Abstract
We identified a nonpathogenic strain of Ustilago maydis by tagging mutagenesis. The affected gene, glucosidase1 (gas1), displays similarity to catalytic α-subunits of endoplasmic reticulum (ER) glucosidase II. We have shown that Gas1 localizes to the ER and complements the temperature-sensitive phenotype of a Saccharomyces cerevisiae mutant lacking ER glucosidase II. gas1 deletion mutants were normal in growth and mating but were more sensitive to calcofluor and tunicamycin. Mutant infection hyphae displayed significant alterations in the distribution of cell wall material and were able to form appressoria and penetrate the plant surface but arrested growth in the epidermal cell layer. Electron microscopy analysis revealed that the plant–fungal interface between mutant hyphae and the plant plasma membrane was altered compared with the interface of penetrating wild-type hyphae. This may indicate that gas1 mutants provoke a plant response.
INTRODUCTION
With the genome sequences of several plant pathogenic fungi being completed or under way (Broad Institute, http://www.broad.mit.edu/seq/), genomic approaches to study interaction of these fungal pathogens with their hosts are becoming feasible (Lorenz, 2002). Such studies will have to be complemented by the isolation and characterization of mutants, either by generating knockouts systematically or by screening for mutants that specifically affect pathogenic development. The Whitehead Institute has recently completed the sequence of the fungus Ustilago maydis, a pathogen of corn, and a user friendly database has been established by the Munich Information Center for Protein Sequences Institute (MUMDB; http://mips.gsf.de/genre/proj/ustilago/). This basidiomycete has become an attractive organism for the study of fungal dimorphism and pathogenicity because of its ease of genetic handling (Bölker, 2001). In its haploid form, U. maydis grows by budding and can be propagated on artificial media. A filamentous dikaryon arises after fusion of two compatible haploid strains. This form is characterized by tip growth and the generation of empty sections in the older parts of the filament. On the plant surface, the filamentous dikaryons can differentiate appressoria and penetrate the leaf. Appressorial development and the penetration step are poorly understood processes. The infecting hyphae initially grow intracellularly with few macroscopic symptoms before starting to proliferate intercellularly during tumor development (Snetselaar and Mims, 1992, 1993; Banuett and Herskowitz, 1996). Even during the early infection stages the fungal cells inside the plant are separated from the host cell cytoplasm through invaginations of the plasma membrane (Snetselaar and Mims, 1992, 1993). The disease culminates in the production of large tumor-like structures that are filled with black teliospores.
Initially, U. maydis did not lend itself to a genetic study of pathogenicity because the pathogenic form is dikaryotic. However, this has changed with the advancement of understanding the underlying molecular processes of mating. Haploid cells communicate through a pheromone/receptor system whose components, the pheromone and receptor genes, are encoded by the a locus (Bölker et al., 1992). In response to activation of this signaling pathway, a number of pheromone-responsive genes are induced and among these are the genes in the a and b loci (Urban et al., 1996). The multiallelic b locus encodes two homeodomain transcription factors, bE and bW, that dimerize when they originate from different alleles (i.e., in the dikaryon) (Gillissen et al., 1992; Kämper et al., 1995). The heterodimer then serves as the central transcription factor for a large number of genes (Brachmann et al., 2001). By building on this information, it has been possible to construct haploid strains that express an active bE/bW heterodimer and are hence pathogenic without prior cell fusion (Bölker et al., 1995a). Using such a solopathogenic strain, CL13, it has been possible to isolate a large number of nonpathogenic mutants by applying the method of restriction enzyme–mediated integration (REMI) for insertional mutagenesis (Bölker et al., 1995b).
In this article, we describe one such nonpathogenic mutant that is affected in a gene, gas1, encoding an endoplasmic reticulum (ER) glucosidase II (α-subunit). Δgas1 mutants are arrested after formation of appressoria and penetration into the epidermis.
RESULTS
Isolation of the gas1 Gene
In a screen for nonpathogenic mutants, the haploid solopathogenic strain CL13 (a1bE1W2) (Bölker et al., 1995a) was subjected to REMI mutagenesis using the plasmid pSMUT in combination with BamHI restriction enzyme as previously described (Bölker et al., 1995b). From the nonpathogenic mutant CL13pat4339 the integrated plasmid plus flanking sequences was recovered by plasmid rescue (p4339). Linkage between insertion of pSMUT in CL13pat4339 and loss of pathogenicity was demonstrated by regenerating the mutation. To this end, MluI-linearized p4339 was transformed in CL13. Of 15 transformants, seven were shown to have integrated the plasmid by homologous recombination, thus recreating the mutant. These seven strains all proved to be nonpathogenic (data not shown).
Sequencing of the regions flanking the pSMUT backbone in p4339 and subsequent BLAST analyses (Altschul et al., 1997) revealed significant sequence similarity on both sides of the integration site to a putative glucosidase II α-subunit from Neurospora crassa (CAD36981). The corresponding genomic region was isolated from a cosmid library, and an 11-kb MluI DNA fragment was subcloned. Sequence analysis of this subclone revealed the presence of an open reading frame of 3183 bp, termed glucosidase1 (gas1). The pSMUT insertion in the nonpathogenic mutant strain CL13pat4339 had occurred in the 3′ region of gas1, at a genomic BamHI site located at base pair 2642 of the open reading frame (Figure 1A). Two partial cDNA clones corresponding to the 5′- and 3′-ends of gas1, respectively (Figure 1A), and together covering the entire open reading frame, were isolated from a cDNA library and sequenced. Comparison of genomic and cDNA sequences did not reveal the presence of introns in gas1.
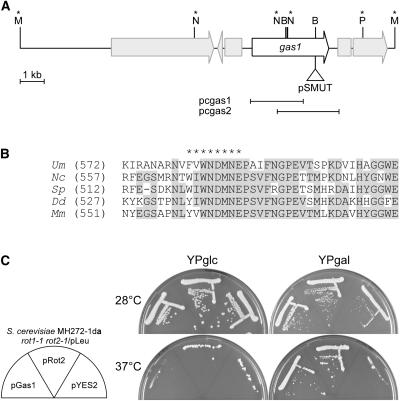
Figure 1.
Identification of gas1 as a Gene Encoding an α-Subunit of Glucosidase II.
(A) Schematic representation of the pSMUT insertion in gas1 in mutant strain CL13pat4339. The open reading frame of Gas1 is indicated by an open arrow. The neighboring predicted open reading frames are indicated by gray arrows, which are interrupted at potential intron positions (MUMDB, http://mips.gsf.de/genre/proj/ustilago/; shown are open reading frames um04407, um10516, um04405, and um04404 from contig 1.157). Relevant restriction enzyme sites are indicated by letters: B, BamHI, M, MluI; N, NcoI, P, PstI. The pSMUT insertion took place in a genomic BamHI site located in the 3′ region of the 3183-bp open reading frame at bp 2642. The restriction sites used for cloning are indicated by asterisks above the respective sites. The relative positions of the two partial cDNA clones pcgas1 and pcgas2 are indicated by black lines below the figure. The pSMUT insertion is not drawn to scale.
(B) Gas1 shares significant sequence similarity with the putative α-glucosidases from N. crassa (Nc, CAD36981), S. pombe (Sc, CAB65603), D. discoideum (Dd, ModA; AAB18921), and M. musculus (Mm, AAC53182). Only the region around the catalytic site is shown. Identical amino acids are highlighted by gray shading. Gas1 is predicted to carry a cleavable N-terminal signal sequence (amino acids 1 to 24; data not shown) and possesses the characteristic catalytic site of glycosyl hydrolases ([(G/F)-(L/I/V/M)-W-X-D-M-N-E], indicated by asterisks above the sequence).
