Abstract
Secondary walls in vessels and fibers of dicotyledonous plants are mainly composed of cellulose, xylan, and lignin. Although genes involved in biosynthesis of cellulose and lignin have been intensively studied, little is known about genes participating in xylan synthesis. We found that Arabidopsis thaliana fragile fiber8 (fra8) is defective in xylan synthesis. The fra8 mutation caused a dramatic reduction in fiber wall thickness and a decrease in stem strength. FRA8 was found to encode a member of glycosyltransferase family 47 and exhibits high sequence similarity to tobacco (Nicotiana plumbaginifolia) pectin glucuronyltransferase. FRA8 is expressed specifically in developing vessels and fiber cells, and FRA8 is targeted to Golgi. Comparative analyses of cell wall polysaccharide fractions from fra8 and wild-type stems showed that the xylan and cellulose contents are drastically reduced in fra8, whereas xyloglucan and pectin are elevated. Further structural analysis of cell walls revealed that although wild-type xylans contain both glucuronic acid and 4-O-methylglucuronic acid residues, xylans from fra8 retain only 4-O-methylglucuronic acid, indicating that the fra8 mutation results in a specific defect in the addition of glucuronic acid residues onto xylans. These findings suggest that FRA8 is a glucuronyltransferase involved in the biosynthesis of glucuronoxylan during secondary wall formation.
INTRODUCTION
Secondary walls are the major constituent of tracheary elements and fibers in wood, which is the most abundant component of the biomass produced by plants. Study of secondary cell wall biosynthesis is not only important in basic plant biology but also has significant economic and agronomic implications because of extensive use of fiber and wood in our daily life. Secondary walls from wood of dicotyledonous plants are mainly composed of cellulose, xylan, and lignin (Carpita and McCann, 2000). The biosynthesis of cellulose and lignin has been studied extensively; genes encoding putative cellulose synthase catalytic subunits (Taylor et al., 2004) and most genes participating in the monolignol biosynthesis (Boerjan et al., 2003) have been isolated and characterized. By contrast, little is known about genes involved in xylan synthesis in secondary walls.
Xylans are the main hemicellulosic polysaccharides found in secondary walls in dicotyledonous plants. They are composed of a backbone polymer of β-(1,4)–linked d-xylosyl (Xyl) residues with α-d-glucuronic acid (GlcA) and/or 4-O-methyl-α-d-glucuronic acid (4-O-Me-GlcA) residues at O-2 of one out of every 6 to 12 xylosyl residues. The xylosyl residues may also be substituted with short side chains containing l-arabinose and may be acetylated on C-2 or C-3 (Ebringerová and Heinze, 2000; Teleman et al., 2000). The type and distribution of substituents are greatly dependent on species and tissues of origin. The specific roles of the xylan substituents are still unknown, but it has been suggested that the acidic substituents contribute to the formation of a helicoidal orientation of cellulose microfibrils during the secondary cell wall assembly (Reis and Vian, 2004). In primary walls of dicotyledons, xylans contain both acidic residues 4-O-Me-GlcA and GlcA and represent ∼5% of the wall (Darvill et al., 1980; Zablackis et al., 1995). On the other hand, arabinoxylans are one of the major hemicellulosic components of the primary walls of grasses (Aspinall, 1980; McNeil et al., 1984; Ebringerová and Heinze, 2000).
Early biochemical analysis demonstrated that microsomal membrane fractions from etiolated pea (Pisum sativum) epicotyls contain both xylosyltransferase and glucuronyltransferase that catalyze the concomitant biosynthesis of xylan backbone and addition of GlcA side chains, respectively (Waldron and Brett, 1983; Baydoun et al., 1989b). Xylosyltransferase activities involved in xylan synthesis were also detected in a number of other sources, including cultured Zinnia elegans mesophyll cells undergoing xylem differentiation (Suzuki et al., 1991), developing xylem cells of sycamore (Acer pseudoplatanus) and poplar (Populus spp) trees (Dalessandro and Northcote, 1981a, 1981b), and wheat (Triticum aestivum) seedlings (Kuroyama and Tsumuraya, 2001). It has been shown that elongation of the xylan backbone and the addition of GlcA residues onto this backbone occur simultaneously and that little GlcA can be added to preformed xylan (Baydoun et al., 1989b). Despite these biochemical studies, genes encoding xylosyltransferases and glucuronyltransferases involved in glucuronoxylan synthesis are still elusive.
Recently, a number of genes involved in the biosynthesis of plant cell wall polysaccharides, including cellulose, xyloglucan, mannan, and pectin, have been identified (Delmer, 1999; Edwards et al., 1999; Keegstra and Raikhel, 2001; Bouton et al., 2002; Iwai et al., 2002; Madson et al., 2003; Dhugga et al., 2004; Scheible and Pauly, 2004; Somerville et al., 2004; Taylor et al., 2004; Liepman et al., 2005). Two of these genes, tobacco (Nicotiana plumbaginifolia) GUT1 (Iwai et al., 2002) and Arabidopsis thaliana MUR3 (Madson et al., 2003), which have been shown to participate in pectin and xyloglucan synthesis, respectively, belong to glycosyltransferase (GT) family 47 (Coutinho et al., 2003; http://afmb.cnrs-mrs.fr/CAZY/).
The biochemical and biological functions of enzymes in GT family 47 were first found in animal exostosins. Exostosins exhibit glucuronyltransferase activities involved in the biosynthesis of the backbones of glycosaminoglycan and heparan sulfate (Lind et al., 1998; Wei et al., 2000). Mutations of the human exostosin genes have been shown to cause the development of hereditary exostoses characterized by multiple cartilaginous tumors (Sugahara and Kitagawa, 2000). In plants, the biological functions of two members of GT family 47 were revealed by studies of the tobacco nolac-H18 and the Arabidopsis mur3 mutants. The nolac-H18 mutation, which occurs in the Np GUT1 gene, causes defective cell wall adhesion, a property conferred by pectin (Iwai et al., 2002). Cell wall structural analysis demonstrated that the nolac-H18 mutation results in a dramatic reduction in GlcA residues in pectin, and this reduction was proven to occur in rhamnogalacturonan II (RGII). Therefore, it was proposed that NpGUT1 is a β-glucuronyltransferase participating in the transfer of GlcA residues onto RGII (Iwai et al., 2002).
The mur3 mutation has been found to result in a reduction in cell wall galactose and fucose (Reiter et al., 1997). Cell wall structural analysis revealed that the mur3 mutation specifically blocks the addition of galactose to O-2 of the xylosyl residue closest to the reducing end of the XXXG core structure of xyloglucan oligosaccharide subunits (Madson et al., 2003). Biochemical study further demonstrated that MUR3 exhibits a β-galactosyltransferase activity that transfers galactose onto xyloglucan, confirming that MUR3 is a xyloglucan β-galactosyltransferase. In the Arabidopsis genome, 39 genes have been proposed to encode GTs belonging to GT family 47 (http://afmb.cnrs-mrs.fr/CAZY/; Zhong and Ye, 2003; Li et al., 2004). However, MUR3 and the tobacco NpGUT1 are the only plant members of GT family 47 whose biological and biochemical functions have been demonstrated. Recently, analysis of 10 genes encoding MUR3 homologs indicated that two of these genes may also encode β-galactosyltransferases involved in cell wall synthesis (Li et al., 2004).
In this report, we show that mutation of another member of the GT family 47 in the Arabidopsis fragile fiber8 (fra8) mutant causes a severe reduction in the secondary wall thickness of fibers and vessels and a dramatic decrease in the amount of cellulose and xylan. We demonstrate that the fra8 mutation results in a specific defect in the addition of GlcA residues onto xylans, indicating that FRA8 is a glucuronyltransferase involved in the transfer of GlcA residues onto xylans. Consistent with a role of FRA8 in glucuronoxylan synthesis, the FRA8 protein is shown to be localized in Golgi. Furthermore, we have found that the FRA8 gene is specifically expressed during secondary wall thickening in fibers and vessels. Taken together, these results suggest that FRA8 is a glucuronyltransferase involved in the biosynthesis of glucuronoxylan during secondary wall formation.
RESULTS
The fra8 Mutation Dramatically Reduces Secondary Wall Thickness of Fibers and Vessels
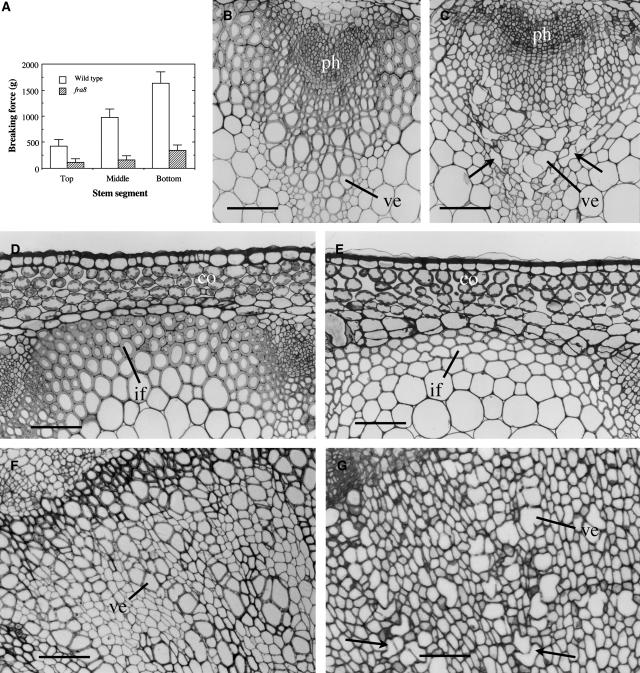
Arabidopsis inflorescence stems develop interfascicular fibers that confer increased mechanical strength to the stem, as demonstrated in the ifl1 mutant (Zhong et al., 1997). To investigate genes involved in secondary wall synthesis, we screened for Arabidopsis mutants defective in fiber wall strength and isolated fra8, a previously unidentified mutant. The fra8 mutation caused a dramatic reduction in the mechanical strength of stems. The force required for breaking apart the bottom parts of mature fra8 stems was 7 times less than that for the wild type (Figure 1A).
Figure 1.
Effects of the fra8 Mutation on Stem Strength and Secondary Wall Thickness of Fibers and Vessels.
Inflorescence stems of 10-week-old plants were used for breaking strength measurements, and the bottom internodes were sectioned for examination of fibers and vessels. co, cortex; if, interfascicular fiber; ph, phloem; ve, vessel. Bars = 78 μm in (B) to (E) and 105 μm in (F) and (G).
(A) Breaking force measurement showing that the force to break the fra8 stems is much lower than that for the wild type. Error bars represent se.
(B) and (C) Cross sections of stems showing vascular bundles of the wild type (B) and fra8 (C). Note that some fra8 vessels were severely deformed (arrows).
(D) and (E) Cross sections of interfascicular regions of stems showing the fra8 fibers with thin walls (E) compared with the wild type (D).
(F) and (G) Cross sections of 1-month-old roots showing secondary xylem of the wild type (F) and fra8 (G). Note that some fra8 vessels had irregular shapes (arrows).
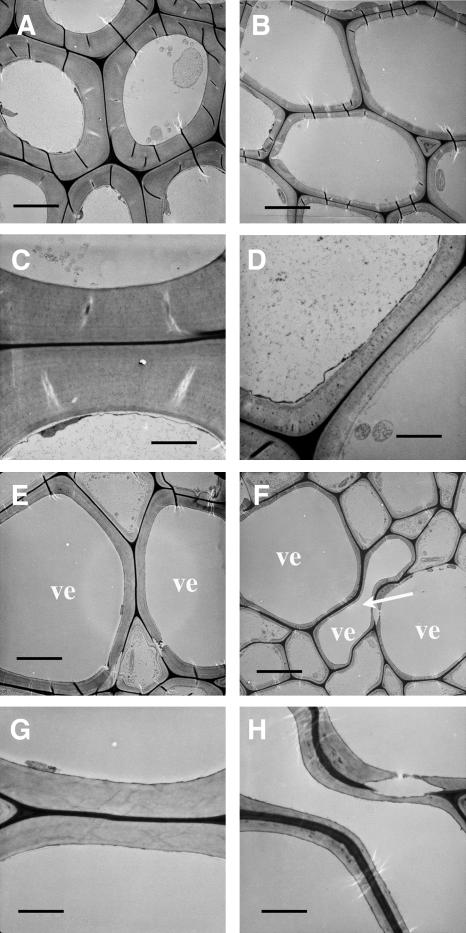
To determine whether the fragile stem phenotype in fra8 was caused by defective fibers, we examined fiber cell morphology and wall thickness. Cross and longitudinal sections of stems showed that although the length of fiber cells was not affected in the fra8 mutant (data not shown), the wall thickness of fra8 fibers was greatly decreased compared with that of the wild type (Figures 1D and 1E). Transmission electron microscopy demonstrated that the wall thickness of fra8 fibers was reduced to 38% of that of the wild type (Figures 2A to 2D, Table 1). Since the staining did not resolve the distinct layers of secondary walls, it is not certain whether the fra8 mutation affected the deposition of specific cell wall layers. It was also found that the fra8 mutation resulted in deformation of xylem vessels in stems and roots (Figures 1B, 1C, 1F, and 1G). In the fra8 mutant, the vessel walls often collapsed inward, most likely due to the reduced wall strength. This phenotype is similar to the collapsed xylem phenotype caused by mutations of cellulose synthase genes involved in secondary wall synthesis (Turner and Somerville, 1997; Taylor et al., 1999, 2000) and by downregulation of lignin biosynthesis in tobacco and Arabidopsis (Chabannes et al., 2001; Jones et al., 2001; Goujon et al., 2003). The wall thickness of fra8 vessels was reduced to 42% of that of the wild type as shown by transmission electron microscopy (Figures 2E to 2H, Table 1). These results demonstrate that the fra8 mutation causes a dramatic decrease in secondary wall thickness in both fibers and vessels.
