Abstract
The Brg1 catalytic subunit of SWI/SNF-related complexes has been implicated in many developmental and physiological processes, but null homozygotes die as blastocysts prior to implantation. To circumvent this early embryonic lethality, we performed an ENU mutagenesis screen and generated a Brg1 hypomorph mutation in the ATPase domain. The mutant Brg1 protein is stable, assembles into SWI/SNF-related complexes, and exhibits normal ATPase activity but is unable to establish DNase I hypersensitivity sites characteristic of open chromatin. Mutant embryos develop normally until midgestation but then exhibit a distinct block in the development of the erythroid lineage, leading to anemia and death. The mutant Brg1 protein is recruited to the β-globin locus, but chromatin remodeling and transcription are perturbed. Histone acetylation and DNA methylation are also affected. To our knowledge, Brg1 is the first chromatin-modifying factor shown to be required for β-globin regulation and erythropoiesis in vivo. Not only does this mutation establish a role for Brg1 during organogenesis, it also demonstrates that ATPase activity can be uncoupled from chromatin remodeling.
Keywords: ATPase, β globin, Brg1, locus-control region, SWI/SNF, chromatin remodeling
Sequence-specific transcription factors are necessary, but usually not sufficient, to regulate the transcription of downstream target genes because the RNA Polymerase II (Pol II) holoenzyme cannot access DNA in the context of nucleosomes or higher-order chromatin structures (Struhl 1999; Lemon and Tjian 2000). However, transcriptional activators bound to enhancers or promoters can recruit chromatin-modifying complexes, which covalently modify N-terminal histone tails or utilize ATPase activity to alter the conformation and/or position of nucleosomes (Fry and Peterson 2001; Narlikar et al. 2002). As a result, these promoters adopt an “open” chromatin configuration capable of binding Pol II so that transcription can be initiated. Conversely, transcriptional repressors can recruit similar or identical complexes to other loci to establish a “closed” chromatin configuration that precludes transcription.
The Saccharomyces cerevisiae SWI/SNF complex, which was one of the first chromatin-modifying complexes to be identified, consists of 11 subunits with a total molecular mass of ∼1 MDa (Sudarsanam and Winston 2000; Smith et al. 2003; Mohrmann and Verrijzer 2005). After being recruited to a specific regulatory element, the SWI2/SNF2 catalytic subunit hydrolyzes tens to hundreds of ATP molecules per minute, thus enabling the complex to break DNA-histone contacts, slide histone octamers to alternate positions, and activate or repress ∼300 genes (Holstege et al. 1998; Sudarsanam et al. 2000). Conserved throughout metazoan evolution, SWI/SNF-related complexes have been characterized in Drosophila melanogaster and mammals. In Drosophila, the catalytic subunit is called brahma (brm), which is the Hindi word meaning “fate”, because mutations perturb the expression of homeotic genes and cause homeotic transformations (Tamkun et al. 1992; Tamkun 1995).
The biochemical composition and function of SWI/SNF-related complexes are more varied in mammals (Mohrmann and Verrijzer 2005). Duplication and divergence events during vertebrate evolution resulted in an increased number of SWI/SNF-related gene products, leading to subunit diversity in various mouse tissues and human cell lines. For example, mammalian complexes utilize either the brahma (Brm) or brahma-related gene 1 (Brg1) catalytic subunits, but not both (Wang et al. 1996). Interestingly, despite having similar sequences (75% identity at the amino acid level), in vitro activities, and widespread expression, gene targeting experiments have demonstrated that Brm is dispensable in mice (Reyes et al. 1998), whereas Brg1 is essential (Bultman et al. 2000). In fact, Brg1 is required for development beyond the peri-implantation stage of early embryogenesis. Null mutations of Baf155 or Baf47/Snf5, both of which encode integral components of Brm- and Brg1-containing complexes, also confer peri-implantation lethality (Klochendler-Yeivin et al. 2000; Roberts et al. 2000; Guidi et al. 2001; Kim et al. 2001). One possible explanation for such a marked difference in phenotypic severity is that Brm- and Brg1-containing complexes might be recruited to different downstream target genes. Indeed, the N termini of Brm and Brg1 are divergent and physically interact with different transcription factors in vitro. Brm associates with ankyrin repeats of factors in the Notch pathway, whereas Brg1 associates with zinc-finger transcription factors (Kadam and Emerson 2003).
To achieve a better understanding of the mechanism and developmental role of mammalian SWI/SNF-related complexes, a number of issues must be addressed. First, although Brg1 ATPase activity is crucial, it is not clear how the energy of ATP hydrolysis is harnessed to remodel nucleosomes. Second, these complexes have been implicated in a number of developmental and physiological processes, but functional data have been lacking because null homozygotes die at such an early stage of embryogenesis. Third, chromatin-remodeling properties have been described using artificial nucleosome arrays in vitro, but mechanistic studies have not been performed at physiologically relevant sites in vivo. Therefore, it is not known whether SWI/SNF-related complexes also affect covalent histone modifications or DNA methylation at these sites during remodeling.
Here we describe the isolation and characterization of an N-ethyl-N-nitrosourea (ENU)-induced Brg1 mutation that addresses all of the issues listed above. This partial loss-of-function mutation changes a single amino acid residue (E1083G) in a highly conserved region of the catalytic ATPase domain. The mutant protein is stable, assembles into SWI/SNF-related complexes, and exhibits normal ATPase activity, but its nucleosome remodeling properties are diminished. Mutant embryos develop normally until midgestation, but then exhibit a distinct block in the development of the erythroid lineage, leading to anemia and death. Mutant Brg1 complexes are recruited to the β-globin locus control region (LCR), but chromatin remodeling and expression are significantly reduced. Both histone acetylation and DNA methylation are also affected in vivo.
Results
A novel Brg1 hypomorph allele
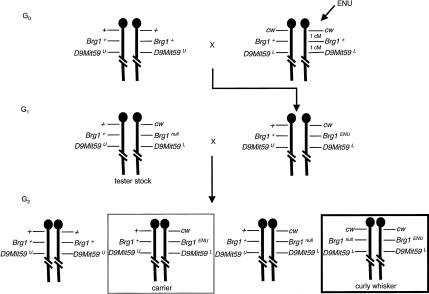
To circumvent the early embryonic lethality of Brg1null homozygotes, we isolated a novel Brg1 hypomorph allele in an ENU mutagenesis screen. We performed this screen by exploiting a visible, recessive marker, called curly whiskers (cw), that is tightly linked to Brg1 and a microsatellite marker (D9Mit59) on proximal chromosome 9 (Fig. 1). cw/cw mice are viable and fertile but have curly vibrissa (Falconer and Isaacson 1966). In the first step of this screen, cw/cw males were mutagenized and bred to wild-type females. Five hundred twenty-five G1 progeny were recovered. Each one inherited a wild-type chromosome 9 from the dam and a mutagenized cw chromosome 9 from the sire. The second step of this screen was designed to identify which, if any, G1 progeny carried a Brg1 mutation (Brg1ENU). Toward this end, every G1 specimen was bred to a tester stock that had a wild-type chromosome 9 balanced with a cw, Brg1null, D9Mit59L haplotype. The G2 progeny were expected to fall into four equally represented genotypic classes unless a Brg1ENU mutation was inherited that failed to complement Brg1null. In this case, the cw/cw class would be underrepresented (<25%) or completely missing. Therefore, we scored whiskers of neonatal pups from all 525 pedigrees. Collectively, 2033 out of 8774 G2 progeny were cw/cw. However, no cw/cw pups were obtained for one pedigree after 83 G2 progeny were scored from eight litters (Supplementary Table 1). This Brg1ENU1 mutation was recovered in carriers, which were identified as normal whiskered G2 mice with a D9Mit59U/L genotype not carrying the Brg1null allele, as determined by PCR genotyping.