(C) Gas1 complements the temperature-sensitive phenotype of a S. cerevisiae rot1-1 rot2-1 double mutant. S. cerevisiae MH272-1da rot1-1 rot2-1/pLeu cells transformed with either empty vector (pYES2) or the pYES2 derivatives containing either the yeast ROT2 gene encoding α-glucosidase II (pRot2) or the U. maydis gas1 gene (pGas1) under the control of a galactose-inducible promoter were streaked on plates containing glucose (YPglc) or galactose (YPgal) and incubated at 28 and 37°C. rot1-1 rot2-1 mutants are viable at 37°C if they express Rot2p or U. maydis Gas1.
gas1 Encodes an ER Glucosidase II α-Subunit
The gas1 gene is predicted to encode a protein of 120.8 kD. The N terminus of Gas1 carries a signal sequence with a predicted cleavage site between amino acids 24 and 25 (GvH score 2.87, PSORT II) (Nakai and Horton, 1999). The central part of Gas1 contains sequences diagnostic of the family-31 glycosyl hydrolases (amino acids 176 to 967; Pfam 10.0) (Bateman et al., 2002). Glycosyl hydrolases are characterized by a conserved sequence motif [(G/F)-(L/I/V/M)-W-X-D-M-N-E] of the catalytic site (Henrissat, 1991), and a perfect match to this consensus is found between amino acids 581 and 588 of Gas1 (Figure 1B). BLAST sequence comparison of the entire Gas1 sequence confirmed sequence similarity to a number of putative glucosidase II α-subunits (GIIαs) from various organisms, including GIIα of Aspergillus fumigatus (EAL85956, 36% identity), Cryptococcus neoformans (AAW41957, 34% identity), Schizosaccharomyces pombe (CAB65603; 33% identity), N. crassa (CAD36981; 32% identity), mouse (Mus musculus; AAC53182; 31% identity), and Dictyostelium discoideum (ModA, AAB18921; 30% identity). ER glucosidase II plays a pivotal role in processing of N-linked oligosaccharides in the ER (Parodi, 2000).
GIIα proteins do not possess characteristic ER retention signals but are retained in the ER through dimerization with a noncatalytic β-subunit carrying an ER retention signal (D'Alessio et al., 1999). As Gas1 also lacks a typical ER retention signal, we determined its subcellular localization. A C-terminal fusion of Gas1 and yellow fluorescent protein (YFP) (Cormack et al., 1996) was generated in plasmid pOGEY, and transcription of the gas1-eYFP gene was placed under the control of the constitutive otef promoter (Spellig et al., 1996) in order to facilitate detection of the fusion protein. Ectopic integration of pOGEY in the gas1 deletion strain HBU15, a derivative of the solopathogenic strain CL13, restored pathogenicity, indicating that the Gas1-YFP fusion was biologically active (data not shown).
For localization studies, plasmid pOGEY was integrated ectopically in FB1 carrying pERCFP that expressed cyan fluorescent protein (CFP) targeted to the ER of U. maydis (Wedlich-Söldner et al., 2002). CFP fluorescence (Figure 2A, left panel) visualized the ER network at the periphery of the cell and the nuclear envelope, as described (Wedlich-Söldner et al., 2002). YFP fluorescence (Figure 2A, middle panel) was found to stain a similar network. Merging the two images showed significant colocalization of Gas-YFP and ER-CFP (Figure 2A, right panel). This suggests that Gas1 localizes to the ER.
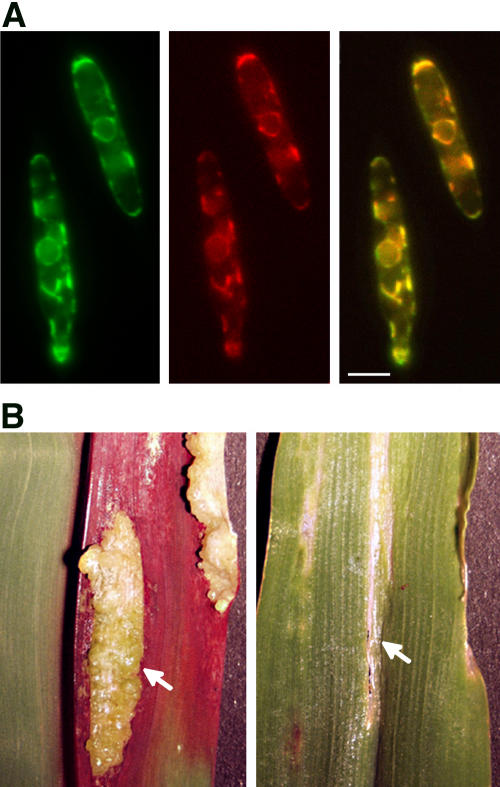
Figure 2.
Gas1-YFP Fusion Protein Localizes to the ER of U. maydis.
(A) Left panel: CFP fluorescence of CFP fusion protein that is targeted to the ER by a calreticulin signal peptide and a C-terminal ER retention signal (Wedlich-Söldner et al., 2002). Middle panel: YFP fluorescence of Gas-YFP fusion protein in the same cell. Since the Gas-YFP signal was faint and required long integration time, ER motility had to be stopped by brief formaldehyde treatment. The overall organization of the ER remained unaffected. Right panel: Merged images of left and middle panels showing colocalization of Gas1-YFP and ER-resident CFP. For clarity, signals emitted by CFP fluorescence were colored in green, while those emitted by YFP fluorescence were colored in red. Merged signals appear yellow. Bar = 3 μm.
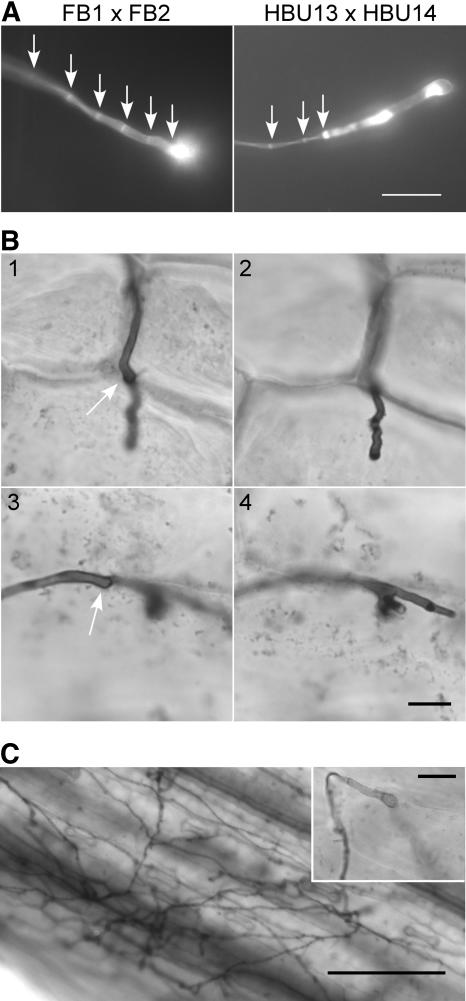
(B) Pathogenicity symptoms on infected plants. Five-day-old maize seedlings were inoculated either with a mixture of haploid wild-type cells (FB1 × FB2) or with a mixture of Δgas1 mutant strains (HBU13 × HBU14). Ten days after infection, tumors are formed on plants infected with the wild type (left panel, arrow), while infection with Δgas1 strains leads to the formation of localized slightly necrotic areas on the leaf (right panel, arrow).
To prove that gas1 encodes an α-glucosidase, we have attempted to complement an ER glucosidase II mutant of Saccharomyces cerevisiae. We took advantage of the temperature-sensitive phenotype of rot1-1 rot2-1 mutant cells (Bickle et al., 1998) and expressed gas1 from a galactose-inducible promoter on pYES2 (Figure 1C). Introduction of pGas1 restored growth at 37°C on galactose-containing medium as did introduction of pRot2 expressing the S. cerevisiae ROT2 gene. This demonstrates that gas1 encodes a functional ER glucosidase II.
