in both LDPs and SDPs (Lin, 2000; Yanovsky and Kay, 2003). An inhibitory effect of phyB on FT expression has been shown in Arabidopsis (Cerdan and Chory, 2003; Halliday et al., 2003; Valverde et al., 2004; Endo et al., 2005), and more recently it was shown that phyB exerts an inhibitory effect on flowering in rice under LD conditions (Takano et al., 2005).
The NB effect on the suppression of Hd3a transcription appears to be mediated by two mechanisms: the major one is Hd1-dependent, and the minor one is Hd1-independent. Therefore, NB may interfere with two transcription complexes on the Hd3a promoter, which can be functionally separated by the requirement of Hd1. It was recently shown that Early heading date1 (Ehd1) encoding the B-type response regulator activates Hd3a expression under SD conditions and that the peak of the Ehd1-dependent Hd3a expression was in the middle of the day (Doi et al., 2004). This expression pattern was similar to that of Hd3a mRNA in the hd1 mutant (Figure 4D). Therefore, it is possible that Ehd1-dependent activation of Hd3a expression is also suppressed by NB. Because it was shown that an Arabidopsis response regulator, ARR4, interacts with the phyB protein (Sweere et al., 2001), it will be of interest to examine whether EHD1 can also interact with phyB and activate Hd3a expression. If that were the case, the EHD1–phyB complex could be a potential target of light during the NB treatment.
One possible mechanism for the suppression of Hd3a mRNA by an NB is either the degradation or destabilization of the transcription machinery acting at the Hd3a promoter by a short light pulse given during the night. It has been shown that phytochrome has a general inhibitory effect on Hd3a mRNA (Izawa et al., 2002). Whether the general inhibition of Hd3a mRNA accumulation by phytochrome and the NB suppression of Hd3a mRNA mediated by phyB are operated by the same molecular mechanism remains to be studied. It was recently shown that the CO protein is destabilized by phyB in the morning but stabilized by phyA and cryptochromes in the morning and evening (Valverde et al., 2004). Furthermore, in the same study, the involvement of the proteasome in the degradation of CO was demonstrated (Valverde et al., 2004). Therefore, it is possible that the NB effect may be regulated by phyB by controlling the stability of the Hd1 protein. In any case, more information on the nature of the transcriptional machinery regulating Hd3a mRNA expression needs to be obtained to better understand the molecular mechanism of the NB effect by phyB on flowering.
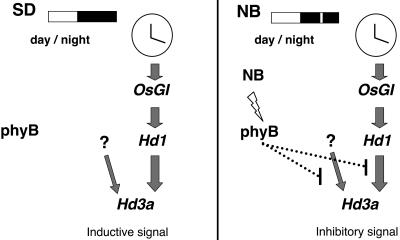
Based on the results we obtained in this study, we propose a model to explain the NB effect on the suppression of flowering in rice (Figure 7). This model is consistent with all of the results we obtained in our NB studies and suggests that Hd3a plays a central role in the NB effect on flowering. This model raises a number of interesting questions regarding the physiology and evolution of the NB effect. First, Hd3a is a key regulator of flowering in rice under both SD and LD conditions, and its expression is suppressed under LD conditions. Therefore, one important issue is whether Hd3a suppression by NB and under LD conditions is caused by the same molecular mechanism. The recent finding that phyB has an inhibitory effect on flowering in rice under LD conditions (Takano et al., 2005) might indicate that with respect to the light quality regulating the photoperiodic flowering they are similar. If this were the case, NB could be used as a tool to study the molecular mechanism of the daylength control of flowering. Second, because the NB effect is highly conserved among SDPs, it would be interesting to determine whether the mRNA of FT orthologs is suppressed by an NB in other SDPs. Third, because many important physiological studies on the NB have been done using morning glory and some genes whose expression patterns are altered by NB treatment have been cloned (O'Neill et al., 1994; Thomas and Vince-Prue, 1997; Sage-Ono et al., 1998b), it would be interesting to determine how the expression of its FT homolog is regulated by the NB. Fourth, the promotion of flowering by NB is also observed in some LDPs, although its effect is not as clear as that in SDPs. Therefore, it would also be interesting to examine whether the expression of FT orthologs is promoted by NB in NB-responsive LDPs. Finally, our results suggest that NB can be used as a tool to study phytochrome signaling in plants.
Figure 7.
Model of the NB Effect on Flowering in Rice.
The NB effect on Hd3a mRNA suppression is mediated by phytochrome B, and its regulation of Hd3a mRNA is possibly at the level of transcription. The NB effect on the suppression of Hd3a transcription may be mediated by two pathways: one is Hd1-dependent, and the other is Hd1-independent. The latter is mediated by an unknown factor. Thus, the phyB-mediated NB effect interferes with two distinct transcription pathways for the Hd3a gene.