Figure 1.
Brg1 ENU mutagenesis screen. The mutagenesis and breeding strategy that was implemented is outlined by showing a schematic of mouse chromosome 9 with the position of curly whiskers (cw), Brg1, and D9Mit59 loci. Genetic distances are indicated in centimorgans (cM) in the top right panel. cw/cw males (top right) were mutagenized (ENU) and bred to wild-type females (top left). (Middle right) G1 progeny inherited one wild-type chromosome 9 from the dam and one mutagenized cw chromosome 9 from the sire. (Middle left) G1 specimens were mated to a tester stock to determine which, if any, carried a Brg1 mutation (Brg1ENU). (Bottom) In the absence of a Brg1ENU mutation, G2 progeny fell into four equally represented genotypic classes. (Bottom right) However, inheritance of a Brg1ENU mutation that failed to complement Brg1null would result in underrepresentation or loss of the curly whisker class (enclosed by a thick box). (Bottom, second from left) In this case, the Brg1ENU mutation could be recovered in carriers (enclosed by a thin box).
Brg1+/ENU1 carriers were bred to the tester stock to confirm that the mutation was heritable and to determine when Brg1null/ENU1 mutants die. The cw marker is not informative before birth so Brg1null/ENU1 mutant embryos were identified by PCR as D9Mit59L/L. We recovered mutants at each stage of development but observed midgestation lethality based on lack of a heartbeat. Lethality started at embryonic day 11.5 (E11.5), nearly half were dead at E12.5, and every mutant was dead by E14.5 (Supplementary Table 1). The orientation of these crosses was not a factor, which is consistent with Brg1 not being imprinted. The fact that Brg1null/ENU1 embryos survived much longer than Brg1null/null embryos (midgestation compared with peri-implantation, respectively) indicated that Brg1ENU1 is a hypomorph (i.e., partial loss-of-function) allele.
Molecular and biochemical nature of the Brg1ENU1 mutation
One advantage of ENU mutagenesis is that point mutations are induced, which provide valuable structure-function correlations. To identify the molecular nature of the Brg1ENU1 lesion, we performed RT-PCR on wild-type (i.e., the C57BL/10 genetic background carrying cw that was mutagenized) and mutant tissues and sequenced full-length Brg1 cDNA products. A single adenosine-to-guanine transition was identified in exon 23 that changes the corresponding amino acid sequence from a glutamic acid (E) to a glycine (G) (Fig. 2A). The fact that this E1083G substitution is deleterious was not surprising because a negatively charged residue in a highly conserved segment of the ATPase domain, between putative helicase motifs IV and V, was converted to an uncharged polar residue (Fig. 2B). The position and hypomorphic nature of this substitution is informative because a K-to-A change two amino acids upstream of this residue in yeast SWI2/SNF2 was shown to be a null, whereas a D-to-A change three positions downstream had little or no phenotypic effect (Richmond and Peterson 1996).
Figure 2.
Molecular nature of the Brg1ENU1 mutation. (A) DNA sequences of wild-type (WT) and Brg1ENU1 (Mut) RT-PCR products are displayed showing an A- to-G point mutation, resulting in a glutamic acid (Glu, E)-to-glycine (Gly, G) amino acid substitution. (B, top) A schematic of the Brg1 protein, indicating the position of the E1083G amino acid substitution between putative helicase motifs IV and V of the catalytic ATPase domain. (ZFP) Zinc-finger protein interaction domain; (Rb) retinoblastoma protein interaction domain. (Bottom) Amino acid sequence alignment showing Homo sapiens and Mus musculus Brg1 E1083 and adjacent residues compared with H. sapien/M. musculus Brm, D. melanogaster BRM, and S. cerevisiae SWI2 and STH1 within the SWI2/SNF2 subfamily, as well as H. sapien/M. musculus Snf2l and Etl1 and S. cerevisiae RAD54 outside the SWI2/SNF2 subfamily. E1083 and corresponding residues are highlighted with a yellow vertical bar. Identities are represented by dots.
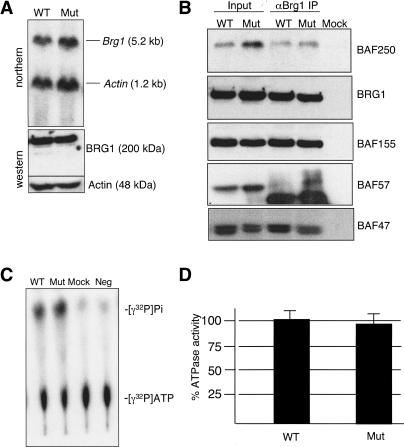
The Brg1ENU1 mutation did not affect mRNA or protein stability based on Northern blot and Western blot analyses, respectively (Fig. 3A). Mutant embryos also expressed normal levels of Brg1-associated factors (BAFs), which are noncatalytic subunits of SWI/SNF-related complexes (see Mut lanes under Input heading of Western blot panels in Fig. 3B). In addition, the Brg1ENU1 protein was shown to physically interact with BAFs in coimmunoprecipitation assays using a BRG1 antibody that does not cross-react with BRM (see Mut lanes under αBrg1 IP heading of Western blot panels in Fig. 3B; see Supplementary Fig. 1 for specificity of BRG1 antibody). This finding indicated that SWI/SNF-related complexes assemble and are stable in mutants. In these experiments, a mock immunoprecipitation was performed, where BRG1 antibody was omitted from wild-type embryo lysates, as a negative control. Complete lack of signal demonstrated that Protein A/G agarose beads did not bind Brg1 or BAFs nonspecifically (see Mock lanes in Western blot panels of Fig. 3B).
Figure 3.
Normal stability and ATPase activity of mutant SWI/SNF-related complexes. (A) Brg1 mRNA and protein expression. (Top) Northern blot of wild-type (WT) and mutant (Mut) E12.5 embryo RNAs hybridized with Brg1 and Actin control probes. (Bottom) Western blot of E12.5 embryo protein lysates probed with BRG1 and Actin control antibodies. (B) Coimmunoprecipitation of BAFs from wild-type (WT) and mutant (Mut) E12.5 embryo protein lysates. Shown are Western blot panels containing input protein (under Input heading), proteins coimmunoprecipitated with a BRG1 antibody (under αBrg1 IP heading), and a mock coimmunoprecipitation where BRG1 antibody was omitted but Protein A/G-conjugated agarose beads were added to wild-type protein lysates and processed (Mock). Western blot panels were probed with antibodies specific for BAF250, BRG1, BAF155, BAF57, and BAF47. IgG heavy-chain bands (∼55 kDa) are visible in IP lanes of the BAF57 panel. (C) ATPase assay. Wild-type (WT) and mutant (Mut) SWI/SNF-related complexes, isolated from E12.5 embryos by coimmunoprecipitation using a BRG1-specific antibody, were incubated with radiolabeled ATP, and reactions were subjected to thin-layer chromatography. Hydrolyzed [γ32P]Pi and unhydrolyzed [γ32P]ATP and are indicated on the right. Two negative controls are shown: (Mock) Mock immunoprecipitation as described above; (Neg) no protein added to radiolabeled ATP reaction mixture. (D) Quantification of ATPase assays. Densitometry was used to measure [γ32P]Pi and [γ32P]ATP values. The ratio of [γ32P]Pi to [γ32P]ATP was calculated and the negative control background signal was subtracted. Wild-type samples were normalized to 100%. The experiment was repeated three times, and error bars are shown.