Δgas1 Mutants Develop Normal Dikaryons
To create a complete deletion of the gas1 gene and allow monitoring of gas1 expression, the entire open reading frame of gas1 was replaced by the enhanced green fluorescent protein (eGFP) reporter gene and a hygromycin resistance cassette in plasmid pΔgas1:eGFP. Following transformation into compatible haploid strains FB1 and FB2, homologous integration of this construct gave rise to the gas1 deletion strains HBU13 and HBU14, respectively. Previous experiments showed that two genes causing pathogenicity defects in CL13 resulted only in reduced pathogenicity when the respective mutations were introduced in compatible haploid strains (Böhnert, 2001). Therefore, we conducted pathogenicity assays with mixtures of haploid strains. Twenty-five of 27 plants infected with a mixture of FB1 and FB2 developed tumors and anthocyanin coloration (Figure 2B, left panel). By contrast, of 48 plants infected with a mixture of the gas1 deletion strains HBU13 and HBU14, none produced these typical pathogenicity symptoms (tumors and/or anthocyanin). The only reaction seen on these plants was a slight necrosis in places where the fungal inoculum had been in direct contact with the leaf (Figure 2B, right panel). This demonstrates that gas1 is required for pathogenicity in the dikaryon.
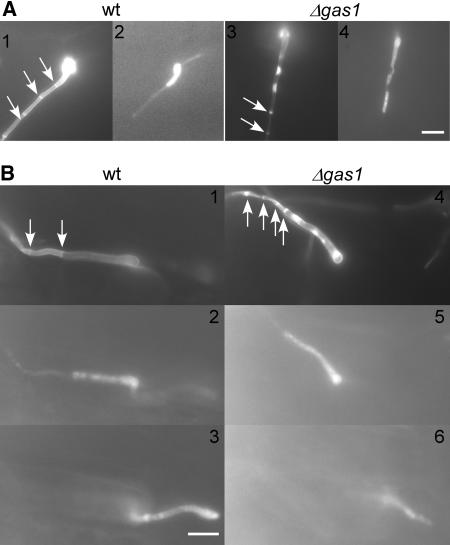
No difference of gas1 deletion strains to wild-type strains could be observed when growth was assessed in axenic culture either in complete medium (CM) or YEPS light or on nitrate minimal plates containing a variety of different carbon sources (see Methods; data not shown). With respect to cell morphology, gas1 deletion strains were also indistinguishable from their wild-type progenitors (Figure 3A, left panel). When stained for chitin with wheat germ agglutinin (WGA), Δgas1 mutants consistently showed increased cell wall staining, suggesting differences in wall composition or accessibility to the dye (Figure 3A, right panel). To assay mating competence, HBU13 and HBU14 were cospotted on CM-charcoal agar plates. Under these conditions, compatible wild-type strains fuse and develop filamentous dikaryotic hyphae (Day and Anagnostakis, 1971). The mixture of HBU13 and HBU14 formed dikaryotic hyphae, which developed more slowly but were comparable to wild-type hyphae after 48 h (Figure 3B). Dikaryons generated by mating of strains HBU13 and HBU14 (Δgas1) were indistinguishable from wild-type hyphae generated by mating of strains FB1 and FB2 (wild type) with respect to distance between nuclei (wild type, 9.1 ± 3.3 μm, n = 21; Δgas1, 10.6 ± 4.8 μm, n = 19), distance between nuclei and tip (wild type, 44.2 ± 8.5 μm, n = 14; Δgas1, 44.6 ± 12.6 μm, n = 13), number of nuclei (wild type, 2.3 ± 0.5, n = 42; Δgas1, 2.2 ± 0.4 μm, n = 17), length of tip cell (wild type, 108.7 ± 19.2 μm, n = 13; Δgas1, 93.2 ± 23.8 μm, n = 15), diameter of hyphae (wild type, 2.1 ± 0.3 μm, n = 20; Δgas1, 2.0 ± 0.3 μm, n = 19), and length of empty hyphal sections (wild type, 13.1 ± 1.9 μm, n = 15; Δgas1, 13.9 ± 3.7 μm, n = 17), and chitin distribution as visualized by WGA staining (Figure 3C). To test for altered cell wall composition, mutant and wild-type cells were resuspended in water for 30 min and the diameter determined. HBU14 cells increased significantly in size, while wild-type cells did not (Figure 4A), suggesting an altered cell wall composition. To test sensitivity to the cell wall–interfering drug calcofluor, FB2 and the Δgas1 mutant HBU14 were serially diluted and spotted on CM plates with and without the addition of calcofluor (50 μM). HBU14 was significantly more sensitive to calcofluor than FB2 (Figure 4B). This further strengthens the notion that the cell wall of the mutant strain is altered. Strains FB2 and HBU14 were also tested for growth on sublethal concentrations of tunicamycin, an inhibitor of N-glycosylation (Tkacz and Lampen, 1975; Lehle and Tanner, 1976). Growth of the Δgas1 strain was significantly reduced when compared with the wild type (Figure 4C). Tunicamycin is a potent inhibitor of UDP-GlcNAc:dolichol phosphate GlcNAc-1-P transferase (GPT) (Tkacz and Lampen, 1975; Lehle and Tanner, 1976). In absence of GPT, the synthesis of glycans for subsequent attachment of nascent polypeptides is prevented, and when GPT activity is reduced, hypersensitivity to tunicamycin results (Kukuruzinska and Lennon, 1995). The increased sensitivity of the Δgas1 mutant to tunicamycin thus indicates already existing defects in glycosylation, which are further enhanced by the drug.
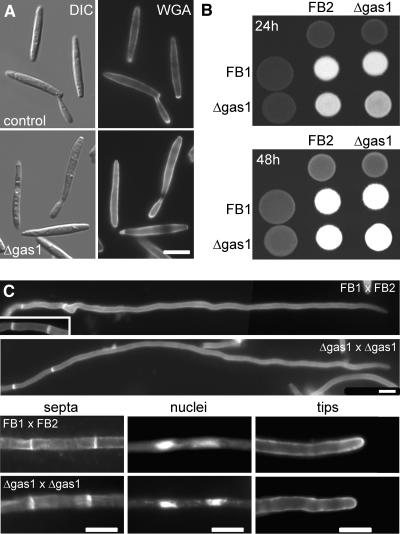
Figure 3.
Morphology of Δgas1 Mutant Strains.
(A) Haploid sporidia of wild-type strain FB2 (top panel) and Δgas1 strain HUB14 (bottom panel) as visualized by differential interference contrast optics (left panel) and after staining for chitin with fluorescein isothiocyanate–conjugated WGA (right panel). The lateral walls of Δgas1 mutant cells are more strongly stained, suggesting that chitin is more accessible to the dye. Bar = 5 μm.
(B) Mating assays of wild-type and mutant strains. Wild-type (FB1 and FB2) and Δgas1 mutant strains (HBU13 and HBU14) were spotted on charcoal-containing agar plates either as pure cultures or as mixtures with the compatible strains and incubated for 24 h (top panel) or 48 h (bottom panel) at 20°C. The appearance of white filaments indicates formation of dikaryotic hyphae. The cross of Δgas1 mutant strains develops dikaryotic hyphae slightly slower.
(C) Dikaryotic hyphae formed by a cross of the wild-type strains FB1 and FB2 or the Δgas1 mutant strains HBU13 and HBU14 after 24 h on charcoal-containing agar plates. Both wild-type and Δgas1 hyphae are straight, grow with a long extended tip cell, and form basal empty sections (top panel). With both strain combinations, these sections are limited by two septa as visualized using WGA (septa, bottom left panel). The tip cells of both strain combinations contain two nuclei as seen by 4′,6-diamidino-2-phenylindole (DAPI) straining of DNA (nuclei, lower middle panel) and carry a cap of chitin at the tip (tips, bottom right panel), indicating growth by tip extension. Bars = 10 μm.
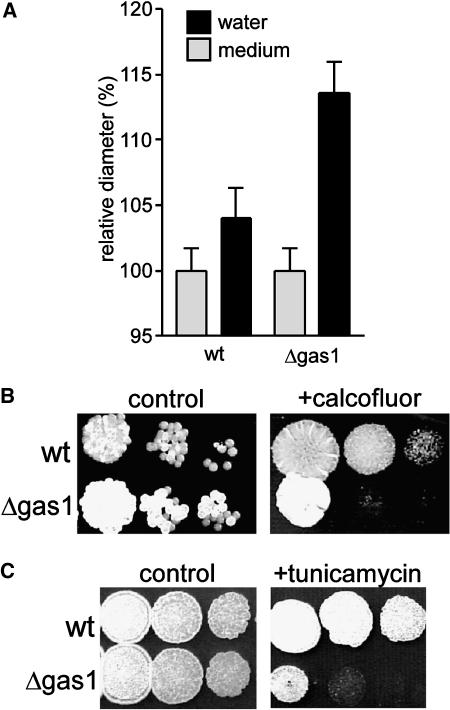
Figure 4.
