Abstract
The prevailing view of the RNA polymerase II (RNAP II) transcription cycle is that RNAP II is recruited to the promoter, transcribes a linear DNA template, then terminates transcription and dissociates from the template. Subsequent rounds of transcription are thought to require de novo recruitment of RNAP II to the promoter. Several recent findings, including physical interaction of 3′-end processing factors with both promoter and terminator regions, challenge this concept. Here we report a physical association of promoter and terminator regions of the yeast BUD3 and SEN1 genes. These interactions are transcription-dependent, require the Ssu72 and Pta1 components of the CPF 3′-end processing complex, and require the phosphatase activity of Ssu72. We propose a model for RNAP II transcription in which promoter and terminator regions are juxtaposed, and that the resulting gene loops facilitate transcription reinitiation by the same molecule of RNAP II in a manner dependent upon Ssu72-mediated CTD dephosphorylation.
Keywords: CPF complex, gene loops, RNA polymerase II, Ssu72, transcription termination
The RNA polymerase II (RNAP II) transcription cycle involves distinct steps that include assembly of a preinitiation complex (PIC), initiation, elongation, termination, and reinitiation. Whereas initiation requires recruitment of RNAP II and a complete set of initiation factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) (Woychik and Hampsey 2002; Hahn 2004), reinitiation has been proposed to occur via a different pathway (Yudkovsky et al. 2000). Following initiation, a subset of the initiation factors and Mediator, which facilitates the interaction between gene-specific regulatory proteins and the general transcription factors, is retained at the promoter, forming a “scaffold” that functions as an intermediate for subsequent rounds of transcription in vitro. Accordingly, reinitiation can facilitate higher levels of transcription by bypassing recruitment of factors retained in the scaffold, requiring de novo recruitment of only TFIIB, TFIIF, and RNAP II (Yudkovsky et al. 2000). Whether transcription reinitiation involves termination-dependent recycling of RNAP II from the terminator to the promoter without release from template DNA is not known, although such facilitated recycling has been reported for yeast RNAP III (Dieci and Sentenac 1996, 2003).
Progression of RNAP II through the transcription cycle is accompanied by changes in the phosphorylation status of the CTD, a reiterated heptapeptide sequence (Y1-S2-P3-T4-S5-P6-S7) present at the C terminus of the Rpb1 subunit of RNAP II (Kobor and Greenblatt 2002). RNAP II is recruited to the promoter in an unphosphorylated form (RNAP IIA) that becomes extensively phosphorylated (RNAP IIO) during transcription. In yeast, phosphorylation of Ser5 of the CTD is catalyzed by the Kin28 subunit of TFIIH, whereas Ser2 is phosphorylated by the Ctk1 subunit of the CTDK-I elongation complex (Cho et al. 2001). Dephosphorylation of Ser5-P and Ser2-P are, in turn, catalyzed by the Ssu72 and Fcp1 phosphatases, respectively (Cho et al. 2001; Krishnamurthy et al. 2004). CTD phosphorylation and dephosphorylation play important and complex roles in recycling of RNAP II and in coordinating RNA processing events during the transcription cycle (Bentley 2002; Proudfoot et al. 2002; Calvo and Manley 2003; Hampsey and Reinberg 2003; Proudfoot 2004).
Cotranscriptional 3′-end processing in yeast requires the CPF and CF-I complexes, which recognize specific sequences within the nascent mRNA and catalyze endonucleolytyic cleavage and poly(A) addition. The Ssu72 phosphatase is a component of the CPF complex and plays an essential role in 3′-end formation of mRNAs and snoRNAs (Ganem et al. 2003; He et al. 2003; Nedea et al. 2003; Steinmetz and Brow 2003; Krishnamurthy et al. 2004). Intriguingly, Ssu72 also interacts physically and functionally with the transcription initiation factor TFIIB (Sun and Hampsey 1996; Wu et al. 1999; Dichtl et al. 2002). Chromatin immunoprecipitation (ChIP) also revealed that Ssu72 occupies the terminator region of RNAP II genes and, to a lesser extent, the promoter region, but not the ORF (Nedea et al. 2003). Thus, even though Ssu72 is an integral component of the CPF complex, it appears to play an important, albeit undefined, role in initiation.
One possibility to account for these results would be the juxtaposition of promoter and terminator regions by gene looping. Indeed, recent experiments in yeast showed that promoter and terminator regions can be juxtaposed in a manner dependent upon the transcriptional status of the gene (O'Sullivan et al. 2004). Here we confirm and extend those results by demonstrating transcription-dependent looping of the BUD3 and SEN1 genes of Saccharomyces cerevisiae. Moreover, we establish that looping is a consequence of terminator contact with the promoter and is dependent upon the Ssu72 and Pta1 components of the CPF complex. We propose that CPF-mediated gene looping is a mechanism to promote transcription reinitiation by facilitating recycling of RNAP II from the terminator to the promoter, and that this event involves Ssu72-catalyzed conversion of RNAP IIO to RNAP IIA.
Results
Modified chromosome conformation capture analysis to detect gene looping
To detect transcription-dependent gene looping, defined here as a physical interaction between the distal ends of genes, we adapted a technique called capturing chromosome conformation (mCCC) (Dekker et al. 2002). The method is summarized in Figure 1A. Briefly, cells are grown under transcriptionally permissive conditions to mid-log phase, and transient protein-protein and protein-DNA interactions are stabilized by formaldehyde cross-linking. Chromatin is extracted, partially purified, and digested with restriction enzymes that cut upstream of the promoter, downstream of the terminator, and at one or more sites within the coding region. DNA ends are then ligated under conditions that minimize intermolecular reactions. If looping has occurred, such that distal regions of the gene (P and T) are covalently cross-linked, then ligation of P1 and T1 ends will occur as a favorable intramolecular reaction. Conversely, in the absence of looping, ligation of P1 and T1 ends will occur only rarely, as an intermolecular event. Following reversal of cross-links, ligation of the distal P1 and T1 ends is assayed by PCR (30 cycles) using divergent P1 and T1 primers. Accordingly, a P1-T1 PCR signal is indicative of gene looping.