Next, we wanted to determine whether mutant complexes exhibit reduced ATPase activity. We isolated native SWI/SNF-related complexes by coimmunoprecipitation using a BRG1 antibody that does not cross-react with BRM as before. Western blot analyses indicated that equivalent amounts of wild-type and mutant Brg1 were recovered (data not shown), and ATPase assays were performed directly on agarose beads. Hydrolyzed [γ32P]Pi was separated from [γ32P]ATP on cellulose plates by thin layer chromatography (Fig. 3C) and quantified by densitometry (Fig. 3D). Wild-type controls and mutants had similar ATPase activities (Fig. 3C,D). ATPase activity was stimulated by DNA as expected for SWI/SNF (data not shown). The high degree of similarity between wild-type and mutant ATPase activities was surprising because Brg1 is the catalytic subunit and the E1083G substitution lies within the ATPase domain. To reconcile this apparent paradox, additional ATPases were included in the amino acid sequence alignment (Fig. 2B). Importantly, although E1083 is absolutely conserved from yeast to human within the SWI2/SNF2 subfamily, it is not conserved in closely related proteins (e.g., Snf2l and Etl1) or more distantly related members of the ATPase superfamily (e.g., RAD54) (Eisen et al. 1995). Based on this observation and X-ray crystal structures of the catalytic domain from related proteins (Durr et al. 2005; Thoma et al. 2005), E1083 cannot be part of the active site. Instead, E1083 is part of a SWI2/SNF2 subfamily signature that might be involved in determining how the energy of ATP hydrolysis is harnessed to change the conformation and position of nucleosomes.
Brg1null/ENU1 erythropoiesis defect
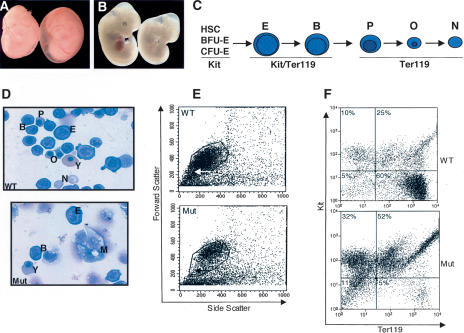
The increased penetrance of Brg1null/ENU1 lethality from E11.5 to E14.5 (Supplementary Table 1) coincides with erythropoiesis switching from the yolk sac to the fetal liver. The main switch occurs at E11.5, but yolk sac-derived erythrocytes persist and continue to circulate for several days. Accordingly, these primitive erythrocytes account for nearly all of the peripheral red blood cells at E10.5 but drop to 75% at E12.5 and 10% by E14.5 (Mucenski et al. 1991; Trimborn et al. 1999). There is normally a concomitant increase in fetal liver-derived definitive erythrocytes in the general circulation from very few at E10.5 to 25% at E12.5 and 90% by E14.5. However, this switch did not occur properly in Brg1null/ENU1 embryos, hereafter referred to as mutants, which led to severe anemia and death. Mutants still alive at E12.5 exhibited signs of normal primitive erythropoiesis in the yolk sac (Fig. 4A) but greatly diminished definitive erythropoiesis in the fetal liver (Fig. 4B). Mutant fetal livers had either a red speckled appearance or were completely pale, indicating a lack of hemoglobin-containing cells. We stained cytospin preparations of fetal liver cells with Wright-Giemsa to determine which cell types were present or absent (Fig. 4C). Wild-type controls had erythroid cells at each stage of the lineage. These cells ranged from proerythroblast progenitors, through a series of increasingly differentiated erythroblasts that become progressively smaller with nuclei that are smaller and more condensed, to enucleated reticulocytes (Fig. 4C,D). Nucleated yolk sac-derived erythrocytes were also present as expected. In contrast, mutants had proerythroblasts and basophilic erythroblasts but lacked more mature polychromatic erythroblasts, orthochromatic erythroblasts, and enucleated reticulocytes (Fig. 4D). An arrest at this stage in the lineage explains the extreme pallor of mutant fetal livers because hemoglobin begins to accumulate at significant levels in polychromatic erythroblasts (Wojda et al. 2002). Nucleated yolk sac-derived erythrocytes and megakaryocytes were present in mutant fetal livers (Fig. 4D), indicating that the defect was restricted to definitive erythropoiesis.
Figure 4.
Brg1null/ENU1 erythroid phenotype. (A,B) Bright-field photographs of wild-type control (left) and mutant (right) E12.5 embryos inside of yolk sacs (A) and with yolk sacs removed (B). (C) Schematic of erythroid lineage. (HSC) Hematopoietic stem cell; (BFU-E) erythroid burst-forming unit; (CFU-E) erythroid colony-forming unit; (E) proerythroblast; (B) basophilic erythroblast; (P) polychromatic erythroblast; (O) orthochromatic erythroblast; (N) enucleated reticulocyte. Expression status of Kit and Ter119 cell-surface markers utilized for flow cytometric anlayses in panel F are indicated below. (D) Cystospin preparations of wild-type (top) and mutant (bottom) fetal liver cells stained with Wright-Giemsa. Abbreviations are the same as for panel C except for two additions: (Y) yolk sac derived erythrocyte; (M) megakaryocyte. Megakaryocytes were present in wild-type fetal livers but none are shown in the upper panel. (E) Flow cytometry of wild-type (top) and mutant (bottom) fetal liver cells showing forward scatter versus side scatter. Arrows point to smaller, more mature population of erythroid cells that were well represented in wild type but underrepresented in mutants. (F) Distribution of Kit and Ter119 wild-type (top) and mutant (bottom) E12.5 fetal liver cells. (Upper left quadrants) Kit-positive cells. (Upper right quadrants) Kit/Ter119 double-positive cells. (Lower right quadrants) Ter119-positive cells. (Lower left quadrants) Double-negative cells. Percentage of cells in each quadrant is indicated.
Flow cytometry was performed to confirm and extend the previous histological findings. Using forward scatter as a measure of cell size, a broad distribution of fetal liver cell sizes was observed in wild-type controls, whereas smaller, more differentiated cells were underrepresented in mutants (Fig. 4E). Kit and Ter119 cell-surface markers were utilized to identify specific classes of cells in a quantitative manner. Kit is a progenitor cell marker (Uoshima et al. 1995), and Ter119 is a pan-erythroid marker (Fig. 4C; Kina et al. 2000). Therefore, Kit-positive cells represent hematopoietic stem cells (HSC), erythroid burst forming units (BFU E), and erythroid colony forming units (CFU E). Kit/Ter119 double-positive cells correspond to proerythroblasts and basophilic erythroblasts. Ter119-positive cells represent terminally differentiated polychromatic erythroblasts, orthochromatic erythroblasts, and enucleated reticulocytes (and nucleated yolk sac-derived erythrocytes). Finally, double-negative cells include nonerythroid hematopoietic cells, fibroblasts, and hepatocytes. The percentage of Kit, Kit/Ter119, Ter119, and double-negative cells in wild-type fetal livers was 10%, 25%, 60%, and 5%, respectively (Fig. 4F). In mutants, the Ter119 class was markedly reduced, from 60% to 5% (Fig. 4F). Moreover, most or all of these cells were probably yolk sac derived, since they also express Ter119. On the other hand, Kit (32%) and Kit/Ter119 progenitor cells (52%) were well represented in Brg1 mutants (Fig. 4F), consistent with the Wright-Giemsa-stained cytospin results. Double-negative cells were also well represented (11%). However, each of these cell types was overrepresented only on a percentage basis as a consequence of mutants having so few Ter119-positive cells. Therefore, mutant progenitor cells were not accumulating or piling up in the lineage upstream of the developmental block. Taken together, the Wright-Giemsa-stained cytospin results and flow cytometry data indicate mutant progenitor cells arrest at the basophilic-to-polychromatic erythroblast transition. Interestingly, EKLF and GATA-1 mutants exhibit a similar arrest at the proerythroblast stage (Fujiwara et al. 1996; McDevitt et al. 1997; Drissen et al. 2005).