The Δgas1 Mutant Displays Increased Sensitivity to Low Osmotic Conditions, Calcofluor, and Tunicamycin.
(A) The Δgas1 strain HUB14 reacts by swelling in hypo-osmotic conditions. Cells of overnight cultures of FB2 (wt) and HUB14 (Δgas1) in CM-containing glucose were washed twice in water and incubated for 30 min either in water (black bars) or in CM-containing glucose (gray bars). FB2 cells show some swelling; however, this is not statistically significant (P = 0.1664, t test, Welsh correction). HBU14 cells show significant swelling (P < 0.0001). Data represent mean + se, n > 50.
(B) The Δgas1 strain HUB14 is more sensitive to calcofluor. Serial dilutions of haploid sporidia of wild-type strain FB2 (wt) and Δgas1 strain HUB14 (Δgas1) were spotted on CM plates containing either glucose (control) or glucose and 50 μM calcofluor (+ calcofluor) and were incubated for 2 d at 28°C.
(C) The Δgas1 strain HUB14 is more sensitive to tunicamycin. Serial dilutions of haploid sporidia of wild-type strain FB2 (wt) and Δgas1 strain HUB14 (Δgas1) were spotted on CM plates containing either glucose (control) or glucose and 2.5 μg/mL tunicamycin (+ tunicamycin) and were incubated for 2 d at 28°C.
gas1 Is Induced during Pathogenesis
Since pathogenic development of gas1 deletion strains HBU13 and HBU14 was arrested shortly after penetration of the plant surface (see below), we examined gas1 expression in axenic culture after growth in YEPS light and during pathogenic development by real-time PCR after infection with wild-type strains. Compared to axenic culture, gas1 was upregulated approximately fivefold 1 d after infection (i.e., at a stage where appressoria have formed) (Brachmann et al., 2003), and expression was detectable throughout the biotrophic phase (see Supplemental Figure 1 online for details). In addition, eGFP fluorescence of strain HBU14, which carried the eGFP gene under control of the gas1 promoter, was studied in axenic culture after growth in CM containing different carbon sources (glucose, fructose, sucrose, rafinose, trehalose, arabinose, and glycerol trioleate). HBU14 showed the same low level of eGFP fluorescence in all of the tested conditions (data not shown). Thus, the observed specific upregulation of gas1 during early biotrophic development coincides with the stage where Δgas1 mutants arrest growth (see below).
Pathogenic Development of gas1 Deletion Strains Is Arrested after Plant Penetration
To establish at which stage pathogenic development of the mutant strains is blocked, we compared fungal development after inoculation of maize (Zea mays) plants with wild-type and Δgas1 strains using calcofluor staining. On the leaf surface, mating of haploid cells, formation and growth of the dikaryon, and development of appressoria were comparable for wild-type and mutant strains (data not shown). In wild-type strains, the final septum visible on the host surface delimited the strongly stained appressorium from the older part of the filamentous dikaryon (Figure 5A, left panel, arrows). This last visible septum was located 7 ± 2 μm (n = 56) from the tip. Hyphae of Δgas1 mutants showed normal septation in the older parts (Figure 5A, right panel, arrows). However, the last septum delimiting the appressorial cell was located 36 ± 9 μm (n = 69) from the tip. Throughout the appressorial cell, Δgas1 mutants displayed irregular deposits strongly stained with calcofluor, while in wild-type cells, strong calcofluor staining was restricted to the appressorial tip (Figure 5A). Of >100 appressoria analyzed, 30 h after infection 94% of the mutant hyphae showed this irregular distribution of cell wall material in the appressorial cell, while none of the wild-type appressoria showed this phenotype. Despite these alterations, Δgas1 appressoria were able to penetrate the plant surface as visualized by Chlorazole Black E staining (Figure 5B). However, while wild-type hyphae had ramified extensively in the host tissue by 2 d after infection (Figure 5C), the Δgas1 mutant hyphae arrested growth after penetration of the plant surface and never (66 infection structures analyzed) extended growth beyond the epidermal cell layer (Figure 5B).
Figure 5.
Δgas1 Strains Arrest Growth after Plant Penetration.
(A) Appressoria visualized by leaf staining with calcofluor 1 d after infection of young maize seedlings with a combination of wild-type (FB1 and FB2) or Δgas1 strains (HBU13 and HBU14). The wild-type appressoria show regular septation up to the appressorial tip (arrows), while in mutant appressoria, regular septation (arrows) does not continue up to the hyphal tip. The remaining elongated cell shows irregular distribution of calcofluor-stainable cell wall material. Bar = 10 μm.
(B) Δgas1 strains arrest growth after plant penetration. Maize seedlings were infected with a mixture of the Δgas1 strains HBU13 and HBU14. Two days after infection, leaf samples were stained with Chlorazole Black E and analyzed by light microscopy. Two different hyphae at their site of penetration are shown (1 and 3), also in a different focal plane (2 and 4), with the arrows indicating the appressorial tip on the plant surface. The invading hyphae grow into the plant cell but arrest growth (2) and may branch (4). Bar = 10 μm.
(C) Wild-type hyphae ramify extensively in the host leaf. Maize seedlings were infected with a mixture of FB1 and FB2. Two days after infection, leaf samples were stained with Chlorazole Black E and analyzed by light microscopy. The inset shows a typical wild-type appressorium with an invading hypha growing into the plant tissue out of focus. Bar = 100 μm (bar in inset = 10 μm).
To find out whether the irregular deposits in Δgas1 strains hide septa, we determined the distribution of cytoplasm in those cells. To this end, a plasmid expressing cytoplasmic GFP was integrated in single copy into the cbx-locus of strains FB1, FB2, HBU13, and HBU14, resulting in strains JSD2, JSF6, JSA8, and JSB11, respectively. Microscopic analysis of appressoria formed after a cross of the Δgas1 mutant strains JSA8 and JSB11 revealed that the elongated appressorial tip cell is completely filled with cytoplasm (Figure 6A). Focusing further down into the leaf revealed that the extended tip cells of JSA8/JSB11 dikaryons reached into the epidermal layer of the plant (Figure 6B, right panel). Comparisons with wild-type cells expressing cytoplasmic GFP (JSD2 and JSF6) showed that at earlier time points (22 h after inoculation) elongated tip cells filled with cytoplasm could also be observed that had already penetrated (Figure 6B, left panel). However, these structures did not show the enhanced calcofluor staining used as marker for detecting appressoria on leaves in previous experiments. At later time points, when strong calcofluor staining of wild-type appressoria was observed, appressoria no longer showed GFP fluorescence, presumably because all cytoplasm was already in hyphal parts within leaf tissue. By contrast, appressoria of the gas1 deletion strains JSA8/JSB11 always displayed GFP fluorescence (35 appressoria analyzed). From this, it follows that the strong calcofluor staining of the wild-type appressorium occurs at a time when most of the cytoplasm has already been translocated to the hyphal parts growing inside the plant. By contrast, the invading hyphae of Δgas1 mutants are unable to translocate their cytoplasm completely and are blocked halfway in.
Figure 6.
U. maydis Penetrates the Plant Surface before the Appressorial Tip Compartment Has Been Sealed by Septum Formation.
(A) A typical appressorium formed by wild-type strains (1 and 2) or by Δgas1 mutant strains (3 and 4). Young maize seedlings were infected with a combination of wild-type (JSD2 and JSF6) or Δgas1 mutant (JSA8 and JSB11) strains expressing cytoplasmic GFP. Appressoria were stained with calcofluor 1 d after inoculation and visualized by fluorescence microscopy (1 and 3) or GFP fluorescence (2 and 4). In wild-type appressoria, regular septation continues up to the appressorial tip (1, arrows), which is filled with cytoplasm (2). In appressoria of Δgas1 strains, the regular septation stops before reaching the tip (3, arrows), leaving an elongated tip cell with irregular wall deposits (3), which is filled with cytoplasm (4).
