Abstract
ABSCISIC ACID–RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1) (i.e., ABF2) is a basic domain/leucine zipper transcription factor that binds to the abscisic acid (ABA)–responsive element (ABRE) motif in the promoter region of ABA-inducible genes. Here, we show that expression of the intact AREB1 gene on its own is insufficient to lead to expression of downstream genes under normal growth conditions. To overcome the masked transactivation activity of AREB1, we created an activated form of AREB1 (AREB1ΔQT). AREB1ΔQT-overexpressing plants showed ABA hypersensitivity and enhanced drought tolerance, and eight genes with two or more ABRE motifs in the promoter regions in two groups were greatly upregulated: late embryogenesis abundant class genes and ABA- and drought stress–inducible regulatory genes. By contrast, an areb1 null mutant and a dominant loss-of-function mutant of AREB1 (AREB1:RD) with a repression domain exhibited ABA insensitivity. Furthermore, AREB1:RD plants displayed reduced survival under dehydration, and three of the eight greatly upregulated genes were downregulated, including genes for linker histone H1 and AAA ATPase, which govern gene expression and multiple cellular activities through protein folding, respectively. Thus, these data suggest that AREB1 regulates novel ABRE-dependent ABA signaling that enhances drought tolerance in vegetative tissues.
INTRODUCTION
Water deficit leads directly to fatal damage in all living things. Hence, the molecular machinery involved in crucial stress responses has developed redundant and complex signal transduction networks that overcome and circumvent fatal injury. However, the inherent complexity and redundancy cause difficulties in the analysis of the key signal transduction pathways. In this study, to overcome this problem, we have used two approaches: (1) an active form of a transcription factor to activate expression of target genes without an originally required posttranscriptional modification and (2) a transcription factor fused to a repression domain (RD) that consists of only 12 amino acids to overcome potential functional redundancy conferred by homologs (Hiratsu et al., 2003).
The plant hormone abscisic acid (ABA) regulates many essential processes, including inhibition of germination, maintenance of seed dormancy, control of stomatal closure, and adaptive responses to a variety of environmental stresses (for review, see Finkelstein et al., 2002). In Arabidopsis thaliana, for example, analyses of ABA-hypersensitive mutants have revealed that ABA is involved in cellular processes such as farnesylation (era1), inositol signaling (fry1), and RNA metabolism (abh1, sad1, and hyl1). In addition, genetic screens for mutations that display a reduced sensitivity to ABA have identified homologous type 2C phosphatases (ABI1 and ABI2) and three different classes of transcription factors (ABI3, ABI4, and ABI5).
ABA is produced under dehydration conditions and plays pivotal roles in response to drought stress (Shinozaki and Yamaguchi-Shinozaki, 2000; Finkelstein et al., 2002; Xiong et al., 2002). Numerous drought stress–inducible genes have been reported in vegetative tissues, and many of them are also activated by ABA (Ingram and Bartels, 1996; Seki et al., 2002). In analyses of the promoters of such ABA-regulated genes, a conserved cis-element designated ABA-responsive element (ABRE; PyACGTGGC), which controls ABA-regulated gene expression, has been identified (Bray, 1994; Giraudat et al., 1994; Busk and Pages, 1998). Various types of ABRE-like sequences have been reported, including the G-box sequence (CACGTG) and coupling element (CGCGTG), CE3, hex3, and motif III (Shen et al., 1996; Busk and Pages, 1998). So far, all isolated interactors with divergent types of ABRE sequences belong to the basic domain/leucine zipper (bZIP) class of transcription factors and can bind to them at least in vitro (Foster et al., 1994; Busk and Pages, 1998).
The RD29B promoter region carries two ABRE sequences, and the drought-inducible expression of RD29B is controlled mainly by ABA, according to analyses in the ABA-deficient and -insensitive mutants aba1 and abi1, respectively (Koornneef et al., 1984, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994). Using yeast one-hybrid screening with the RD29B promoter including the ABRE sequences, we cloned three different cDNAs encoding ABRE binding proteins (AREB1, AREB2, and AREB3) of Arabidopsis (Uno et al., 2000). Expression of AREB1 and AREB2 is upregulated by ABA and by drought and high-salinity stresses. Both AREB1 and AREB2 function as trans-acting activators, as identified by transient expression analysis in protoplasts (Uno et al., 2000). In the Arabidopsis genome, to date, nine AREB homologs have been identified: AREB1/ABF2, AREB2/ABF4, AREB3/DPBF3, ABF1, ABF3/DPBF5, ABI5/DPBF1, EEL/DPBF4, DPBF2, and AT5G42910 (Choi et al., 2000; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000; Bensmihen et al., 2002; Jakoby et al., 2002; Kim et al., 2002; Suzuki et al., 2003). In this study, we show that expression of only three members, AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5, is induced by ABA, drought, and high salinity in vegetative tissues. Because AREB1 has the highest ABA and drought inducibility in RNA gel blot, histochemical, and transient analyses, we focus on AREB1/ABF2 in order to elucidate its role in ABA-dependent responses to drought in vegetative tissues.
Here, we report that AREB1 is a key positive regulator of ABA signaling in vegetative tissues under drought stress. Our results indicate that AREB1 directs expression of ABA- and dehydration-inducible regulatory genes such as linker histone H1-3 and AAA ATPase genes, as well as late embryogenesis abundant (LEA) class genes, which are thought be involved in alleviation of water stress. Also, we demonstrate that two powerful tools, a constitutive active form and a dominant loss-of-function mutant with an RD, are useful for dissecting important transcription factors that seem to be required for posttranscriptional modification or in which there is expected to be phenotypic masking due to potential functional redundancy.
RESULTS
Three bZIP Proteins Are Involved in the ABA-Mediated Signal Transduction Pathway under Drought and High-Salinity Conditions
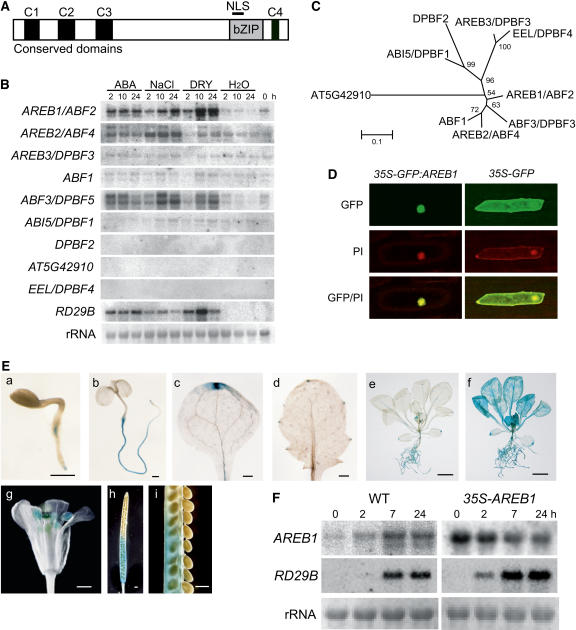
The Arabidopsis AREB/ABF/ABI5/DPBF bZIP subfamily consists of nine members that contain three N-terminal (C1, C2, and C3) and one C-terminal (C4) conserved domains (Bensmihen et al., 2002; Jakoby et al., 2002; Kim et al., 2002; Suzuki et al., 2003) (Figure 1A). To identify members that are involved in an ABA signal transduction pathway under drought, we conducted RNA gel blot analysis of the nine genes. The expression of three genes, AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5, was induced by dehydration, high-salt, and exogenous ABA treatments (Figure 1B). These three members are in the same clade in the phylogenetic tree of the nine members of the subfamily (Figure 1C).
Figure 1.
Expression of the AREB1 Gene and Subcellular Localization of the AREB1 Protein.
(A) Structure of AREB1 family proteins. NLS, nuclear localization signal. C1 to C4 indicate conserved domains within the family.
(B) Expression profiles of AREB family genes in response to dehydration, high salt, or ABA. Each lane was loaded with 20 μg of total RNA from 3-week-old Arabidopsis plants that had been dehydrated (DRY), transferred to hydroponic growth in 250 mM NaCl (NaCl), transferred to hydroponic growth in 100 μM ABA (ABA), or transferred to water (H2O). rRNAs are shown as equal loading controls. A band located in the center of each column indicates a transcript that corresponds to each gene.
(C) Phylogenetic tree of AREB family proteins. Proteins were aligned using ClustalX software, and the tree was constructed using MEGA software.
(D) Nuclear localization of AREB1 protein in onion epidermal cells: fluorescent images of GFP, fluorescent images stained with propidium iodide (PI), and merged images (GFP/PI).
(E) Patterns of AREB1 promoter-driven GUS expression in seedlings at different ages or in different tissues: (a) 2-d-old seedling, (b) 5-d-old seedling, (c) cotyledon, (d) primary leaf, (e) 2-week-old seedling, (f) 2-week-old seedling treated with 50 μM ABA, (g) flower, (h) immature silique, (i) seeds from (h). Bars = 0.5 mm in (a) to (d) and (g) to (i) and 5.0 mm in (e) and (f).
(F) Expression of AREB1 and RD29B in wild-type and 35S-AREB1 plants (line 6) induced by 50 μM ABA treatment. Representative data are shown. Each lane was loaded with 15 μg of total RNA from 2-week-old Arabidopsis plants. rRNAs are shown as equal loading controls.
More AREB1/ABF2 mRNA accumulated under dehydration stress than under high-salt treatment, but the reverse was true for AREB2/ABF4 and ABF3/DPBF5 mRNAs (Figure 1B; Uno et al., 2000). Furthermore, in transactivation experiments using Arabidopsis protoplasts, the ABA-induced transactivation ability of AREB1 was higher than that of AREB2 (Uno et al., 2000). Thus, these data suggest that AREB1/ABF2 is a key regulator of ABA signaling under drought stress. Therefore, we focused on AREB1/ABF2. AREB1/ABF2, AREB2/ABF4, and ABF3/DPBF5 are referred to hereafter as AREB1, AREB2, and ABF3, respectively.
AREB1 Is Localized in the Nucleus
The AREB1 nuclear localization signal is located in the basic region of the bZIP DNA binding domain (Figure 1A). To examine the subcellular localization of the AREB1 protein in plant cells, we fused the AREB1 coding region in frame to the coding region for the C-terminal side of green fluorescent protein (GFP), and the fusion gene was expressed under the control of the 35S promoter of Cauliflower mosaic virus (CaMV). Onion epidermal cells transformed with an expression plasmid for the GFP:AREB1 fusion protein exhibited GFP fluorescence in the nucleus (Figure 1D). By contrast, GFP fluorescence was observed in the entire region of the cell when intact GFP was expressed. These results show that AREB1 is localized in the nucleus.
AREB1 Is Expressed Constitutively in Roots, Leaf Vascular Tissues, and Hydathodes or in All Tissues under Stress Conditions
Previously, we detected weak basal AREB1 expression in roots and leaves but not in seeds in an RNA gel blot analysis (Uno et al., 2000). Here, to determine the temporal and spatial expression patterns of AREB1 in more detail, we analyzed transgenic Arabidopsis plants expressing an AREB1 promoter–β-glucuronidase (GUS) transgene. GUS expression was observed in roots at all developmental stages that we assessed (Figure 1E, a, b, e, and f). In unstressed plants, leaf vascular tissues and hydathodes also exhibited AREB1 promoter activity (Figure 1E, c to e). By contrast, ABA or drought treatment of seedlings enhanced the AREB1 promoter activity in all tissues (Figure 1E, f; data not shown). In mature plants, GUS activity was observed also in anthers, stigma, and siliques (abscission zone and carpels) (Figure 1E, g and h). The false septum and carpels were stained gradually from the abscission zone to the remains of the stigma with maturity (Figure 1E, h and i).