METHODS
Plant Materials
Japonica rice (Oryza sativa) varieties Norin 8 and Nipponbare were used as wild-type rice. The se5 mutant was described previously (Izawa et al., 2000). A Tos17-induced mutant of Hd1 (NG6082, Nipponbare) was obtained from the National Institute of Agrobiological Sciences of Japan (Tos17 mutant panel project; http://tos.nias.affrc.go.jp/∼miyao/pub/tos17/). The Tos17 insertion into Hd1 was confirmed by PCR using genomic DNA with the primers shown in Supplemental Table 1 online. Descriptions and some phenotypic characterization of phyA-2, phyB-1, and phyC-1 mutants of rice are provided by Takano et al. (2001, 2005). Flowering time was defined as the time when the first panicle appeared from the node.
Growth Conditions and NB Treatment
Plants were grown in climate chambers at 70% humidity under SD conditions with daily cycles of 10 h of light at 30°C and 14 h of dark at 25°C. Light was provided by fluorescent white light tubes (400 to 700 nm, 100 μmol·m−2·s−1). NB experiments were performed in the climate chamber with fluence rates equivalent to those used on subjective days. The temperature and humidity for NB experiments were the same as those on subjective nights.
Hd3a-gus Transgenic Plants
The Hd3a-gus gene was described previously (Hayama et al., 2003). It was introduced into Agrobacterium tumefaciens for rice transformation. Agrobacterium-mediated transformation of rice was performed according to a published protocol (Hiei et al., 1994). Transformed calli were selected by hygromycin resistance, and plants were regenerated from transformed calli.
RNA Extraction and Real-Time PCR Analysis
Leaves were harvested at the indicated times, and total RNA was extracted using the RNeasy plant mini kit (Qiagen) and treated with DNase I (Invitrogen). The cDNA was synthesized from 1 μg of total RNA using Superscript II reverse transcriptase (Invitrogen). One microliter of cDNA was used for the quantitative analysis of gene expression performed with SYBR Green PCR master mix (Applied Biosystems) with the gene-specific primers listed in Supplemental Table 1 online. Data were collected using the ABI PRISM 7000 sequence detection system in accordance with the instruction manual.
Phylogenetic Analysis
The predicted peptides from FTL/FT-L1, Hd3a/FT-L2, RFT/FT-L3, FT-L4, FT-L5, FT-L6, FT-L7, FT-L8, FT-L9, FT-L10, and FT-L11 are aligned based on analysis using the ClustalW method.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under the following accession numbers: FTL/FT-L1 (AP002861), Hd3a/FT-L2 (AP005828), RFT/FT-L3 (AP005858), FT-L4 (AP005828), FT-L5 (AP004124), FT-L6 (OJ991113-30), FT-L7 (AL954160), FT-L8 (AP003105), FT-L9 (AP003076), FT-L10 (AP004070), and FT-L11 (AP003682).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Primers Used in This Study.
Supplemental Figure 1. Multiple Alignments of 11 FT-L Genes in Rice.
Supplemental Figure 2. The Phytochrome Pathway Is Required for the NB Effect.
Supplemental Figure 3. OsGI and Hd1 mRNA Levels under SD Conditions and with a 30-min NB in the wild type (Nipponbare) and phyA-2, phyB-1, and phyC-1 Mutants.
Supplemental Figure 4. Quantification of gus mRNA under SD Conditions and with a 2-h NB in Transgenic Line 8.
Supplementary Material
Acknowledgments
We thank Toru Ishizuka (Hitachi Central Research Laboratory) for his earlier work of NB experiments on phytochrome mutants and Sawako Kohashi (Nara Institute of Science and Technology) for her technical assistance. We also thank the members of the Plant Molecular Genetics Laboratory at the Nara Institute of Science and Technology for their participation in discussions. This research was supported by Grants-in-Aid for Scientific Research on Priority Areas (Grant 10182102 to K.S.) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Rice Genome Programs (Grant IP-1006 to T.S. and M.T.). S.T. and R.I. were supported by fellowships from the Japanese Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ko Shimamoto (simamoto@bs.naist.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037028.