(B) Wild-type appressoria penetrate the plant surface before septation has reached the appressorial tip. A penetrating early appressorium of a wild-type dikaryon (left column) and a Δgas1 dikaryon (right column) expressing cytoplasmic GFP as visualized by calcofluor staining (1 and 4), GFP fluorescence (2 and 5), and GFP fluorescence in a different focal plane (3 and 6). At this early stage, both wild-type and Δgas1 hyphae show an elongated appressorial cell on the plant surface that is filled with cytoplasm. The cytoplasm reaches into the plant tissue. Bars = 10 μm.
Δgas1 Mutants Fail to Establish a Normal Host–Pathogen Interface
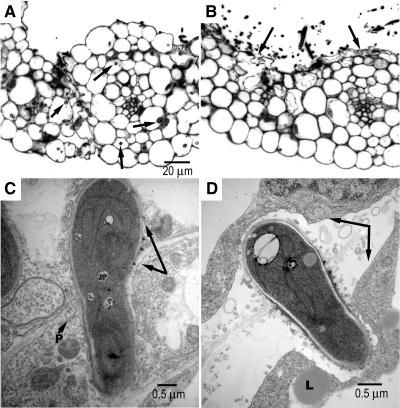
While the Δgas1 dikaryon is able to produce appressoria and penetrate, it is unable to proliferate extensively in host tissue. To characterize the host–pathogen interface, inoculated leaves were examined in section, both with light and electron microscopy. In sections of maize seedlings inoculated with compatible wild-type cells (FB1 and FB2), many hyphal profiles were seen (Figure 7A), showing that the dikaryotic filaments had invaded the leaf. By contrast, leaf sections of plants inoculated with compatible Δgas1 strains (HBU13 and HBU14) showed prominent hyphal growth only on the leaf surface (Figure 7B). Some epidermal plant cells appeared to be collapsed after infections with Δgas1 strains, while such collapsed cells were not detected after infections with wild-type strains (Figure 7B, arrows). Using transmission electron microscopy, the host–pathogen interface of invading wild-type hyphae was visualized (Figure 7C). As shown before for a variety of smut fungi, the interaction zone is electron opaque (Bauer et al., 1997), can be seen between the host plasma membrane and the fungal cell wall (Figure 7C, asterisks), and has also previously been described for U. maydis (Snetselaar and Mims, 1994). The wild-type hyphae were typically surrounded by host cells containing normal-appearing organelles (Figure 7C). By contrast, Δgas1 hyphae were often separated from the host plasma membrane by a wide, electron-transparent zone (Figure 7D, arrows), although the fungal wall and electron-dense interaction zone appeared normal (Figure 7D, asterisks). We speculate that these alterations in host tissue relate to the observed arrest of Δgas1 hyphae.
Figure 7.
Δgas 1 Mutants Fail to Establish a Normal Host–Parasite Interface.
(A) Section of a young maize leave 40 h after inoculation with a mixture of haploid wild-type cells (FB1 and FB2) visualized by light microscopy. Numerous invading hyphae can be detected (arrows) but the host cells or tissue organization seems undisturbed.
(B) Section of a young maize leave 40 h after inoculation with a mixture of haploid Δgas1 mutant cells (HBU13 and HBU14) visualized by light microscopy. Most hyphal profiles can be observed on the leaf surface. Some epidermal cells are collapsed (arrows). The bar in (A) is for (A) and (B).
(C) Invading dikaryotic hypha of a wild-type cross (FB1 and FB2) as visualized by transmission electron microscopy. The fungal hypha is in close contact with the plant cells. Arrows point to the plasma membranes of host cells, which delimit a characteristic interaction zone (asterisks) in close contact with the fungal wall. In the host wall, plasmodesmata (P) can be seen.
(D) Invading dikaryotic hypha of a Δgas1 mutant cross (HBU13 and HBU14) as visualized by transmission electron microscopy. The fungal hypha itself does not differ greatly in appearance from the wild-type filament seen in (C). However, there is a notable difference with respect to the host–fungus interface. The fungal filament is separated from the host plasma membrane by a large space (arrows) containing small vesicles. Host lipid deposits (L) can be observed in the vicinity of invading mutant hyphae. The fungus–plant interaction zone (asterisks) seems intact.
DISCUSSION
In this study, we have identified a gene, gas1, which is essential for pathogenicity in U. maydis. Gas1 is the catalytic α-subunit of a glucosidase II and localizes to the ER. Δgas1 deletion mutants are not affected in growth on artificial media, cell fusion, or dikaryon formation and on the leaves form appressoria that are able to penetrate the surface. However, pathogenic development of Δgas1 mutants is arrested soon after penetration. At this stage, mutant hyphae are unable to continue ordered septum formation and display irregular deposits of cell wall material as well as alterations in the host–fungus interface.
The gas1 gene disrupted in the nonpathogenic REMI mutant CL13pat4339 encodes a predicted polypeptide of 1061 amino acids that displays the highest degrees of similarity to GIIαs from various eukaryotic organisms, including S. pombe, mouse, and human. The notion that Gas1 is the catalytic subunit of glucosidase II and has a function in the ER is validated by demonstrating that gas1 complements a yeast mutant defective in the ER GIIα gene. Like ER-localized GIIα subunits from other systems, Gas1 does not possess an ER retrieval sequence. The observed ER localization is probably mediated by dimerization with a noncatalytic β-subunit, as has been shown for GIIα of S. pombe (D'Alessio et al., 1999). The genome of U. maydis contains one gene (Broad Institute, http://www.broad.mit.edu/seq/; contig 1.249, position 17485-15649) with significant similarity to glucodisase II β-subunits from mouse (AAC53183, 25% amino acid identity) and man (P14314, 23% amino acid identity). Glucosidase II enzymes play a central role in the processing of N-linked oligosaccharides in the ER, where they excise both remaining glucose units after glucosidase I has removed the terminal glucose from newly synthesized glycoproteins. UDP-Glc:glycoprotein glucosyltransferase then reglucosylates glucose-free oligosaccharides, thus establishing, together with GIIα, a deglucosylation-reglucosylation cycle (Trombetta and Helenius, 1998). Glucosidases are proposed to be crucial for ER quality control of glycoprotein folding and exit from the ER (Helenius and Aebi, 2001). In all systems studied to date, GIIα is encoded by a unique gene (Trombetta et al., 1996; Freeze et al., 1997; Simons et al., 1998). In the available U. maydis sequence, we have detected one gene with significant similarity to gas1 whose product shares with Gas1 the catalytic site motif of family-31 glycosyl hydrolases (Henrissat, 1991) (Broad Institute, http://www.broad.mit.edu/seq/; contig 1.92, position 7939-4835). It is feasible that this protein related to Gas1 provides for GIIα function primarily in haploid cells and during mating.