Expression of AREB1 on Its Own Is Insufficient to Induce the Expression of the Downstream Gene RD29B
To assess the in vivo function of AREB1, we generated transgenic plants expressing the AREB1 cDNA under the control of the CaMV 35S promoter (35S-AREB1). Thirty-six T3 homozygous lines were obtained, and eight transgenic lines with higher expression levels of the transgene were selected for further analysis by RNA gel blot analysis.
No obvious difference was observed in growth phenotypes between the wild-type and 35S-AREB1 plants growing on GM agar plates containing 1 or 3% sucrose, implying the possibility that constitutive overexpression of intact AREB1 on its own is not sufficient to activate downstream genes such as RD29B. To examine this possibility, we monitored RD29B mRNA levels in the 35S-AREB1 transgenic plants at several time points with or without exogenous ABA. As shown in Figure 1F, constitutive overexpression of AREB1 did not activate expression of the downstream RD29B in the absence of exogenous ABA (time point 0). Within 2 h after the addition of ABA, RD29B was expressed in the 35S-AREB1 plants but not in the wild-type plants. These results indicate that not only the presence of AREB1 in plants but also the exogenous addition of ABA is required for the expression of downstream genes such as RD29B. These data also suggest that the preexistence of AREB1 before the addition of ABA contributed to earlier accumulation of RD29B mRNA in the 35S-AREB1 plants than in the wild-type plants. Taken together with our previous report of ABA-dependent phosphorylation of the N-terminal region of AREB1 and suppression of the activation of AREB1 by protein kinase inhibitors in a transient assay using protoplasts (Uno et al., 2000), in addition to the ABA-dependent expression of AREB1, these results indicate that the ABA-induced modification of the AREB1 protein seems to be also required for the expression of its downstream genes.
The N-Terminal Conserved Region of AREB1 Has Transactivation Activity in Protoplasts
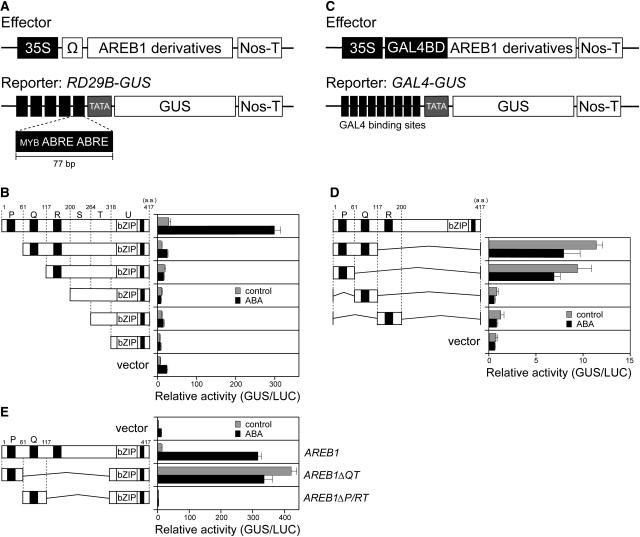
In a previous study, we showed that the AREB1 protein transactivates the RD29B promoter–GUS fusion gene (RD29B-GUS) in Arabidopsis protoplasts (Uno et al., 2000). To identify the transcriptional activation domain of AREB1, we constructed a series of effector plasmids bearing N-terminal deletion mutants of AREB1 under the control of the constitutive CaMV 35S promoter (Figure 2). The effector plasmids were cotransfected into protoplasts prepared from Arabidopsis T87 cultured cells, with a reporter plasmid, RD29B-GUS, carrying a GUS reporter gene fused to five tandem copies of a 77-bp fragment of the RD29B promoter containing two ABRE motifs (Figure 2A). A small deletion of 60 amino acids (region P) from the N terminus of AREB1 produced a significant decrease in the transactivation of the reporter gene in protoplasts treated either with or without 100 μM ABA, suggesting the presence of a positive regulatory domain in this region (Figure 2B).
Figure 2.
The N-Terminal Conserved Region of AREB1 Functions as a Transcriptional Activation Domain in Protoplasts Derived from Arabidopsis T87 Cultured Cells.
All transactivation experiments were performed 3 to 10 times, and results from one representative experiment are shown. Bars indicate standard deviation; n = 3 to 5.
(A) Scheme of the effector and reporter constructs used in the transactivation analysis with AREB1 bZIP DNA binding domain. The effector constructs contain the CaMV 35S promoter and TMV Ω sequence fused to AREB1 cDNA fragments encoding different portions of AREB1. The reporter construct, RD29B-GUS, contains 77-bp fragments of the RD29B promoter connected tandemly five times. The promoters were fused to the −51 RD29B minimal TATA promoter–GUS construct. Nos-T, nopaline synthase terminator.
(B) Transactivation domain analysis of AREB1 using N-terminal deletion constructs. Protoplasts were cotransfected with the RD29B-GUS reporter and the effector construct (shown on the left) carrying an N-terminal truncated form of AREB1 cDNA or pBI-35SΩ (vector). To normalize for transfection efficiency, the pBI35SΩ-LUC reporter was cotransfected as a control in each experiment. Bars indicate standard deviation of three replicates. “Relative activity” indicates the multiples of expression compared with the value obtained with the pBI221-35SΩ vector control. Top numbers indicate amino acid numbers of AREB1. P, Q, R, S, T, and U indicate the partial region of the AREB1 cDNA. The region P, Q, R, or U includes a conserved domain (solid rectangle), C1, C2, C3, or C4, respectively, in Figure 1A.
(C) Schematic diagram of the effector and reporter constructs used in the transactivation analysis with the GAL4 DNA binding domain. The effector plasmids encoding the GAL4 DNA binding domain are fused to AREB1 cDNA fragments encoding different portions of AREB1. The GUS reporter construct, GAL4-GUS, containing GAL4 binding sites, is fused to the minimal promoter of CaMV 35S.
(D) N-terminal conserved P region of AREB1 contains sufficient domain for transcriptional activation. Protoplasts were cotransfected with the GAL4-GUS reporter and the effector construct expressing a portion of AREB1 or the vector DNA.
(E) AREB1ΔQT is a constitutive active form of AREB1. Protoplasts were cotransfected with the RD29B-GUS reporter and the effector construct expressing intact AREB1, AREB1ΔQT, or AREB1ΔP/RT, or vector DNA.
To determine whether the N-terminal P region of AREB1 functions as a transcriptional activation domain in combination with the other DNA binding domain, we constructed a series of effector plasmids that were driven by the CaMV 35S promoter, carrying fusion genes that consisted of the DNA binding domain of the yeast transcriptional activator GAL4 and the PQ (amino acids 1 to 116), P (1 to 60), Q (61 to 116), or R (117 to 199) regions of AREB1 (Figures 2C and 2D). These plasmids were cotransfected into Arabidopsis protoplasts with a reporter plasmid, GAL4-GUS, that contains nine copies of a GAL4 binding site fused to the minimal promoter of CaMV 35S and GUS. The effector plasmids encoding the GAL4-PQ and GAL4-P fusion proteins transactivated the reporter gene, demonstrating that the N-terminal P region of AREB1 functions as a transcriptional activation domain even in combination with a non-native DNA binding domain (Figure 2D).
We further analyzed whether an AREB1 mutant protein containing the P region and its native binding domains transactivates the RD29B-GUS reporter gene without exogenous ABA treatment. AREB1ΔQT and AREB1ΔP/RT were constructed as effector plasmids, carrying the AREB1 internal deletion mutants containing the bZIP DNA binding domain of AREB1 and region P or Q, respectively (Figure 2E). Cotransfection of AREB1ΔQT together with RD29B-GUS resulted in a significant activation of the GUS reporter gene even in the absence of ABA, but that of AREB1ΔP/RT did not activate the reporter gene either with or without ABA. These results indicate that the N-terminal P region of AREB1 contains a transcriptional activation domain, and AREB1ΔQT is a constitutive active form of AREB1 in protoplasts (Figure 2E). Also, significant reduction of activation in the AREB1ΔP/RT deletion mutants or most of the N-terminal deletion mutants of AREB1 compared with the vector control in the presence of ABA suggests that overexpression of the AREB1ΔP/RT or N-terminal deletion mutants dominantly inhibits ABA-induced binding of endogenous transcription factors to the ABRE motifs of the reporter plasmids (Figures 2B and 2E). Among N-terminal deletion mutants of AREB1, however, only the deletion mutant lacking the P region does not seem to repress the activation of the reporter gene in the presence of ABA, suggesting that the deletion mutant lacking the P region may not dominantly inhibit such ABA-induced binding owing to the sequence-specific effect of the deletion mutant (Figure 2B).
AREB1ΔQT Is a Constitutive Active Form of AREB1 in Planta
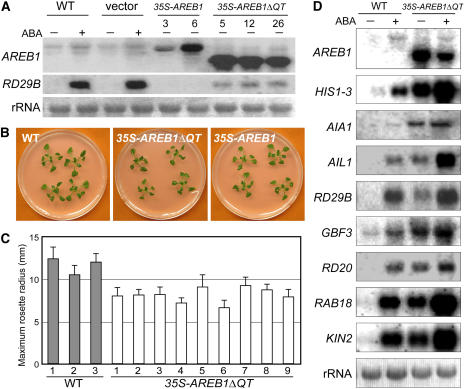
Our previous study showed that the AREB1 protein binds to two ABRE sequences in the RD29B promoter and activates expression of the gene (Uno et al., 2000). As described above, overexpression of intact AREB1 does not activate the expression of the downstream RD29B gene in the absence of exogenous ABA. We then analyzed whether AREB1ΔQT overexpression in Arabidopsis plants activates the transcription of downstream genes such as RD29B in the absence of exogenous ABA. Transgenic plants expressing the AREB1ΔQT cDNA under the control of the CaMV 35S promoter were generated, and expression of the AREB1ΔQT transgene and the downstream RD29B gene was analyzed in 33 independent transgenic lines under unstressed conditions. The accumulation levels of both AREB1ΔQT and RD29B mRNA were elevated in all examined lines in the absence of exogenous ABA (data not shown); so, we selected eight transgenic lines with higher AREB1ΔQT expression levels for phenotypic analysis. Because the eight transgenic lines behaved in a similar manner (data not shown) and because the number of seeds in each line was sometimes not enough to allow statistical analysis, in some cases, we used different lines in different experiments. Overexpression of AREB1ΔQT in plants activated the expression of the downstream RD29B gene in the absence of exogenous ABA (Figure 3A). This result shows that AREB1ΔQT is a constitutive active form of AREB1 even in whole plants and that the N-terminal P region of AREB1 functions as a transcriptional activation domain in plants as well as in protoplasts. At 3 weeks after stratification, the maximum rosette radius of the 35S-AREB1ΔQT plants averaged 70% of that of the wild-type plants (Figures 3B and 3C). The 35S-AREB1ΔQT plants were slightly smaller than the wild type throughout their life (data not shown). By contrast, the 35S-AREB1 transgenic plants showed a similar growth phenotype to that of the wild type (Figure 3B).
Figure 3.
AREB1ΔQT Is a Constitutive Active Form of AREB1 in Planta.
(A) RNA gel blot analysis of AREB1 and RD29B expression in wild-type, vector control (vector), 35S-AREB1, and 35S-AREB1ΔQT plants. Two-week-old seedlings were either not treated (−) or treated (+) with ABA for 7 h. Each lane contained 10 μg of total RNA. Two lines of the 35S-AREB1 plants (3 and 6) and three lines of the 35S-AREB1ΔQT plants (5, 12, and 26) are shown. rRNAs on ethidium bromide–stained gel are shown as equal loading controls.