Figure 1.
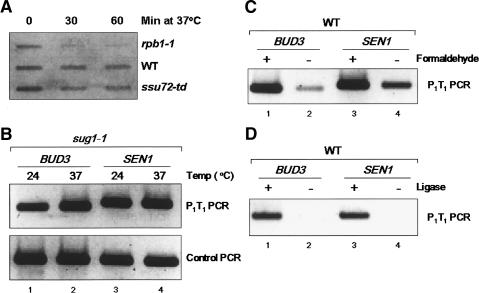
Gene looping at the BUD3 and SEN1 genes. (A) Schematic depiction of mCCC analysis to detect gene looping. Under transcriptionally permissive conditions, formaldehyde is used to cross-link transiently interacting chromatin regions, followed by restriction digestion to cut DNA upstream of the promoter, within the ORF, and downstream of the terminator. DNA ends are then ligated in dilute solution to minimize intermolecular ligation. Cross-links are reversed, and PCR is performed with divergent primers P1 and T1. Accordingly, P1-T1 PCR products represent ligation of distal, divergent regions of the gene of interest and are therefore indicative of gene looping. If looping does not occur, then ligation of the DNA ends adjacent to P1 and T1 should occur infrequently, and the P1-T1 PCR products should be diminished relative to the signal observed under conditions that favor looping. (B) Schematic depiction of the BUD3 and SEN1 genes showing the positions of the XhoI or HindIII restriction sites and the P1 and T1 PCR primers used in mCCC analysis. (C) The rpb1-1 mutation diminishes loop formation. Chromatin was isolated from the rpb1-1 strain grown at 24°C (permissive temperature, transcription ON) or following a 1-h temperature shift to 37°C (non-permissive temperature, transcription OFF). The P1-T1 PCR products correspond to ligation of distal, divergent ends of the BUD3 and SEN1 genes that result from cross-linking of the respective promoter and terminator regions as depicted in A. Control PCR represents an intergenic region of chromosome V to ascertain that equal amounts of template DNA were present in all reactions. (D) Identical to C, except using the isogenic RPB1 wild-type strain.
Transcription-dependent gene looping at the BUD3 and SEN1 genes
The BUD3 (4.9 kb) and SEN1 (6.7 kb) genes were chosen for mCCC analysis because of their exceptionally long open reading frames, thereby maximizing the distance between the promoter and terminator regions. mCCC analysis of BUD3 was done using XhoI, which cuts once within the ORF; SEN1 was analyzed with HindIII, which cuts at three sites within the ORF (Fig. 1B).
To assess whether loop formation occurs at the BUD3 gene and if loop formation is transcription-dependent, we used an isogenic RPB1 wild-type and rpb1-1 strain pair. The rpb1-1 mutant encodes an altered form of RNAP II that rapidly ceases transcription upon a temperature shift from 24°C to 37°C (Nonet et al. 1987). mCCC analysis of the BUD3 gene yielded a distinct P1-T1 PCR product from the rpb1-1 mutant grown at 24°C (Fig. 1C, lane 1). This signal was markedly diminished when cells were shifted to 37°C for 1 h prior to formaldehyde cross-linking (Fig. 1C, lane 2). This effect can be directly attributed to the RNAP II defect, as there was no diminution in the P1-T1 PCR signal associated with the 37°C shift for the isogenic RPB1 wild-type strain (Fig. 1D, lanes 1,2). Gene looping is not specific to the BUD3 gene, as similar results were obtained by mCCC analysis of the SEN1 gene (Fig. 1C,D, lanes 3,4).
Several control experiments were done to validate these results. To confirm that RNA synthesis is diminished in the rpb1-1 mutant, we assayed total poly(A) RNA in the rpb1-1 and wild-type strains following the 37°C temperature shift. Consistent with earlier results (Nonet et al. 1987), mRNA levels were markedly diminished 30 and 60 min after the temperature shift, whereas there was no appreciable change in the mRNA levels in the wild-type strain (Fig. 2A). To ascertain that the rpb1-1 effect is due to the transcriptional defect and is not simply a consequence of cell growth inhibition at the restrictive temperature, we repeated the mCCC assay using a sug1-1 mutant, which exhibits rapid cell growth inhibition at 37°C, comparable to that of the rpb1-1 mutant, but does not affect global transcription (Russell et al. 1996). In this case, no effect on the P1-T1 PCR signal was observed for either BUD3 or SEN1 (Fig. 2B). If the mCCC reaction products represent facilitated interaction between physically distinct promoter and terminator DNA fragments, then the P1-T1 PCR products should be dependent upon both cross-linking and subsequent ligation reactions (Fig. 1A). Indeed, the P1-T1 PCR products were significantly diminished in the absence of formaldehyde cross-linking (Fig. 2C). In the absence of DNA ligase, no P1-T1 PCR products were observed (Fig. 2D), as expected if the P1-T1 PCR products reflect ligation of the distal, divergent ends of the BUD3 and SEN1 genes. Thus, the P1-T1 PCR products are diagnostic for physical interaction between distal fragments of BUD3 and SEN1. We conclude from these experiments that DNA looping occurs at the BUD3 and SEN1 genes and that looping is transcription-dependent.
Figure 2.