Despite the pivotal function of glucosidase II in glycoprotein processing and maturation, mutants in GIIα genes in S. cerevisiae and S. pombe are also viable and display no apparent growth defects (Fanchiotti et al., 1998; D'Alessio et al., 1999). In S. pombe, mutants lacking GIIα accumulate misfolded glycoproteins in the ER under nonstressed conditions (D'Alessio et al., 1999). Nevertheless, such cells do not show a discernible phenotype, and this is explained by the upregulation of BiP and probably other proteins that assist folding in the ER (Simons et al., 1998; D'Alessio et al., 1999). The only other case where mutation in an α-glucosidase leads to a developmental defect is D. discoideum, where modA mutants are perfectly viable but produce stunted fruiting bodies with a short, thickened stalk. For this system, it has been speculated that correctly trimmed sugar chain structures in glycoproteins may be required for correct stalk assembly (Freeze et al., 1997). N-linked sugars of glycoproteins have been implicated in correct protein folding, secretion, and protein stability (Rademacher et al., 1988; Varki, 1993). With respect to growth on a variety of substrates (see Methods), including some requiring the action of secreted proteins, Δgas1 mutant strains showed no differences with wild-type cells (data not shown). This could indicate that gas1 is specifically required during infection-related fungal development where the structure of sugar chains of the fungal cell wall may be crucial for establishment of a functioning interface with the host plant. Cytological analysis of the penetration process revealed that U. maydis does not accumulate its entire cytoplasm inside the appressorial head before penetrating the leaf. Instead, the infection hypha gradually progresses through the plant cuticle. The appressorium simply marks the point at which growth direction changes. This penetration mechanism sets U. maydis apart from other phytopathogenic fungi such as Magnaporthe grisea, Colletotrichum species, and rust fungi (Staples, 1985; Bechinger et al., 1999; Talbot, 2003). In these fungi, appressorium development is characterized by the deposition of melanin and the generation of high turgor pressure due to the accumulation of osmolytes (Bechinger et al., 1999; Talbot, 2003). When such a dome-shaped appressorium has fully matured, penetration occurs primarily by mechanical force through a penetration peg with a much smaller diameter than the appressorium. By contrast, appressoria of U. maydis are unmelanized and only slightly swollen relative to hyphae and form penetration structures that are much less constricted (Snetselaar and Mims, 1992, 1993; Snetselaar et al., 2001), making it unlikely that entry of U. maydis hyphae occurs by mechanical force. Infection hyphae of Δgas1 mutants form appressoria and penetrate the leaf surface. However, growth becomes arrested in the epidermal cell layer. At this stage, the tip cell extends from the surface to the underlying plant tissue, and no new septa are formed at the posterior end of the cytoplasm. Based on electron microscopy sectioning, gas1 mutant hyphae inside the plant tissue appeared normal, but the space between fungal cell wall and host membrane appeared widened, and mutant hyphae were surrounded by vesicular material. To address the possibility of an enhancement of plant defense reactions, invading hyphae of wild-type and Δgas1 mutants were stained with diaminobenzidine or dichlorofluoresceine diacetate to visualize reactive oxygen species. However, no differences in staining could be detected nor were there differences in callose accumulation (data not shown). Since diaminobenzidine staining could successfully be applied in mutants defective in H2O2 signaling (L. Molina and R. Kahmann, unpublished data), we consider it likely to indicate that Gas1 may be required for the production of infection-specific wall components. Specific modifications in hyphal wall structure during infection have been reported for several phytopathogenic fungi. For M. grisea (Howard et al., 1991), Colletotrichum lindemuthianum (O'Connell, 1991), and various rust fungi (Deising et al., 1996), infection hyphae displayed a dramatically reduced lectin affinity when compared to germ tubes. The strong uniform staining of wild-type appressoria by calcofluor, which is absent in the Δgas1 mutants, might be indicative of such cell wall alterations. Through the modified cell wall structure, the fungus might be protected from host defense systems. Alternatively, improper glycosylation of one or more proteins could prevent establishment of the appropriate host–parasite interface required for development of U. maydis in planta. Unlike many other biotrophic fungi, U. maydis does not form regular haustoria; nevertheless, plant cell membranes invaginate around the invading hyphae as they grow through the cell and must provide for the passage of nutrients from the plant to the fungus (Snetselaar and Mims, 1994; Mendgen and Hahn, 2002). At present, we are unable to distinguish between these possibilities (i.e., whether Δgas1 mutants are arrested due to lack of protection against host defense mechanisms, insufficient supply of nutrients, or both). In either case, the defect of the Δgas1 mutants is highly specific and manifests itself in the growing tip that has invaded the plant. It will be a challenge to find other U. maydis mutants that arrest growth in the epidermal cell layer, as this would allow attribution of the defects seen in the Δgas1 mutants to one specific gene.
METHODS
Strains, Growth Conditions, Cloning Procedures, and Constructs
Ustilago maydis strains were grown at 28°C in 2.5% (w/v) potato dextrose, YEPS (Tsukuda et al., 1988), or YEPS light (1% yeast extract [w/v], 0.4% Bacto-pepton [w/v], and 0.4% sucrose [w/v]). Solid media contained 2% (w/v) bacto-agar. Mating assays and plant infections were carried out as described (Gillissen et al., 1992). Strains were classified as nonpathogenic if neither tumor formation nor anthocyanin coloration of the infected plant was observed.
For cloning purposes, the Escherichia coli K12 strain DH5α (Bethesda Research Laboratories) was used and molecular methods followed described protocols (Sambrook et al., 1989). DNA isolation from U. maydis and transformation procedures were carried out as described (Schulz et al., 1990). Sequencing and cloning was performed in plasmids pUC19 (Yanisch-Perron et al., 1985), pBluescript II KS+ (Stratagene), and pSL1180 (Pharmacia).
When testing for drug sensitivity, strains were grown on CM plates containing glucose (50 mM) and either calcofluor (50 μM) or tunicamycin (2.5 μg/mL), as indicated for the individual experiments.
U. maydis strains FB1 (a1b1) and FB2 (a2b2), CL13 (a1bE1W2), and FB1EG (a1b1/pERGFP) have been described previously (Banuett and Herskowitz, 1989; Bölker et al., 1995a; Wedlich-Söldner et al., 2000). CL13pat4339 is a nonpathogenic REMI mutant obtained from CL13 by REMI using BamHI and the plasmid pSMUT (Bölker et al., 1995b). HBU13 (a1b1 Δgas1:eGFP) and HBU14 (a1b1 Δgas1:eGFP) were created by transformation of FB1 and FB2, respectively, with pΔgas1:eGFP. Homologous recombination led to the replacement of the open reading frame of gas1 by the eGFP gene, a nopaline synthase terminator, and a hygromycin resistance cassette. In these strains, eGFP expression is under the control of the gas1 promoter. HBU15 (a1bE1W2Δgas1) is a derivative of CL13, in which gas1 has been replaced by the phleomycin resistance cassette of pΔgas1. LAH1 (a1b1/pERGFP/pOGEY) was created by ectopic integration of pOGEY into strain FB1EG. This strain expresses the gas1-eYFP fusion protein in addition to a CFP fusion protein that is targeted to the ER by a calreticulin signal peptide and a C-terminal ER retention signal (Wedlich-Söldner et al., 2002). Strains JSD2 (a1b1 cbx::Potef-eGFP), JSF6 (a2b2 cbx::Potef-eGFP), JSA8 (a1b1 Δgas1:eGFP cbx::Potef-eGFP), and JSB11 (a2b2 Δgas1:eGFP cbx::Potef-eGFP) are derivatives of strains FB1, FB2, HBU13, and HBU14, respectively, and were obtained by homologous integration of the eGFP gene under the control of the strong constitutive otef promoter (Spellig et al., 1996) from p123 (C. Aichinger, unpublished data) at the cbx locus in single copy as described (Loubradou et al., 2001). All strains generated by homologous integration were verified by DNA gel blot analysis. To test growth on different carbon sources, FB2 and HBU14 were grown on nitrate minimal plates (Holliday, 1974) containing one of the following carbon sources (w/v): glucose (1%), fructose (1%), arabinose (1%), rafinose (1%), sucrose (1%), trehalose (1%), maltose (1%), sorbitol (1%), BSA (0.08%), glycerol trioleate (0.08%), oleic acid (0.08%), glycogen (0.08%), pectin from apple (Malus domestica; 0.08%), amylopectin from potato (Solanum tuberosum; 0.08%), or starch from maize (Zea mays; 0.08%).