(B) Growth phenotype of 35S-AREB1ΔQT (line 5) and 35S-AREB1 (line 6) plants that were grown for 3 weeks on GM agar plates containing 1% sucrose.
(C) Maximum rosette radius (i.e., length of the longest rosette leaf) of each plant on a GM agar plate containing 3% sucrose was measured 3 weeks after stratification. Three independent lines of wild-type plants and nine independent lines of 35S-AREB1ΔQT plants were used. Bars indicate standard deviation; n = 7.
(D) Expression profile of downstream genes identified by microarray analysis (Table 1) in 35S-AREB1ΔQT plants (line 5). Two-week-old seedlings were either not treated (−) or treated (+) with ABA for 7 h. Each lane contained 7 μg of total RNA. Three to eight independent lines were used, and results from one representative experiment are shown. rRNAs on ethidium bromide–stained gel are shown as equal loading controls.
Overexpression of AREB1ΔQT Leads to Expression of LEA Proteins and Activation of ABRE-Dependent Signal Transduction Pathways
Recent studies using genome-wide location analysis, which combines chromatin immunoprecipitation with DNA microarray technology, have demonstrated that the selection of target genes is determined at the level of DNA binding rather than by signaling events after DNA binding (Ren et al., 2000; Zeitlinger et al., 2003). Furthermore, DNA microarray analysis in combination with active forms of transcription factors carrying activation and DNA binding domains revealed that the DNA binding domain alone is sufficient to confer target gene specificity of the native transcription factors (Devaux et al., 2001). On the basis of these reports, we used 35S-AREB1ΔQT plants, which express a constitutive active form of AREB1 carrying the activation and DNA binding domains of AREB1, to identify target genes of AREB1. We compared the expression profiles in two independent lines (numbers 8 and 12) of 2-week-old 35S-AREB1ΔQT plants under unstressed conditions with that of wild-type plants using Agilent near-full-genome gene chips (Arabidopsis 22K) with Agilent's propagated error method (http://www.chem.agilent.com/scripts/generic.asp?lpage=11617&indcol=Nandprodcol=Y). Among the 22,000 genes represented on the array, only 31 showed more than a threefold increase in transcript accumulation in the 35S-AREB1ΔQT plants (P < 0.001; Table 1). As shown in Table 1, many genes with higher changes in expression seem to respond to multiple stresses. Among the top 11 genes with greatest increase in expression, eight were induced by both dehydration and exogenous ABA treatment and carry two or more ABRE motifs in the promoter regions (Table 1). Moreover, using RNA gel blot analysis, we confirmed that the all eight of these genes were ABA inducible and were expressed even in the absence of ABA in the 35S-AREB1ΔQT transgenic plants, suggesting that these eight genes are candidates for direct target genes of AREB1 (Figure 3D). These eight genes are divided into two groups. Four genes encode LEA or LEA-like proteins: At3g17520 (encoding a group 3 LEA class protein; Wise, 2003), RD29B/LTI65 (Nordin et al., 1993; Yamaguchi-Shinozaki and Shinozaki, 1994), RAB18 (Lang and Palva, 1992), and KIN2/COR6.6 (Gilmour et al., 1992; Kurkela and Borg-Franck, 1992). The other four genes encode regulatory proteins: HIS1-3 (encoding a linker histone H1; Ascenzi and Gantt, 1997), At1g64110 (encoding an AAA ATPase), GBF3 (Schindler et al., 1992), and RD20 (Yamaguchi-Shinozaki et al., 1992). Two novel candidate target genes, At3g17520 and At1g64110, were named AIL1 (for ABA-inducible LEA class gene) and AIA1 (for ABA-inducible AAA ATPase gene), respectively. These data suggest that AREB1 plays an important role in the ABRE-dependent ABA signal transduction pathway. For convenience, we refer to the RD29B/LTI65 gene as RD29B and to KIN2/COR6.6 as KIN2 in this report.
Table 1.
Genes Upregulated in Plants Overexpressing AREB1ΔQT, Identified by Microarray Analysis
| Experiment 1 (Transgenic Line 8)a |
Experiment 2 (Transgenic Line 12)a |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inducibilityb | Genec | Descriptionc | RAFL Clone No.d | AGI Codee | Median of Fold Changef | Fold Change 1 | P Value 1g | Fold Change 2 | P Value 2g | Fold Change 1 | P Value 1g | Fold Change 2 | P Value 2g | No. of ABREsh | |||
| D | – | A | – | HIS1-3 | Linker histone H1 | RAFL05-20-P13 | AT2G18050 | 15.5 | 17.6 | 5.6E-32 | 20.9 | 1.5E-32 | 12.1 | 3.2E-30 | 13.3 | 1.1E-30 | 2 |
| D | S | A | – | AIA1 | AAA family ATPase | RAFL08-19-A04 | AT1G64110 | 13.3 | 12.8 | 9.1E-30 | 8.6 | 7.1E-26 | 13.9 | 1.1E-30 | 16.1 | 1.2E-30 | 2 |
| D | S | A | – | AIL1 | LEA class protein | RAFL15-06-F04 | AT3G17520 | 13.0 | 9.8 | 1.2E-28 | 8.5 | 4.0E-27 | 28.0 | 2.2E-33 | 16.2 | 1.5E-31 | 2 |
| – | – | – | – | Dihydroflavonol 4-reductase | AT5G42800 | 7.9 | 15.1 | 8.2E-30 | 7.5 | 3.2E-24 | 6.4 | 3.6E-22 | 8.3 | 4.9E-21 | 2 | ||
| D | S | A | – | RD29B | Transcription factor | RAFL05-11-I09 | AT5G52300 | 6.6 | 9.8 | 2.0E-27 | 4.6 | 2.1E-18 | 7.3 | 6.5E-25 | 6.0 | 7.2E-21 | 5 |
| D | S | A | – | GBF3 | Transcription factor | RAFL05-09-G15 | AT2G46270 | 5.7 | 5.2 | 1.7E-22 | 4.0 | 1.2E-18 | 6.9 | 6.7E-26 | 6.2 | 2.6E-24 | 8 |
| D | S | A | C | RD20 | Ca2+ binding EF-hand protein | RAFL08-16-M12 | AT2G33380 | 5.5 | 6.5 | 1.3E-25 | 7.3 | 1.6E-26 | 4.5 | 1.9E-21 | 4.5 | 3.1E-21 | 3 |
| D | S | A | C | RAB18 | LEA class protein | RAFL05-18-M18 | AT5G66400 | 5.5 | 5.7 | 3.2E-24 | 4.1 | 5.0E-20 | 6.0 | 8.0E-25 | 5.2 | 4.0E-23 | 5 |
| – | – | A | – | Protease inhibitor/seed storage/LTP family | RAFL05-12-N10 | AT2G37870 | 5.4 | 4.6 | 2.3E-21 | 6.9 | 1.3E-25 | 5.6 | 5.2E-24 | 5.1 | 8.6E-23 | 1 | |
| D | S | – | C | Φ-1-like protein | RAFL03-02-F02 | AT5G64260 | 4.8 | 6.9 | 4.5E-26 | 5.6 | 6.9E-24 | 4.1 | 6.8E-20 | 4.1 | 9.1E-20 | 3 | |
| D | S | A | C | KIN2 | LEA class protein | RAFL04-17-B12 | AT5G15970 | 4.8 | 4.0 | 7.3E-20 | 5.8 | 2.4E-24 | 4.5 | 2.0E-21 | 5.1 | 5.2E-23 | 6 |
| – | – | – | – | Putative isocitrate lyase | RAFL06-07-C24 | AT3G21720 | 4.7 | 5.0 | 1.5E-22 | 5.1 | 2.2E-22 | 4.5 | 3.6E-21 | 3.4 | 3.3E-17 | 1 | |
| D | – | – | C | WD 40 repeat protein | RAFL05-19-N20 | AT1G24530 | 4.3 | 4.1 | 1.9E-19 | 4.5 | 7.5E-20 | 2.3 | 6.9E-11 | 6.0 | 7.6E-24 | 1 | |
| – | – | – | – | ORG2 | bHLH proteini | AT3G56970 | 4.2 | 2.0 | 4.7E-07 | 2.3 | 1.8E-06 | 7.6 | 1.6E-26 | 6.2 | 3.2E-24 | 0 | |
| – | – | – | – | Jasmonate inducible protein, putative | RAFL16-34-L18 | AT1G52100 | 4.2 | 3.5 | 3.8E-17 | 3.5 | 1.7E-16 | 5.1 | 1.4E-22 | 4.9 | 6.8E-22 | 3 | |
| D | – | – | – | Senescence-associated protein | RAFL02-09-F24 | AT5G66170 | 4.2 | 4.2 | 1.5E-19 | 4.1 | 5.6E-18 | 4.5 | 1.2E-20 | 3.6 | 3.5E-17 | 3 | |
| – | – | A | – | GBF2 | Transcription factor | RAFL08-16-I23 | AT4G01120 | 3.9 | 4.3 | 2.5E-20 | 4.2 | 1.3E-19 | 3.6 | 3.8E-18 | 3.0 | 8.0E-15 | 7 |
| D | S | A | – | SAG29 | Senescence-associated protein | RAFL05-19-F21 | AT5G13170 | 3.9 | 4.3 | 1.1E-20 | 3.8 | 7.1E-19 | 4.0 | 1.3E-19 | 3.6 | 5.0E-18 | 4 |
| – | – | – | – | Invertase-like protein | RAFL08-08-F02 | AT4G34860 | 3.6 | 3.0 | 3.2E-14 | 2.3 | 7.3E-10 | 4.3 | 4.4E-20 | 4.8 | 5.6E-20 | 2 | |
| – | – | A | – | Lipase class 3 family protein | RAFL05-18-O21 | AT2G30550 | 3.5 | 3.0 | 8.7E-15 | 3.1 | 4.2E-15 | 3.9 | 3.2E-19 | 4.0 | 7.0E-19 | 1 | |
| D | – | – | – | Dormancy/auxin-associated family protein | RAFL05-13-B18 | AT1G56220 | 3.5 | 4.0 | 1.3E-19 | 5.3 | 1.7E-23 | 2.7 | 1.7E-13 | 3.0 | 2.1E-15 | 2 | |
| D | – | A | – | Glycosyl hydrolase family 36 | RAFL05-19-C02 | AT3G57520 | 3.4 | 4.4 | 5.8E-21 | 3.9 | 6.7E-19 | 3.0 | 4.3E-15 | 2.5 | 9.0E-12 | 3 | |
| – | – | – | – | ROC1 | Cyclosporin A binding protein | RAFL05-21-A06 | AT4G38740 | 3.4 | 3.2 | 2.9E-16 | 2.9 | 7.1E-15 | 3.7 | 2.0E-18 | 3.8 | 6.4E-19 | 2 |
| – | – | – | – | ELIP2 | Early light-induced protein, putative | RAFL04-12-M20 | AT4G14690 | 3.4 | 3.5 | 2.3E-17 | 2.9 | 1.9E-14 | 3.9 | 1.8E-19 | 3.2 | 2.3E-16 | 3 |
| – | – | – | – | FLS1 | Flavonol synthase 1 | RAFL09-32-C09 | AT5G08640 | 3.3 | 2.7 | 4.6E-13 | 2.6 | 1.2E-11 | 4.0 | 1.9E-19 | 3.9 | 5.5E-19 | 2 |
| – | – | – | – | ORG3 | bHLH protein | AT3G56980 | 3.3 | 1.7 | 1.3E-04 | 2.6 | 6.4E-07 | 4.0 | 7.8E-19 | 6.4 | 4.2E-22 | 0 | |
| – | – | – | – | ENTH domain-containing protein | AT4G32285 | 3.2 | 3.8 | 2.2E-18 | 2.6 | 1.6E-11 | 4.6 | 2.8E-21 | 2.7 | 7.0E-13 | 0 | ||
| – | – | – | – | Glutathione S-transferase, putative | RAFL14-52-N20 | AT1G17190 | 3.2 | 2.2 | 1.5E-09 | 3.2 | 6.9E-14 | 3.1 | 1.5E-15 | 4.2 | 4.1E-19 | 0 | |
| – | – | – | – | Aldose reductase, putative | RAFL19-21-M03 | AT5G01670 | 3.1 | 2.1 | 9.0E-09 | 3.2 | 2.2E-14 | 5.4 | 5.4E-23 | 3.0 | 1.1E-14 | 1 | |
| – | – | – | – | LTP4 | Lipid transfer protein 4 | RAFL05-08-P24 | AT5G59310 | 3.1 | 3.6 | 7.5E-18 | 4.9 | 2.0E-20 | 2.3 | 1.4E-10 | 2.5 | 1.3E-11 | 1 |
| – | S | – | C | Lipid-associated protein family | RAFL14-89-N02 | AT2G22170 | 3.1 | 2.9 | 6.3E-15 | 3.3 | 6.9E-17 | 3.0 | 2.7E-15 | 3.1 | 7.6E-16 | 0 | |
Upregulated genes in untreated 35S-AREB1ΔQT transgenic plants (AREB1ΔQT/control). A complete data set is available at arraExpress (http://www.ebi.ac.uk/arrayex/query/entry) under accession number E-MEXP-397.