Figure 2.
Dramatic Reduction in the Thickness of Secondary Walls of Fibers and Vessels by the fra8 Mutation.
Fibers and vessels were examined for their wall thickness under a transmission electron microscope. ve, vessel. Bars = 5.8 μm in (A), (B), (E), and (F) and 2.1 μm in (C), (D), (G), and (H).
(A) and (B) Interfascicular fiber cells showing thin walls in fra8 (B) compared with the wild type (A).
(C) and (D) High magnification of fiber walls of the wild type (C) and fra8 (D).
(E) and (F) Xylem vessels showing thin walls in fra8 (F) compared with the wild type (E). Note the presence of a deformed vessel (arrow).
(G) and (H) High magnification of vessel walls of the wild type (G) and fra8 (H).
Table 1.
Wall Thickness of Fibers and Vessels in the Stems of Wild-Type and fra8 Mutant Plants
| Sample | Fiber Cells | Vessels |
|---|---|---|
| Wild type | 2.29 ± 0.21 | 1.41 ± 0.22 |
| fra8 | 0.86 ± 0.20 | 0.59 ± 0.13 |
Wall thickness was measured from transmission electron micrographs of fibers and vessels. Data are means (μm) ± se from 20 cells.
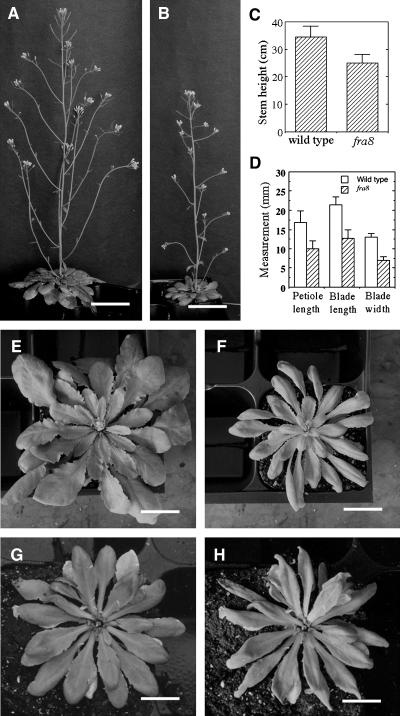
In addition to the defective cell wall phenotypes, we found that the fra8 mutation affected plant growth. The fra8 inflorescence stems were shorter in height (Figures 3A to 3C), and its rosette leaves were smaller in size (Figures 3D to 3F), as compared with the wild type. It was also noted that fra8 leaves exhibited a mild wilty phenotype on sunny afternoons under greenhouse conditions even when the plants were well watered (Figures 3G and 3H). It is likely that the deformed vessels in fra8 impede the transport of water and minerals, which in turn results in the wilty phenotype and the reduced plant growth. This wilty phenotype is similar, albeit to a lesser extent, to the reduced water transport and wilty phenotype of the tomato (Lycopersicon esculentum) wilty-dwarf mutant, in which the vessels have compound perforation plates instead of simple perforation plates as seen in the wild type (Rick, 1952; Alldridge, 1964).
Figure 3.
Alteration of Plant Growth by the fra8 Mutation.
(A) and (B) Eight-week-old plants showing shorter stems in fra8 (B) compared with the wild type (A). Bars = 4.9 cm.
(C) Quantitative measurement of stem heights of 8-week-old wild-type and fra8 plants. Error bars represent se.
(D) Quantitative measurement of the sizes of the fifth rosette leaves of 4-week-old wild-type and fra8 plants.
(E) and (F) Four-week-old plants showing smaller rosette leaves in fra8 (F) compared with the wild type (E). Bars = 15 mm.
(G) and (H) Rosette leaves of a fra8 plant were well expanded in the early morning (G) but were slightly wilted in the sunny afternoon (H) under the greenhouse conditions. Bars = 9 mm.
Positional Cloning of the FRA8 Gene
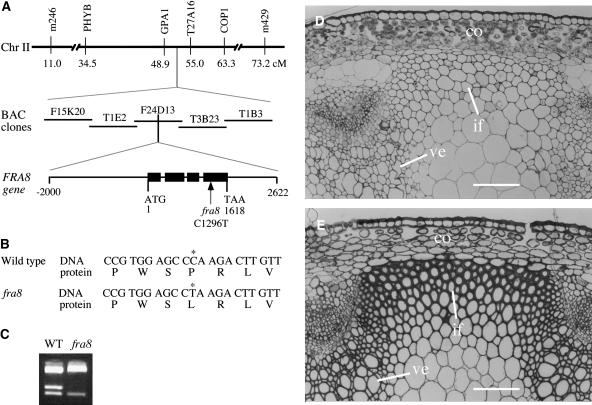
To further investigate the function of FRA8, we undertook the positional cloning of the FRA8 gene. Using F2 mapping plants generated by crossing the fra8 mutant and the wild-type ecotype Landsberg erecta, we mapped the fra8 locus to a region near the COP1 marker on chromosome 2 (Figure 4A). Further mapping with adjacent markers revealed that the fra8 locus was located between GPA1 and T27A16 markers. Based on the sequence information of BAC clones between these two markers, we developed additional mapping markers and used them to gradually narrow the fra8 locus to a 42-kb region within the BAC clone F24D13 (Figure 4A).
Figure 4.
Positional Cloning of the FRA8 Gene.
(A) The fra8 locus was mapped to a small region in the BAC clone F24D13 on chromosome 2. The FRA8 gene is composed of four exons and three introns. The fra8 mutation results in a C-to-T transition in the fourth exon. Black boxes in the FRA8 gene diagram indicate exons, and lines between exons indicate introns. Chr, chromsome; cM, centimorgan.
(B) Nucleotide and amino acid sequences around the fra8 mutation site. The fra8 mutation changes a wild-type codon encoding Pro into a codon encoding Leu.
(C) Elimination of a BanII site in the fra8 mutant gene. The single nucleotide mutation in fra8 occurs at a BanII restriction endonuclease cleavage site. This is revealed by digestion of the PCR-amplified DNA fragments with BanII, which shows that a BanII site is missing in the fra8 mutant DNA compared with the wild type.
(D) and (E) Complementation of the fra8 mutant by the wild-type FRA8 gene. The thin fiber wall and collapsed vessel phenotypes exhibited by the fra8 mutant (D) were rescued by introduction of the wild-type FRA8 gene into the fra8 mutant (E). co, cortex; if, interfascicular fiber; ve, vessel. Bars = 116 μm.
According to the genome annotations of chromosome 2 from the Arabidopsis genome database, the 42-kb region in which the fra8 locus is located encompasses nine putative genes. In order to determine which of these genes carried the fra8 mutation, we sequenced all nine genes from the fra8 mutant. Comparison of the gene sequences from the fra8 mutant with those from the wild type revealed a point mutation (C-to-T) in one of these genes, F24D13.10 (At2g28110) (Figure 4B). The C-to-T mutation resulted in loss of a BanII site in the F24D13.10 gene (Figure 4C).
To confirm that the C-to-T mutation in F24D13.10 was responsible for the phenotypes observed in the fra8 mutant, we transformed a 4.6-kb genomic DNA fragment containing the wild-type F24D13.10 gene (Figure 4A) into the fra8 mutant and examined the phenotypes of the transgenic plants. It was found that the wild-type F24D13.10 gene completely rescued the fra8 phenotypes, including the fiber wall thickness and vessel morphology (Figures 4D and 4E), along with stem mechanical strength and plant growth (data not shown). These results unequivocally demonstrate that the C-to-T mutation in the F24D13.10 gene is responsible for the fra8 mutant phenotypes; therefore, F24D13.10 represents the FRA8 gene.
Sequence Analysis of the FRA8 Gene and Its Encoded Protein
To perform molecular characterization of the FRA8 gene, we isolated the full-length FRA8 cDNA. Comparison of the sequences of FRA8 gene and its cDNA revealed that the FRA8 gene consists of four exons and three introns with a length of 1618 bp from the start codon to the stop codon (Figure 4A). The longest open reading frame in the FRA8 cDNA is 1347 bp long, and it encodes a protein of 448 amino acid residues with a predicted molecular mass of 51,694 D and a predicted pI of 9.3. The fra8 mutation occurred in the 4th exon of the FRA8 gene. Comparison of the wild-type and fra8 cDNAs and their deduced amino acid sequences showed that the fra8 missense mutation changes a Pro codon CCA to a Leu codon CTA (Figure 4B).
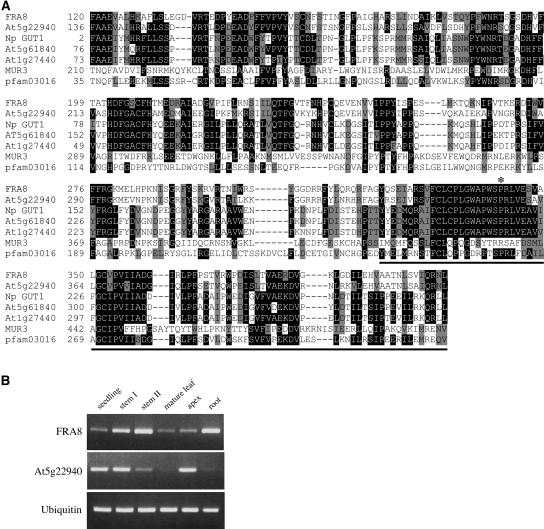
A search of the GenBank conserved domain database revealed that the deduced FRA8 protein contains a domain that shares significant sequence similarity with the pfam03016 domain (Figure 5A). The pfam03016 domain was first defined based on the β-glucuronyltransferase domain of animal exostosins, and putative GTs containing this domain are grouped as GT family 47 (Coutinho et al., 2003). In the Arabidopsis genome, 39 genes, including FRA8, have been identified as members of GT family 47 (http://afmb.cnrs-mrs.fr/CAZY/; Zhong et al., 2003; Li et al., 2004). Of these, the At1g27440 (AtGUT1) and At5g61840 (AtGUT2) genes, two putative orthologs of tobacco NpGUT1, were thought to encode β-glucuronyltransferases involved in pectin synthesis (Iwai et al., 2002), and the MUR3 gene was proven to encode a xyloglucan β-galactosyltransferase (Madson et al., 2003). FRA8, together with a FRA8 homolog (At5g22940), show the highest sequence similarity to tobacco NpGUT1 and its putative Arabidopsis orthologs, At1g27440 and At5g61840 (Figure 5A, Table 2).
Figure 5.
Sequence Analysis of FRA8 and Gene Expression Analysis of the FRA8 Gene in Arabidopsis Organs.
(A) Alignment of FRA8 and other GT family 47 members, including tobacco NpGUT1 and four Arabidopsis members, with the conserved pfam03016 domain. The numbers shown at the left of each sequence are the positions of amino acid residues in the corresponding proteins. Gaps (marked with dashes) were introduced to maximize the sequence alignment. Identical and similar amino acid residues are shaded with black and gray, respectively. The location of the fra8 missense mutation is indicated by an asterisk. The sequence corresponding to the GT signature motif is underlined.