References
- Araki, T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4, 63–68. [DOI] [PubMed] [Google Scholar]
- Borthwick, H.A., Hendricks, S.B., and Parker, M.W. (1952). The reaction controlling floral initiation. Proc. Natl. Acad. Sci. USA 38, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.), S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünning, E. (1936). Die endogene Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber. Dtsch. Bot. Ges. 54, 590–607. [Google Scholar]
- Carr, D.J. (1952). The photoperiodic behaviour of short-day plants. Physiol. Plant. 5, 70–84. [Google Scholar]
- Cerdan, P.D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. [DOI] [PubMed] [Google Scholar]
- Corbesier, L., and Coupland, G. (2005). Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 28, 54–66. [Google Scholar]
- Coulter, M.W., and Hamner, K.C. (1964). Photoperiodic flowering responses of Biloxi soybean in 72-hour cycles. Physiol. Plant. 39, 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., Yano, M., and Yoshimura, A. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M., Nakamura, S., Araki, T., Mochizuki, N., and Nagatani, A. (2005). Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17, 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, N., Kumagai, T., and Koornneef, M. (1991). Flowering responses to night-breaks in photomorphogenesis mutants of Arabidopsis thaliana, a long-day plant. Physiol. Plant. 83, 209–215. [Google Scholar]
- Halliday, K.J., Salter, M.G., Thingnaes, E., and Whitelam, G.C. (2003). Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J. 33, 875–885. [DOI] [PubMed] [Google Scholar]
- Hamner, K.C., and Bonner, J. (1938). Photoperiodism in regulation to hormones as factors in floral initiation and development. Bot. Gaz. 100, 388–431. [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., Izawa, T., and Shimamoto, K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43, 494–504. [DOI] [PubMed] [Google Scholar]
- Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (2001). Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 4, 118–122. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Tran, H.G., Swartz, T.E., Briggs, W.R., and Kay, S.A. (2003). FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16, 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22, 391–399. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Takahashi, Y., and Yano, M. (2003). Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 6, 113–120. [DOI] [PubMed] [Google Scholar]
- Jack, T. (2004). Molecular and genetic mechanisms of floral control. Plant Cell 16 (suppl.), S1–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Lin, C. (2000). Photoreceptors and regulation of flowering time. Plant Physiol. 123, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Yu, J., McIntosh, L., Kende, H., and Zeevaart, J.A. (2001). Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 125, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden, P.J., and Furuya, M. (1986). Evidence for two actions of light in the photoperiodic induction of flowering in Pharbitis nil. Plant Cell Physiol. 27, 1541–1551. [Google Scholar]
- Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., Mouradov, A., Fowler, S., Kamada, H., Putterill, J., and Coupland, G. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, S.D., and Zheng, C.C. (1998). Abundance of mRNAs encoding HMG1/HMG2 class high-mobility-group DNA-binding proteins are differentially regulated in cotyledons of Pharbitis nil. Plant Mol. Biol. 37, 235–241. [DOI] [PubMed] [Google Scholar]
- O'Neill, S.D., Zhang, X.S., and Zheng, C.C. (1994). Dark and circadian regulation of mRNA accumulation in the short-day plant Pharbitis nil. Plant Physiol. 104, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Pittendrigh, C.S., and Minis, D.H. (1964). The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am. Nat. 108, 261–295. [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Sage-Ono, K., Ono, M., Harada, H., and Kamada, H. (1998. a). Dark-induced accumulation of mRNA for a homolog of translationally controlled tumor protein (TCTP) in Pharbitis. Plant Cell Physiol. 39, 357–360. [DOI] [PubMed] [Google Scholar]
- Sage-Ono, K., Ono, M., Harada, H., and Kamada, H. (1998. b). Accumulation of a clock-regulated transcript during flower-inductive darkness in Pharbitis nil. Plant Physiol. 116, 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle, I., and Coupland, G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23, 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G. (2003). Evolution of flowering in response to day length: Flipping the CONSTANS switch. Bioessays 25, 829–832. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sugiyama, N., Izawa, T., Oikawa, T., and Shimamoto, K. (2001). Light regulation of circadian clock-controlled gene expression in rice. Plant J. 26, 607–615. [DOI] [PubMed] [Google Scholar]
- Sweere, U., Eichenberg, K., Lohrmann, J., Mira-Rodado, V., Baurle, I., Kudla, J., Nagy, F., Schafer, E., and Harter, K. (2001). Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science 294, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Takano, M., Inagaki, N., Xie, X., Yuzurihara, N., Hihara, F., Ishizuka, T., Yano, M., Nishimura, M., Miyao, A., Hirochika, H., and Shinomura, T. (2005). Distinct and cooperative functions of phytochromes A, B and C in the control of de-etiolation and flowering in rice. Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- Takano, M., Kanegae, H., Shinomura, T., Miyao, A., Hirochika, H., and Furuya, M. (2001). Isolation and characterization of rice phytochrome A mutants. Plant Cell 13, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto, A., and Hamner, K.C. (1964). Effect of temperature and preconditioning on photoperiodic response of Pharbitis nil. Plant Physiol. 39, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B., and Vince-Prue, D. (1997). Photoperiodism in Plants. (London: Academic Press).
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Kojima, S., Takahashi, Y., Lin, H., and Sasaki, T. (2001). Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 127, 1425–1429. [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2003). Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 4, 265–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.