Data on inducibility were based on microarray analysis (Seki et al., 2002; K. Maruyama and K. Yamaguchi-Shinozaki, unpublished data). D, drought; S, high salinity; A, ABA; C, cold.
Description as given by The Institute for Genomic Research database.
RAFL, RIKEN Arabidopsis full-length cDNA.
AGI, Arabidopsis Genome Initiative.
Genes with median of fold change (untreated 35S-AREB1ΔQT plants/untreated control plants) of >3 are listed.
P values < 0.001 were studied.
Number of ABRE core sequences in 1000 bp of the sequence upstream of the gene.
bHLH, basic helix-loop-helix.
The expressions of RD29B and RAB18 in the 35S-AREB1ΔQT plants without exogenous ABA were weak compared with the expressions in the wild-type plants in response to ABA treatment, whereas the mRNA accumulation levels of HIS1-3, AIA1, and GBF3 in the 35S-AREB1ΔQT plants without exogenous ABA were higher than those in wild-type plants treated with exogenous ABA (Figure 3D). Expressions of AIL1, RD20, and KIN2 were similar between the 35S-AREB1ΔQT plants without exogenous ABA and the wild-type plants with ABA (Figure 3D). The difference in these expression patterns between the 35S-AREB1ΔQT plants without exogenous ABA and wild-type plants with exogenous ABA may reflect some difference in the composition of the transcriptional regulatory complex of the cis-element in the promoter region of the target genes.
Transgenic Plants Overexpressing AREB1ΔQT Are Hypersensitive to ABA
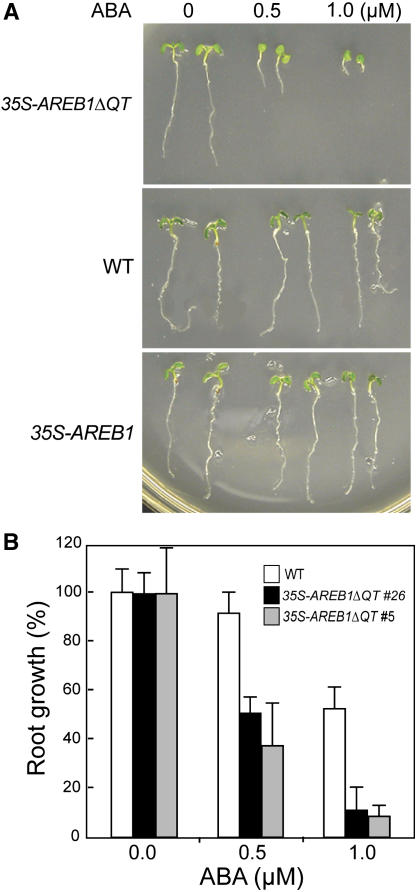
To evaluate the effect of AREB1ΔQT overexpression in transgenic plants on ABA sensitivity, we germinated 35S-AREB1ΔQT seeds and grew the seedlings on growth medium (GM) containing various concentrations of ABA. During the germination process, no obvious difference was observed between the 35S-AREB1ΔQT and wild-type plants (data not shown). However, the 35S-AREB1ΔQT seedling growth, including root growth, and cotyledon greening and expansion were severely inhibited when the ABA concentration was 0.5 μM or higher (Figures 4A and 4B). By contrast, seeds of the wild-type plants germinated and the seedlings grew normally, although at a slower rate than those on ABA-free GM (Figures 4A and 4B). In addition, we found no differences in germination or seedling growth between the 35S-AREB1 and wild-type plants by 6 d after stratification, whereas at 2 weeks after stratification, the seedling growth of 35S-AREB1 plants was severely inhibited when the ABA concentration was 0.5 μM or higher compared with the wild-type plants (data not shown).
Figure 4.
35S-AREB1ΔQT Plants Are Hypersensitive to ABA.
(A) Growth of 35S-AREB1ΔQT (line 5) and 35S-AREB1 (line 6) plants on GM agar plates containing 0, 0.5, or 1.0 μM ABA. Seeds were germinated and grown on GM agar plates for 6 d; representative plants are shown.
(B) ABA dose response of root growth. Seeds were germinated and grown on GM agar plates containing various concentrations of ABA and 1% sucrose. Root elongation was measured 6 d after stratification, and relative growth compared with that on ABA-free medium is indicated. Bars indicate standard deviation; n = 26 to 38. The experiments were performed three or more times, sometimes using different transgenic lines, and the results were consistent.
Transgenic Plants Overexpressing AREB1ΔQT Display Enhanced Drought Tolerance
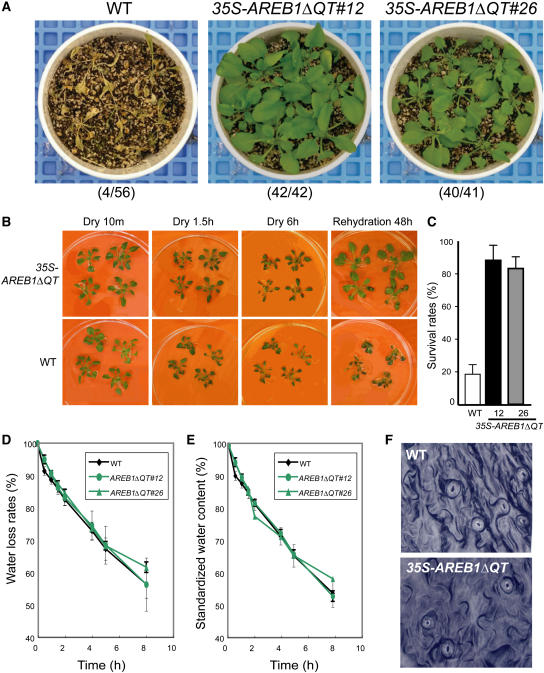
Stress-responsive genes that encode LEA class proteins are thought to be involved in dehydration tolerance (Ingram and Bartels, 1996; Thomashow, 1999). Four of the eight candidate target genes in the 35S-AREB1ΔQT plants under unstressed conditions were LEA class genes. Therefore, the 35S-AREB1ΔQT plants could be expected to have enhanced tolerance to drought. To examine this possibility, we examined whether overexpression of AREB1ΔQT affects tolerance to drought stress. Because several independent transgenic lines behaved in a similar manner (data not shown), we performed detailed analysis on transgenic lines 12 and 26. Almost all the wild-type plants withered completely when water was withdrawn for 12 d, whereas nearly all the transgenic plants of both AREB1ΔQT lines survived to maturity when rewatered afterward (Figure 5A). During the drought stress experiment, soil water content differed by <5% among all pots (data not shown). To exclude growth-dependent effects in the drought tolerance test, we further tried to explore the difference in recovery after dehydration using plants grown on agar plates. Three-week-old wild-type and transgenic plants were removed from agar plates and kept on plastic plates for 6 h (20% ± 10% relative humidity) and then rehydrated. During the first 1.5 h of dehydration, all wild-type plants had flopped, while the main stems of all transgenic plants remained standing. By 6 h, the wild-type plants had withered almost completely, while the 35S-AREB1ΔQT plants withered only slowly. Two days after rehydration, the wild-type plants had wilted and crinkled leaves, whereas the transgenic plants had standing main stems and green leaves that were spread out (Figure 5B). More than 80% of the 35S-AREB1ΔQT plants survived, whereas <20% of the wild-type plants did (Figure 5C). Thus, 35S-AREB1ΔQT plants survived dehydration better than did the wild-type plants (Figures 5B and 5C) and showed enhanced tolerance to drought stress (Figure 5A).
Figure 5.
Enhanced Drought Tolerance in 35S-AREB1ΔQT Plants.
(A) Enhanced tolerance to drought in the 35S-AREB1ΔQT plants (lines 12 and 26). Watering was withheld from 3-week-old plants for 12 d, then rewatering for 10 d, before the photograph was taken. Number codes indicate number of surviving plants out of total number.
(B) Enhanced ability of 35S-AREB1ΔQT plants (line 12) to survive the dehydration condition. Three-week-old transgenic and wild-type plants were grown on GM agar plates, transferred to Petri dishes, left unwatered for 6 h, and then rewatered.
(C) Increased survival rates of the 35S-AREB1ΔQT plants (lines 12 and 26) under dehydration. Water was withheld for 5 to 6 h from 3-week-old plants and then survival rates were counted. Surviving plants were scored on the second day. Survival rates and standard deviations (bars) were calculated from results of three independent experiments.
(D) Water loss rates of 35S-AREB1ΔQT (lines 12 and 26) plants. Each data point represents the mean of duplicate measurements (n = 7 each). Error bars represent standard deviation.
(E) Standardized water content of 35S-AREB1ΔQT (lines 12 and 26) plants. Details as in (D). Some error bars are smaller than the symbols.
(F) Stomatal aperture of 35S-AREB1ΔQT plants (line 12). Stomatal guard cells were observed in the middle of the watering cycle.
Stomatal closure under dehydration is one of the crucial ABA-regulated processes (Leung and Giraudat, 1998). Since the 35S-AREB1ΔQT transgenic plants are hypersensitive to ABA, we expected the overexpression of AREB1ΔQT to cause constitutive stomatal closure, thereby minimizing water loss and enhancing survival under dehydration. To assess whether altered transpiration rates contribute to the better survival of the 35S-AREB1ΔQT plants, we measured water loss rates and standardized water contents of whole plants under dehydration. As shown in Figures 5D and 5E, the water loss rates and standardized water contents of 35S-AREB1ΔQT were similar to those of control plants. Moreover, no remarkable difference was observed in the status of the stomatal opening between the 35S-AREB1ΔQT and wild-type plants grown on soil (Figure 5F) or on plates for 30 min after excision of the leaves (data not shown). These data together suggest that the enhanced tolerance of the 35S-AREB1ΔQT plants can be attributed to the AREB1ΔQT-dependent overexpression of the downstream genes, including LEA class genes, rather than to ABA-mediated stomatal closure. Overall, the overexpression of AREB1ΔQT resulted in the expression of downstream genes that are thought to protect the plants from water deficit stress and enhance their tolerance to drought.