Brg1 versus Brm expression in the erythroid lineage
We were curious why most tissues and organ systems developed properly in Brg1null/ENU1 mutants, considering that Brg1 encodes a chromatin-modifying factor expressed in a widespread manner. One possible explanation was that the Brm paralog could functionally compensate everywhere except in the erythroid lineage. Brm is 75% identical to Brg1 at the amino acid level, has identical activities in several in vitro assays, and is also broadly expressed. Expression of these two genes has not been compared in the erythroid lineage, so we performed RT-PCR on Ter119-positive flow sorted cells. Although both Brg1 and Brm were expressed in whole embryos, which served as a positive control, only Brg1 was expressed in Ter119-positive cells (cf. Brg1 and Brm panels for Ter119 lane in Fig. 5). Gapdh was amplified to demonstrate that the Ter119-positive cells and other RT templates were of good quality and present in equivalent amounts. An Epor control amplicon confirmed that Ter119-positive cells were, in fact, erythroid. These results support the notion that Brm may compensate in many cell types but cannot do so in the erythroid lineage because it is not expressed. We propose that Brg1 and SWI/SNF-related complexes are particularly important at the basophilic-to-polychromatic erythroblast transition because the genome is becoming highly condensed but certain genes must be maintained in an open chromatin configuration and continue to be expressed. A similar proposal has been made regarding the role of SWI/SNF complexes regulating gene expression while the chromosomes are highly condensed during mitosis in yeast (Krebs et al. 2000).
Figure 5.
Brg1 and Brm expression in erythroid cells and other hematopoietic lineages. Shown are negative images of ethidium bromide-stained agarose gels of Brg1 and Brm RT-PCR products amplified from the following flow-sorted cell populations: (CD4+) T helper lymphocytes; (CD8+) cytotoxic T lymphocytes; (CD4+ CD8+) immature double-positive T lymphocytes; (Mac1+) macrophages; (Gr1+) granulocytes; (Ter119+) erythroid (highlighted with a vertical arrowhead); (B220+) B lymphocytes. Two controls were also performed: (Embryo) E12.5 positive control; (Neg) no template negative control. In addition to Brg1 and Brm, two control amplicons were included: (Gapdh) glyceraldehyde 3-phosphate dehydrogenase; (Epor) erythropoietin receptor, to control for specificity of Ter119 erythroid marker.
We also compared Brg1 and Brm expression in several other hematopoietic lineages (Fig. 5). Brg1 was robustly expressed in all of the lineages that were analyzed. In contrast, Brm was differentially expressed with moderate to relatively high levels of mRNA in T helper cells (CD4+), cytotoxic T cells (CD8+), macrophages (Mac+), and granulocytes (Gr+) but very low levels of mRNA in immature, double-positive T cells (CD4+ CD8+) and in B cells (B220+). The Brg1null/ENU1 mutant phenotype occurs too early to assess either type of lymphocyte, but Cre-loxP strategies have already demonstrated a role for Brg1 in T-cell development (Chi et al. 2003; Gebuhr et al. 2003). In these studies, a developmental block was shown to occur at the double-negative-to-double-positive transition, which coincides with lack of Brm expression in this lineage.
A Brg1 downstream target is the β-globin locus
Gene expression profiling of Kit-positive cells flow sorted from wild-type and mutant fetal livers identified β globin as a downstream target gene (data not shown), and this result was confirmed by RT-PCR. The expression of both fetal/adult β-globin genes, βmaj (Hbb-b1) and βmin (Hbb-b2), was reduced in mutants (Fig. 6). In contrast, we did not observe any difference in the expression of either embryonic β-globin gene (εY or βH1) (Fig. 6). These results are consistent with the Brg1 mutant phenotype (i.e., E11.5-E14.5 lethality). εY and βH1 are expressed by yolk sac-derived primitive erythrocytes (but not fetal liver-derived definitive erythrocytes), and primitive erythropoiesis defects usually result in lethality by E10.5. βmaj and βmin are expressed by fetal liver-derived definitive erythrocytes (but not yolk sac-derived primitive erythrocytes), and definitive erythropoiesis defects usually result in lethality at E11.5 or later in development. These results indicate that globin switching occurs normally in mutants, but definitive erythroblasts express fetal/adult β globins at reduced levels.
Figure 6.
Expression of β-globin genes. (A) Shown are images of ethidium bromide-stained agarose gels containing Actin and β-globin RT-PCR products amplified from wild-type (WT) or mutant (Mut) E12.5 fetal livers. For each panel, the upper band is Actin and the lower band is a β-globin (the positions of which are indicated with arrowheads). The individual β globin amplicons are, from left to right, βmaj, βmin, εY, and βH1. For the εY and βH1 panels, two templates were used: (1) Fetal livers; (2) Kit-positive cells from fetal livers that were isolated by flow sorting. Expression of both embryonic globins can be observed in wild-type and mutant fetal livers because of yolk sac-derived primitive erythocytes still in the general circulation (see 1s). In contrast, neither embryonic globin is ectopically expressed in mutant Kit-positive definitive erythroblasts (see 2s). (MW) One-kilobase molecular weight standard. (B) Quantification of β-globin expression levels. For wild-type samples (black bars), ratios of β globin to actin were normalized to 100%. Mutant samples (gray bars) were expressed as a percentage relative to wild type. εY and βH1 values are based on fetal liver data (1s from A).
Reduced β-globin transcription is consistent with previous in vitro data. The zinc-finger transcription factors EKLF and GATA-1 bind directly to Brg1 (but not Brm), and EKLF can recruit SWI/SNF-related complexes to the β-globin locus in erythroleukemia cell lines (Armstrong et al. 1998; O'Neill et al. 1999; Kadam and Emerson 2003). This recruitment step and Brg1 ATPase activity are necessary to establish DNase I hypersensitive sites (HSs) characteristic of open chromatin and stimulate transcription of a β-globin reporter construct (Armstrong et al. 1998).
Perturbed chromatin remodeling at the β-globin locus
We performed a series of experiments to investigate the role of Brg1 and SWI/SNF-related complexes in the regulation of the β-globin locus in vivo. The mouse β-globin LCR consists of a series of tissue-specific DNase I HSs that have enhancer and possibly insulator or boundary element properties in the erythroid lineage. The LCR directs transcription of four downstream β-globin genes arranged in the order of their developmental expression (Fig. 7A). EKLF and GATA-1 bind to their consensus sequences (CACCC and A/TGATAA/G, respectively) in HS2 and HS3 (Lee et al. 2000; Duan et al. 2001; Horak et al. 2002; Levings and Bungert 2002). Therefore, we first wanted to determine whether Brg1 is recruited to these sites in vivo as expected. We performed chromatin immunoprecipitation (ChIP) assays on E12.5 fetal liver cells with a BRG1 antibody. Robust PCR products indicated that Brg1 was localized to both HS2 and HS3 in wild-type and mutant samples (see ChIP lanes for HS2 and HS3 panels in Fig. 7B). The finding that mutant Brg1 also localized to HS2 and HS3 is consistent with the point mutation (E1083G) not disrupting the N-terminal EKLF/GATA-1 interaction domain (corresponding to ZFP in Fig. 2B) but rather lying in the ATPase domain. Mock immunoprecipitations were also performed, where BRG1 antibody was omitted, to serve as a negative control. Complete lack of HS2 and HS3 PCR products demonstrated that Protein A/G agarose beads were not binding Brg1 or HS2/HS3 nonspecifically (see M lanes in Fig. 7B). As an additional negative control, we attempted to amplify the promoter of a gene (Necdin) not regulated by Brg1 from the same ChIP templates. Unlike the HS2 and HS3 results, Brg1 was not localized to this promoter (see ChIP lanes for Necdin promoter panel in Fig. 7B).
Figure 7.