(B) Expression of the FRA8 gene and its homolog, At5g22940, in various Arabidopsis organs. The expression level of a ubiquitin gene was used as an internal control. The seedlings used were 2 weeks old. Mature leaves were from 6-week-old plants. Inflorescence apices and mature roots were from 8-week-old plants. Stems I and II were from 4- and 8-week-old plants, respectively.
Table 2.
Identity and Similarity of the Conserved Domains of FRA8 and Other GT Family 47 Proteins
| Identity/Similarity (%)
|
|||||||
|---|---|---|---|---|---|---|---|
| Protein | FRA8 | At5g22940 | NpGUT1 | At5g61840 | At1g27440 | MUR3 | pfam03016 |
| FRA8 | – | 91.6 | 71.0 | 70.6 | 72.0 | 51.5 | 58.4 |
| At5g22940 | 72.0 | – | 75.0 | 73.6 | 75.0 | 50.5 | 60.4 |
| Np GUT1 | 41.3 | 42.6 | – | 97.5 | 97.8 | 49.2 | 59.4 |
| At5g61840 | 40.6 | 42.3 | 95.0 | – | 96.4 | 47.8 | 58.4 |
| At1g27440 | 39.9 | 43.2 | 91.1 | 90.4 | – | 50.5 | 58.0 |
| MUR3 | 20.2 | 20.6 | 20.0 | 22.0 | 18.9 | – | 65.3 |
| pfam03016 | 30.3 | 29.8 | 32.4 | 33.1 | 32.0 | 39.7 | – |
Values in the lower left portion represent identity, and values in the upper right portion represent similarity.
The most significant sequence similarity among the 39 Arabidopsis GT family 47 proteins and between Arabidopsis GT family 47 proteins and animal exostosins resides within the GT signature motif (Zhong et al., 2003; Li et al., 2004). The fra8 missense mutation occurred at an amino acid residue Pro within the GT signature motif. It is noteworthy that the GT signature motifs of FRA8 and its homolog, At5g22940, show much higher sequence similarity with those of NpGUT1 and its Arabidopsis orthologs than with MUR3 (Figure 5A). This is consistent with the phylogenetic data showing that FRA8, At5g22940, At1g27440, At5g61840, and NpGUT1 are grouped as closely related members (Zhong et al., 2003; Li et al., 2004). In addition, the Pro residue that is mutated in fra8 is completely conserved among FRA8 and NpGUT1 and its Arabidopsis orthologs but not among MUR3 and its homologs (Figure 5A). These findings suggest that FRA8 and NpGUT1 might share a similar (i.e., glucuronyltransferase) biochemical activity and that the Pro residue is essential for their functions.
Expression Pattern of the FRA8 Gene
To investigate the expression pattern of the FRA8 gene, we first used the semiquantitative RT-PCR method to examine its expression in various organs. It was found that although the FRA8 gene was expressed in all of the organs examined, it showed the highest expression in developing stems and 8-week-old roots (Figure 5B), both of which had a large number of developing vessels and fibers (data not shown). Interestingly, the FRA8 homolog, At5g22940, was mainly expressed in seedlings, young stems, and inflorescence apices (Figure 5B).
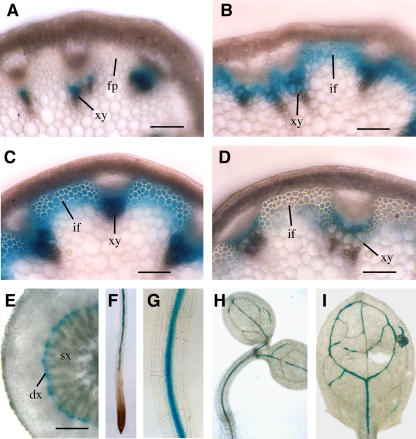
We next used the β-glucuronidase (GUS) reporter gene to examine the tissue-level expression pattern of the FRA8 gene. To do this, we employed the 4.6-kb genomic DNA fragment containing the wild-type FRA8 gene that was used for the complementation study (Figure 4A). Because this genomic DNA fragment was sufficient to complement the fra8 mutant phenotypes, it should contain all elements responsible for the expression of the endogenous FRA8 gene. The GUS reporter gene was inserted in frame just before the stop codon of the FRA8 gene in a binary vector to create the FRA8-GUS construct. Transformation of this construct into the fra8 mutant plants completely rescued the mutant phenotypes (data not shown). Examination of GUS activity in the inflorescence stems of the transgenic plants revealed that in early elongating internodes where no interfascicular fibers were visible, the GUS signals were only present in developing xylem but absent in interfascicular regions (Figure 6A). In internodes near cessation of elongation when fiber cells are clearly visible and in nonelongating internodes in which fiber cells undergo massive secondary wall deposition (Ye et al., 2002), the GUS staining was intensive in both interfascicular fibers and developing xylem cells (Figures 6B and 6C). In mature internodes in which interfascicular fibers had thick secondary walls, little GUS staining was seen in interfascicular fibers. However, intensive GUS staining was still present in developing xylem cells but absent in mature ones (Figure 6D).
Figure 6.
Expression Pattern of the FRA8 Gene Revealed by the GUS Reporter Gene.
The GUS reporter gene was inserted just before the stop codon of the FRA8 gene, which includes a 2-kb 5′ upstream sequence, the entire exon and intron sequence, and a 1-kb 3′ downstream sequence. The construct was introduced into Arabidopsis plants, and various organs of the transgenic plants were examined for GUS activity, which is indicated by the blue color. dx, developing xylem; fp, fiber precursor; if, interfascicular fiber; sx, secondary xylem; xy, xylem. Bars = 350 μm in (A) to (D) and 162 μm in (E).
(A) Cross section of a young elongating internode showing GUS staining in xylem.
(B) Cross section of an internode near the end of elongation showing GUS staining in both interfascicular fibers and xylem.
(C) Cross section of a nonelongating internode showing intensive GUS staining in both interfascicular fibers and xylem.
(D) Cross section of a mature internode showing high GUS staining in developing xylem but little GUS staining in interfascicular fibers.
(E) Cross section of a root from a 4-week-old plant showing GUS staining in developing secondary xylem.
(F) Primary root of a 4-d-old seedling showing GUS staining in the vascular cylinder in the maturation zone but absent in the apex.
(G) High magnification of (F) showing intensive GUS staining in the vascular cylinder.
(H) Four-day-old seedling showing GUS staining in the vascular strands of the cotyledons and hypocotyl.
(I) Leaf from a 2-week-old plant showing GUS staining in veins.
A similar GUS expression pattern was also observed in roots that underwent secondary growth. The GUS staining was evident only in developing root xylem cells but absent in mature ones (Figure 6E). In the seedling stage, the GUS staining was confined in the vascular strands of roots, cotyledons, hypocotyls, and leaves (Figures 6F to 6I). These results demonstrate that the FRA8 gene is specifically expressed in cells undergoing secondary wall deposition. The tight association of FRA8 gene expression with secondary wall synthesis in fibers and vessels is consistent with the defective fiber and vessel phenotypes seen in the fra8 mutant.
Subcellular Localization of the FRA8 Protein
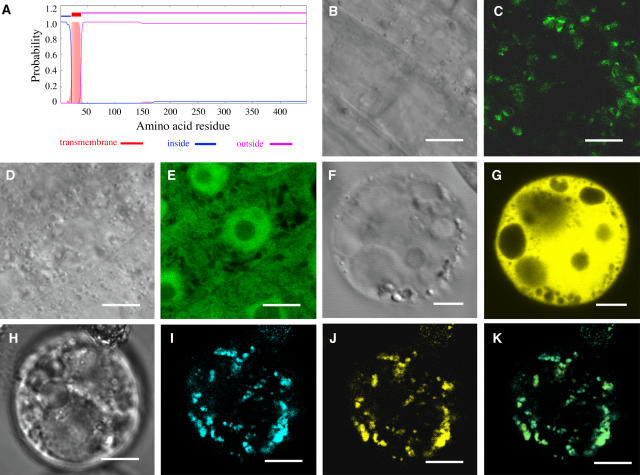
Sequence analysis using the TMHMM2.0 program for prediction of transmembrane helices in proteins (http://www.cbs.dtu.dk/services/TMHMM-2.0/) predicted that FRA8 is a type II membrane protein that contains a short cytoplasmic N terminus followed by a single transmembrane helix and a long noncytoplasmic C terminus (Figure 7A). Further sequence analysis predicted FRA8 to be a Golgi-localized protein using the Golgi predictor program (http://ccb.imb.uq.edu.au/golgi/golgi_predictor.shtml). To determine the actual subcellular location of FRA8, we expressed green fluorescent protein (GFP)–tagged FRA8 in Arabidopsis plants. Examination of the root cells of the transgenic plants showed that the FRA8-GFP signal displayed a punctate pattern (Figures 7B and 7C), whereas the GFP control protein had signals throughout the cytoplasm and the nucleus (Figures 7D and 7E). This observation indicates that FRA8 is targeted to certain subcellular organelles.
Figure 7.
Subcellular Localization of Fluorescent Protein-Tagged FRA8.
Fluorescent protein–tagged FRA8 was expressed in Arabidopsis plants and carrot protoplasts, and its subcellular location was examined with a laser confocal microscope. Bars = 9 μm in (B) to (E) and 14 μm in (F) to (K).
(A) FRA8 is a type II membrane protein as predicted by the TMHMM2.0 program. FRA8 is predicted to have a transmembrane helix between amino acid residues 21 to 38, a short N-terminal region located on the cytoplasmic side of the membrane (inside), and a long stretch of C-terminal region located on the noncytoplasmic side of the membrane (outside).
(B) and (C) Differential interference contrast (DIC) image (B) and the corresponding fluorescent signals (C) of Arabidopsis root epidermal cells expressing FRA8-GFP. Note that the FRA8-GFP signals show a punctate pattern.
(D) and (E) DIC image (D) and the corresponding fluorescent signals (E) of Arabidopsis root epidermal cells expressing GFP alone. Note the presence of GFP signals throughout the cytoplasm.
(F) and (G) DIC image (F) and the corresponding fluorescent signals (G) of a carrot cell expressing EYFP alone.
(H) to (K) DIC image (H) and the corresponding FRA8-ECFP signals (I), MUR4-EYFP signals (J), and a merged image (K) of a carrot cell expressing FRA8-ECFP and the Golgi-localized MUR4-EYFP. It is evident that the FRA8-ECFP signals are identical to MUR4-EYFP signals.
To discern the exact subcellular location of FRA8, we performed colocalization experiments in carrot (Daucus carota) protoplasts. As with the FRA8-GFP protein expressed in Arabidopsis root cells, enhanced cyan fluorescent protein (ECFP)–tagged FRA8 exhibited a punctate localization pattern in carrot protoplasts (Figures 7H and 7I). Colocalization experiments (Figures 7H to 7K) revealed that the pattern of FRA8-ECFP was identical to that of enhanced yellow fluorescent protein (EYFP)–tagged MUR4, a UDP-d-xylose 4-epimerase previously shown to be localized in Golgi (Burget et al., 2003). The EYFP control protein was seen to be distributed throughout the cytoplasm and the nucleus (Figures 7F and 7G). Together, these results demonstrate that FRA8 is a Golgi-localized protein.
Compositional Analysis of Total Cell Walls from fra8 Stems
Because FRA8 is a member of GT family 47 and it is localized in Golgi, we reasoned that FRA8 may be involved in the biosynthesis of a noncellulosic polysaccharide in the cell wall. To ascertain whether the fra8 mutation caused an overall reduction in cell wall synthesis or a specific effect on the synthesis of a particular polysaccharide, we examined the neutral sugar composition of cell walls isolated from fra8 stems. This analysis revealed that xylose and glucose, which are the main components of xylan and cellulose, respectively, were profoundly affected in fra8. The amount of cell wall xylose and glucose in fra8 stems was reduced by 56 and 25%, respectively, compared with the wild type (Table 3). By contrast, the amount of several other wall neutral sugars, including mannose, galactose, and arabinose, was significantly increased in the fra8 mutant.
Table 3.
Neutral Monosaccharide Composition of Cell Walls from the Stems of Wild-Type and fra8 Plants
| Sample | Glucose | Xylose | Mannose | Galactose | Arabinose | Rhamnose | Fucose |
|---|---|---|---|---|---|---|---|
| Wild type | 345 ± 29 | 125 ± 4 | 20 ± 1 | 11.5 ± 0.7 | 8.7 ± 0.5 | 4.7 ± 0.4 | 2.7 ± 0.1 |
| fra8 | 260 ± 12 | 55 ± 2 | 32 ± 1 | 22.0 | 17.0 ± 4.2 | 4.6 ± 0.4 | 3.6 ± 0.7 |
Cell wall residues used for composition analysis were prepared from stems of 10-week-old plants. Data are means (mg/g dry cell wall) ± se of three independent assays.