Loss-of-Function Mutants of AREB1 Display Opposite Phenotypes to That Conferred by Overexpression of AREB1ΔQT
The constitutive expression of AREB1ΔQT in transgenic plants resulted in significant changes in ABA-associated phenotypes, such as ABA sensitivity and drought tolerance, and altered expression of ABA/stress-responsive genes, suggesting that AREB1 functions in the ABA-mediated stress signaling pathway. The overexpression of the active form of AREB1, however, might have caused unnatural conditions; so, the overexpression phenotypes need to be interpreted with caution. For example, a high level of the active form of AREB1 may result in altered specificity of interaction with the target proteins, leading to an altered function in the cell. In addition, overexpression of the active form of AREB1 also may have other, nonspecific effects on gene expression. However, the fact that overexpression of the active form of AREB1 conferred a positive effect on ABA-mediated drought stress response suggests that AREB1 function is specific even after overexpression. Nevertheless, we cannot exclude the possibility that a gain of function occurred as a result of the overproduction of the active form of AREB1.
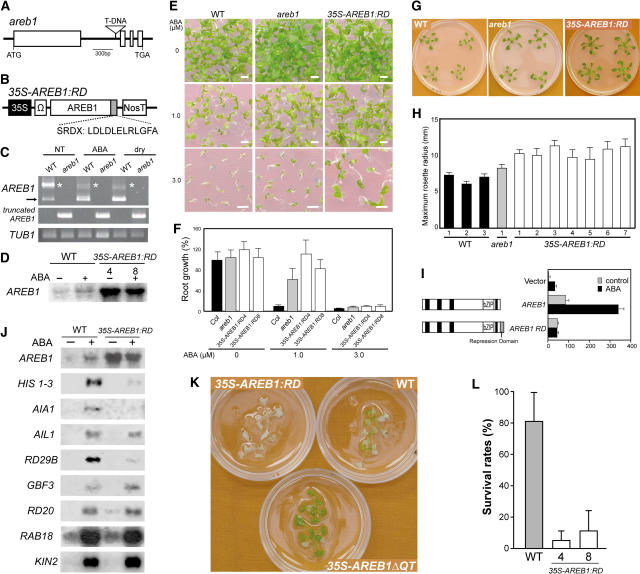
To further investigate the function of the AREB1 gene, we assessed two types of loss-of-function mutants: a T-DNA insertion mutant of AREB1, areb1, and transgenic plants overexpressing AREB1 fused to a fragment of the EAR motif RD, SRDX, under the control of the CaMV 35S promoter, 35S-AREB1:RD (Figures 6A to 6D). The mutant areb1 has a T-DNA insertion in the first intron of AREB1 (Figure 6A). RT-PCR analysis indicated that the knockout mutant plants did not produce AREB1 transcripts even after the application of exogenous ABA or during dehydration, while AREB1 and a larger mRNA band were observed in the wild-type plants (Figure 6C). This larger band seems to correspond to the estimated size of the unspliced form of AREB1 mRNA and was also detected using the other AREB1-specific primer pairs, suggesting that this may indeed be the unspliced form of AREB1 mRNA (Figure 6C; data not shown). By contrast, in the areb1 knockout mutant, a truncated form of AREB1 mRNA was detected using primers carrying AREB1- and T-DNA–specific binding sequences (Figure 6C). Growth of wild-type seedlings was inhibited gradually with increasing amounts of ABA, and growth of areb1 seedlings was also inhibited, but to a lower degree, especially at 3.0 μM ABA on the GM agar plates 2 weeks after stratification (Figure 6E). Furthermore, for more detailed analysis, because it is difficult to get intact roots from GM agar plates >2 weeks after stratification, we used GM plates containing 2.5% Gelrite (Merck), which is soft enough to release them even later than 2 weeks after stratification. Increased concentrations of ABA also resulted in greater growth retardation of both wild-type and areb1 primary roots, although the effect of ABA on primary root growth in the Gelrite plates seems to have been severer than in the agar plates (Figures 6E and 6F). At 1.0 μM ABA, primary root growth of wild-type plants was 7% of that in the medium without ABA, while that of the areb1 plants was 61% of that in the medium without ABA (Figure 6F). Thus, these data demonstrate insensitivity of the areb1 plants to ABA (Figures 6E and 6F) despite no obvious difference in ABA sensitivity having been observed between areb1 and wild-type plants in the germination process (data not shown). At 3 weeks after stratification, the maximum rosette radius of areb1 plants averaged 23% larger than that of the wild-type plants (Figures 6G and 6H). Thus, before the bolting stage, the areb1 plants are slightly larger than wild-type plants, but after that, the size difference gradually decreased, and then eventually the sizes were indistinguishable (data not shown), indicating that the areb1 plants grew slightly faster than the wild-type plants during the vegetative phase.
Figure 6.
Analysis of AREB1 Loss-of-Function Mutants.
(A) Scheme of the Arabidopsis AREB1 gene. Exons (open boxes) and introns (lines) are indicated. The position of the T-DNA insertion is shown (not to scale).
(B) Schematic representation of the 35S-AREB1:RD construct used for expression of the chimeric repressor with a modified version of the EAR-motif RD (SRDX), consisting of 12 peptides.
(C) Expression levels of AREB1 in the areb1 knockout mutant were determined by RT-PCR using total RNAs isolated from 2-week-old plants with or without 6-h treatment of 100 μM ABA or dehydration and grown on GM agar plates. Primers for detection of a truncated form of AREB1 mRNA, generated by T-DNA insertion into the first intron of AREB1, have T-DNA– and AREB1-specific binding sequences. The arrow and asterisks indicate expression of AREB1 and a larger band, respectively. TUB1, β-1 tubulin transcript as a control.
(D) RNA gel blot analysis of AREB1 mRNA in wild-type and 35S-AREB1:RD plants (lines 4 and 8) in the absence or presence of 50 μM ABA for 7 h. Eight micrograms of total RNA from 3-week-old seedlings was probed with AREB1 cDNA.
(E) Growth of the mutant areb1 and 35S-AREB1:RD plants (line 4) on GM agar plates containing 0, 1.0, or 3.0 μM ABA, supplemented with 1% sucrose. Seeds were germinated and grown on the medium for 2 weeks. Bars = 25 mm.
(F) ABA dose response of primary root growth. Seeds were geminated and grown on GM plates containing 0.25% Gelrite, 1% sucrose, and various concentrations of ABA. Primary root elongation was measured 19 d after stratification, and relative growth compared with that on ABA-free medium is indicated. Bars indicate standard deviation; n = 15 to 31.
(G) Growth phenotypes of areb1 and 35S-AREB1:RD (line 4) plants that were grown for 3 weeks on GM agar plates supplemented with 1% sucrose.
(H) Maximum rosette radius (i.e., length of the longest rosette leaf) of each plant on a GM agar plate containing 3% sucrose was measured 3 weeks after stratification. Three independent lines of wild-type plants, one line of the areb1 T-DNA insertion mutant, and seven independent lines of 35S-AREB1:RD plants were used. Bars indicate standard deviation; n = 7.
(I) The fusion of the RD to AREB1 creates a repressor. Arabidopsis protoplasts were cotransfected with the RD29B-GUS reporter and the effector construct expressing AREB1 or AREB1:RD, or vector DNA. The RD29B-GUS reporter plasmid and the transient assay system are described in the legend of Figure 2.
(J) Expression profile of downstream genes identified by microarray analysis (Table 1) in 35S-AREB1:RD plants (line 4). Two-week-old seedlings were either not treated (−) or treated (+) with ABA for 7 h. Each lane contained 7 μg of total RNA. Three to eight independent lines were used; results from one representative experiment are shown.
(K) Difference in recovery after rehydration among 35S-AREB1ΔQT (line 12), 35S-AREB1:RD (line 4), and wild-type plants. Transgenic and wild-type plants were grown on GM agar plates for 2 weeks, transferred to Petri dishes, left unwatered for 4 h, and then rewatered. The photograph was taken 2 d after rewatering.
(L) Quantification of the survival rates of the wild-type and 35S-AREB1:RD plants (lines 4 and 8) after rehydration. Water was withheld for 5 h from 3-week-old plants and then survival rates were counted. Surviving plants were scored on the second day. Survival rates and standard deviations were calculated from the results of four independent experiments (n = 10 each). Bars indicate standard deviations.
The areb1 plants were more resistant to ABA after germination and grew slightly faster than the wild-type plants (Figures 6E to 6H). Considering the potential functional compensation, however, due to the redundancy of the AREB family and other bZIP members (see Introduction for details), we could not exclude the possibility that the knockout plants would not work sufficiently as loss-of-function mutants. Recently, Hiratsu et al. (2003) clearly demonstrated that expression of specific target genes was suppressed dominantly by a chimeric transcription factor fused to an RD derived from the EAR motif of SUPERMAN, a TFIIIA-type zinc finger repressor, even in the presence of redundant transcription factors. This technology, using the SRDX RD, which consists of only 12 amino acids (LDLDLELRLGFA; Figure 6B), enabled us to see loss-of-function phenotypes that we have not yet observed in the other loss-of-function mutants (Hiratsu et al., 2003). Before creating AREB1 transgenic plants with the RD, first we confirmed the fusional AREB1:RD-dependent repression of the transactivation of the reporter gene in the transient assay system using protoplasts from T87 cells (Figure 6I).
To minimize the effects of phenotypic masking due to functional redundancy, we used this technology to generate 26 35S-AREB1:RD transgenic lines overexpressing AREB1 fused to the SRDX RD under the control of the CaMV 35S promoter. Expression levels of the transgene in the 26 independent transgenic lines were analyzed by RNA gel blot analysis using an AREB1-specific probe; we selected eight transgenic lines with higher AREB1:RD expression levels for phenotypic analysis. Because the eight transgenic lines behaved in a similar manner (Figures 6D to 6H; data not shown), we performed detailed analysis on representative lines 4 and 8. RNA gel blot analysis of the 35S-AREB1:RD plants showed that in the 35S-AREB1ΔQT plants under unstressed condition, three of the eight candidate target genes, HIS1-3, AIA1, and RD29B, were downregulated significantly in the presence of ABA (Figure 6J). By contrast, expression of another four candidate target genes, AIL1, RD20, RAB18, and KIN2, was not altered, and only GBF3 expression seemed to be only slightly upregulated (Figure 6J). Thus, although ABA-dependent secondary gene expression could be induced, approximately half of the genes downstream of AREB1 were suppressed even in the presence of exogenous ABA, indicating that the 35S-AREB1:RD plants act as loss-of-function mutants. These data also suggest that the three genes that are upregulated by the overexpression of AREB1ΔQT and downregulated in the loss-of-function mutant of AREB1 with the RD (HIS1-3, AIA1, and RD29B) are target genes regulated mainly by AREB1. Since upregulated genes such as RD20, RAB18, and KIN2 are well-known stress-inducible markers (Seki et al., 2002), we expect the expression of these genes to be also mediated by transcription factors other than AREB1.