Chromatin structure is altered at the β-globin locus. (A) Schematic of mouse β-globin locus. The LCR consists of six HSs (vertical arrows), although HS5 is a weak site in the mouse (denoted by thin vertical arrow). EKLF and GATA-1 bind to HS2 and HS3. All four β-globin genes consist of three exons (black boxes) that are transcribed in the same orientation and arranged in the order of their developmental expression. (B) Recruitment of Brg1 to the β-globin LCR. ChIP assays demonstrating that wild-type and mutant Brg1 are recruited to HS2 and HS3. Shown are images of ethidium bromide-stained agarose gels of HS2, HS3, and a negative control (Necdin promoter) amplified from wild-type (WT) or mutant (Mut) E12.5 fetal liver cells. (MW) One-kilobase molecular weight standard (the 1- and 0.5-kb bands are visible); (Pos) wild-type genomic DNA served as a positive control for PCR; (Neg) no template negative control; (Input) total chromatin, sheared but not immunoprecipitated; (ChIP) chromatin immunoprecipitated with a BRG1 antibody; (M) mock immunoprecipitation, which served as a negative control, where BRG1 antibody was omitted from wild-type chromatin but processed in a manner identical to ChIP samples. (C) DNase I hypersensitivity assays of β-globin LCR. Nuclei from wild-type (WT) and mutant (Mut) E12.5 fetal liver cells were digested with increasing amounts of DNase I (from left to right: 0.01, 0.10, 0.50, 1.00, and 2.50 U of enzyme per microgram of DNA). Genomic DNA was subsequently prepared, and HSs 1, 2, 3, 4, and 6 were PCR amplified. To correct for small differences in the amount of DNA that might have been recovered for each sample, an Mit marker (D9Mit59) was amplified as a control (see CNTRL panel). (Pos) Wild-type genomic DNA served as a positive control for each amplicon; (Neg), no template was used as a negative control for each amplicon. Each panel is an image of PCR products run on an ethidium bromide-stained agarose gel. (MW) One-kilobase Plus molecular weight standard (bands from 1 kb to 100 bp are visible).
Considering that mutant SWI/SNF-related complexes localize to the β-globin locus, we wanted to test whether chromatin structure was affected. Therefore, DNase I hypersensitivity assays were performed to compare the chromatin structure of the LCR in wild-type and mutant embryos. Fetal liver cells were harvested, nuclei were digested with DNase I, DNA was subsequently purified, and five out of the six HSs were amplified by PCR. HS5 was excluded from this analysis because it is a relatively weak site in the mouse (Bender et al. 1998). As wild-type nuclei were digested with increasing concentrations of DNase I, more DNA molecules were cleaved, fewer PCR templates remained intact, and PCR product yields declined (Fig. 7C). These data indicate the LCR was in an open chromatin configuration as expected. In mutant nuclei, increasing DNase I concentrations had less of an effect and PCR product yields were greater than the corresponding controls for each HS amplicon (Fig. 7C). These data indicate that the LCR in mutants was not as accessible to DNase I because of unusually condensed chromatin. To rule out the possibility that different amounts of wild-type and mutant DNAs were recovered from DNase I-treated nuclei, all of the samples had an Mit marker that is relatively resistant to DNase I amplified as a control (Fig. 7C).
It has been known for a long time that Brg1 ATPase activity is required for SWI/SNF function, but it still is not clear how the energy of ATP hydrolysis is harnessed to change the conformation and position of nucleosomes. The Brg1ENU1 mutation may provide insight into this mechanism since ATPase activity (which is normal, see Fig. 3C) is uncoupled from chromatin remodeling (which is abnormal, see Fig. 7C). It is interesting to note that deletion of putative helicase motif V in yeast SWI2/SNF2 (72 residues downstream) also uncouples ATPase activity from chromatin remodeling (even though a lysine residue two amino acids upstream of E1083 is required for ATPase activity) (Smith and Peterson 2005). Point mutations in putative helicase motif V of BRG1 seem to be overrepresented in primary tumors and tumor cell lines, which lends credence to the importance of this energy transduction mechanism (Wong et al. 2000; Medina et al. 2004; Smith and Peterson 2005).
Altered histone acetylation and DNA methylation at the LCR
More distantly related SNF2-like proteins, which are presumably the catalytic subunits of other ATPase remodeling complexes, have been shown to influence covalent histone modifications or DNA methylation (Gibbons et al. 2000; Dennis et al. 2001; Meehan et al. 2001; Pray-Grant et al. 2005). Less is known about Brg1 and SWI/SNF-related complexes in this regard, so we evaluated both types of epigenetic marks. First, we performed ChIP assays to measure histone 3 (H3) acetylation levels at the β-globin LCR. We cross-linked and sonicated chromatin from fetal liver and head cells, immunoprecipitated with an anti-pan-acetyl H3 antibody, reversed the cross-links, prepared genomic DNAs, and amplified HSs 1, 2, 3, 4, and 6. In wild-type embryos, each site was acetylated in a tissue-specific manner. Robust acetylation was detected in erythroid cells of the fetal liver with little or no background signal detected in nonerythroid cells of the head (cf. FL and H ChIP lanes in WT panels of Fig. 8A). Mock immunoprecipitations were performed, where acetyl H3 antibody was omitted from fetal liver ChIP assays, to serve as a negative control. Complete lack of signal at each HS demonstrated that Protein A/G agarose beads were not binding acetyl H3 or any of the HSs nonspecifically (see Neg. lanes in Fig. 8A). In mutant embryos, none of the HSs were acetylated at normal levels. Instead, mutant fetal livers had either no signals or faint signals comparable to background levels observed in head tissue (cf. FL and H ChIP lanes in Mut panels in Fig. 8A). Chromatin prepared from head tissue was of good quality (cf. FL and H Input lanes in Fig. 8A) and capable of being H3 acetylated based on enrichment of this mark at the promoter of a gene (Necdin) expressed in neurons and other cell types (see Necdin panel in Fig. 8A; Uetsuki et al. 1996; Forsberg et al. 2000).
Figure 8.
Aberrant epigenetic modifications at the β-globin locus. (A) ChIP assays showing histone 3 (H3) acetylation status at the β-globin LCR. Shown at the left are images of ethidium bromide-stained agarose gels containing HSs 1, 2, 3, 4, and 6 amplified from wild-type (WT) or mutant (Mut) E12.5 tissues. Necdin promoter was also amplified as a control because it corresponds to the promoter of a gene (Necdin) that is acetylated and expressed in neurons and other cell types. (Pos) Wild-type genomic DNA served as a positive control for PCR; (Neg) negative control, mock immunoprecipitation where no antibody was added to wild-type FL chromatin but processed in a manner identical to ChIP samples; (Input) total chromatin, sheared but not immunoprecipitated, prepared from fetal liver (FL) or head (H) cells; (ChIP) FL or H chromatin immunoprecipitated with an anti-acetyl H3 antibody. Shown at the right is quantification of the ChIP data utilizing digitized images of the gels. ChIP band intensities were divided by Input band intensities after subtracting out background levels. Values were normalized to 100% for wild-type samples (black bars). Mutant samples (gray bars) were expressed as a percentage of wild-type values. (B) Genome-wide DNA methylation analysis. Image of an ethidium bromide-stained agarose gel of wild-type (WT) and mutant (Mut) genomic DNAs digested with Hpa II (H) or Msp I (M). (MW) One-kilobase ladder molecular weight standard. (B) Sodium bisulfite mutagenesis analysis of HS1 and HS2 within β-globin LCR of wild-type and mutant fetal liver cells. (Unfilled circles) Unmethylated CpGs; (filled circles) methylated CpGs.
Second, we analyzed DNA methylation at a genome-wide level and at the β-globin LCR. To evaluate genome-wide DNA methylation, we prepared genomic DNA from wild-type and mutant embryos, digested with Hpa II or Msp I restriction enzymes, and separated the DNA fragments by size on agarose gels. Hpa II and Msp I recognize the same DNA sequence (CCGG) but are isoschizomers. Hpa II is methylation sensitive and cuts relatively infrequently, whereas Msp I is methylation insensitive and cuts much more often, giving rise to smearing of lower molecular weight fragments. We did not observe any difference between the wild-type and mutant digests (Fig. 8B), suggesting that genome-wide methylation is normal in mutants.