(A) RNA slot blot analysis of total poly(A) RNA in rpb1-1, wild-type (WT), and ssu72-td strains following incubation of cells at 37°C for the indicated periods of time. (B) The sug1-1 mutant (KMY1171) does not affect loop formation. Identical to Figure 1C, except the mCCC assay was performed using a sug1-1 mutant, which exhibits rapid cell growth inhibition at 37°C, comparable to rpb1-1, but does not affect global transcription. In this case no effect on the P1-T1 PCR signal was observed for either BUD3 or SEN1. (C,D) Transcription-induced gene looping at BUD3 and SEN1 is dependent on formaldehyde cross-linking and ligation. Chromatin was isolated from the wild-type strain (FY23) and grown at 24°C, and mCCC analysis was performed as depicted in Figure 1, except without formaldehyde cross-linking (C) or ligation (D).
Gene looping is associated with transcriptional activation
The deleterious effect of rpb1-1 on promoter-terminator interactions is an important result because it establishes that DNA looping is dependent upon the transcriptional state of the cell. However, these results do not distinguish between indirect effects of global rpb1-1 transcription shut-off versus defective transcription specifically of BUD3 and SEN1. To address this issue, we repeated the mCCC analysis of the BUD3 and SEN1 genes that had been placed under control of the GAL1 promoter (Fig. 3A). Results demonstrate that when cells are grown under transcriptionally repressing conditions (dextrose), the P1-T1 PCR products for both BUD3 and SEN1 are markedly diminished relative to the readout under transcriptionally inducing conditions (galactose) (Fig. 3B). Thus, either global inhibition of transcription by the rpb1-1 mutation or gene-specific repression from the GAL1 promoter demonstrates that loop formation at the BUD3 and SEN1 genes is transcription-dependent. Moreover, these results demonstrate that transcriptional activation is associated with enhanced loop formation.
Figure 3.
Galactose-induced transcription of GAL1p-BUD3 and GAL1p-SEN1 results in gene looping. (A) The GAL1 promoter (GAL1p) was cloned upstream of the BUD3 and SEN1 genes at their native chromosomal loci. (B) Strains were grown in dextrose (transcription repressed) or galactose (transcription induced), and gene looping was assayed by mCCC analysis as depicted in Figure 1.
Gene looping is dependent upon the Ssu72 and Pta1 components of the CPF complex
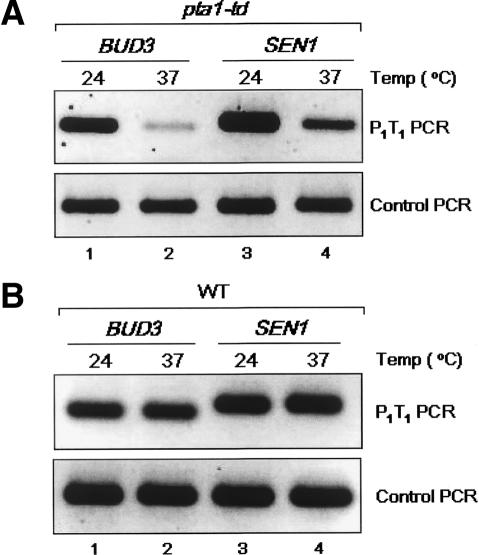
To identify factors that affect loop formation, we first asked whether the Ssu72 protein, which interacts with both the initiation and termination machineries, is required. We repeated mCCC analysis of the BUD3 and SEN1 genes using a degron-tagged ssu72-td strain that results in rapid depletion of Ssu72 upon a temperature shift from 24°C to 37°C (He et al. 2003; Krishnamurthy et al. 2004). Following a 1-h incubation of cells at 37°C, the P1-T1 PCR signal was dramatically diminished for BUD3 and SEN1 (Fig. 4A). These are not indirect effects due to impaired transcription, as neither depletion nor inactivation of Ssu72 blocks transcription within 1 h, as defined by retention of total poly(A) RNA following the 37°C temperature shift (Fig. 2A, cf. ssu72-td and rpb1-1 strains). Earlier transcription run-on (TRO) assays also indicated that RNAP II is actively engaged in transcription in ssu72-td and ssu72-2 mutants 1 h after the 37°C temperature shift (Dichtl et al. 2002; He et al. 2003). Diminished looping was directly attributed to loss of Ssu72, as plasmid-borne wild-type SSU72 fully rescued the P1-T1 PCR signals for both genes (Fig. 4B). In contrast, plasmid-borne ssu72 (C15S), which encodes a catalytically inactive Ssu72 phosphatase (Krishnamurthy et al. 2004), is unable to rescue the ssu72-td defect (Fig. 4C). Thus, the Ssu72 component of the CPF complex is required for loop formation in a manner dependent upon its catalytic activity.
Figure 4.
Gene looping at BUD3 and SEN1 is dependent on the RNAP II CTD phosphatase activity of Ssu72. (A) The Ssu72 protein was depleted from the ssu72-td degron strain (XH-24) following a 1-h temperature shift from 24°C to 37°C as described previously (Krishnamurthy et al. 2004). Chromatin was isolated from cells and subjected to mCCC analysis as depicted in Figure 1. (B) Identical to A except that the ssu72-td strain carries the SSU72 wild-type plasmid (pM712), which restores the normal level of Ssu72 protein and its CTD phosphatase activity (Krishnamurthy et al. 2004). (C) Identical to A except that the ssu72-td strain carries the ssu72-C15S plasmid (pM698), which encodes a catalytically inactive form of the Ssu72 phosphatase (Krishnamurthy et al. 2004).