At 3 weeks after stratification, the maximum rosette radius of 35S-AREB1:RD plants averaged 56% larger than that of wild-type plants (Figures 6G and 6H). Compared with the wild-type plants, petioles of the 35S-AREB1:RD plants were longer and thicker. This enhanced growth phenotype contrasted with the growth retardation phenotype of the 35S-AREB1ΔQT plants during the vegetative phase. By flowering time, however, the growth phenotype of the 35S-AREB1:RD plants had become similar to that of the wild-type plants (data not shown). We also tested the ABA sensitivity of the 35S-AREB1:RD transgenic plants. The 35S-AREB1:RD plants were more resistant to ABA than the other loss-of-function mutant areb1 plants or the wild-type plants >2 weeks after stratification (Figures 6E and 6F). In the germination process, however, no obvious ABA insensitivity was observed in the 35S-AREB1:RD plants or in the areb1 plants (data not shown).
As described above, the 35S-AREB1:RD plants were insensitive to ABA, and at least three stress-inducible genes were downregulated in them (Figures 6E, 6F, and 6J). Therefore, compared with wild-type plants, the 35S-AREB1:RD plants could be expected to survive less well under dehydration. To test this, we examined the differences in recovery after dehydration using plants grown on GM agar plates. All 35S-AREB1:RD plants were dead, and many wild-type plants were partially dead, but most 35S-AREB1ΔQT plants survived when they were rewatered afterwards (Figure 6K). To clarify this difference between the 35S-AREB1:RD and wild-type plants, we performed further tests. Compared with the wild-type plants, the survival rates of the 35S-AREB1:RD plants were reduced, confirming that the 35S-AREB1:RD plants survived less well under dehydration (Figure 6L). These phenotypes of the 35S-AREB1:RD plants also support the notion that AREB1 plays a pivotal role in the ABA-mediated stress signaling pathway in vegetative tissues. However, because such ABA-dependent phenotypes were due to overexpression of the gene, we need to interpret them with the cautions described above. Nevertheless, when we consider the two types of loss-of-function mutants, these loss-of-function phenotypes in growth, ABA sensitivity, or survival under dehydration were the opposite of the gain-of-function phenotypes conferred by the overexpression of AREB1ΔQT, demonstrating that AREB1 is involved in the ABA-mediated tolerance to drought through regulation of the ABA-dependent expression of novel downstream genes.
DISCUSSION
Here, we show that expression of the intact AREB1 gene alone is insufficient to upregulate its downstream genes under normal growth conditions. Taken together with data from our previous in-gel kinase assays and protoplast transient assays (Uno et al., 2000), the data may support the notion that not only ABA-induced transcription but also ABA-induced modification of AREB1 is required to activate expression of ABRE-dependent downstream genes. On the basis of recent reports that AREB1 and its homologs are phosphorylated in vitro or in vivo (Uno et al., 2000; Lopez-Molina et al., 2001; Kagaya et al., 2002), phosphorylation of AREB1 may be involved in the modification. We have shown that AREB1 is expressed constitutively in specific tissues (roots, hydathodes, and some vascular systems) and is induced by drought and high salt in all vegetative tissues. Therefore, activation of AREB1 by such modification without de novo protein synthesis allows ABA-dependent gene expression to respond more rapidly to stress conditions in these specific tissues. Since overexpression of intact AREB2 or ABF3, unlike AREB1, affects ABA sensitivity and stress tolerance in normal medium containing 1% sucrose (Kang et al., 2002; Kim et al., 2004; data not shown), this two-step signal perception system containing transcriptional and posttranscriptional regulation is more important in AREB1 than that in the other two homologs. Furthermore, the addition of ABA or drought stress dramatically activated AREB1 promoter activity in all tissues (Figure 1E), whereas very little induction of AREB2 or ABF3 promoter activity was observed under stress conditions in histochemical analyses (Uno et al., 2000; Kang et al., 2002). Our results show that AREB1, rather than the other bZIP homologs, plays an important role in vegetative tissues under drought stress conditions. In this scenario, the recent finding that ABF2 (AREB1) is a positive component of glucose signaling implies the possibility that AREB1-mediated stress response is involved in glucose signal transduction (Kim et al., 2004).
By differential expression analyses with microarray and RNA gel blot analyses, in combination with information on stress inducibility and cis-elements in the promoter region, we identified two groups of candidate target genes of AREB1: (1) four LEA class genes (AIL1, RD29B, KIN2, and RAB18) and (2) four regulatory genes (HIS1-3, AIA1, GBF3, and RD20). Interestingly, promoter regions of all these genes carry two or more ABRE motifs. This is consistent with recent findings that two ABRE motifs are required for activation of gene expression by AREB1 (Choi et al., 2000; Uno et al., 2000) and suggests that these genes are candidates for direct targets of AREB1. In particular, three of the eight genes (HIS1-3, AIA1, and RD29B) were upregulated in 35S-AREB1ΔQT plants even in the absence of exogenous ABA and downregulated in 35S-AREB1:RD plants even in the presence of exogenous ABA, suggesting that these genes are directly and mainly regulated by AREB1. The remaining five upregulated genes that were not downregulated in the 35S-AREB1:RD plants even in the presence of exogenous ABA also could be regulated by cis-elements other than the ABRE sequence and dedicated transcription factors other than AREB1 because at least three genes (KIN2, RAB18, and RD20) are also induced by cold stress and are known to be multiple stress marker genes (Table 1). For example, RD20 is upregulated by overexpression of RD26, which is a transcription factor with a NAC domain, which is induced by dehydration, high salinity, or ABA (Fujita et al., 2004).
Among the four regulatory genes identified as candidate target genes of AREB1, we expect two genes, HIS1-3 and AIA1, to be direct target genes of AREB1. HIS1-3, encoding a linker histone H1-3 protein, showed the highest increase of expression in the 35S-AREB1ΔQT plants under unstressed conditions (Table 1), and HIS1-3 expression was suppressed in the 35S-AREB1:RD plants in the presence of exogenous ABA (Figure 6J). Linker histone H1, unlike core histones (H2A, H2B, H3, and H4), is the most variable histone in eukaryotes and regulates specific gene expression, but not global transcription, throughout all tissues (Shen and Gorovsky, 1996; for review, see Jerzmanowski et al., 2000). HIS1-3 (Ascenzi and Gantt, 1997) in Arabidopsis and H1-D (Wei and O'Connell, 1996) and H1-S (Scippa et al., 2000) in tomato (Lycopersicon esculentum) have been reported as drought-inducible variants of histone H1 genes. These findings suggest that HIS1-3 plays an important role in drought-responsive gene expression mediated by chromatin remodeling. Previous results of the expression of HIS1-3 in RNA gel blot and histochemical analyses showed that the stress inducibility and location of expression are very similar to those of AREB1 (Ascenzi and Gantt, 1997, 1999). In particular, HIS1-3 expression is characteristically observed around the transition zone (the area at the junction of root and hypocotyl) and in hydathodes (Ascenzi and Gantt, 1999). AREB1 expression was also characteristically detected in the same region (Figure 1E), but expression patterns of AREB2 or ABF3, two members of the bZIP family, were distinct from that of HIS1-3 (Ascenzi and Gantt, 1999; Kang et al., 2002). Also, interestingly, expressions of AREB1, HIS1-3, RD29B, and RAB18 were not induced by ABA or dehydration treatments in the abi1 mutant (Ascenzi and Gantt, 1997; Uno et al., 2000; Y. Uno and K. Yamaguchi-Shinozaki, unpublished data), implying that the ABA-dependent expression via AREB1 is mediated by the ABI1 protein.
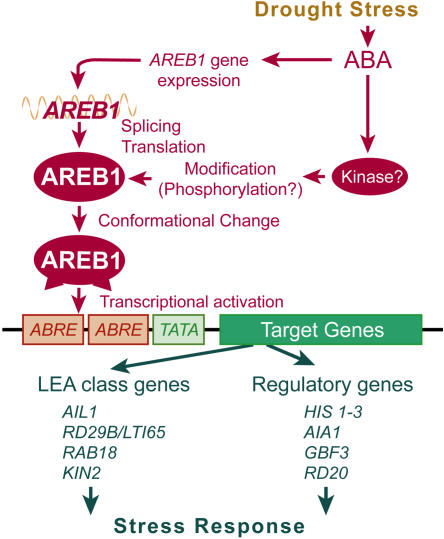
AIA1 encodes an AAA ATPase with chaperone-like activity (Neuwald et al., 1999). AAA ATPases form a large protein family with manifold cellular activities, including proteolysis, protein folding, and cytoskeletal regulation (Vale, 2000). In many cases, AAA domains assemble into hexameric rings that are likely to change their shape during the ATPase cycle (Vale, 2000). Although recent reports have shown that AAA ATPase is involved in multiple cellular functions via chaperone-like activity, the role of AAA ATPase in plants is still unknown. Our findings suggest that AIA1 is involved in drought response via its chaperone-like activity. Since all these candidate target genes were ABA and stress inducible, it is likely that the genes downstream of AREB1 enhance drought stress tolerance and increase fundamental cellular activities under stress conditions (Figure 7). Further dissection of such target genes will give us new insights into the ABA signal transduction network under stress conditions.
Figure 7.
A Model of the Regulation of ABA Signaling by AREB1.
AREB1 is postulated to mainly regulate the expression of stress-responsive genes via ABRE sequences in vegetative tissues.
LEA class proteins contain hydrophilic LEA-like or LEA proteins that typically accumulate during the late stage of embryogenesis or in response to dehydration (Ramanjulu and Bartels, 2002). According to several reports describing the classification of LEA class proteins (Bray, 1994; Cuming, 1999; Wise, 2003), the dehydrin RAB18 is a group 2 LEA protein, and the novel polypeptide AIL1 has a high degree of sequence similarity to group 3 LEA proteins. Also, the polypeptides RD29B and KIN2 share the characteristic biased amino acid compositions but not the canonical consensus motif of the groups 1 and 2 LEA proteins. Although the precise function of these hydrophilic LEA class proteins is yet unknown, several reports have suggested that LEA class proteins play a role in counteracting crystallization of cellular components or the irreversibly damaging effects of increasing ionic strength, which is induced by water deficit (Ingram and Bartels, 1996; Thomashow, 1999; Zhu, 2001). Because the enhanced tolerance to drought in the 35S-AREB1ΔQT plants is not associated with the altered transpiration rates mediated by ABA-dependent stomatal closure, the difference in the degree of wilting between the 35S-AREB1ΔQT and wild-type plants may be attributed to the upregulated LEA class proteins. These data imply that the LEA class proteins function in the detoxification and alleviation of such damage rather than the suppression of the loss of water.
All tested mutants of AREB1 showed no obvious phenotypes in the germination process compared with the wild-type plants (data not shown). Also, histochemical analysis detected very little AREB1 promoter activity in newly germinated seedlings (Figure 1E). These data suggest that AREB1 does not function during germination. This is also consistent with the finding that AREB1, unlike its bZIP homolog ABI5, has not been isolated to date in extensive genetic screening during germination (Leung and Giraudat, 1998; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). By contrast, overexpression of AREB2 or ABF3 caused ABA hypersensitivity to some extent during germination, indicating that the role of AREB1 is distinct from those of AREB2 and ABF3 (Kang et al., 2002; data not shown). This finding may be related to the observation that expression of AREB1, unlike AREB2 or ABF3, was inhibited by overexpression of VP1, a key determinant of seed-specific gene expression, in the vegetative tissues (Suzuki et al., 2003). Thus, AREB1 appears to play a specific role only in the vegetative phase. Moreover, interestingly, in the AREB1- or AREB1ΔQT-overexpressing plants, the mRNA decreased over time after ABA treatment of the plants (Figures 1F, 3D, and 6J). Although this phenomenon may be specific to the AREB1 sequence and may play some role in the regulation of AREB1, the role and mechanism are still unknown.