To evaluate DNA methylation at the β-globin LCR, we performed sodium bisulfite mutagenesis on wild-type and mutant fetal liver DNAs, PCR amplified HS1 and HS2, subcloned the PCR products, and sequenced individual subclones. HS1 and HS2 each contain two CpGs. All four CpGs were unmethylated in wild-type controls but partially methylated in mutants (Fig. 8C). Therefore, although mutant tissues apparently had normal genome-wide methylation levels, methylation of specific, physiologically relevant targets were altered. Similarly, BRG1-deficient tumor cells display aberrant methylation at a specific locus (but not the entire genome) that can be rescued by transfection of a BRG1 expression vector (Banine et al. 2005). Although it is not clear how the energy of ATP hydrolysis is harnessed to change the conformation and position of nucleosomes, histone acetylation and DNA methylation might be part of the mechanism since both are affected as a consequence of the E1083G substitution.
Discussion
Here, we present the characterization of a novel, ENU-induced Brg1 mutation (Brg1ENU1) that results in a single amino acid substitution (E1083G) in the catalytic ATPase domain. Not only is the mutant protein stable and capable of assembling into SWI/SNF-related complexes, it also exhibits normal ATPase activity. The notion that E1083 does not comprise part of the ATPase-active site is supported by the fact that this residue is not conserved in proteins outside of the SWI2/SNF2 subfamily as well as its predicted position in X-ray crystal structures (Durr et al. 2005; Thoma et al. 2005).
Brg1null/ENU1 embryos develop normally until midgestation when erythropoiesis switches from the yolk sac (i.e., primitive erythropoiesis) to the fetal liver (i.e., definitive erythropoiesis). A defect in definitive erythropoiesis leads to anemia and death between E11.5 and E14.5. In mutant fetal livers, erythroid progenitors develop normally until a block occurs at the basophilic-to-polychromatic erythroblast transition. Brg1 and SWI/SNF-related complexes might be particularly important at this stage in the lineage because the nucleus is becoming increasingly condensed but certain genes must be maintained in an open chromatin configuration and continue to be expressed.
Although the erythroid lineage is severely affected, most other tissues/organs develop normally in Brg1null/ENU1 mutants. This finding can possibly be explained by our finding that although Brm is expressed in a widespread manner, it is not expressed in the erythroid lineage. Brm can therefore be expected to compensate in most tissues but not the erythroid lineage.
At a mechanistic level, the zinc-finger transcription factors EKLF and GATA-1 bind Brg1 (Kadam and Emerson 2003) and can recruit wild-type as well as mutant SWI/SNF-related complexes to the β-globin LCR. Once recruited, Brg1 is required for the LCR to adopt an open chromatin structure. Although Brg1 ATPase activity is necessary for function, our results indicate that it is not sufficient. The E1083G substitution may be revealing a segment of the ATPase domain involved in harnessing the energy derived from ATP hydrolysis to alter the conformation and position of nucleosomes. In Brg1 mutants, the LCR is not H3 acetylated and CpGs are methylated. The LCR adopts a closed chromatin configuration, β-globin transcription is reduced, and definitive erythroblasts undergo developmental arrest instead of terminal differentiation. This arrest occurs at a stage when β-globin transcription is normally peaking (Wojda et al. 2002) despite the onset of nuclear condensation.
Materials and methods
ENU mutagenesis
One gram of ENU (Sigma) was dissolved in 10 mL of 95% ethanol and mixed with 90 mL of phosphate/citrate buffer (0.1 M dibasic sodium phosphate, 0.05 M citrate at pH 5.0) while keeping glassware wrapped in aluminum foil to minimize light exposure. To determine concentration, 200 μL of ENU was diluted to 1 mL with phosphate/citrate buffer (1:5 dilution), transferred to disposable cuvettes, and measured in a spectrophotometer at OD398. (OD398/0.72) (5) = X mg/mL. Eight-week-old to 12-wk-old male mice received three weekly intraperitoneal (IP) injections at a dose of 100 mg/kg body weight.
Blots
Northern and Western blots were performed according to standard procedures (Ausubel et al. 1992). The Brg1 Western blots utilized a mouse monoclonal raised against the N-terminal portion of human BRG1 that does not cross-react with BRM (a gift from P. Chambon, Institut Clinique de la Souris [ICS], Ippkirch, France). The BAF antibodies were rabbit polyclonals (a gift from G. Crabtree [Howard Hughes Medical Institute, Department of Developmental Biology, Stanford University, Stanford, CA] and W. Wang [Laboratory of Genetics, National Institute on Aging, National Institute of Health, Geontology Research Center, Baltimore, MD]). Primary antibodies were detected using goat anti-mouse or anti-rabbit secondary antibodies conjugated to horse-radish peroxidase.
Immunoprecipitations
E12.5 embryos were placed in RIPA buffer (100 mM NaCl, 20 mM Tris at pH 7.6, 0.2% deoxycholate, 0.2% Triton X-100, 0.2% NP40) containing DTT (1 mM), β-mercaptoethanol (2 mM), and protease inhibitors. Tissues were sonicated twice at 70% amplitude on wet ice and spun at 3500 rpm for 30 min at 4°C to pellet debris. The concentrations of protein lysates were determined by Bradford assays. Approximately 1 mg of protein was immunoprecipitated with 4 μg of a mouse monoclonal antibody raised against the N-terminal portion of human BRG1 that does not cross-react with BRM (Santa Cruz). This step went overnight at 4°C while rocking on a nutator. Fifty microliters of preblocked Protein G (Oncogene) or Protein A/G (Santa Cruz) agarose beads were added, and the incubation continued for three more hours. Samples were washed two times in IP0.2 buffer (20 mM HEPES at pH 7.6, 10% glycerol, 12.5 mM MgCl2, 0.1 mM EDTA at pH 8.0, 0.2% NP40, 0.1 mM DTT, 0.6 M KOAc) and processed immediately or stored at -80°C.
ATPase assays
Twenty-microliter reactions were carried out directly on agarose beads in 10 mM Tris (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 20% glycerol in the presence of BSA (1 mg/mL), PMSF (0.5 mM), DTT (1mM), ATP (0.5mM), plasmid DNA (20 nM), and 1 μCi [γ32P]-ATP at 37°C. Reactions were stopped by adding 1 μL of 0.5 μM EDTA and placing on wet ice. One microliter of each reaction was spotted onto polyethyleneimine cellulose TLC plates (Sigma), which had been prerun in distilled water and dried. Inorganic phosphate was separated from uncleaved ATP by running the TLC plates in 1 M formic acid and 0.5 M LiCl in a glass TLC developing tank. The ratio of inorganic phosphate to ATP was quantified using a Molecular Dynamics Phosphorimager (Amersham Biosciences) and ImageQuant software.
Cytospin preparations
Fetal livers were triturated in PBS containing 2% FBS using 1-cc syringes fitted with needles having progressively smaller apertures (18, 22, and 25 gauges). Two hundred microliters of the single-cell suspensions were added to cuvettes assembled with paper pads and microscope slides (that were pretreated with PBS/FBS) in metal holders. Samples were spun at 700 rpm for 10 min, air dried, fixed in methanol for 2 min, stained in Wright's stain (200 mL of Wright's stain from Sigma mixed with 200 mL methanol) for 4 min, stained in Giemsa buffer (11 mL of Giemsa stain from Sigma freshly mixed with 50 mL distilled water) for 12 min, and dipped five times in 20% methanol followed by five times in distilled water.