Compositional Analysis of Individual Polysaccharide Fractions Isolated from fra8 Cell Walls
In order to determine which specific cell wall polysaccharides are affected by the fra8 mutation, we isolated individual polysaccharide fractions for compositional analysis. Cell walls, in the form of alcohol insoluble residues (AIRs), were prepared from the stems of wild-type and fra8 plants, and they were subjected to sequential extraction with specific polysaccharide-cleaving enzymes and chemical reagents that disrupt the cell wall, yielding five water-soluble fractions plus cellulose (see Methods). These fractions, in turn, were subjected to glycosyl composition analysis using techniques that permit the quantitation of both neutral and acidic monosaccharides. As shown in Table 4, the overall monosaccharide yields were lower for the fra8 mutant than for the wild type. Enzyme-extracted fractions were obtained by treatment with endo-polygalacturonase (EPG) to generate the pectin fraction and subsequent treatment with a xyloglucan-specific endo-glucanase (XEG) to generate the XEG fraction. Although these enzyme-extracted fractions constitute a greater portion of the cell wall in fra8 plants than in wild-type plants, the ratios of individual monosaccharides within these two fractions are not significantly different when fra8 and wild-type cell walls are compared (Table 4).
Table 4.
Glycosyl Compositions of Fractions from Wild-Type and fra8 Cell Walls
| Cell Wall Fraction | Rha | Fuc | Ara | Xyl | Man | 4-O-Me-GlcA | Gal | Glc | GalA | GlcA | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pectin | WT | 7.94 | 0.87 | 8.36 | 0.87 | 0.25 | 0.59 | 10.59 | 0.34 | 11.23 | 0.97 | 42.02 |
| fra8 | 11.09 | 1.11 | 12.13 | 1.07 | 0.33 | 0.90 | 15.92 | 0.53 | 15.26 | 1.12 | 59.47 | |
| XEG | WT | 0 | 0.22 | 0.04 | 1.17 | 0.05 | nd | 0.65 | 2.05 | nd | nd | 4.19 |
| fra8 | 0 | 0.38 | 0.07 | 1.89 | 0.16 | nd | 1.10 | 3.55 | nd | nd | 7.14 | |
| 1 N KOH | WT | 1.04 | 0.30 | 1.39 | 58.51 | 0.38 | 5.00 | 1.83 | 3.27 | 1.89 | 3.25 | 76.86 |
| fra8 | 1.38 | 0.92 | 2.75 | 15.49 | 1.37 | 2.37 | 4.80 | 9.16 | 3.55 | 0.60 | 42.40 | |
| 4 N KOH | WT | 0.83 | 0.78 | 1.33 | 37.97 | 2.17 | 2.55 | 2.75 | 6.22 | 2.88 | 1.73 | 59.21 |
| fra8 | 1.01 | 1.76 | 2.06 | 19.26 | 5.74 | 2.92 | 4.20 | 10.12 | 4.68 | 0.34 | 52.09 | |
| TFA | WT | 1.81 | 1.10 | 2.30 | 20.23 | 5.78 | 4.05 | 7.45 | 14.56 | 6.47 | 2.62 | 66.37 |
| fra8 | 1.74 | 0.72 | 2.76 | 15.80 | 6.64 | 3.04 | 5.47 | 10.02 | 8.46 | 1.04 | 55.69 | |
| Cellulose | WT | 331.65 | ||||||||||
| fra8 | 243.45 | |||||||||||
| Total | WT | 11.62 | 3.28 | 13.42 | 118.75 | 8.64 | 12.18 | 23.29 | 358.09 | 22.46 | 8.57 | 580.29 |
| fra8 | 15.22 | 4.89 | 19.76 | 53.50 | 14.24 | 9.24 | 31.50 | 276.83 | 31.96 | 3.10 | 460.23 |
Data are means (mg of sugar/g of AIR) of two duplications of two independent reductions. AIR was fractioned sequentially with a combination of EPG and pectin methylesterase (PME) (pectin fraction), a xyloglucan-specific endoglucanase (XEG fraction), and 1 and 4 N KOH (KOH fractions). The resulting residue was hydrolyzed with TFA (TFA fraction) or with sulfuric acid to determine cellulose content (see Methods). Uronosyl residues in the fraction were reduced to their correspondent 6-6 dideutered sugars before sugar analysis (as alditol acetates). The 3-0-methyl glucose (Biochemical) was used as standard to determine the amount of 4-O-methyl glucuronic acid after reduction. Gas chromatography–mass spectrometry and gas chromatography with flame-ionization detection were used for identification and quantification of the monosaccharides. Bold numbers highlight significant changes between the wild type and fra8. For clarity, the variance is not shown but is <10%. nd, not determined.
Xyl, GlcA, and 4-O-Me-GlcA are the main components of xylans, which are a major component of the 1 and 4 N KOH-extracted fractions (Table 4). It was found that the Xyl content of the 1 and 4 N KOH fractions extracted from fra8 walls was decreased to ∼25 and 50%, respectively, of the wild-type amounts, and the GlcA content of these fractions was <20% of the wild-type amounts. Interestingly, the ratio of 4-O-Me-GlcA to Xyl was higher in fra8 than in the wild type. The amount of Xyl that remained associated in the cell wall after the KOH extraction (TFA fraction in Table 4) was also reduced in the fra8 mutant in comparison with the wild type. The fra8 mutation did not lead to any apparent reduction in the base extractability of xylan, as this would lead to an increase in the xylose content of the trifluoroacetic acid (TFA) fraction or the residual α-cellulose fraction, but this was not observed. In addition, the cellulose content of the fra8 cell walls was drastically reduced compared with that of wild-type walls (Table 4).
Analysis of Pectins from fra8 Cell Walls
Of the GT family 47 enzymes whose specific biochemical function is supported by experimental evidence, the one most closely related to FRA8 is NpGUT1 (Iwai et al., 2002). Based on sequence homology to exostosins and the fact that mutation of the NpGUT1 gene causes a decrease in the GlcA content of a complex pectic polysaccharide known as RGII, it was concluded that NpGUT1 encodes a pectin β-glucuronyltransferase. Mutation of NpGUT1 also leads to observable cell wall abnormalities that are likely to stem from a decreased capacity of the modified RGII to dimerize via the formation of borate-diester cross-links. The fact that the fra8 mutation has no significant effect on the GlcA content of the pectin fraction makes it unlikely that FRA8 is a β-glucuronyltransferase involved in pectin biosynthesis. However, in order to rule out the possibility that FRA8 and NpGUT1 have the same activity and that the fra8 phenotype is due to a change in RGII structure, the dimerization of RGII from fra8 and wild-type cell walls was analyzed chromatographically.
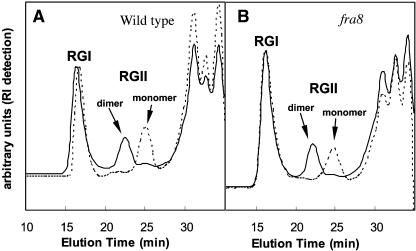
Figure 8 shows the main components of the pectin fractions obtained from the wild-type and fra8 mutant cell walls separated by size-exclusion chromatography. The RGII dimer was significantly more abundant than the monomer in the pectic fractions from both wild-type and fra8 plants. Treatment of either pectin fraction with 0.1 N HCl, which hydrolyzes the RGII dimer to the monomer form, produced the same effect in both cases, confirming the identity of the RGII dimer peak. These results demonstrate that the fra8 mutation has no effect on RGII dimerization, indicating that FRA8 and NpGUT1 have distinct biochemical activities. They also suggest that the physiological effects of the fra8 mutation have a different origin than those observed when the NpGUT1 gene is mutated in tobacco.
Figure 8.
Size-Exclusion Chromatography, Using Refractive Index Detection, of Pectin Fractions Obtained from Stems of Wild-Type and fra8 Plants.
Pectin fractions were treated with 0.1 M HCl for 1 h to dissociate the borate-diester cross-linked RGII dimers into monomers. RI, refractive index.
(A) Pectin fractions from wild-type plants before (solid lines) and after (dashed lines) mild-acid treatment.
(B) Pectin fractions from fra8 plants before (solid lines) and after (dashed lines) mild-acid treatment.
Structural Analysis of Xyloglucans from fra8 Cell Walls
We also analyzed the structure of xyloglucan oligosaccharides isolated from the XEG-extracted fraction from the cell walls of fra8 and wild-type plants. Matrix-assisted laser-desorption ionization time of flight (MALDI-TOF) and NMR analyses of these oligosaccharides showed no significant structural differences (data not shown).
Structural Analyses of Xylans from fra8 Cell Walls
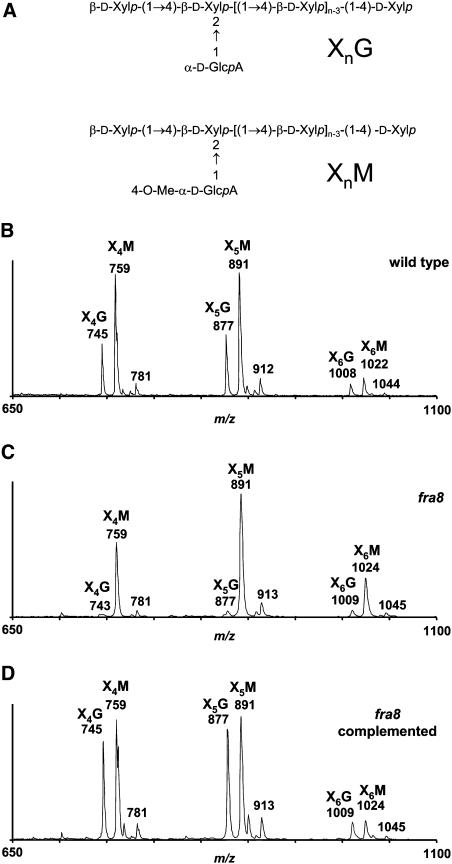
As the fra8 mutation has significant effects on the Xyl and GlcA content of the cell wall (Table 4), the xylose-rich 1 and 4 N KOH fractions from the cell walls of the wild-type, fra8, and fra8 plants complemented with the wild-type FRA8 gene were analyzed. Xyloglucan and xylan are released from the cell wall with 1 and 4 N KOH, and both polymers contain high amounts of xylose. To identify the polysaccharide responsible for the observed loss of xylose, these fractions were incubated with a β-endoxylanase, which digests glucuronoxylans but is not able to degrade xyloglucan, as confirmed by our experiments. The resulting digestion products were separated by size-exclusion chromatography. Endoxylanase treatment converted ∼78% of the 1 and 4 N KOH fractions from wild-type stems into oligosaccharides. By contrast, this treatment of the 1 and 4 N KOH fractions from fra8 stems converted only 53 and 44%, respectively, of these samples into oligosaccharides. The undigested (polymeric) materials were identified by NMR as mainly xyloglucan (data not shown). The acidic oligosaccharides produced by digestion were partially purified and analyzed by MALDI-TOF mass spectrometry (MS), as described in the next paragraph. Neutral xylan oligosaccharide products were also detected. The yields of both the acidic and neutral oligosaccharides were lower when fra8 extracts were digested than when wild-type extracts were digested. Analysis of the 1 and 4 N KOH fractions gave similar results, and only the spectra of the oligosaccharides from the 4 N KOH fraction are presented (Figure 9).
Figure 9.
MALDI-TOF Mass Spectra of Acidic Oligoxylan Products Generated by β-Endoxylanase Digestion of the 4 N KOH Fractions from Wild-Type and fra8 Stems.
(A) General structures of the oligoxylans.
(B) Oligosaccharides from wild-type plants.
(C) Oligosaccharides from fra8 plants.
(D) Oligosaccharides from fra8 plants complemented with the wild-type FRA8 gene.
Two series of ions showing a mass increment of 132 D (corresponding to one xylosyl residue) were observed in all the spectra. These series correspond to oligosaccharides composed of four to six Xyl residues bearing a 4-O-Me-GlcA residue (X4M, X5M, and X6M) or a nonmethylated GlcA residue (X4G, X5G, and X6G). The abundance of XnG oligosaccharides with a nonmethylated glucuronic acid was reduced drastically in the oligosaccharides isolated from fra8 plants (C) compared with the wild type (B). Complementation of the fra8 mutant with the wild-type FRA8 gene (D) restored the production of XnG oligosaccharides with a nonmethylated glucuronic acid residue.