Saccharomyces cerevisiae strains MH272-1da and MH272-1da rot1-1 rot2-1 (Bickle et al., 1998) were obtained from M. Hall. Plasmid-containing strains were grown on CM lacking the amino acid required for plasmid maintenance (Treco and Lundblad, 1993). Rich media contained yeast extract (10 g/L), peptone (10 g/L), and either glucose (20 g/L; YPglc) or galactose (20 g/L) and rafinose (10g/L; YPgal). Solid media contained agar (20 g/L). Transformation of yeast strains was carried out as described (Becker and Lundblad, 1994).
pΔgas1:eGFP
Plasmid pΔgas1:eGFP contains the eGFP gene fused to 2 kb of gas1 promoter region. The gas1 gene, including 2-kb promoter region and 1-kb terminator region, was cloned as a 3.8-kb NcoI-NcoI fragment plus a 3.1-kb NcoI-PstI fragment in pUC19. The promoter and terminator regions, including plasmid backbone, were amplified using two primers, HB1 (5′-CATGCCATGGTGACGTATATGCTGAGATCTACACTG-3′) and HB2 (5′-AGTTGCCATGGCTTGGGATCCTGATCATCAAAGATCCAGCTGTTGCCATTA-3′), generating an NcoI site at the 5′-end and a BamHI site at the 3′-end of gas1, respectively. The resulting PCR product was digested with NcoI and BamHI and ligated with the 700-bp NcoI-BamHI eGFP fragment from peGFP (Clontech), giving rise to pPgas1eGFP. The hygromycin resistance cassette from pTZhyg (R. Kahmann, unpublished data) was excised as a 3-kb BamHI fragment and inserted at the corresponding site of pPgas1eGFP.
pΔgas1
pΔgas1 is like pΔgas1:eGFP, but the eGFP gene and the hygromycin resistance cassette are replaced by a phleomycin resistance cassette. The 3-kb BamHI-BspHI phleomycin resistance cassette from pNEBble+ (New England Biolabs; A. Brachmann, unpublished data) was fused with the NcoI-BamHI–digested PCR product generated for the construction of pΔgas1:eGFP.
pOGEY
The plasmid pOGEY contains a C-terminal Gas1-eYFP fusion under the control of the otef promoter and was generated in three steps. (1) Fusion of the 5′-end of gas1 to the otef promoter: base pair 1 to 1329 (AatII site) of the gas1 open reading frame was amplified by PCR with HB3 (5′-CCACATGACATCCATGGG-3′) and HB4 (5′-CGTCACCATGGTCAGCAAGCATCGCTGGTCATGC-3′), generating an NcoI site at the start codon of gas1. The resulting PCR product was digested with NcoI and AatII. The otef promoter was recovered as a 900-bp HindIII-NcoI fragment from p123 (C. Aichinger, unpublished data). Both fragments were cloned in HindIII-AatII–digested pSL1180, giving rise to pOG1329. Subsequently, a 1.2-kb AatII-NotI fragment from the cDNA clone pgas1c14, corresponding to the gas1 open reading frame from base pair 1330 to 2524, was inserted in the corresponding sites of pOG1329, producing pOG2524. (2) Fusion of the 3′-end of gas1 to eYFP: base pair 2642 (BamHI site) to 3183 was PCR amplified with HB5 (5′-CCAAGGCTCTGAACGCGG-3′) and HB6 (5′-ATGTCCATGGACTCGAACTCGATGGTCCAGTCCG-3′), introducing an NcoI site at the 3′-end of gas1. The PCR product was digested with BamHI and NcoI and introduced in the corresponding sites of pEYFP (Clontech) to produce pGEY2641. A 700-bp SalI-BamHI fragment corresponding to the gas1 open reading frame from base pair 1939 to 2641 was recovered from the cDNA clone pgas1c14 and cloned in the corresponding sites of pGEY2641, giving rise to pGEY1939. (3) Assembly of pOGEY: the 3.4-kb HindIII-SspI fragment from pOG2524, carrying the otef promoter and gas1 up to base pair 2389, and the 2-kb SspI fragment from pGEY1939, carrying the 3′-end of the gas1 gene from base pair 2390, fused to the eYFP gene, were transferred to HindIII-SspI–digested plasmid pSMUT (Bölker et al., 1995b).
pRot2
For the construction of pRot2, the yeast ROT2 gene was amplified by PCR from genomic DNA of strain MH272-1da, using primers oJS122 (5′-TATTCCATGGTCCTTTTGAAATGGCTCGTATGC-3′) and oJS123 (5′-ATACGGCCGTCAAAAAATAACTTCCCAATCTTCAGTTATGTC-3′). The 3-kb PCR product was cloned into pCR2.1-TOPO (Invitrogen) and introduced as a 3-kb HindIII-XhoI fragment into the respective sites of pYES2.
pGas1
pGas1 is a derivative of pYESGasYFP, which contains the gas1-eyfp fusion from pOGEY in the PvuII and XbaI sites of pYES2. To generate pGas1, the eyfp part of pYESGasYFP was exchanged by a PCR product generated using genomic DNA of U. maydis FB1 and primers oJS118 (5′-TCCTGCGGTAGACAAAGACG-3′) and oJS119 (5′-CTACTCGAACTCGATGGTCCAGTC-3′), restoring the original stop codon of the gas1 gene. This exchange took several steps. First, the 760-bp PCR product was cloned into pCR2.1-TOPO to generate pTOPO-PCR. Second, a 676-bp EcoRV-NotI fragment of pTOPO-PCR was ligated to the 8279-bp XbaI-NotI fragment of pYESGasYFP. The ligation product was blunted using Klenow polymerase and self-ligated.
pLeu
pLeu is a derivative of pYES2, where the URA3 marker of pYES2 was exchanged by the LEU2 marker of pRS415Met25 (Mumberg et al., 1994). The LEU2 gene was amplified using primers oJS246 (5′-TATAAGCTAGCATGCCGATTTCGGCCTATTGG-3′) and oJS247 (5′-TATCAGGGCCCAACTATGCGGCATCAGAGC-3′) and introduced as a 2.4-kb fragment into the ApaI-NheI sites of pYES2.
Plasmid Rescue
Three micrograms of genomic DNA of mutant strain CL13pat4339 was digested with MluI, ligated in a total volume of 100 μL to achieve circularization of individual fragments, and transformed in E. coli by electroporation. The resulting plasmid p4339 carries the genomic regions flanking the insertion site cloned in pSMUT (Bölker et al., 1995b).
Sequence Analysis
Predicted amino acid sequences were analyzed using the programs BLAST (Altschul et al., 1997), Pfam (Bateman et al., 2002), and PSORT (Nakai and Horton, 1999).
Staining Procedures, Light and Electron Microscopy, and Image Processing
For staining of nuclei and chitin in haploid sporidia and dikaryotic filaments, cells were fixed for 30 min in 4% formaldehyde in PBS (Sambrook et al., 1989), followed by two washes. Cells were mechanically disrupted and attached to poly-l-Lys–treated cover slips. The DAPI staining and WGA staining were done as previously described (Wedlich-Söldner et al., 2000). Calcofluor staining using Fluorescent Brightener 28 (Sigma-Aldrich) for the microscopy of prepenetration stages of pathogenic development, and Chlorazole Black E staining for microscopy of post-penetration stages were performed as described (Brachmann et al., 2003). Staining for callose deposition was done by placing the infected leaves into a solution of 0.005% aniline blue in 50% ethanol for 16 h. Reactive oxygen species detection was done either by staining with a solution of diaminobenzidine (1 mg/mL) for 8 h followed by clearing in a solution of acetic acid:glycerol:ethanol (1:3:3) at 80°C for 5 min and subsequent storage of the samples in 60% glycerol or by staining with 2′, 7′-dichlorofluorescein diacetate (10 μM) in a solution of KCl (30 mM) and MES-KOH (10 mM) at pH 6.15 for 1 h at 22°C.
For sectioning, leaves were prepared by fixation in 2% glutaraldehyde/3% formaldehyde in phosphate buffer. Dehydration in ethanol was followed by infiltration and embedment in Spurr's epoxy resin. For light microscopy, 2-μm sections were cut with glass knives, dried onto glass slides, and stained with toluidine blue. For transmission electron microscopy, 80-nm sections were cut with a diamond knife, stained with uranyl acetate and lead citrate, and observed with a JEOL 1010 electron microscope.
Microscopic analysis was performed using a Zeiss Axiophot microscope. Frames were taken with a CoolSNAP-HQ CCD camera (Photometrics) or a SIT camera (Hamamatsu; C2400-08). Samples were observed either with differential interference contrast optics or by fluorescence microscopy using the standard fluorescein isothiocyanate, DAPI, or rhodamine filter sets or a specific filter set (excitation band-pass filter BP 470/20, beam splitter FT 493, emission band-pass filter BP 505-503) for eGFP fluorescence. Processing of images was carried out using Photoshop 6.0 (Adobe Systems).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AJ619756.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Kinetics of Expression of gas1 during the Biotrophic Phase of U. maydis by Real-Time PCR.