Ssu72 is a component of the APT subcomplex of CPF, whereas Pta1 is a component of the CPF core complex (Nedea et al. 2003). Pta1 physically interacts with Ssu72 (He et al. 2003) and is required for Ssu72 CTD phosphatase activity in vivo (Krishnamurthy et al. 2004). We therefore asked whether Pta1 is also required for loop formation. Analogous to the effect of ssu72-td, degron-mediated depletion of Pta1 resulted in a marked diminution of the P1-T1 PCR products for BUD3 and SEN1 (Fig. 5A). This effect was attributed specifically to loss of Pta1, as no diminution of the P1-T1 PCR signal was observed following the temperature shift with the isogenic wild-type strain (Fig. 5B). Thus, the Ssu72 component of the APT subcomplex and the Pta1 component of the CPF core complex are required for loop formation, implicating the CPF complex and RNAP II CTD phosphorylation/dephosphorylation in transcription-induced changes in gene configuration.
Figure 5.
Gene looping at BUD3 and SEN1 is dependent upon the Pta1 subunit of the CPF 3′-end processing complex. (A) Pta1 protein was depleted in the pta1-td degron strain (XH-23) following a 1-h temperature shift from 24°C to 37°C as described previously (Krishnamurthy et al. 2004). Chromatin was isolated from cells and subjected to mCCC analysis as depicted in Figure 1. (B) Identical to A except that the experiment was done using an isogenic wild-type strain.
DNA looping occurs between promoter and terminator regions
The P1 and T1 primers used for mCCC analysis of the BUD3 and SEN1 genes were designed to detect looping between distal regions of the two genes. To map the juxtaposed regions more precisely, we took advantage of the multiple HindIII sites within and flanking the SEN1 gene. We repeated mCCC analysis as outlined in Figure 1A using the divergent primer pairs depicted in Figure 6A. Results of the PCR analyses are shown in Figure 6B.
Figure 6.
DNA looping juxtaposes the promoter and terminator regions of SEN1. mCCC analysis was performed as depicted in Figure 1 with modifications described in the Materials and Methods. (A) Schematic depiction of the SEN1 gene, showing HindIII sites (HIII) and the P, C, and T primers used in mCCC analysis. The relative positions of the HindIII sites are drawn approximately to scale. (B) PCR products derived from the indicated primer pairs. PCR signals are expected for the P1-C1 and T1-C6 primer pairs, as these correspond to intramolecular ligation products. As the number of HindIII sites between primer pairs increases, the PCR signals decrease (cf. P1-C1, P1-C3, P1-C5; and T1-C6, T1-C4, T1-C2). These results stand in marked contrast to P1-T1, which produces a strong PCR signal despite the presence of three intervening HindIII sites, indicating that DNA looping juxtaposes restriction fragments encompassing the promoter and terminator regions.
As expected, a P1-C1 PCR signal was detected (Fig. 6B, lane 2), reflecting intramolecular ligation of the HindIII fragment encompassing the SEN1 promoter. A P1-C3 PCR signal was also detected (Fig. 6B, lane 3), albeit diminished relative to the P1-C1 signal; and no P1-C5 signal was observed (Fig. 6B, lane 4). Accordingly, as the probability increases that template DNA is cut at least once between the P1 and C primer pairs (no HindIII sites between P1-C1, one site between P1-C3, and two sites between P1-C5), the PCR signal decreases. Similar results were obtained with respect to the DNA fragment encompassing the SEN1 terminator. The T1-C6 PCR signal (Fig. 6B, lane 8) was comparable to the P1-C1 signal (Fig. 6B, lane 2), whereas the T1-C4 (Fig. 6B, lane 7) and T1-C2 (Fig. 6B, lane 6) signals diminished with increasing probability of HindIII digestion between the primer pairs. These are precisely the results expected if these HindIII fragments are not tethered during the ligation reaction. Yet a strong P1-T1 PCR signal was observed (Fig. 6B, lane 5), despite the presence of three HindIII sites located between the P1 and T1 primers. Furthermore, neither P3-T1 (Fig. 6B, lane 1) nor P1-T3 (Fig. 6B, lane 9) PCR signals were detected, indicating that the promoter DNA fragment is not juxtaposed to DNA downstream of the terminator, nor is the terminator DNA fragment juxtaposed to DNA upstream of the promoter. These results (1) validate mCCC analysis for detection of gene looping by showing that cross-linking is not uniform along the length of the gene; (2) map the juxtaposed ends of the loop to DNA fragments encompassing the promoter and terminator regions of SEN1; and (3) demonstrate that the promoter region does not cross-link to DNA downstream of the terminator, nor does the terminator region cross-link to DNA upstream of the promoter.
Ligation-facilitated ChIP to detect promoter-terminator interactions
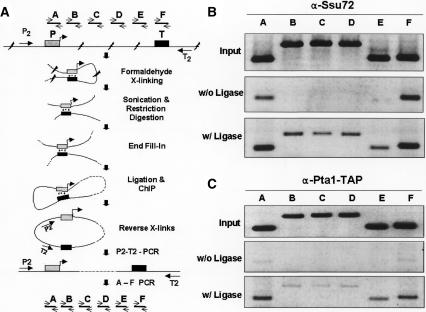
As an alternative method to mCCC analysis for analyzing gene looping, we developed a technique called ligation-facilitated ChIP (LF-ChIP). This method is described schematically in Figure 7A. Transient DNA loops are first stabilized by formaldehyde cross-linking. Chromatin is then extracted from cells and sheared into DNA fragments of ∼500 base pairs (bp) by extensive sonication and restriction digestion. DNA ends are filled in with T4 DNA polymerase and ligated in dilute solution to minimize intermolecular ligation. DNA that has been cross-linked to a specific protein is immunoprecipitated, cross-links are reversed, and DNA is amplified by PCR (30 cycles) using convergent primer pairs P2 and T2 that flank the gene of interest. The amplified DNA is subsequently assayed for the presence of specific regions of the gene (A through F) by a second round of PCR. P2-T2 amplification of immunoprecipitated DNA assures that subsequent amplification of regions A through F occurs only from template DNA that has been religated. As in the case of mCCC analysis, ligation reactions are performed in dilute solution such that distal ends of the gene (proximal to the P2 and T2 primers) would be ligated to each other only if these DNA fragments are tethered by cross-linking. To assure that P2-T2-amplified DNA does not correspond to DNA that had not been cut between P2 and T2, PCR products should be ligation-dependent. Furthermore, we should detect equal amounts of fragments A and F, as these regions are proximal to the P2 and T2 primers, but we should detect diminishing amounts of fragments B through E with increasing distance toward the middle of the gene, due to a higher probability of multiple cut sites with increasing distance from P2 and T2.