The most abundant ions in the spectra of all acidic xylan oligosaccharide fractions occur at mass-to-charge ratios (m/z) 759, 891, and 1022, a series with an incremental mass of 132 D, which is consistent with the sequential addition of a pentosyl residue to the oligosaccharide. These fractions are rich in Xyl and contain very little Ara (Table 4), indicating that this pentosyl residue is Xyl. Therefore, this series of ions was attributed to oligosaccharides with four, five, and six Xyl residues substituted with one 4-O-Me-GlcA (XnM). The ions at m/z 781, 912, and 1044 were attributed to the doubly sodiated species [M-H + 2Na]+ of the same series of oligosaccharides (Reis et al., 2003). Assignment of these ions was supported by observation of the same series of ions in the MALDI-TOF spectrum of oligosaccharides obtained by endoxylanase digestion of a 4-O-methyl-glucuronoxylan purchased from Sigma-Aldrich (data not shown). This commercially available polysaccharide has a backbone of 1,4-linked d-Xylp residues, some of which are substituted at O-2 with 4-O-Me-α-d-GlcpA residues. Comparison of the spectra of the wild type and fra8 showed no alteration in the abundance of ions at m/z 759, 891, and 1022 (Figures 9B to 9D), indicating that the fra8 mutation does not affect the addition of 4-O-Me-GlcA onto xylan.
Another series of ions at m/z 745, 877, and 1008 also showed a mass increment of 132 D with a difference of 14 D relative to the XnM series (Figure 9B). These ions were attributed to oligosaccharides with four, five, and six Xyl residues substituted with one GlcA (XnG). The spectrum for the fra8 mutant showed a drastic reduction in the abundance of these ions compared with the wild type (Figures 9B and 9C), indicating that the fra8 mutation causes a specific defect in the addition of GlcA onto xylan. Xylan oligosaccharide fractions obtained from the fra8 mutant complemented with the wild-type FRA8 gene gave MALDI-TOF spectra similar to the wild type with high abundance ions in the XnG series (Figure 9D), confirming that the defect in the addition of GlcA onto xylan in the mutant is caused by mutation of the FRA8 gene.
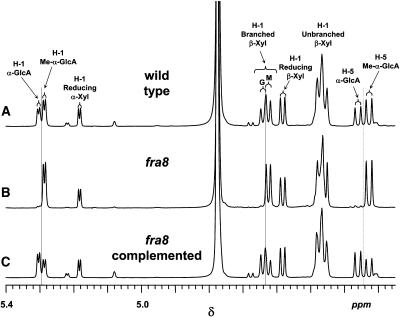
To further prove the structural defect of xylan in the fra8 mutant, 1H-NMR spectra of the purified xylan oligosaccharides obtained from the cell walls of wild-type, fra8, and fra8 complemented plants were recorded (Figure 10). All the spectra included anomeric resonances at δ 5.183 and 4.583, characteristic for the α- and β-configurations, respectively, of the reducing d-Xylp residue. In the spectrum of the oligosaccharides from wild-type plants (Figure 10A), two additional anomeric resonances (δ 5.301 and 5.282) were attributed to α-d-GlcpA and 4-O-Me-α-d-GlcpA residues, respectively, in glucuronoxylan oligosaccharides. The scalar coupling (J1,2) of ∼3.9 Hz for these H-1 signals indicates an α-configuration, consistent with this assignment. Two other diagnostic resonances (δ 5.301 and 5.282) were assigned to H-5 of α-d-GlcpA and 4-O-Me-α-d-GlcpA residues, respectively. Substitution (at O-2) of β-d-Xylp residues with an α-d-GlcpA or 4-O-Me-α-d-GlcpA residue caused an downfield shift of H-1 (relative to the unsubstituted β-d-Xylp residues) from δ 4.45 to 4.48 to δ 4.638 and 4.624, respectively.
Figure 10.
Partial 600-MHz 1H NMR Spectra of Acidic Oligoxylan Products Generated by β-Endoxylanase Digestion of the 4 N KOH Fractions from the Stems of Wild-Type and fra8 Plants.
(A) Wild-type plants.
(B) fra8 plants.
(C) fra8 plants complemented with the wild-type FRA8 gene.
Resonances are labeled with the position of the assigned proton (i.e., H-1 or H-5) and the identity of the residue containing that proton. The resonances due to H-1 of branched β-Xyl residues are labeled with a G if the β-Xyl residue bears an α-GlcA side chain or M if the β-Xyl residue bears a 4-O-Me-α-GlcA side chain. All spectra include resonances due to H-1 of α-Xyl and β-Xyl residues at the reducing end of the oligosaccharides and H-1 of unbranched β-Xyl residues, which do not bear a uronic acid side chain residue. Resonances due to H-1 and H-5 of 4-O-Me-GlcA residues are also found in all spectra, along with H-1 resonances of branched β-Xyl residues bearing a 4-O-Me-GlcA residue at O-2. However, resonances assigned as H-1 and H-5 of nonmethylated GlcA residues and H-1 resonances of branched β-Xyl residues bearing a nonmethylated GlcA residue at O-2 are strong in the spectra of oligosaccharides from wild-type (A) and fra8 complemented stems (C) but virtually absent in the spectrum of oligosaccharides from fra8 stems (B).
The resonances assigned to 4-O-Me-α-d-GlcpA residues and β-d-Xylp residues with a 4-O-Me-α-d-GlcpA residue at O-2 were present in the 1H-NMR spectrum of the oligosaccharides obtained by digestion of the commercial 4-O-methyl glucuronoxylan (Sigma-Aldrich). However, the 1H-NMR spectra of the oligosaccharides from this commercial source did not contain resonances assigned to α-d-GlcpA residues or β-d-Xylp residues with an α-d-GlcpA residue at O-2 (data not shown). The 1H-NMR assignments were realized by comparison with the data in the literature for sorghum (Sorghum bicolor) glucuronoarabinoxylan (Verbruggen et al., 1998) and rigorously confirmed by two-dimensional NMR experiments (gCOSY, gHSQC, and HMBC; data not shown) (Jia et al., 2003). These experiments also established O-2 of the β-d-Xylp residues as the location of the α-d-GlcpA and 4-O-Me-α-d-GlcpA substituents, as previously reported for other acidic xylans (Ebringerová and Heinze, 2000; Teleman et al., 2000; Habibi et al., 2002).
Examination of the 1H-NMR spectra of the acidic xylan oligosaccharides from fra8 and wild-type plants showed that the wild type and fra8 contain similar resonances assigned to H-1 and H-5 of 4-O-Me-α-d-GlcpA residues and to H-1 of unbranched β-d-Xylp residues and β-d-Xylp residues substituted at O-2 with 4-O-Me-α-d-GlcpA (Figures 10A and 10B). By contrast, whereas resonances assigned to H-1 and H-5 of α-d-GlcpA residues and to H-1 of β-d-Xylp residues substituted with α-d-GlcpA are evident in the spectrum of the wild-type oligosaccharides, they are virtually absent in fra8 (Figures 10A and 10B). Complementation of the fra8 mutant with the wild-type FRA8 gene resulted in the production of acidic xylan oligosaccharides with an 1H-NMR spectrum similar to the wild type (Figures 10A and 10C), in which the resonances diagnostic for the presence of α-d-GlcpA residues are restored.
These spectroscopic results lead to the following conclusions. (1) Acidic xylans in the stems of wild-type Arabidopsis plants contain both α-d-GlcpA and 4-O-Me-α-d-GlcpA residues, with the latter being the most abundant. (2) Acidic xylans in the stems of fra8 plants retain 4-O-Me-α-d-GlcpA residues but are practically devoid of α-d-GlcpA residues. (3) Complementation of fra8 plants with the wild-type FRA8 gene restores the production of acidic xylans containing both α-d-GlcpA and 4-O-Me-α-d-GlcpA residues.
DISCUSSION
Secondary cell walls are the most abundant components of many economically important products such as wood and paper. Understanding how secondary walls are synthesized is a long-sought goal, with the hope of genetic manipulation of wood formation. Although cellulose synthase genes participating in cellulose synthesis during secondary wall formation have been characterized (Taylor et al., 2004), genes involved in the biosynthesis of xylan, another major polysaccharide in secondary walls, have not been identified. Our finding that FRA8, a member of the GT family 47, is essential for glucuronoxylan synthesis during secondary wall formation marks an important step toward further understanding of how plants make secondary walls.
The FRA8 GT Is Essential for Secondary Wall Synthesis
Sequence analyses have shown that the FRA8 gene encodes a member of GT family 47. The biological functions of two plant members of GT family 47 have been characterized previously. Biochemical evidence supports the ideas that NpGUT1 is a β-glucuronyltransferase important in pectin synthesis in tobacco (Iwai et al., 2002) and that Arabidopsis MUR3 is a xyloglucan β-galactosyltransferase (Madson et al., 2003). FRA8 and its homolog, At5g22940, show the highest sequence similarity to tobacco NpGUT1 and its Arabidopsis orthologs, suggesting that FRA8 might have a biochemical function similar to that of NpGUT1 (i.e., glucuronyltransferase activity). This hypothesis is further substantiated by the fact that the biological activity of FRA8 is disrupted by a missense mutation at a Pro residue conserved between FRA8 and NpGUT1 but not among MUR3 and its homologs.
The FRA8 gene was found to be specifically expressed in developing fibers and vessels, the cell types that undergo secondary wall synthesis. Consistent with its expression pattern, mutation of the FRA8 gene resulted in a severe defect in secondary wall synthesis. Cell wall composition analysis demonstrated that the fra8 mutation caused a dramatic reduction in the levels of xylan and cellulose in cell walls, indicating that FRA8 is most likely involved in the synthesis of xylan or cellulose during secondary wall formation. Because the cellulose synthesizing machineries reside in the plasma membrane (Delmer, 1999) and FRA8 is located in Golgi, it is unlikely that FRA8 is directly involved in cellulose synthesis. These results strongly suggest that FRA8 most likely participates in xylan synthesis since it is known that noncellulosic polysaccharides are synthesized in Golgi (Levy and Staehelin, 1992). This is consistent with previous observations that a xylan synthase-associated polypeptide is specifically localized in the Golgi apparatus in xylem cells of French bean (Phaseolus vulgaris) (Gregory et al., 2002).
Role of FRA8 in Xylan Synthesis
Thorough analysis of the polysaccharide components of cell walls isolated from the stems of fra8 plants indicate that FRA8 is involved in glucuronoxylan synthesis. Mutation of the FRA8 gene was found to result in a specific defect in the addition of GlcA residues onto xylan. Although xylan isolated from wild-type Arabidopsis stems contains both GlcA and 4-O-Me-GlcA side chains, xylan from the fra8 mutant is devoid of nonmethylated GlcA residues. This finding suggests that FRA8 is a glucuronyltransferase involved in the transfer of GlcA onto xylan, which is supported by the sequence analysis showing that FRA8 is closely related to the putative tobacco pectin glucuronyltransferase NpGUT1. Both xylose and GlcA are significantly reduced in the xylan-enriched cell wall fractions from fra8, suggesting that addition of GlcA side chains is essential for the normal synthesis of xylan. It is not known how a defect in the addition of GlcA side chains could have such a profound effect on the synthesis of the xylan backbone. Because the synthesis of xylan backbone and GlcA side chains is thought to be a coupled process (Baydoun et al., 1989b), it is possible that the fra8 mutation may disrupt the efficient synthesis of glucuronoxylan. The coordination of enzymatic activities involved in the synthesis of other cell wall polysaccharides has also been documented. The biosynthesis of galactomannan has been shown to be mediated by the coordinated activities of a mannan synthase and a galactosyltransferase that catalyze the synthesis of the mannan backbone and the transfer of galactose side chains, respectively (Edwards et al., 1989, 1999). The apparent cooperativity of the glucuronosyl and xylosyl transferase activities may stem from a decrease in xylan solubility that is likely to accompany a deficit in the transfer of glucuronosyl side chains, hindering the efficient synthesis of xylan chains.