Flow cytometry
Fetal liver cell suspensions were prepared as described for cytospins. Approximately 1 × 106 cells were incubated with anti-Ter119-PE (Pharmingen) and anti-Kit-FITC (Pharmingen) on wet ice in the dark for 1 h. Cells were washed several times, fixed in 200 μL 2% PFA, and analyzed using a FACScan instrument (Bectin Dickinson). For each experiment, wild-type cells were also incubated with Ter119-PE only, Kit-FITC only, or no antibody to serve as compensation controls.
RT-PCR
RNA was prepared from E12.5 embryos, fetal livers, or flow-sorted cell populations using Trizol reagent (Invitrogen) and reverse transcribed using random hexamers or Oligo d(T) and SuperScript II RT (Invitrogen) according to standard procedures (Ausubel et al. 1992). The Brg1 and Brm primers and amplification conditions have already been described (Bultman et al. 2000). The Epor primers were 5′-TGAGCTTCCTGAAGCTAGGG-3′ and 5′-TCGCTAGCGGTCAGTAGTGA-3′. The 765-base-pair (bp) Epor product was amplified at 1 mM MgCl2 and a 55°C annealing temperature. For the β-globin expression analyses, the βmaj primers were 5′-CGTTTGCTTCTGATTCTGTTG-3′ and 5′-CTAGATGCCCAAAGGTCTTC-3′, and the corresponding product was 590 bp. The βmin primers were 5′-AAAGGTGAACCCCGATGAAG-3′ and 5′-TGTGCATAGACAATAGCAGA-3′, and the corresponding product was 411 bp. The εY primers were 5′-TGACACTCCTGTGATCACCA-3′ and 5′-AAAGGAGGCATAGCGGACAC-3′, and the corresponding product was 535 bp. The βH1 primers were 5′-TCTCCAAGCTTCTATACCTC-3′ and 5′-CATGGGATTGCCAGTGTACT-3′, and the corresponding product was 498 bp. All of the β-globin amplicons were amplified for 30 cycles at a 1 mM MgCl2 concentration. The βmaj and βmin had a 61°C annealing temperature, whereas the εY and βH1 had a 55°C annealing temperature. The actin control amplicon (5′-CAAGGTGTGATGGTGGGAAT-3′ and 5′-GGTGTAAAACGCAGCTCAGT-3′) was 1038 bp and worked at both 61°C and 55°C.
ChIP assays
The ChIP protocol was adapted from Upstate (http://www.upstate.com) and Forsberg et al. (2000). Single cell suspensions were prepared from E12.5 fetal livers as described above for cytospin preparations. Protein-protein and protein-DNA interactions were cross-linked in 0.4% formaldehyde at room temperature for 10 min and stopped by adding glycine to a final concentration of 0.125 M. Aliquots of 2 × 106 cells were lysed in 200 μL SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris at pH 8.1, protease inhibitors), and chromatin was sonicated with four 10-sec pulses at 30% of maximum power. Samples were diluted to 2 mL with IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris at pH 8.1, 167 mM NaCl, protease inhibitors). One hundred microliters was removed as input, and the remainder was processed for immunoprecipitations. J1 antibody (a gift from G. Crabtree and W. Wang) was used for Brg1, and anti-pan-acetyl H3 (Upstate) was used for histone acetylation. Following overnight incubation at 4°C, protein A/G agarose beads (Santa Cruz) were added and incubated for at least 2 h at 4°C. Samples were washed in low salt buffer, high salt buffer, LiCl buffer, and TE and then eluted in elution buffer (Forsberg et al. 2000). Cross-links were reversed with 0.2 M NaCl overnight at 65°C. DNA was purified by Proteinase K treatment, phenol/chloroform extractions, and ethanol precipitations. DNAs were resuspended in 25 μL and PCR amplified. The HS 1, 2, 3, 4, and 6 oligonucleotide primers that were used have already been described (McArthur et al. 2001). The Necdin promoter oligonucleotide primers have also been described (Forsberg et al. 2000).
DNase I hypersensitivity assays
Single cell suspensions were prepared from E12.5 fetal livers as described above for cytospin preparations. The DNase I hypersensitivity assays were performed as described (McArthur et al. 2001). The same HS 1, 2, 3, 4, and 6 oligonucleotide primers were used as described (McArthur et al. 2001). The D9Mit59 control primers were 5′-CAGCCAGAGGCAGTGTTTTA-3′ and 5′-TAGGCTTCAGCTGCAACTCA-3′.
Sodium bisulfite mutagenesis
Genomic DNA (2.5 μg) was digested with the Sph I restriction enzyme, extracted with phenol and chloroform, ethanol precipitated, and resuspended in 50 μL TE. DNA was denatured in 0.3 N NaOH at 42°C for 30 min and then mutagenized in sodium bisulfite (2.9 M) and hydroquinone (0.5 mM) at 55°C for ∼19 h with minimal light exposure. DNA was purified using a genomic DNA purification kit (Promega), desulfonated with 0.3 N NaOH treatment at 37°C for 15 min, neutralized with NH4OAc, and ethanol precipitated. DNAs were ligated into the pGem-T Easy vector (Promega) and transformed. Plasmid DNA was prepared from individual ampicillin-resistant colonies (Qiagen miniprep kit) and sequenced.
Acknowledgments
We thank P. Chambon, G. Crabtree, and W. Wang for Brg1 antibodies; G. Crabtree and W. Wang for BAF antibodies; and S. Kirby for Wright-Giemsa staining protocol. G. Rossen at University of North Caroloina-Chapel Hill kindly provided the Western blot data in Supplementary Figure 1. We also thank members of the Magnuson laboratory for helpful discussions, and J. Brennan, D. Ciavatta, and G. Narlikar for comments on this manuscript. This work was supported, in part, by a grant from the NIH.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1364105.
Supplemental material is available at http://www.genesdev.org.
References
- Armstrong, J.A., Bieker, J.J., and Emerson, B.M. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95: 93-104. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. 1992. Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York.