Supplementary Material
Acknowledgments
We thank K.H. Braun and J. Görl for performing the REMI screen and the isolation of numerous nonpathogenic mutants, H.-U. Mösch for supply of pRS415Met25, and E. Mayer for technical assistance. We are indebted to M. Hall for supply of S. cerevisiae strains. This work was in part supported by the German Research Foundation program Molecular Phytopathology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Regine Kahmann (kahmann@staff.uni-marburg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036285.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1989). Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F., and Herskowitz, I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, R., Oberwinkler, F., and Vanky, K. (1997). Ultrastructural markers and systematics in smut fungi and allied taxa. Can. J. Bot. 75, 1273–1314. [Google Scholar]
- Bechinger, C., Giebel, K.F., Schnell, M., Leiderer, P., Deising, H.B., and Bastmeyer, M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Becker, D.M., and Lundblad, V. (1994). Introduction of DNA into yeast cells. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.7.1–13.7.10.
- Bickle, M., Delley, P.A., Schmidt, A., and Hall, M.N. (1998). Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17, 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnert, H.U. (2001). Charakterisierung Apathogener REMI-Mutanten in Ustilago maydis. PhD dissertation (Munich, Germany: Ludwigs-Maximilians-Universität).
- Bölker, M. (2001). Ustilago maydis–A valuable model system for the study of fungal dimorphism and virulence. Microbiology 147, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Böhnert, H.U., Braun, K.H., Görl, J., and Kahmann, R. (1995. b). Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 248, 547–552. [DOI] [PubMed] [Google Scholar]
- Bölker, M., Genin, S., Lehmler, C., and Kahmann, R. (1995. a). Genetic regulation of mating, and dimorphism in Ustilago maydis. Can. J. Bot. 73, 320–325. [Google Scholar]
- Bölker, M., Urban, M., and Kahmann, R. (1992). The a mating type locus of Ustilago maydis specifies cell signaling components. Cell 68, 441–450. [DOI] [PubMed] [Google Scholar]
- Brachmann, A., Schirawski, J., Müller, P., and Kahmann, R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, A., Weinzierl, G., Kämper, J., and Kahmann, R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42, 1047–1063. [DOI] [PubMed] [Google Scholar]
- Cormack, B.P., Valdivia, R.H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- D'Alessio, C., Fernandez, F., Trombetta, E.S., and Parodi, A.J. (1999). Genetic evidence for the heterodimeric structure of glucosidase II. The effect of disrupting the subunit-encoding genes on glycoprotein folding. J. Biol. Chem. 274, 25899–25905. [DOI] [PubMed] [Google Scholar]
- Day, P.R., and Anagnostakis, S.L. (1971). Corn smut dikaryon in culture. Nat. New Biol. 231, 19–20. [DOI] [PubMed] [Google Scholar]
- Deising, H., Heiler, S., Rauscher, M., Xu, H., and Mendgen, K. (1996). Cellular Aspects of Rust Infection Structure Differentiation: Spore Adhesion and Fungal Morphogenesis. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Fanchiotti, S., Fernandez, F., D'Alessio, C., and Parodi, A.J. (1998). The UDP-Glc:glycoprotein glucosyltransferase is essential for Schizosaccharomyces pombe viability under conditions of extreme endoplasmic reticulum stress. J. Cell Biol. 143, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze, H.H., Lammertz, M., Iranfar, N., Fuller, D., Panneerselvam, K., and Loomis, W.F. (1997). Consequences of disrupting the gene that encodes alpha-glucosidase II in the N-linked oligosaccharide biosynthesis pathway of Dictyostelium discoideum. Dev. Genet. 21, 177–186. [DOI] [PubMed] [Google Scholar]
- Gillissen, B., Bergemann, J., Sandmann, C., Schröer, B., Bölker, M., and Kahmann, R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68, 647–657. [DOI] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2001). Intracellular functions of N-linked glycans. Science 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday, R. (1974). Ustilago maydis. In Handbook of Genetics, R.C. King, ed (New York: Plenum Press), pp. 575–595.
- Howard, R.J., Ferrari, M.A., Roach, D.H., and Money, N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88, 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: Nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell 81, 73–83. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska, M.A., and Lennon, K. (1995). Diminished activity of the first N-glycosylation enzyme, dolichol-P-dependent N-acetylglucosamine-1-P transferase (GPT), gives rise to mutant phenotypes in yeast. Biochim. Biophys. Acta 1247, 51–59. [DOI] [PubMed] [Google Scholar]
- Lehle, L., and Tanner, W. (1976). The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 72, 167–170. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C. (2002). Genomic approaches to fungal pathogenicity. Curr. Opin. Microbiol. 5, 372–378. [DOI] [PubMed] [Google Scholar]
- Loubradou, G., Brachmann, A., Feldbrügge, M., and Kahmann, R. (2001). A homolog of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40, 719–730. [DOI] [PubMed] [Google Scholar]
- Mendgen, K., and Hahn, M. (2002). Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7, 352–356. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Müller, R., and Funk, M. (1994). Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- O'Connell, R.J. (1991). Cytochemical analysis of infection structures of Colletotrichum lindemuthianum using fluorochrome-labelled lectins. Physiol. Mol. Plant Pathol. 39, 189–200. [Google Scholar]
- Parodi, A.J. (2000). Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 69, 69–93. [DOI] [PubMed] [Google Scholar]
- Rademacher, T.W., Parekh, R.B., and Dwek, R.A. (1988). Glycobiology. Annu. Rev. Biochem. 57, 785–838. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Frisch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schulz, B., Banuett, F., Dahl, M., Schlesinger, R., Schäfer, W., Martin, T., Herskowitz, I., and Kahmann, R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60, 295–306. [DOI] [PubMed] [Google Scholar]
- Simons, J.F., Ebersold, M., and Helenius, A. (1998). Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17, 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snetselaar, K., and Mims, C. (1994). Light and electron microscopy of Ustilago maydis hyphae in maize. Mycol. Res. 98, 347–355. [Google Scholar]
- Snetselaar, K.M., Carfioli, M.A., and Cordisco, K.M. (2001). Pollination can protect maize ovaries from infection by Ustilago maydis, the corn smut fungus. Can. J. Bot. 79, 1390–1399. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84, 193–203. [Google Scholar]
- Snetselaar, K.M., and Mims, C.W. (1993). Infection of maize stigmas by Ustilago maydis: Light and electron microscopy. Phytopathology 83, 843–850. [Google Scholar]
- Spellig, T., Bottin, A., and Kahmann, R. (1996). Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252, 503–509. [DOI] [PubMed] [Google Scholar]
- Staples, R.C. (1985). The development of infection structures by the rusts and other fungi. Microbiol. Sci. 2, 193–194, 197–198. [PubMed] [Google Scholar]
- Talbot, N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. [DOI] [PubMed] [Google Scholar]
- Tkacz, J.S., and Lampen, O. (1975). Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem. Biophys. Res. Commun. 65, 248–257. [DOI] [PubMed] [Google Scholar]
- Treco, D.A., and Lundblad, V. (1993). Preparation of yeast media. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.1.1-13.1.7.
- Trombetta, E.S., and Helenius, A. (1998). Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol. 8, 587–592. [DOI] [PubMed] [Google Scholar]
- Trombetta, E.S., Simons, J.F., and Helenius, A. (1996). Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271, 27509–27516. [DOI] [PubMed] [Google Scholar]
- Tsukuda, T., Carleton, S., Fotheringham, S., and Holloman, W.K. (1988). Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8, 3703–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M., Kahmann, R., and Bölker, M. (1996). The biallelic a mating type locus of Ustilago maydis: Remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 250, 414–420. [DOI] [PubMed] [Google Scholar]
- Varki, A. (1993). Biological roles of oligosaccharides: All of the theories are correct. Glycobiology 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner, R., Bölker, M., Kahmann, R., and Steinberg, G. (2000). A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J. 19, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner, R., Schulz, I., Straube, A., and Steinberg, G. (2002). Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron, C., Vieira, J., and Messing, J. (1985). Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.