Previous biochemical studies indicate that 4-O-methylation of GlcA occurs after transfer of this residue to the polymeric glucuronoxylan and that a nucleotide-linked precursor containing 4-O-methylated GlcA is not involved (Kauss and Hassid, 1967; Baydoun et al., 1989a). Therefore, it is intriguing to find that the fra8 mutation specifically affects the addition of GlcA but not 4-O-Me-GlcA residues onto xylan. One possible explanation is that a limited amount of glucuronoxylan methyltransferase activity is available and that this activity is able to methylate the GlcA residues in the fra8 mutant to a level comparable to that of the 4-O-Me-GlcA residues in the wild type; therefore, the total amount of 4-O-Me-GlcA is not altered in fra8 stems. It is also possible that another glucuronyltransferase is involved in the addition of 4-O-Me-GlcA residues onto xylan. We hypothesize that this other putative glucuronyltransferase might form a complex with a glucuronoxylan methyltransferase and that the cooperation of both enzymes is required for the addition of 4-O-Me-GlcA side chains to the xylan. Since At5g22940 shows the highest sequence similarity to FRA8, it will be interesting to investigate whether At5g22940 is a glucuronyltransferase involved in the addition of 4-O-Me-GlcA residues onto xylan. Alternatively, At5g22940 may have the same biochemical function as FRA8 but play a minor role in secondary wall synthesis; thus, it could not compensate for the loss of FRA8 activity in the fra8 mutant.
It is important to note that, although the available evidence indicates that FRA8 is a glucuronyltransferase that adds GlcA side chains to the xylan backbone, the data do not allow us to unambiguously exclude the possibility that FRA8 is a xylosyltransferase directly involved in the synthesis of xylan backbone, considering the fact that the amount of Xyl in the xylan-containing fractions is also dramatically reduced in the fra8 mutant. However, this possibility is not consistent with the current hypothesis that such β-xylosyltransferases are encoded by cellulose synthase-like genes (Hazen et al., 2002; Dhugga et al., 2004; Liepman et al., 2005) in the cellulose synthase superfamily (Richmond and Somerville, 2000). Furthermore, if FRA8 were a xylosyltransferase, one might expect to see a proportional reduction in the GlcA and 4-O-Me-GlcA residues in xylans prepared from fra8 plants. However, an overall decrease in the rate of xylan synthesis might result in an increase in its overall degree of methylation if the rate of methylation remained constant, as suggested above.
Definitive proof that FRA8 is a xylan glucuronyltransferase would depend on demonstration of its biochemical activity in vitro. We have attempted to detect the transfer of GlcA to xylan oligomers by recombinant FRA8 protein expressed in yeast but failed to observe any glucuronyltransferase activity (data not shown). This could be due to the lack of xylosyltransferase activity in the assay reaction, as it has been suggested that glucuronyltransferase and xylosyltransferase act cooperatively during xylan synthesis. It has been shown that glucuronyltransferases catalyze the addition of GlcA residues onto xylan chains when xylan chains are being synthesized but exhibit little activity toward preformed xylan chains (Baydoun et al., 1989b). It will be important to identify xylosyltransferase genes involved in xylan synthesis and then use both glucuronyltransferase and xylosyltransferase to investigate their cooperative role in the synthesis of glucuronoxylans.
It is interesting to note that GlcA and 4-O-Me-GlcA residues are linked to the xylan backbone via an α-glycosidic linkage (Ebringerová and Heinze, 2000; Teleman et al., 2000; Habibi et al., 2002). However, the members of GT family 47 with known biochemical functions all possess β-GT activities with a catalytic mechanism that inverts the anomeric configuration of the donor substrate to generate β-linked products from α-linked donor substrates (Coutinho et al., 2003; http://afmb.cnrs-mrs.fr/CAZY/). This implies that FRA8 might be an exception to the rule that all GT family 47 enzymes have inverting catalytic mechanisms. It might also be possible that FRA8 is a glucuronyltransferase involved in the production and/or utilization of an intermediate donor substrate that contains a β-linked GlcA. There exists a precedent for this type of mechanism. Most members of GT family 2 use α-linked precursors (e.g., UDP-Glc) to catalyze the addition of β-linked glycosidic residues to polysaccharides such as β-linked glucans (Coutinho et al., 2003; http://afmb.cnrs-mrs.fr/CAZY/GT_2.html). However, this family also includes an enzyme (EC 2.4.1.83) that uses α-linked GDP-Man to generate β-linked dolichol-P-mannose, which can be used as a donor substrate by another enzyme (EC 2.4.1.199) in GT family 2 to generate α-linked mannosyl oligosaccharides. If FRA8 is involved in the transfer of GlcA residues by a similar mechanism, in vitro assays to detect the catalytic activity of heterologously expressed protein will require addition of the appropriate intermediate donor or acceptor substrates. The exact biochemical mechanisms of how FRA8 is involved in glucuronoxylan synthesis remain to be investigated.
Changes in the Amount of Other Cell Wall Polysaccharides in the fra8 Mutant
The only cell wall polysaccharides that are dramatically reduced in the fra8 mutant are xylan and cellulose. FRA8 is unlikely to be a part of cellulose synthase complex due to its localization in Golgi. We did not detect any changes in other cell wall polysaccharides that could account for a possible indirect effect on xylan synthesis in the fra8 mutant. For example, thorough analyses of the cell walls of fra8 and wild-type stems did not reveal any compositional or structural alterations in their pectin and xyloglucan components. Although all available evidence indicates that FRA8 is a GT involved in glucuronoxylan synthesis, we could not rule out the possibility that the fra8 mutation affects the synthesis of other cell wall polymers, which may result in an indirect defect in glucuronoxylan and cellulose synthesis. It has been known that an alteration in the biosynthesis of one cell wall polysaccharide may cause indirect changes in the biosynthesis of another cell wall polysaccharide. One of the best-documented examples is that a reduction in cellulose synthesis in several Arabidopsis mutants, such as kor (His et al., 2001) and prc1 (Fagard et al., 2000), leads to a compensatory increase in pectin content. It has been shown that mutation of the QUASIMODO1 gene, which encodes a putative α-1,4-d-galacturonosyltransferase involved in homogalacturonan synthesis, results in a significant decrease in xylan synthase activity, although its effect on cell wall xylose content is mild (Orfila et al., 2005). The tobacco nolac-H18 mutation, which is defective in the transfer of GlcA onto pectin RGII, was found to cause a reduction in several other cell wall sugars, including glucose, xylose, galactose, fucose, and arabinose (Iwai et al., 2002). In tobacco and Arabidopsis plants with downregulation of lignin biosynthesis, the assembly of cellulose microfibrils in the secondary walls was profoundly affected (Pincon et al., 2001; Goujon et al., 2003), and the cellulose content was decreased (Jones et al., 2001).
It is intriguing that, in addition to the dramatic reduction in xylan synthesis, the fra8 mutation also results in a severe effect on cellulose synthesis, suggesting that a reduction in xylan synthesis affects cellulose synthesis. Xylan is a matrix polysaccharide that spontaneously binds to the surface of cellulose microfibirils and may thereby play a crucial role in the normal assembly of cellulose microfibrils during secondary wall synthesis. The acidic substituents of xylan have been proposed to play an important role in the formation of a helicoidal orientation of cellulose microfibrils during the secondary wall assembly (Reis and Vian, 2004). A reduction in the amount of xylan and/or its GlcA side chains might result in aberrant assembly of nascent cellulose microfibrils at the plasma membrane, which could, in turn, impede the further synthesis of cellulose micrifibrils.
Although the amount of cellulose is reduced in the fra8 mutant, the amount of pectin and xyloglucan is elevated. The relative increase in the levels of these polysaccharides could be due simply to the decrease in the amounts of xylan and cellulose or due to a compensatory effect in response to the reduction in cellulose, as suggested for other cellulose-deficient mutants, such as kor (His et al., 2001), prc1 (Fagard et al., 2000), and fra5 (Zhong et al., 2003).
The identification of FRA8 as a putative xylan glucuronyltransferase provides an important tool for further studies of secondary wall synthesis in wood. In this regard, it should be noted that a putative GT belonging to GT family 47 that is expressed during wood formation in poplar (Aspeborg et al., 2005) shows high sequence identity to FRA8 (88% similarity within the pfam03016 domain), indicating that this GT is likely involved in the synthesis of glucuronoxylans in poplar wood.
METHODS
Mutant Isolation
M2 Arabidopsis thaliana (ecotype Columbia) plants generated from ethyl methanesulfonate mutagenization were grown in a greenhouse, and their inflorescence stems were screened for mutants with reduced breaking strength. Stems were divided into three equal segments, and each segment was measured for its breaking force using a digital force/length tester (model DHT4-50; Larson System). The breaking force was calculated as the force needed to break apart a stem segment (Zhong et al., 1997). Putative mutants with reduced stem breaking strength were selected and backcrossed with wild-type Columbia three times before analysis.
Microscopy
Stem samples were fixed in 2% (v/v) glutaraldehyde in PEMT buffer (50 mM PIPES, 2 mM EGTA, 2 mM MgSO4, and 0.05% [v/v] Triton X-100, pH 7.2) at 4°C overnight. After being washed in phosphate buffer (50 mM, pH 7.2), samples were postfixed in 2% (v/v) OsO4 for 2 h and then dehydrated through a gradient of ethanol, incubated in propylene oxide, and embedded in low viscosity (Spurr's) resin (Electron Microscopy Sciences; Taylor and Mims, 1991). One-micrometer-thick sections were cut, stained with toluidine blue, and viewed under a light microscope. For transmission electron microscopy, 85-nm ultrathin sections were cut, picked up on slot grids, and allowed to dry down onto formvar-coated aluminum racks according to the procedure of Rowley and Moran (1975). After post-staining with uranyl acetate and lead citrate, the specimens were observed under a Zeiss EM 902A transmission electron microscope (Carl Zeiss).
Map-Based Cloning
The fra8 mutant (ecotype Columbia) was crossed with Arabidopsis ecotype Landsberg erecta to generate 1650 F2 mapping plants. Fine mapping of the fra8 locus was done with codominant amplified polymorphic sequence markers according to Konieczny and Ausubel (1993). Codominant amplified polymorphic sequence markers were developed based on sequence information from the Cereon Arabidopsis polymorphic database (http://www.arabidopsis.org/cereon).
For complementation analysis, a 4.6-kb genomic DNA fragment containing the wild-type FRA8 gene, including a 2-kb 5′ upstream sequence, the entire exon and intron regions, and a 1-kb 3′ downstream sequence, was PCR amplified with high-fidelity DNA polymerase with gene-specific primers (5′-GGTTGACGAGTCGTTTTCTAAGTG-3′ and 5′-GGGTTTTAGGTTTAGCAGCTGTTA-3′), confirmed by sequencing, and cloned into the binary vector pBI101 (BD Biosciences Clontech). The construct was introduced into the fra8 mutant by the Agrobacterium tumefaciens–mediated transformation (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and grown to maturity for analysis of their ability to complement the mutant phenotypes.
Gene Expression Analysis
Total RNA was isolated from leaves, stems, roots, flowers, and seedlings using a Qiagen RNA isolation kit. One microgram of the purified RNA was treated with DNase I to remove any potential genomic DNA contamination and then used for first-strand cDNA synthesis. One-twentieth of the first-strand cDNA was used for PCR amplification of FRA8 with gene-specific primers (5′-AAGGACGGAAGATCCGTATGAAGC-3′ and 5′-TTACAAGAAAGAGTTTGACCTTCT-3′). The primers used for RT-PCR span two introns so that any potential amplification from genomic DNA could be easily identified based on its larger size. No genomic DNA was amplified in the RT-PCR reactions. The PCR was performed for variable cycles to determine the logarithmic phase of amplifications for the samples. The RT-PCR reactions were repeated three times, and identical results were obtained. The expression of a ubiquitin gene was used as an internal control for determining the RT-PCR amplification efficiency among different samples.
The tissue-level expression pattern of the FRA8 gene was studied using the GUS reporter gene. To do this, we used the 4.6-kb genomic DNA fragment containing the wild-type FRA8 gene that was used for the complementation study. The GUS reporter gene was inserted in frame right before the stop codon of the FRA8 gene in the binary vector pBI101 to create the FRA8-GUS construct. The construct was transformed into wild-type and fra8 mutant plants by the Agrobacterium-mediated transformation procedure (Bechtold and Bouchez, 1994). Transgenic plants were selected on kanamycin and used for expression analysis of the GUS reporter gene. Tissues were first immersed in 90% ice-cold acetone for 20 min and then incubated in the GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocynide, and 1 mM 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid) at 37°C. After cleared in 70% ethanol, the tissues were observed for the GUS staining under a light microscope (Gallagher, 1992).
Localization of Fluorescent Protein–Tagged FRA8
The wild-type FRA8 cDNA was PCR amplified, confirmed by sequencing, and then fused in frame with the GFP cDNA (ABRC, Columbus, OH; developed by S.J. Davis and R.D. Vierstra). The FRA8-GFP fusion cDNA was cloned downstream of the cauliflower mosaic virus 35S promoter in the binary vector pBI121. The FRA8-GFP construct was introduced into Arabidopsis plants by Agrobacterium-mediated transformation. Transgenic plants were selected on kanamycin, and the T2 progeny were used for GFP localization.