Our results show that the overexpression of the AREB1 mutation carrying an internal deletion between the N-terminal activation and bZIP DNA binding domains enables the constitutive activation of transcription of downstream genes in the absence of ABA. This suggests that the region of the internal deletion functions as a regulatory domain in response to ABA. Because it appears that AREB1 mutations lacking the N-terminal activation domain have a dominant negative effect on binding of endogenous transcription factors to the ABRE motifs in the promoter region of the reporter plasmid (Figures 2B and 2E), the altered function of the activation domain, rather than the change in binding activity of AREB1 to ABRE motifs, seems to help overcome the problem in some modifications. Hence, conformational change generated by the internal deletion may be associated with exposure of an activation domain masked in the normal folded complex (Figure 7). Furthermore, in the protoplast transient assay, an effector plasmid producing fusion proteins consisting of the DNA binding domain of GAL4 and the P region of AREB1 (which has a single amino acid substitution of Ser-26 to Ala in the R-x-x-S/T phosphorylation target site in the N-terminal conserved region of AREB1) did not decrease expression of the GUS reporter gene, suggesting that phosphorylation of the target site in the N-terminal conserved domain in the P region does not affect transactivation activity itself (data not shown). Taken together, these results indicate that ABA-induced modification, such as phosphorylation, might contribute to enabling the function of the N-terminal transactivation domain via conformational change of AREB1 rather than to activating the N-terminal transactivation domain itself.
Considering the potential functional redundancy of AREB family proteins and many bZIP factors interacting with ABREs, traditional loss-of-function approaches would not be appropriate for studying such bZIP factors (Foster et al., 1994; Kang et al., 2002). Although an areb1 T-DNA insertion mutant exhibited a milder phenotype, we could not determine whether this was an AREB1-specific phenotype or if it was compensated functionally by the other redundant bZIPs. To minimize the effects of phenotypic masking due to functional redundancy, we used the recently established chimeric repressor silencing technology (Hiratsu et al., 2003) in loss-of-function analysis of AREB1. Using several well-studied transcription factors, such as CUC1 and EIN3, they demonstrated that the chimeric repressors, which express transcription factors with an RD, SRDX, exhibited dominant loss-of-function phenotypes even in the presence of functionally redundant transcription factors (Hiratsu et al., 2003). In the case of AREB1, we show here that the phenotype conferred by the overexpression of the AREB1 mutant with the RD is opposite to that rendered by the overexpression of the constitutive active form of AREB1. Gain-of-function phenotypes in the AREB1 homolog are very different from each other (Kang et al., 2002), and loss-of-function phenotypes of the AREB1 mutant with the RD are also different from those of the homologs AREB2 and ABF3 (Y. Fujita and K. Yamaguchi-Shinozaki, unpublished data). These data suggest that phenotypes conferred by either type of mutant (with an internal deletion or the RD) are AREB1 specific. Thus, in the study of important transcription factors that have redundant homologs and are modulated by several means, the creation of constitutive active and chimeric repression forms of transcription factors are attractive approaches for revealing the mechanism of such transcription factors.
Here, we show that expression of the intact AREB1 gene alone is insufficient to lead to expression of the downstream genes under normal growth conditions. Furthermore, we identified the N-terminal region of AREB1 as a transcriptional activation domain and then created a constitutive active form of AREB1 carrying the N-terminal activation and bZIP DNA binding domains. Using the constitutive active form of AREB1, a T-DNA insertion knockout mutant, and a dominant loss-of-function mutant with the SRDX RD, we further demonstrate that AREB1 plays a key role in vegetative tissues under drought stress and mediates novel ABRE-dependent ABA signaling that enhances drought stress tolerance. Overall, these findings may contribute to our understanding of several important mechanisms underlying AREB1 functions in a novel ABRE-dependent ABA signal transduction pathway in vegetative tissues under drought stress.
METHODS
Plant Materials and Growth Conditions
Plants (Arabidopsis thaliana ecotype Columbia) were grown on GM agar plates for 2 to 3 weeks as described (Osakabe et al., 2005) under a 16-h-light/8-h-dark regime (40 ± 10 μmol photons/m2/s). The GM agar plates were supplemented with 1 or 3% sucrose and, as described in Results, with ABA as needed. T87 cultured cells, derived from Arabidopsis, were maintained, grown, and treated as described (Satoh et al., 2004). An Arabidopsis AREB1 T-DNA insertion line (SALK_002984; Col-0 ecotype) was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Insertion mutant information was obtained from the Salk Institute Genomic Analysis Laboratory's website (http://signal.salk.edu). The T-DNA insertion sites were confirmed by PCR using T-DNA left-border primer 5′-GCGTGGACCGCTTGCTGCAACT-3′ and AREB1-specific primer 5′-TCAAGCTCCACGGTGTAAGCC-3′. We confirmed that the intact AREB1 gene was not expressed in the areb1 mutant using RT-PCR analysis as described (Ito and Shinozaki, 2002).
RNA Gel Blot Analysis
Total RNA extraction and RNA gel blot analysis were conducted as described (Satoh et al., 2004) using a Shakemaster sonicator (BioMedical Science) for disruption of the cells. Probes for RNA gel blot analysis were prepared as described (Maruyama et al., 2004).
Transient Expression in Onion Epidermal Cells
The 35S-GFP:AREB1 plasmid was constructed by subcloning a full-length cDNA of AREB1 into the pGFP3BX vector (Fujita et al., 2004). The constructs were introduced into onion epidermal cells with a pneumatic particle gun (PDS-1000/He; Bio-Rad Laboratory) as described (Ito and Shinozaki, 2002). After incubation at 22°C for 8 to 12 h, the tissues were stained with propidium iodide (10 μg/mL), and GFP fluorescence was observed in whole mounts under a confocal laser scanning microscope (LSM510; Zeiss) as described (Ito and Shinozaki, 2002).
Histochemical GUS Staining
The AREB1 promoter–GUS reporter plasmid was constructed by cloning a PCR-amplified DNA fragment containing an AREB1 5′ sequence (−1123 to −1) into the GUS reporter plasmid pBI101.1 (Clontech). Histochemical localization of GUS activity was determined as described (Satoh et al., 2002). Whole plants were immersed in X-Gluc for 20 h at 37°C.
Transient Expression Assay Using Arabidopsis Protoplasts
Transient expression assay using protoplasts derived from Arabidopsis T87 cultured cells was performed as described (Fujita et al., 2004; Satoh et al., 2004) with minor modifications. The protoplasts were isolated and transformed at room temperature (25 to 28°C). The transformed protoplasts were incubated at 22°C for 16 to 20 h in the dark. Enzyme solution (0.4 M mannitol, 1.5% [w/v] cellulase Onozuka R-10 [Yakult], 0.3% [w/v] macerozyme R-10 [Yakult], 0.1% [w/v] BSA, 10 mM CaCl2, 20 mM KCl, and 20 mM MES, pH 5.7) was prepared according to J. Sheen (http://genetics.mgh.harvard.edu/sheenweb/).
Effector plasmids used in the transient transactivation experiment with the AREB1 bZIP DNA binding domain were constructed with PCR-amplified DNA fragments containing a partial or whole AREB1 cDNA, which were cloned into NotI sites of the expression vector pBI35SΩ (Abe et al., 1997). pBI35SΩ-AREB1 was partially digested with EcoT14I and then self-ligated to remove the 0.64-kb EcoT14I partially digested fragment. The resultant plasmid, pBI35SΩ-AREB1ΔQT, had an internal deletion (amino acids 65 to 277) spanning the Q to T region. Two DNA fragments containing a portion of AREB1 cDNA were generated by PCR with the following two pairs of primers: forward primer A, 5′-GGGGCGGCCGCATGACACAAGCCATGGCTAGTG-3′; reverse primer A, 5′-GCAGAAGCACCTTGACTTCCCCCTACTCCAC-3′; forward primer B, 5′-GTAGGGGGAAGTCAAGGTGCTTCTGCTGC-3′; reverse primer B, 5′-GGGGAGCTCTCACCAAGGTCCCGACTCTG-3′. The resulting purified fragments A and B were mixed in a tube for PCR, denatured at 94°C for 10 min, annealed, and polymerized at 72°C for 3 min. Then, a DNA fragment amplified in the tube with forward primer A and reverse primer B was digested with NotI and SacI and cloned into pBI35SΩ. The resultant plasmid, pBI35SΩ-AREB1ΔP/RT, has two internal deletions (amino acids 1 to 60 and 117 to 277).
The effector plasmid expressing the GAL4 DNA binding domain fused to the GAL4 activation domain (p35S-562) and the GAL4-GUS reporter plasmid (pGUS-558) were kindly provided by T. Hattori (Nagoya University, Japan). Effector plasmids used in the transient transactivation experiment with the GAL4 binding domain were constructed with PCR-amplified DNA fragments containing a portion of AREB1 cDNA, which were cloned into BamHI-SacI sites of the expression vector p35S-562. The 0.9-kb HindIII-BamHI fragment of pBI-35SLUC (Urao et al., 1996) was inserted into the HindIII and BamHI sites of the plant expression vector Japan International Research Center for Agricultural Sciences, Tsukuba, pBI221(−46/Ω)LUC, which was kindly provided by T. Urao (Japan International Research Center for Agricultural Sciences, Tsukuba, Japan). The resulting pBI35SΩ-LUC reporter plasmid was used as an internal control in each transactivation experiment.
Construction of Transgenic Plants
To construct the pBE2113Not-AREB1 plasmid, the entire coding region of AREB1 was amplified by PCR with NotI linker primers and cloned into the binary vector pBE2113Not (Liu et al., 1998) in the sense orientation. To create pBE2113Not-AREB1ΔQT, the AREB1ΔQT coding region was PCR amplified with XbaI-BamHI linker primers and cloned into the XbaI and BamHI sites of pBE2113Not in the sense orientation.
The plasmid pBI101.1 (Clontech) was modified for Gateway technology by cloning the Gateway vector conversion cassette (reading frame A; Invitrogen) into the EcoRI and HindIII sites to create the pBCKK vector. A DNA fragment containing AREB1 cDNA was generated by PCR with the following pair of 5′-phosphorylated primers: forward primer, 5′-GGGATGGATGGTAGTATGAATTTG-3′; reverse primer, 5′-CCAAGGTCCCGACTCTGTCCTCC-3′. To generate a Gateway entry clone (p35S-AREB1:RD), the resulting PCR product was cloned into the dephosphorylated SmaI site of p35S-SRDXG, which contains two Gateway recombination sites (attL1 and attL2; Invitrogen), the CaMV 35S promoter, an Ω translation enhancer sequence, the SRDX (LDLDLELRLGFA) RD sequence (Hiratsu et al., 2003), and a NOS terminator in the pUC19 vector. pBCKK-35S-AREB1:RD was formed from a destination vector, pBCKK, and an entry clone, p35S-AREB1:RD, using the Gateway LR clonase reaction (Invitrogen).
To create 35S-AREB1, 35S-AREB1ΔQT, and 35S-AREB1:RD transgenic plants, the plant transformation vectors described above (pBE2113Not-AREB1, pBE2113Not-AREB1ΔQT, and pBCKK-35S-AREB1:RD) were transformed into Arabidopsis plants (Columbia) by the vacuum infiltration method using Agrobacterium tumefaciens strain C58 (Osakabe et al., 2005).
Drought Tolerance Assays
Drought tolerance assays were performed as described (Sakamoto et al., 2004) with minor modifications. The plants were grown under 16-h illumination of 50 ± 10 μmol photons/s/m2 at 22°C ± 1°C and 35% ± 5% relative humidity. Drought stress was imposed by withholding water for 12 d.