Figure 7.
Detection and mapping of gene looping at SEN1 by LF-ChIP. (A) Schematic depiction of LF-ChIP to detect and map gene looping. Transiently interacting regions of chromatin are cross-linked in whole cells using formaldehyde. Chromatin is extracted, extensively sonicated, and digested with HindIII, which cuts SEN1 upstream of the promoter, downstream of the terminator, and at three sites within the ORF (Fig. 1B). Following fill-in of overhanging DNA ends, samples are ligated in dilute solution to minimize intermolecular ligation products, followed either by immunoprecipitation with anti-Ssu72 antibodies or by Pta1-TAP pull-down, and the DNA is amplified with the P2-T2 primer pair that flanks the SEN1 ORF. Ligated DNA is then assayed using primer pairs that define regions A-F. A critical control in this procedure is omission of the DNA ligation step, in which case PCR products simply reflect Ssu72 and Pta1 occupancy of the respective DNA fragments (A-F) by ChIP. (B) Promoter and terminator regions of SEN1 are physically linked during transcription. LF-ChIP was performed as described in A. The indicated PCR products for regions A-F of SEN1 represent input DNA prior to ChIP (upper panel), Ssu72 ChIP DNA products obtained without ligation of fragmented DNA (middle panel), and PCR products representing regions of DNA brought together by gene looping (bottom panel). (C) Identical to B, except ChIP was performed using Pta1-TAP.
As Ssu72 and Pta1 are required for loop formation, we preformed LF-ChIP by immunoprecipitation of these two proteins; the target gene was SEN1. Results from immunoprecipitation of Ssu72 are shown in Figure 7B. The top panel (input) of Figure 7B indicates that regions A through F are represented equally in the chromatin samples prior to ChIP. Following ChIP and P2-T2 PCR amplification of DNA that had not been ligated prior to immunoprecipitation, the second round of PCR generated fragments A and F, but not fragments B through E (Fig. 7B, middle panel). This result demonstrates that Ssu72 occupies both the promoter and terminator regions of SEN1 and is consistent with earlier ChIP analysis of Ssu72 (Nedea et al. 2003); it does not distinguish between occupancy of promoter and terminator regions that are juxtaposed versus physically distinct. Little or no amplification of regions B through E was detected, indicating that SEN1 chromosomal DNA had been extensively cut prior to ChIP. If detection of regions A and F by ChIP is a consequence of Ssu72 occupancy of unlinked promoter and terminator regions, then there should be no amplification of Ssu72-immunoprecipitated DNA by the P2 and T2 PCR primers, and no subsequent amplification of regions A through F. Instead, we observed ligation-dependent P2-T2 PCR products that were enriched for regions A and F (Fig. 7B, lower panel). We also detected fragments B through E, albeit at lower levels than A and F. Detection of lower levels of B through D is an important result because it confirms that P2-T2-amplified DNA is derived from DNA that had been cut and religated, rather than residual DNA that had not been cut between P2 and T2. Similar results were obtained when LF-ChIP was performed using TAP-tagged Pta1 (Fig. 7C). These results (1) provide independent experimental evidence for DNA looping at the SEN1 gene; (2) refine the mCCC mapping of the juxtaposed regions to the promoter and terminator; and (3) demonstrate that the Ssu72 and Pta1 components of the CPF complex are components of the loop.
Discussion
The results presented in this paper demonstrate that DNA looping occurs at the BUD3 and SEN1 genes of yeast. Three lines of evidence argue that loop formation is a physiological consequence of transcription and not due simply to DNA bending or cross-linking of distal nucleosomes. First, looping is transcription-dependent. Global inhibition of RNAP II transcription dramatically diminished loop formation, and activation of GAL1p-BUD3 and GAL1p-SEN1 stimulated loop formation. Second, looping is dependent upon the Ssu72 and Pta1 components of the pre-mRNA 3′-end processing machinery. Cellular depletion of either of these components of the CPF complex, or inactivation of the Ssu72 CTD phosphatase, diminished loop formation. These are direct effects of the CPF complex on looping, and not indirect effects due to impaired transcription, as demonstrated (1) by retention of total poly(A) RNA in the ssu72-td strain following the 37°C temperature shift; and (2) by earlier transcription run-on experiments showing that neither Ssu72 inactivation (ssu72-2) nor depletion (ssu72-td) diminished the density of elongating RNAP II on a CYC1 reporter gene (Dichtl et al. 2002; He et al. 2003). Third, looping is due to interaction of the promoter and terminator regions of BUD3 and SEN1. mCCC and LF-ChIP mapped the DNA regions responsible for looping to within ∼500 bp of the SEN1 promoter and terminator regions. Although our evidence for gene looping is limited to the BUD3 and SEN1 genes, which were chosen for this analysis simply because of the technical advantages offered by these exceptionally long genes, we do not think that loop formation is a characteristic unique to long genes. Rather, we propose that gene looping is widespread and facilitates transcription by RNAP II.