The GFP signals from roots of 3-d-old transgenic seedlings were viewed under a Leica TCs SP2 spectral confocal microscope (Leica Microsystems). Images were saved and processed with Adobe Photoshop version 7.0.
To determine the exact subcellular location of FRA8, the FRA8 cDNA was fused in frame with an ECFP and ligated between the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator in a high copy vector. The FRA8-ECFP expression construct together with another expression construct containing the Golgi-localized MUR4 tagged with an EYFP were cotransfected into carrot (Daucus carota) protoplasts according to Liu et al. (1994). The transfected protoplasts were incubated in darkness for 20 h before examination under a Leica TCs SP2 spectral confocal microscope. Images from single optical sections were collected and processed with Adobe Photoshop version 7.0.
Expression of Recombinant FRA8 Protein in Yeast
The full-length FRA8 cDNA was PCR amplified using high-fidelity DNA polymerase from cDNAs synthesized from wild-type stems. The FRA8 cDNA was confirmed by sequencing and ligated in frame into the yeast expression vector pYES2/NT, which is tagged with six histidines and the Xpress epitope (DLYDDDDK) at the N terminus (Invitrogen). The construct was transformed into the yeast strain INVSc1 (Invitrogen). The expression of recombinant FRA8 protein was induced in the presence of 2% galactose for 24 h. After induction, yeast cells were broken using glass beads in the extraction buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM PMSF, and 1% Triton X-100). After centrifugation at 12,000 rpm for 20 min, the protein extracts were separated on a 12.5% SDS gel and transferred onto a nitrocellulose membrane. The recombinant FRA8 protein was detected by incubation with a monoclonal antibody against the Xpress epitope (Invitrogen) and horseradish peroxidase–conjugated secondary antibodies. The recombinant FRA8 protein with the expected molecular mass was confirmed to be expressed in yeast cells.
Glucuronyltransferase Activity Assay
Yeast protein extract with expression of recombinant FRA8 protein was assayed for the glucuronyltransferase activity according to Waldron and Brett (1983). The control used was protein extract with expression of β-galactosidase prepared under the same conditions. The protein extract (100 μg) was mixed with the reaction solution in a total volume of 100 μL. The reaction solution contains 50 mM Tris-HCl, pH 7.5, 10 mM MnCl2, 0.1 μCi UDP-D-[U-14C]glucuronic acid (American Radiolabeled Chemicals) and xylan oligomers (ranging from 5 to 100 xylose residues) prepared from xylanase digestion of larchwood xylan and 4-O-methyl glucuronoxylan (Sigma-Aldrich). After incubation at 25°C for 4 h, the reaction product was precipitated with 70% ethanol, and the incorporation of radioactivity into 70% ethanol-insoluble materials was determined by counting in a scintillation counter.
Neutral Glycosyl Composition Analysis of Whole Cell Walls
Inflorescence stems of 10-week-old plants were collected for cell wall isolation. Stems were ground into fine powder in liquid nitrogen with a mortar and pestle, homogenized with a polytron, and extracted in 70% ethanol at 70°C. The resulting cell wall residues were dried in a vacuum oven at 60°C and used for analysis of total sugar composition. Cell wall sugars (as alditol acetates) were determined following the procedure described by Hoebler et al. (1989). Briefly, cell walls were incubated with 70% sulfuric acid at 37°C for 60 min followed by the addition of inositol as the internal standard and dilution with water to 2 N sulfuric acid. After heating for 120 min at 100°C, the solution was cooled and treated with 25% ammonium solution. After reduction with sodium borohydride in dimethyl sulfoxide, the solution was heated for 90 min at 40°C, followed by sequential treatment with glacial acetic acid, acetic anhydride, 1-methylimidazole, dichloromethane, and water. The organic layer containing the alditol acetates of the hydrolyzed cell wall sugars was washed three times with water, and sugars were analyzed on an Agilent 6890N gas–liquid chromatograph) equipped with a 30 m × 0.25 mm (i.d.) silica capillary column DB 225 (Alltech Associates). All samples were run in triplicate.
Preparation of Alcohol Insoluble Cell Wall Residues
AIR was prepared as described previously (Perrin et al., 2003). In brief, frozen mature inflorescence stems of 10-week-old fra8 and wild-type Arabidopsis plants were ground into fine power under liquid nitrogen. The ground material was suspended in 80% (v/v) ethanol and further disrupted using a Polytron homogenizer. The AIR was collected on nylon mesh and washed with 80% (v/v) ethanol and then absolute ethanol. The washed AIR was suspended in methanol:chloroform (1:1, v/v), stirred for 1 h at room temperature, collected by filtration through glass-fiber filter (GF/A; Whatman), washed with acetone, and air dried. The AIR preparations were sequentially extracted with enzymes and chemical agents that disrupt the cell wall to isolate polysaccharide fractions as described below.
Pectin Isolation and Analysis
AIR preparations from wild-type and fra8 stems were suspended in 50 mM NaOAc buffer, pH 5.0 (10 mg/mL), containing 0.01% (w/v) thimerosal and incubated with endo-polygalacturonase (20 units) from Aspergillus niger (provided by Carl Bergmann, Complex Carbohydrate Research Center) and pectin methylesterase (20 units) from A. oryzae (Novozymes). The suspensions were incubated at 24°C for 24 h in a shaking incubator and then filtered, and the solid residues were subjected to a second incubation under the same conditions. For each plant source, the filtrates were combined and labeled the EPG and PME soluble fractions. Aliquots of the EPG and PME soluble fractions were incubated with 1 M HCl solution for 1 h at 25°C, conditions which cleave the RGII dimer to form RGII monomer. The resulting changes in molecular weight were monitored by size-exclusion chromatography on a Superdex-75 HR10/30 column (Amersham Biosciences) using refractive index detection. Samples of the original fraction without acid treatment were used as controls.
XEG Treatment of Cell Walls
The partial depectinated AIR were suspended in 20 mM NaOAc buffer, pH 5, containing 0.01% (w/v) thimerosal and incubated with XEG (supplied by Novozymes and purified as described by Pauly et al., 1999). The incubation was performed at 37°C for 24 h in a shaking incubator. The suspension was passed through a glass-fiber filter, and the solid residues were treated a second time with XEG. Three volumes of 95% ethanol were added to the filtrates, and the resulting precipitate was removed by centrifugation (10 min at 3000g). The supernatant containing the oligosaccharides was concentrated by rotary evaporation to remove ethanol and desalted on Sephadex G-25. Xyloglucan oligosaccharides were detected in the G-25 eluant by the anthrone assay for hexoses (Dische, 1962). The salt-free fractions were pooled and lyophilized. The resulting purified xyloglucan oligosaccharides were analyzed by MALDI-TOF MS. The structural features of the oligosaccharides were determined by analysis of diagnostic resonances in their one-dimensional 1H-NMR spectra (http://www.ccrc.uga.edu/xgdb.html).
1 and 4 N KOH Treatments
The remaining insoluble residues after XEG treatment were suspended in 1 N KOH containing 1% (w/v) NaBH4 and stirred at room temperature for 24 h. The suspensions were passed through a glass-fiber filter, and the insoluble residues were collected and suspended in 4 N KOH containing 1% (w/v) NaBH4. The suspensions were stirred at room temperature for 24 h and then filtered. The 1 and 4 N KOH extracts were chilled in an ice bath, acidified, pH 5, with glacial acetic acid, extensively dialyzed (3500 Mr cutoff tubing; Spectrum Laboratories) against running deionized water, and lyophilized.
Glycosyl Composition Analysis of Cell Wall Fractions
Uronic acids in the cell walls were activated with N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide methyl-p-toluenesulfonate powder and reduced with NaBD4 to their respective 6,6-didueterio sugars (Kim and Carpita, 1992; as modified by Carpita and McCann, 1996). Uronosyl-reduced wall materials (1 to 2 mg) were converted into alditol acetates as described previously (Carpita and Shea, 1989). The glycosyl-residue composition was determined by gas chromatography–electron impact mass spectrometry. The proportion of 6,6-dideuteriogalactosyl was calculated using ion abundance ratios at m/z 187/189, 217/219, and 289/291 according to the equation described by Kim and Carpita (1992). The ion abundances of interfering isotopomers (at M+2 D) were determined empirically with nondeuterated samples and subtracted. Total monosaccharide abundances were quantified by GC-FID. The 4-O-methyl glucuronic acid was reduced to 4-O-methyl glucose that was quantified using 3-O-methyl glucose (Calbiochem) as standard that has the identical molar-response factor (Sweet et al., 1975).
Cellulose Determination
Samples (2 to 4 mg) of the residues obtained after KOH treatment and uronosyl reduction were suspended at room temperature for 3 h in concentrated H2SO4 containing 1 μmol of myo-inositol as internal standard. After dilution to 1 M H2SO4 by addition of water, the reaction was heated to 120°C for 1 h. The sample was cooled, neutralized with BaCO3, centrifuged, filtered through glass-fiber filter (GF/A; Whatman), and dried under a stream of air. Similar samples (1 to 2 mg) were hydrolyzed with 1 mL of 2 M TFA containing 1 μmol of myo-inositol as internal standard at 120°C for 1.5 h. The materials released in both hydrolysis were converted to alditol acetates and analyzed as described above.
β-Endoxylanase Treatment
The 1 and 4 N KOH fractions were incubated with a β-endoxylanase from Trichoderma viride (catalog number 70502; Megazyme) at 37°C for 24 h in a shaking incubator. The digested products were desalted and separated by size-exclusion chromatography on a Sephadex G-25 column (Sigma-Aldrich). Carbohydrate in the eluant was monitored by the phenol-sulfuric assay (DuBois et al., 1956). Fractions containing the oligosaccharides were pooled and lyophilized.
MALDI-TOF MS
Xylan oligosaccharides obtained from endo-β-xylanase digestion were analyzed by MALDI-TOF MS using a Hewlett-Packard LDI 1700 XP spectrometer operated in the positive-ion mode with an accelerating voltage of 30 kV, an extractor voltage of 9 kV, and a source pressure of ∼8 × 10−7 torr. The matrix was prepared by mixing (1:1 v/v) 2,5-dihydroxbenzoic acid (0.2 M) and 1-hydroxyisoquinoline (0.06 M), both in 50% aqueous MeCN. The typical spectra shown represent the sums of 200 such laser shots.
NMR Spectroscopy
Purified xylan oligosaccharides were dissolved in D2O (0.6 mL, 99.9%; Cambridge Isotope Laboratories) and transferred to a 5-mm NMR tube. NMR spectra were recorded at 298 K with an Inova-600 MHz NMR spectrometer. Chemical shifts were measured relative to internal acetone at δ 2.225. Two-dimensional gCOSY, HSQC, and HMBC spectra (Kessler et al., 1988) were recorded using standard pulse sequences provided by Varian.
Accession Numbers
Sequence data for the sequences shown in Figure 5 can be found in the GenBank/EMBL data libraries under the following accession numbers: FRA8 (DQ182567 and DQ182568), At5g22940 (BT011629), At1g27440 (BT022053), At5g61840 (AY054180), NpGUT1 (AB080676), and MUR3 (AY195743).
Acknowledgments
This work was supported by grants from the U.S. Department of Energy-Bioscience Division (DE-FG02-03ER15415 to Z.-H.Y. and DE-FG05-93ER20097 and DE-FG02-96ER20220 to M.J.P., A.G.D., and W.S.Y.). We would like to thank Malcolm O'Neill and Carl Bergmann of the Complex Carbohydrate Research Center for helpful discussions and for providing the pectolytic enzymes used in this research and the editor and reviewers for their constructive suggestions.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: William S. York (will@ccrc.uga.edu) and Zheng-Hua Ye (zhye@plantbio.uga.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035501.
References
- Alldridge, N.A. (1964). Anomalous vessel elements in wilty-dwarf tomato. Bot. Gaz. 125, 138–142. [Google Scholar]
- Aspeborg, H., et al. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137, 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinall, G.O. (1980). Chemistry of cell wall polysaccharides. In The Biochemistry of Plants, Vol. 3, J. Preiss, ed (London: Academic Press), pp. 473–500.
- Baydoun, E.A.-H., Usta, J.A.-R., Waldron, K.W., and Brett, C.T. (1989. a). A methyltransferase involved in the biosynthesis of 4-O-methylglucuronoxylan in etiolated pea epicotyls. J. Plant Physiol. 135, 81–85. [Google Scholar]
- Baydoun, E.A.-H., Waldron, K.W., and Brett, C.T. (1989. b). The interaction of xylosyltransferase and glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem. J. 257, 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Bouchez, D. (1994). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants, I. Potrykus and G. Spangenberg, eds (Berlin: Springer-Verlag), pp. 19–23.
- Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M.-T., Talbotec, J., Grainier, F., Lahaye, M., Höfte, H., and Truong, H.-N. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14, 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget, E.G., Verma, R., Molhoj, M., and Reiter, W.-D. (2003). The biosynthesis of l-Arabinose in plants: Molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and McCann, M.C. (1996). Some new methods to study plant polyuronic acids and their esters. In Progress in Glycobiology, R. Townsend, and A. Hotchkiss, eds (New York: Marcel Dekker), pp. 595–611.
- Carpita, N.C., and McCann, M.C. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Carpita, N.C., and Shea, E.M. (1989). Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GC-MS) of partially methylated alditol acetates. In Analysis of Carbohydrates by GLC and MS. C.J. Biermann and G.D. MacGinnis, eds (Boca Raton, FL: CRC Press), pp. 157–216.
- Chabannes, M., Barakate, A., Lapierre, C., Marita, J.M., Ralph, J., Pean, M., Danoun, S., Halpin, C., Grima-Pettenati, J., and Boudet, A.M. (2001). Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 28, 257–270. [DOI] [PubMed] [Google Scholar]
- Coutinho, P.M., Deleury, E., Davies, G.J., and Henrissat, B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317. [DOI] [PubMed] [Google Scholar]
- Dalessandro, G., and Northcote, D.H. (1981. a). Xylan synthetase activity in differentiated xylem cells of sycamore trees (Acer pseudoplatanus). Planta 151, 53–60. [DOI] [PubMed] [Google Scholar]
- Dalessandro, G., and Northcote, D.H. (1981. b). Increase of xylan synthetase activity during xylem differentiation of the vascular cambium of sycamore and poplar trees. Planta 151, 61–67. [DOI] [PubMed] [Google Scholar]
- Darvill, J.E., McNeil, M., Darvill, A.G., and Albersheim, P. (1980). The structure of plant cell walls. 11. Glucuronoarabinoxylan, a second hemicellulose in the primary cell wall of suspension-cultured sycamore cells. Plant Physiol. 66, 1135–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- Dhugga, K.S., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G.S., Dolan, M., Kinney, A.J., Tomes, D., Nichols, S., and Anderson, P. (2004). Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303, 363–366. [DOI] [PubMed] [Google Scholar]
- Dische, Z. (1962).General color reactions: Color reactions of carbohydrates. In Methods in Carbohydrate Chemistry, R.L. Whistler and M.L. Wolfrom, eds (New York: Academic Press), pp. 478–481.
- DuBois, M., Gilles, K.A., Hamilton, J.K., Rebers, P.A., and Smith, F. (1956). Colorimtric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. [Google Scholar]
- Ebringerová, A., and Heinze, T. (2000). Xylan and xylan derivatives-biopolymers with valuable properties, 1. Naturally occurring xylans structures, isolation procedured and properties. Macromol. Rapid Commun. 21, 542–556. [Google Scholar]
- Edwards, M., Bulpin, P.V., Dea, I.C.M., and Reid, J.S.G. (1989). Biosynthesis of legume-seed galactomannans in vitro. Planta 178, 41–51. [DOI] [PubMed] [Google Scholar]
- Edwards, M.E., Dickson, C.A., Chengappa, S., Sidebottom, C., Gidley, M.J., and Reid, J.S.G. (1999). Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19, 691–697. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Höfte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, S.R. (1992). GUS Protocols. (San Diego, CA: Harcourt Brace Jovanovich).
- Goujon, T., Ferret, V., Mila, I., Pollet, B., Ruel, K., Burlat, V., Joseleau, J.P., Barriere, Y., Lapierre, C., and Jouanin, L. (2003). Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217, 218–228. [DOI] [PubMed] [Google Scholar]
- Gregory, A.C.E., Smith, C., Kerry, M.E., Wheatley, E.R., and Bolwell, G.P. (2002). Comparative subcellular immunolocation of polypeptides associated with xylan and callose synthases in French bean (Phaseolus vulgaris) during secondary wall formation. Phytochemistry 59, 249–259. [DOI] [PubMed] [Google Scholar]
- Habibi, Y., Mahrouz, M., and Vignon, M.R. (2002). Isolation and structure of D-xylans from pericarp seeds of Opuntia ficus-indica prickly pear fruits. Carbohydr. Res. 337, 1593–1598. [DOI] [PubMed] [Google Scholar]
- Hazen, S.P., Scott-Craig, J.S., and Walton, J.D. (2002). Cellulose synthase-like genes of rice. Plant Physiol. 128, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- His, I., Driouich, A., Nicol, F., Jauneau, A., and Höfte, H. (2001). Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-β-glucanase. Planta 212, 348–358. [DOI] [PubMed] [Google Scholar]
- Hoebler, C., Barry, J.L., David, A., and Delort-Laval, J. (1989). Rapid acid-hydrolysis of plant cell wall polysaccharides and simplified quantitative determination of their neutral monosaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Iwai, H., Masaoka, N., Ishii, T., and Satoh, S. (2002). A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc. Natl. Acad. Sci. USA 99, 16319–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Z., Qin, Q., Darvill, A.G., and York, W.S. (2003). Structure of the xyloglucan produced by suspension-cultured tomato cells. Carbohydr. Res. 338, 1197–1208. [DOI] [PubMed] [Google Scholar]
- Jones, L., Ennos, A.R., and Turner, S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- Kauss, H., and Hassid, W.Z. (1967). Biosynthesis of the 4-O-methyl-D-glucuronic acid unit of hemicellulose B by transmethylation from S-adenosyl-L-methionine. J. Biol. Chem. 242, 1680–1684. [PubMed] [Google Scholar]
- Keegstra, K., and Raikhel, N. (2001). Plant glycosyltransferases. Curr. Opin. Plant Biol. 4, 219–224. [DOI] [PubMed] [Google Scholar]
- Kessler, H., Gehrke, M., and Griesinger, C. (1988). Two-dimensional NMR spectroscopy: Background and overview of the experiments. Angew. Chem. Int. Ed. Engl. 27, 490–536. [Google Scholar]
- Kim, J.-B., and Carpita, N.C. (1992). Changes in esterification of the uronic acids groups of cell wall polysaccharides during elongation of maize coleoptiles. Plant Physiol. 98, 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kuroyama, H., and Tsumuraya, Y. (2001). A xylosyltransferase that synthesizes β-(1->4)-xylans in wheat (Triticum aestivum L.) seedlings. Planta 213, 231–240. [DOI] [PubMed] [Google Scholar]
- Levy, S., and Staehelin, L.A. (1992). Synthesis, assembly and function of plant cell wall macromolecules. Curr. Opin. Cell Biol. 4, 856–862. [DOI] [PubMed] [Google Scholar]
- Li, X., Cordero, I., Caplan, J., Molhoj, M., and Reiter, W.D. (2004). Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol. 134, 940–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A.H., Wilkerson, C.G., and Keegstra, K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 102, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, T., Tufaro, F., McCormick, C., Lindahl, U., and Lidholt, K. (1998). The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 273, 26265–26268. [DOI] [PubMed] [Google Scholar]
- Liu, Z.B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G.F., Caplan, J., Shoue, D.A., Carpita, N.C., and Reiter, W.D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, M., Darvill, A.G., Fry, S.C., and Albersheim, P. (1984). Structure and function of the primary walls of plants. Annu. Rev. Biochem. 53, 625–663. [DOI] [PubMed] [Google Scholar]
- Orfila, C., Sorensen, S.O., Harholt, J., Geshi, N., Crombie, H., Truong, H.-N., Reid, J.S.G., Knox, J.P., and Scheller, H.V. (July 30, 2005). QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta http://dx.doi.org/10.1007/s00425-005-0008-z. [DOI] [PubMed]
- Pauly, M., Andersen, L.N., Kauppinen, S., Kofod, L.V., York, W.S., Albersheim, P., and Darvill, A.G. (1999). A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: Expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology 9, 93–100. [DOI] [PubMed] [Google Scholar]
- Perrin, R.M., Jia, Z., Wagner, T.A., O'Neil, M.A., Sarria, R., York, W.S., Raikhel, N.V., and Keegstra, K. (2003). Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol. 132, 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincon, G., Chabannes, M., Lapierre, C., Pollet, B., Ruel, K., Joseleau, J.P., Boudet, A.M., and Legrand, M. (2001). Simultaneous down-regulation of caffeic/5-hydroxy ferulic acid-O-methyltransferase I and cinnamoyl-coenzyme A reductase in the progeny from a cross between tobacco lines homozygous for each transgene. Consequences for plant development and lignin synthesis. Plant Physiol. 126, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis, A., Domingues, M.R.M., Ferrer-Correia, A.J., and Coimbra, M.A. (2003). Structural characterisation by MALDI-MS of olive xylo-oligosaccharides obtained by partial acid hydrolysis. Carbohydr. Polym. 53, 101–107. [Google Scholar]
- Reis, D., and Vian, B. (2004). Helicoidal pattern in secondary cell walls and possible role of xylans in their construction. C. R. Biol. 327, 785–790. [DOI] [PubMed] [Google Scholar]
- Reiter, W.-D., Chapple, C.C.S., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335–345. [DOI] [PubMed] [Google Scholar]
- Richmond, T.A., and Somerville, C.R. (2000). The cellulose synthase superfamily. Plant Physiol. 124, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C.M. (1952). The grafting relations of wilty dwarf, a new tomato mutant. Am. Nat. 86, 173–184. [Google Scholar]
- Rowley, J.C., and Moran, D.T. (1975). A simple technique for mounting wrinkle-free sections on formvar-coated slot grids. Ultramicroscopy 1, 151–155. [DOI] [PubMed] [Google Scholar]
- Scheible, W.-R., and Pauly, M. (2004). Glycosyltransferases and cell wallbiosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 7, 285–295. [DOI] [PubMed] [Google Scholar]
- Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., Osborne, E., Paredez, A., Persson, S., Raab, T., Vorwerk, S., and Youngs, H. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sugahara, K., and Kitagawa, H. (2000). Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 10, 518–527. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Ingold, E., Sugiyama, M., and Komamine, A. (1991). Xylan synthase activity in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements. Plant Cell Physiol. 32, 303–306. [Google Scholar]
- Sweet, D.P., Shapiro, R.H., and Albersheim, P. (1975). Quantitative analysis by various g.l.c. response-factor theories for partially methylated and partially ethylated alditol acetates. Carbohydr. Res. 40, 217–225. [Google Scholar]
- Taylor, J., and Mims, C.W. (1991). Fungal development and host cell responses to the rust fungus Puccinia substriata var. indica in seedlings and mature leaves of susceptible and resistant pearl millet. Can. J. Bot. 69, 1207–1219. [Google Scholar]
- Taylor, N.G., Gardiner, J.C., Whiteman, R., and Turner, S.R. (2004). Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11, 329–338. [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman, A., Lundqvist, J., Tjerneld, F., Stålbrand, H., and Dahlman, O. (2000). Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr. Res. 329, 807–815. [DOI] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen, M.A., Spronk, B.A., Schols, H.A., Beldman, G., Voragen, A.G.J., Thomas, J.R., Kamerling, J.P., and Vliegenthart, J.F.G. (1998). Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr. Res. 306, 265–274. [DOI] [PubMed] [Google Scholar]
- Waldron, K.W., and Brett, C.T. (1983). A glucuronyltransferase involved in glucuronoxylan synthesis in pea (Pisum sativum) epicotyls. Biochem. J. 213, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, G., Bai, X., Gabb, M.M.G., Bame, K.J., Koshy, T.I., Spear, P.G., and Esko, J.D. (2000). Location of the glucuronyltransferase domain in the heparan sulfate copolymerase EXT1 by analysis of Chinese hamster ovary cell mutants. J. Biol. Chem. 275, 27733–27740. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H., Freshour, G., Hahn, M.G., Burk, D.H., and Zhong, R. (2002). Vascular development in Arabidopsis. Int. Rev. Cytol. 220, 225–256. [DOI] [PubMed] [Google Scholar]
- Zablackis, E., Huang, J., Müller, B., Darvill, A.G., and Albersheim, P. (1995). Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Morrison III, W.H., Freshour, G., Hahn, M., and Ye, Z.-H. (2003). Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132, 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2003). Unraveling the functions of glycosyltransferase family 47 in plants. Trends Plant Sci. 8, 565–568. [DOI] [PubMed] [Google Scholar]