In survivability tests in the dehydration conditions, transgenic and wild-type plants were germinated and grown on GM agar plates for 2 to 3 weeks, transferred to Petri dishes, left unwatered for specific times, and then rewatered. A survivability test in the dehydration condition was conducted at 25°C ± 2°C and 20% ± 10% relative humidity under an illumination of 9 ± 1 μmol photons/s/m2. After the rewatering, the Petri dishes were transferred to a plant incubation room and incubated at 22°C ± 2°C under continuous light (50 ± 5 μmol photons/s/m2) for 1 to 3 d so that we could recognize whether the plants were dead or alive by their coloring. Plants that were green on >50% of their tissue were counted as surviving plants. To minimize any size-dependent effect, plants of similar size were used. All experiments were repeated at least five times, and >40 plants from at least three lines were used in each comparison.
Stomatal Aperture Measurement
Detached rosette leaves from 4-week-old soil-grown plants in the middle of the watering cycle (2 d after watering) were incubated for 2 h in 20 mM KCl, 1 mM CaCl2, and 5 mM MES-KOH, pH 6.15 (Pei et al., 1998). Leaves were placed immediately on slides, abaxial side up, and observed intermittently for 30 min after excision. Photographs of the guard cells were taken through a color laser three-dimensional profile microscope (Keyence).
Microarray Analysis
Two-week-old seedlings of 35S-AREB1ΔQT and vector control plants grown on GM agar plates were harvested directly or after ABA treatment for 7 h and were tested in an Agilent Arabidopsis 2 Oligo Microarray (Agilent Technologies). For each biological replicate, material from eight plants was pooled to make a single sample for RNA purification. Two independent transgenic lines were used for each experiment. Total RNA was isolated with Trizol reagent (Invitrogen) and used for the preparation of Cy5- and Cy3-labeled cDNA probes. All microarray experiments, including the data analysis, were performed according to the manufacturer's manual (http://www.chem.agilent.com/scripts/generic.asp?lpage=11617&indcol=Nandprodcol=Y). The reproducibility of microarray analysis was assayed by a dye swap in each experiment. On the basis of our empirical findings, expression of genes showing average signal intensity values of <500 to ∼1000 in either the Cy3 or Cy5 channel of the control plants was not always detected reproducibly by RNA gel blot analysis. Thus, under our experimental conditions, genes showing a signal value <1000 in both Cy3 and Cy5 channels of the control plants were not considered for the analysis. We studied genes with P values < 0.001. Our previous data also show that most genes with changes in expression of >3 are clearly and reproducibly confirmed by RNA gel blot or real-time quantitative RT-PCR analyses (for example, Rabbani et al., 2003; Fujita et al., 2004; Maruyama et al., 2004). Feature extraction and image analysis software (version A.6.1.1; Agilent Technologies) was used to locate and delineate every spot in the array and to integrate each spot's intensity, filtering, and normalization by the Lowess method. Gene clustering analysis was performed with Genespring 6.1 software (Silicon Genetics). Because the nucleotide sequences of RD29B and At4g25580 are very similar in the Arabidopsis genome, we confirmed only that RD29B was upregulated in the 35S-AREB1ΔQT plants using quantitative real-time PCR and RNA gel blot analysis with several RD29B-specific sequences under our experimental conditions.
Analysis of Plant Water Relations
Water loss and standardized water content were measured as described (Yoshida et al., 2002; Ma et al., 2004), with minor modifications. Aerial parts from 4-week-old soil-grown plants were excised and weighed for fresh weight over time. Detached aerial parts were then dried at 180°C for 3.5 h to determine dry weight. To eliminate variability resulting from plant size or dry weight, water content was standardized as a percentage relative to the initial water content of aerial parts of the plant; it was calculated as [(FWi – DW)/(FW0 – DW)] × 100, where FWi and FW0 are fresh weight for any given interval and original fresh weight, respectively, and DW is dry weight. These tests were conducted on the laboratory bench at 24°C ± 1°C and 65% ± 5% relative humidity under an illumination of 9 ± 1 μmol photons/s/m2.
Phylogenetic Analysis
Three N-terminal conserved (C1, C2, and C3, Figure 1A) and bZIP domain sequences were aligned using the ClustalX program (version 1.83) with the following parameter sets: gap open penalty = 5.00, gap extension penalty = 0.05 (see Supplemental Figure 1 online). The alignment was finally adjusted manually. A phylogenetic tree was constructed by the neighbor-joining method using MEGA software (version 3) as described previously (Fujita et al., 2004). The confidence level of monophyletic groups was estimated by bootstrap analysis of 1000 replicates.
Accession Numbers
The microarray data were submitted in MIAME-compliant (minimum information about a microarray experiment) format to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) and have been assigned the accession number E-MEXP-397. Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers in Table 1 or as follows: AREB1/ABF2, At1g45249; AREB2/ABF4, At3g19290; AREB3/DPBF3, At3g56850; ABF1, At1g49720; ABF3/DPBF5, At4g34000; ABI5/DPBF1, At2g36270; EEL/DPBF4, At2g41070; DPBF2, At3g44460.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Alignment of Three N-Terminal Conserved and bZIP Domain Sequences.
Supplementary Material
Acknowledgments
We thank T. Urao and T. Hattori for providing the vector pBI221(−46/Ω)LUC and yeast GAL4 fusion constructs, respectively; Y. Niwa for letting us use the sGFP gene; E. Ohgawara, K. Amano, H. Sado, and K. Yoshiwara for their excellent technical assistance; M. Toyoshima for skillful editorial assistance; and the Rice Genome Resource Center at the National Institute of Agrobiological Sciences for the use of the Arabidopsis microarray analysis system. This work was also supported in part by a project grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan and the Genome Project in RIKEN, Bio-oriented Technology Research Advancement Institution, and Core Research for Evolutional Science and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuko Yamaguchi-Shinozaki (kazukoys@jircas.affrc.go.jp).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035659.
References
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi, R., and Gantt, J.S. (1997). A drought-inducible histone gene in Arabidopsis is a member of a distinct class of plant linker histone variants. Plant Mol. Biol. 34 629–641. [DOI] [PubMed] [Google Scholar]
- Ascenzi, R., and Gantt, J.S. (1999). Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol. Biol. 41 159–169. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., Rippa, S., Lambert, G., Jublot, D., Pautot, V., Granier, F., Giraudat, J., and Parcy, F. (2002). The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1994). Alterations in gene expression in response to water deficit. In Stress-Induced Gene Expression in Plants, A.S. Basra, ed (Amsterdam: Harwood Academic), pp. 1–23.
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37 425–435. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. [DOI] [PubMed] [Google Scholar]
- Cuming, A.C. (1999). LEA proteins. In Seed Proteins, P.R. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 753–780.
- Devaux, F., Marc, P., Bouchoux, C., Delaveau, T., Hikkel, I., Potier, M.-C., and Jacq, C. (2001). An artificial transcription activator mimics the genome-wide properties of the yeast Pdr1 transcription factor. EMBO Rep. 2 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Gampala, S.S., and Rock, C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, R., Izawa, T., and Chua, N.-H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8 192–200. [DOI] [PubMed] [Google Scholar]
- Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., Tran, L.P., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 863–876. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Artus, N.N., and Thomashow, M.F. (1992). cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 18 13–21. [DOI] [PubMed] [Google Scholar]
- Giraudat, J., Parcy, F., Bertauche, N., Gosti, F., Leung, J., Morris, P.C., Bouvier-Durand, M., and Vartanian, N. (1994). Current advances in abscisic acid action and signalling. Plant Mol. Biol. 5 1557–1577. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Ito, T., and Shinozaki, K. (2002). The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 43 1285–1292. [DOI] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F.; bZIP Research Group. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski, A., Przewloka, M.R., and Grasser, K.D. (2000). Linker histones and HMG1 proteins of higher plants. Plant Biol. 2 586–597. [Google Scholar]
- Kagaya, Y., Hobo, T., Murata, M., Ban, A., and Hattori, T. (2002). Abscisic acid–induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14 3177–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.Y., Choi, H.I., Im, M.Y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Kang, J.Y., Cho, D.I., Park, J.H., and Kim, S.Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40 75–87. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Ma, J., Perret, P., Li, Z., and Thomas, T.L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1992). The isolation of abscisic acid (ABA)-deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant 61 377–383. [Google Scholar]
- Kurkela, S., and Borg-Franck, M. (1992). Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 19 689–692. [DOI] [PubMed] [Google Scholar]
- Lang, V., and Palva, E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20 951–962. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 199–222. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.-H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A post-germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S., Quist, T.M., Ulanov, A., Joly, R., and Bohnert, H.J. (2004). Loss of TIP1;1 aquaporin in Arabidopsis leads to cell and plant death. Plant J. 40 845–859. [DOI] [PubMed] [Google Scholar]
- Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., Shimada, Y., Yoshida, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38 982–993. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9 27–43. [PubMed] [Google Scholar]
- Nordin, K., Vahala, T., and Palva, E.T. (1993). Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 21 641–653. [DOI] [PubMed] [Google Scholar]
- Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyl transferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290. [DOI] [PubMed] [Google Scholar]
- Rabbani, M.A., Maruyama, K., Abe, H., Khan, M.A., Katsura, K., Ito, Y., Yoshiwara, K., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanjulu, S., and Bartels, D. (2002). Drought and desiccation-induced modulation of gene expression in plants. Plant Cell Environ. 25 141–151. [DOI] [PubMed] [Google Scholar]
- Ren, B., et al. (2000). Genome-wide location and function of DNA-binding proteins. Science 290 2306–2309. [DOI] [PubMed] [Google Scholar]
- Sakamoto, H., Maruyama, K., Sakuma, Y., Meshi, T., Iwabuchi, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Arabidopsis Cys2/His2-type zinc finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, R., Fujita, Y., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45 309–317. [DOI] [PubMed] [Google Scholar]
- Satoh, R., Nakashima, K., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiol. 130 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler, U., Beckmann, H., and Cashmore, A.R. (1992). Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis thaliana GBF bZIP proteins. EMBO J. 4 1261–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scippa, G.S., Griffiths, A., Chiatante, D., and Bray, E.A. (2000). The H1 histone variant of tomato, H1-S, is targeted to the nucleus and accumulates in chromatin in response to water-deficit stress. Planta 211 173–181. [DOI] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genomics 2 282–291. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Zhang, P., and Ho, T.-H.D. (1996). Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., and Gorovsky, M.A. (1996). Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell 86 475–483. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Suzuki, M., Ketterling, M.G., Li, Q.B., and McCarty, D.R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation, freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. [DOI] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Noji, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1996). A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. 10 1145–1148. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. (2000). AAA proteins. Lords of the ring. J. Cell Biol. 150 F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T., and O'Connell, M.A. (1996). Structure and characterization of a putative drought-inducible H1 histone gene. Plant Mol. Biol. 30 255–268. [DOI] [PubMed] [Google Scholar]
- Wise, M.J. (2003). LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 29 52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K., and Zhu, J.-K. (2002). Cell signaling during cold, drought and salt stress. Plant Cell 14 (suppl.), S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., Koizumi, M., Urao, S., and Shinozaki, K. (1992). Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: Sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 33 217–224. [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., Ecker, J.R., and Shinozaki, K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43 1473–1483. [DOI] [PubMed] [Google Scholar]
- Zeitlinger, J., Simon, I., Harbison, C.T., Hannett, N.M., Volkert, T.L., Fink, G.R., and Young, R.A. (2003). Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113 395–404. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2001). Plant salt tolerance. Trends Plant Sci. 6 66–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.