Our results are generally consistent with those of O'Sullivan et al. (2004), although they observed DNA looping at the GALp-FMP27 gene under noninducing (raffinose) conditions, and not under repressing (glucose) or inducing (galactose) conditions. However, they also reported different results under inducing conditions depending upon the cross-linking method. In our experiments, we consistently observed looping at BUD3 and SEN1 under transcriptionally permissive conditions and at GAL1p-BUD3 and GAL1p-SEN1 under inducing conditions. Based on these results, and the CPF requirement for looping, we do not believe that loop formation is associated with early transcriptional activation, but instead we favor a model in which juxtaposition of the terminator and promoter regions requires an initial round of transcription.
Our working model (Fig. 8) is that promoter-terminator interaction facilitates transcription reinitiation by RNAP II following a pioneer round of transcription on a linear DNA template. Accordingly, the CPF complex would not only catalyze 3′-end processing, but would mediate loop formation via physical contact with components of the initiation complex. Consistent with this model, direct protein-protein contact between Ssu72 and TFIIB has already been demonstrated (Wu et al. 1999; Dichtl et al. 2002). We propose that CPF interacts with the “scaffold” that is retained at the promoter during the transcription cycle (Yudkovsky et al. 2000). Conceivably, juxtaposition of the terminator with the scaffold might allow RNAP II to be translocated from the terminator to the promoter, thereby accounting for the higher rates of transcription reinitiation relative to de novo initiation (Yudkovsky et al. 2000). We envision that DNA looping is a transient and dynamic process facilitated by promoter-terminator contacts during termination and is released upon promoter clearance. Ssu72 would play a key role in this process (1) by bridging the interaction between CPF and the scaffold via direct contact with TFIIB; and (2) by catalyzing dephosphorylation of Ser5-P of the CTD, thereby converting the elongation-competent IIO form of RNAP II to the initiation-competent IIA form. Consistent with this model, (1) the RNAP II CTD is hyperphosphorylated on Ser5 at both the promoter and terminator region of the GAL1p-FMP27 gene, but not within the ORF (O'Sullivan et al. 2004); and (2) Ssu72 is specific for Ser5-P dephosphorylation in vivo and in vitro (Krishnamurthy et al. 2004; Hausmann et al. 2005).
Figure 8.
A model for CPF-mediated promoter-terminator interactions. We propose that transient DNA looping occurs following a pioneer round of transcription and facilitates transcription reinitiation by RNAP II (see Discussion). Physical interaction between the promoter (P) and terminator (T) regions would be mediated, in part, by direct contact between Ssu72 and TFIIB. (Black line) Double-stranded DNA; (green lines) mRNA.
A prediction of our model is that inactivation of Ssu72 would block loop formation and impair translocation of RNAP II from terminator to promoter. If this were the case, then RNAP II should be depleted near the promoter and accumulate near the terminator in an ssu72 mutant. This is indeed the case. A time-course experiment using the ssu72-2 mutant revealed a pronounced (fivefold) increase in RNAP II density over the terminator-proximal region of CYC1 relative to the promoter-proximal region following inactivation of Ssu72 (Dichtl et al. 2002). Moreover, RNAP II density diminished over the promoter-proximal region, with no increase in RNAP II density downstream of the terminator. Although the authors interpreted their results to mean that ssu72-2 affects elongation (Dichtl et al. 2002), we suggest instead that ssu72-2 impairs reinitiation rather than elongation, resulting in terminator-proximal accumulation of RNAP II.
Although there are no reports of direct transfer of RNAP II from the terminator to the promoter of the same gene in vitro, there is evidence for such “facilitated recycling” in the case of RNAP III (Dieci and Sentenac 1996). Direct translocation of RNAP III from terminator to promoter bypasses several steps in the de novo assembly of the PIC, leading to accelerated reinitiation (Dieci and Sentenac 1996). Although the mechanism by which RNAP III recycling occurs has not been defined, it might involve DNA looping between the terminator and the stable TFIIIB-DNA promoter complex, in a manner analogous to the juxtaposition of the promoter-terminator regions described here for RNAP II genes.
Transcription-dependent loop formation and facilitated recycling of RNAP II makes a number of predictions. For one, direct transfer of RNAP II from the terminator to the scaffold should enhance the efficiency of transcription. This is already borne out for the Gal1p-driven BUD3 and SEN1 genes, in which we observed marked enhancement of looping associated with transcriptional induction (Fig. 3B). It will now be interesting to see whether terminator-dependent transcription is more efficient than run-off transcription in vitro. Standard RNAP II in vitro transcription systems are terminator-independent, based on run-off transcription from a linear DNA template. We anticipate that in a terminator-dependent system that is proficient for both transcription and 3′-end processing, a higher yield of transcripts will be generated than from a run-off template. Although poly(A) site-dependent termination has been demonstrated in vitro (Hyman and Moore 1993; Yonaha and Proudfoot 2000; Park et al. 2004), to our knowledge there are no reports of terminator-dependent effects on transcription initiation. A second prediction of our model is that 3′-end processing and termination factors should occupy both promoter and terminator regions. Indeed, several polyadenylation factors have already been found at the promoter (Calvo and Manley 2003). Finally, components of the 3′-end processing and termination machineries might affect transcription initiation. This is borne out by the ssu72-1 allele that genetically interacts with TFIIB and affects start site selection in vivo (Sun and Hampsey 1996). It will now be interesting to see if other genetic interactions between components of the transcription initiation and 3′-end processing machineries can be uncovered.