- Banine, F., Bartlett, C., Gunawardena, R., Muchardt, C., Yaniv, M., Knudsen, E.S., Weissman, B.E., and Sherman, L.S. 2005. SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res. 65: 3542-3547. [DOI] [PubMed] [Google Scholar]
- Bender, M.A., Reik, A., Close, J., Telling, A., Epner, E., Fiering, S., Hardison, R., and Groudine, M. 1998. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse β-globin locus control region. Blood 92: 4394-4403. [PubMed] [Google Scholar]
- Bultman, S., Gebuhr, T., Yee, D., La Mantia, C., Nicholson, J., Gilliam, A., Randazzo, F., Metzger, D., Chambon, P., Crabtree, G., et al. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6: 1287-1295. [DOI] [PubMed] [Google Scholar]
- Chi, T.H., Wan, M., Lee, P.P., Akashi, K., Metzger, D., Chambon, P., Wilson, C.B., and Crabtree, G.R. 2003. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19: 169-182. [DOI] [PubMed] [Google Scholar]
- Dennis, K., Fan, T., Geiman, T., Yan, Q., and Muegge, K. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes & Dev. 15: 2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen, R., von Lindern, M., Kolbus, A., Driegen, S., Steinlein, P., Beug, H., Grosveld, F., and Philipsen, S. 2005. The erythroid phenotype of EKLF-null mice: Defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25: 5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Z., Stamatoyannopoulos, G., and Li, Q. 2001. Role of NF-Y in in vivo regulation of the γ-globin gene. Mol. Cell. Biol. 21: 3083-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr, H., Korner, C., Muller, M., Hickmann, V., and Hopfner, K.P. 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363-373. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A., Sweder, K.S., and Hanawalt, P.C. 1995. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 23: 2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D.S. and Isaacson, J.H. 1966. Curly-whiskers and its linkage with tail-kinks in linkage group II of the mouse. Genet. Res. 8: 111-113. [DOI] [PubMed] [Google Scholar]
- Forsberg, E.C., Downs, K.M., Christensen, H.M., Im, H., Nuzzi, P.A., and Bresnick, E.H. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. 97: 14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, C.J. and Peterson, C.L. 2001. Chromatin remodeling enzymes: Who's on first? Curr. Biol. 11: R185-R197. [DOI] [PubMed] [Google Scholar]
- Fujiwara, Y., Browne, C.P., Cunniff, K., Goff, S.C., and Orkin, S.H. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. 93: 12355-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebuhr, T.C., Kovalev, G.I., Bultman, S., Godfrey, V., Su, L., and Magnuson, T. 2003. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 198: 1937-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, R.J., McDowell, T.L., Raman, S., O'Rourke, D.M., Garrick, D., Ayyub, H., and Higgs, D.R. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24: 368-371. [DOI] [PubMed] [Google Scholar]
- Guidi, C.J., Sands, A.T., Zambrowicz, B.P., Turner, T.K., Demers, D.A, Webster, W., Smith, T.W., Imbalzano, A.N., and Jones, S.N. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21: 3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege, F.C., Jennings, E.G., Wyrick, J.J., Lee, T.I., Hengartner, C.J., Green, M.R., Golub, T.R., Lander, E.S., and Young, R.A. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717-728. [DOI] [PubMed] [Google Scholar]
- Horak, C.E., Mahajan, M.C., Luscombe, N.M., Gerstein, M., Weissman, S.M., and Snyder, M. 2002. GATA-1 binding sites mapped in the β-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. 99: 2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam, S. and Emerson, B.M. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11: 377-389. [DOI] [PubMed] [Google Scholar]
- Kim, J.K., Huh, S.O., Choi, H., Lee, K.S., Shin, D., Lee, C., Nam, J.S., Kim, H., Chung, H., Lee, H.W., et al. 2001. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 21: 7787-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kina, T., Ikuta, K., Takayama, E., Wada, K., Majumdar, A.S., Weissman, I.L., and Katsura, Y. 2000. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109: 280-287. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin, A., Fiette, L., Barra, J., Muchardt, C., Babinet, C., and Yaniv, M. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1: 500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, J.E., Fry, C.J., Samuels, M.L., and Peterson, C.L. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102: 587-598. [DOI] [PubMed] [Google Scholar]
- Lee, J.S., Ngo, H., Kim, D., and Chung, J.H. 2000. Erythroid Kruppel-like factor is recruited to the CACCC box in the β-globin promoter but not to the CACCC box in the γ-globin promoter: The role of the neighboring promoter elements. Proc. Natl. Acad. Sci. 97: 2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon, B. and Tjian, R. 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 14: 2551-2569. [DOI] [PubMed] [Google Scholar]
- Levings, P.P. and Bungert, J. 2002. The human β-globin locus control region. Eur. J. Biochem. 269: 1589-1599. [DOI] [PubMed] [Google Scholar]
- McArthur, M., Gerum, S., and Stamatoyannopoulos, G. 2001. Quantification of DNaseI-sensitivity by real-time PCR: Quantitative analysis of DNaseI-hypersensitivity of the mouse β-globin LCR. J. Mol. Biol. 313: 27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt, M.A., Shivdasani, R.A., Fujiwara, Y., Yang, H., and Orkin, S.H. 1997. A `knockdown' mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. 94: 6781-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, P.P., Carretero, J., Fraga, M.F., Esteller, M., Sidransky, D., and Sanchez-Cespedes, M. 2004. Genetic and epigenetic screening for gene alterations of the chromatin-remodeling factor, SMARCA4/BRG1, in lung tumors. Genes Chromosomes Cancer 41: 170-177. [DOI] [PubMed] [Google Scholar]
- Meehan, R.R., Pennings, S., and Stancheva, I. 2001. Lashings of DNA methylation, forkfuls of chromatin remodeling. Genes & Dev. 15: 3231-3236. [DOI] [PubMed] [Google Scholar]
- Mohrmann, L. and Verrijzer, C.P. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681: 59-73. [DOI] [PubMed] [Google Scholar]
- Mucenski, M.L., McLain, K., Kier, A.B., Swerdlow, S.H., Schreiner, C.M., Miller, T.A., Pietryga, D.W., Scott Jr., W.J., and Potter, S.S. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677-689. [DOI] [PubMed] [Google Scholar]
- Narlikar, G.J., Fan, H.Y., and Kingston, R.E. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475-487. [DOI] [PubMed] [Google Scholar]
- O'Neill, D., Yang, J., Erdjument-Bromage, H., Bornschlegel, K., Tempst, P., and Bank, A. 1999. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc. Natl. Acad. Sci. 96: 349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant, M.G., Daniel, J.A., Schieltz, D., Yates III, J.R., and Grant, P.A. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434-438. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C., Barra, J., Muchardt, C., Camus, A., Babinet, C., and Yaniv, M. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17: 6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, E. and Peterson, C.L. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24: 3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.W., Galusha, S.A., McMenamin, M.E., Fletcher, C.D., and Orkin, S.H. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. 97: 13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.L. and Peterson, C.L. 2005. A conserved swi2/snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol. 25: 5880-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.L., Horowitz-Scherer, R., Flanagan, J.F., Woodcock, C.L., and Peterson, C.L. 2003. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat. Struct. Biol. 10: 141-145. [DOI] [PubMed] [Google Scholar]
- Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1-4. [DOI] [PubMed] [Google Scholar]
- Sudarsanam, P. and Winston, F. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16: 345-351. [DOI] [PubMed] [Google Scholar]
- Sudarsanam, P., Iyer, V.R., Brown, P.O., and Winston, F. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 97: 3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun, J.W. 1995. The role of brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev. 5: 473-477. [DOI] [PubMed] [Google Scholar]
- Tamkun, J.W., Deuring, R., Scott, M.P., Kissinger, M., Pattatucci, A.M., Kaufman, T.C., and Kennison, J.A. 1992. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561-572. [DOI] [PubMed] [Google Scholar]
- Thoma, N.H., Czyzewski, B.K., Alexeev, A.A., Mazin, A.V., Kowalczykowski, S.C., and Pavletich, N.P. 2005. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12: 350-356. [DOI] [PubMed] [Google Scholar]
- Trimborn, T., Gribnau, J., Grosveld, F., and Fraser, P. 1999. Mechanisms of developmental control of transcription in the murine α- and β-globin loci. Genes & Dev. 13: 112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetsuki, T., Takagi, K., Sugiura, H., and Yoshikawa, K. 1996. Structure and expression of the mouse necdin gene. Identification of a postmitotic neuron-restrictive core promoter. J. Biol. Chem. 271: 918-924. [DOI] [PubMed] [Google Scholar]
- Uoshima, N., Ozawa, M., Kimura, S., Tanaka, K., Wada, K., Kobayashi, Y., and Kondo, M. 1995. Changes in c-Kit expression and effects of SCF during differentiation of human erythroid progenitor cells. Br. J. Haematol. 91: 30-36. [DOI] [PubMed] [Google Scholar]
- Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P.A., Biggar, S.R., Muchardt, C., Kalpana, G.V., Goff, S.P., Yaniv, M., et al. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15: 5370-5382. [PMC free article] [PubMed] [Google Scholar]
- Wojda, U., Noel, P., and Miller, J.L. 2002. Fetal and adult hemoglobin production during adult erythropoiesis: Coordinate expression correlates with cell proliferation. Blood 99: 3005-3013. [PubMed] [Google Scholar]
- Wong, A.K., Shanahan, F., Chen, Y., Lian, L., Ha, P., Hendricks, K., Ghaffari, S., Iliev, D., Penn, B., Woodland, A.M., et al. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60: 6171-6177. [PubMed] [Google Scholar]