Materials and methods
Yeast strains and plasmids
The rpb1-1 (RS420) (Nonet et al. 1987), ssu72-td (XH-24) (He et al. 2003), pta1-td (XH-23) (Krishnamurthy et al. 2004), and FY23 (Winston et al. 1995) strains were described previously. Isogenic wild-type (KMY1170) and sug1-1 (KMY1171) strains were obtained from Kiran Madura (UMDNJ). Strains YMH867 and YMH926 were constructed by integrating the GAL1 promoter upstream of BUD3 and SEN1 genes, respectively, by one-step PCR-based chromosome modification (Longtine et al. 1998), using W303-1A as the host strain. Plasmids pM712 (SSU72+) and pM698 (ssu72-C15S), which encodes the catalytically inactive form of the Ssu72 phosphatase, were described previously (Sun and Hampsey 1996; Krishnamurthy et al. 2004).
mCCC assay
The interaction of promoter and terminator regions during transcription was analyzed using mCCC methodology. This technique was originally described for analyzing long-range chromosomal interactions in yeast (Dekker et al. 2002). To detect short-range interactions between promoter and terminator regions during transcription, the mCCC assay was modified as follows. Since transcription-dependent conformational changes are transient, formaldehyde cross-linking was performed in actively growing cells and not with isolated nuclei. Cells were grown in appropriate medium to A600∼0.7-1.0 and cross-linked with 1% formaldehyde for 10-15 min at 25°C. The reaction was stopped by addition of glycine to 125 mM, and cultures were incubated for an additional 5 min at room temperature. The cell pellet obtained from the 25-mL culture was washed with 10 mL of 1× TBS buffer (10 mM Tris-HCl at pH 7.5, 200 mM NaCl) containing 1% Triton X-100 and resuspended in 400 μL of FA-lysis buffer (50 mM HEPES-KOH at pH 7.9, 140 mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 1 mM PMSF). Approximately 400 μL of acid-washed glass beads were added, and cells were lysed by vigorous shaking for 40 min at 4°C. Cell lysates were collected by puncturing the bottom of the tube with a 22-gauge needle and collecting the filtrate in a 15-mL tube. The filtrate was transferred into a 1.5-mL microfuge tube and spun for 15 min at 4°C in a microfuge. The crude chromatin pellet was washed with 500 μL of FA-lysis buffer and resuspended in 500 μL of chromatin digestion buffer (10 mM Tris-HCl at pH 7.9, 5 mM MgCl2, and 0.1% Triton X-100).
Cross-linked, crude chromatin preparations were digested with a restriction endonuclease (XhoI or HindIII) that cuts at least once within the coding sequence and at sites flanking the promoter and terminator regions, as shown in Figure 1A. Typically, 10 μL of chromatin preparation was digested with 10 μL of restriction enzyme in a 100-μL reaction volume. The restriction digestion was carried out for 90 min with occasional mixing. The reaction was stopped by adding 10 μL of 10% SDS and incubating for 20 min at 65°C. To sequester SDS and allow the subsequent ligation reaction, 75 μL of 10% Triton X-100 was added, followed by 7.5× dilution to a final volume of 750 μL. Ligation reactions were performed for 1 h at 25°C using Quick Ligase (New England Biolabs). The cross-links were reversed for 8 h at 65°C in the presence 100 μg proteinase K (Roche). To ensure complete removal of RNA, 10 μg of DNase-free RNase (Roche) was added to the reaction mixture, and the incubation was carried out for 15 min at 37°C. The samples were extracted with phenol-chloroform three times and ethanol precipitated in the presence of glycogen. DNA concentration was determined spectrophotometrically, and 250 ng of DNA was used as the template in each PCR reaction.
PCR reactions (30 cycles) to detect transcription-induced chromatin looping were performed using divergent primers P1 and T1, as shown in Figure 1A. The BUD3 gene spans coordinates 96280-101190 on chromosome III; the primers P1 and T1 hybridized to regions upstream and downstream of the BUD3 coding region near coordinates 90781 and 118302, respectively. SEN1 spans coordinates 993430-1000125 on chromosome XII; the divergent primers P1 and T1 hybridized to coordinates 993414 and 1000410, respectively. Control PCR reactions were carried out using the same template DNA with primers corresponding to a chromosome V intergenic region. PCR products were fractionated on a 1.5% agarose gel and visualized by ethidium bromide staining using an AlphaImager 2000.
Mapping the interacting regions of SEN1 by mCCC analysis
The juxtaposed regions of SEN1 were mapped by mCCC analysis with the following modifications. The chromatin pellet prepared from 50 mL of formaldehyde cross-linked culture was resuspended in 400 μL of 20 mM Tris-HCl (pH 7.5). In each reaction, 80 μL of the chromatin preparation was used for restriction digestion and subsequent ligation. HindIII restriction digestions were performed in 100-μL reaction volume for 5 h with gentle shaking at 37°C. Reactions were clarified by centrifugation for 5 min at room temperature, and the supernatant fraction was used for subsequent analyses. Ligation and reversal of cross-links were carried out as described in the mCCC assay. Following deproteinization, PCR reactions were carried out using 250 ng of DNA and the primer pairs shown in Figure 6. To confirm the compatibility of primer pairs, yeast genomic DNA was digested with HindIII and ligated as described previously (Dekker et al. 2002). Following deproteinization and ethanol precipitation of DNA, PCR was performed with all primer pairs used in the mapping analysis.
Ligation-facilitated ChIP
Cross-linking and isolation of chromatin was performed as described above. In this case, chromatin preparations were sonicated 12 times for 30 sec with 5 min cooling after each round of sonication. Sonication was performed at the 30% duty cycle in a Branson digital sonifier. Following sonication, samples were spun at 14,000 rpm in a refrigerated microfuge. The supernatant was digested with HindIII, followed by end fill-in using T4 DNA polymerase for 15 min at 25°C. Enzymes were inactivated by adding SDS to a final concentration of 1% and incubating the reaction mixture for 20 min at 65°C. The reaction was diluted twofold using ligation buffer, and Triton X-100 was added to a 1.5% final concentration. DNA was ligated for 1 h at 25°C using Quick Ligase. Approximately 10 μg of affinity-purified anti-Ssu72 polyclonal antibody was added to the ligated chromatin preparation and allowed to bind for 4-12 h at 4°C with gentle shaking. The antigen-antibody complex was adsorbed on 100 μL of Protein A-Sepharose beads and washed successively with 1 mL each of FA-lysis buffer, FA-lysis buffer containing 500 mM NaCl, ChIP wash buffer (10 mM Tris-HCl at pH 8.0, 250 mM LiCl2, 0.5% Nonidet-P40, 0.5% sodium deoxycholate, and 1 mM EDTA), and TE buffer. The beads were resuspended in 100 μL TE and incubated with 10 μg of DNase-free RNase, 20 μg proteinase K, and 2.5 μL of 10% SDS for 15 min at 37°C. Cross-links were reversed by overnight incubation at 65°C. Samples were extracted with phenol-chloroform at least two times followed by ethanol precipitation of DNA in the presence of glycogen. DNA pellets were resuspended in 50 μL TE and used as template for further PCR reaction. The first PCR was performed using 5 μL of immunoprecipitated DNA as template and primer pairs P2 and T2, which hybridize upstream and downstream of the SEN1 promoter and terminator sequences, as depicted in Figure 7. The second PCR was performed using the product from the first PCR as template and primer pairs A, B, C, D, E, and F, which hybridize to the indicated regions of SEN1. PCR products were resolved in a 1.5% agarose gel and visualized as described above.
Acknowledgments
We are grateful to Claire Moore (Tufts University Medical School), Marc Gartenberg, and Danny Reinberg for helpful advice during the course of this work and for critical reading of the manuscript; to B.N. Singh for RNA analyses; to Meredith Prysak, Mariela Reyes, Krishnamurthy Shankarling, and B.N. Singh for many fruitful discussions; and to Marc Gartenberg for suggesting the term ligation-facilitated ChIP. This work was supported by NIH grants GM-39484 and GM-68887.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1362305.
References
- Bentley D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336-342. [DOI] [PubMed] [Google Scholar]
- Calvo O. and Manley, J.L. 2003. Strange bedfellows: Polyadenylation factors at the promoter. Genes & Dev. 17: 1321-1327. [DOI] [PubMed] [Google Scholar]
- Cho E.J., Kobor, M.S., Kim, M., Greenblatt, J., and Buratowski, S. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 15: 3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe, K., Dekker, M., and Kleckner, N. 2002. Capturing chromosome conformation. Science 295: 1306-1311. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank, D., Ohnacker, M., Friedlein, A., Roeder, D., Langen, H., and Keller, W. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10: 1139-1150. [DOI] [PubMed] [Google Scholar]
- Dieci G. and Sentenac, A. 1996. Facilitated recycling pathway for RNA polymerase III. Cell 84: 245-252. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 28: 202-209. [DOI] [PubMed] [Google Scholar]
- Ganem C., Devaux, F., Torchet, C., Jacq, C., Quevillon-Cheruel, S., Labesse, G., Facca, C., and Faye, G. 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22: 1588-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11: 394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M. and Reinberg, D. 2003. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell 113: 429-432. [DOI] [PubMed] [Google Scholar]
- Hausmann S., Koiwa, H., Krishnamurthy, S., Hampsey, M., and Shuman, S. 2005. Different strategies for CTD recognition by serine5-specific CTD phosphatases. J. Biol. Chem. 280: 37681-37688. [DOI] [PubMed] [Google Scholar]
- He X., Khan, A.U., Cheng, H., Pappas Jr., D.L., Hampsey, M., and Moore, C.L. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes & Dev. 17: 1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L.E. and Moore, C.L. 1993. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol. Cell. Biol. 13: 5159-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor M.S. and Greenblatt, J. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577: 261-275. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S., He, X., Reyes-Reyes, M., Moore, C., and Hampsey, M. 2004. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell 14: 387-394. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- Nedea E., He, X., Kim, M., Pootoolal, J., Zhong, G., Canadien, V., Hughes, T., Buratowski, S., Moore, C.L., and Greenblatt, J. 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278: 33000-33010. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe, C., Sexton, J., and Young, R. 1987. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7: 1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J.M., Tan-Wong, S.M., Morillon, A., Lee, B., Coles, J., Mellor, J., and Proudfoot, N.J. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 36: 1014-1018. [DOI] [PubMed] [Google Scholar]
- Park S.J., Tsao, D.C., and Martinson, H.G. 2004. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol. Cell. Biol. 24: 4092-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16: 272-278. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Russell S.J., Sathyanarayana, U.G., and Johnston, S.A. 1996. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J. Biol. Chem. 271: 32810-32817. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J. and Brow, D.A. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23: 6339-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.-W. and Hampsey, M. 1996. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 16: 1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard, C., and Ricuperohovasse, S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53-55. [DOI] [PubMed] [Google Scholar]
- Woychik N.A. and Hampsey, M. 2002. The RNA polymerase II machinery: Structure illuminates function. Cell 108: 453-463. [DOI] [PubMed] [Google Scholar]
- Wu W.-H., Pinto, I., Chen, B.-S., and Hampsey, M. 1999. Mutational analysis of yeast TFIIB: A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M. and Proudfoot, N.J. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19: 3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish, J.A., and Hahn, S. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225-229. [DOI] [PubMed] [Google Scholar]