Abstract
The stepwise loss of cohesins, the complexes that hold sister chromatids together, is required for faithful meiotic chromosome segregation. Cohesins are removed from chromosome arms during meiosis I but are maintained around centromeres until meiosis II. Here we show that Sgo1, a protein required for protecting centromeric cohesins from removal during meiosis I, localizes to cohesin-associated regions (CARs) at the centromere and the 50-kb region surrounding it. Establishment of this Sgo1-binding domain requires the 120-base-pair (bp) core centromere, the kinetochore component Bub1, and the meiosis-specific factor Spo13. Interestingly, cohesins and the kinetochore proteins Iml3 and Chl4 are necessary for Sgo1 to associate with pericentric regions but less so for Sgo1 to associate with the core centromeric regions. Finally, we show that the 50-kb Sgo1-binding domain is the chromosomal region where cohesins are protected from removal during meiosis I. Our results identify the portions of chromosomes where cohesins are protected from removal during meiosis I and show that kinetochore components and cohesins themselves are required to establish this cohesin protective domain.
Keywords: Centromere, cohesion, meiosis, pericentromere, Rec8, Sgo1
The meiotic cell division cycle is a specialized cell cycle that creates haploid gametes in sexually reproducing organisms. In contrast to the mitotic cell divisions, where DNA replication and chromosome segregation alternate, two consecutive segregation phases follow a single round of DNA replication during the meiotic cell division. Homologous chromosomes segregate from each other during meiosis I, and sister chromatids are partitioned during meiosis II.
Protein complexes known as cohesins play an important role in both mitotic and meiotic chromosome segregation (for review, see Nasmyth 2001). They hold sister chromatids together until the onset of chromosome segregation. In the budding yeast Saccharomyces cerevisiae, the mitotic cohesin complex consists of Smc1, Smc3, Scc1/Mcd1, and Scc3 and associates with chromosomes in a nonuniform manner with peaks of binding enriched at kinetochores, AT-rich sequences, and convergent intergenic regions (Glynn et al. 2004; Lengronne et al. 2004; Weber et al. 2004). The removal of cohesin complexes from chromosomes marks the onset of anaphase and requires the cleavage of the Scc1/Mcd1 subunit by the protease Separase (for review, see Nasmyth 2001).
Meiotic cohesin complexes in yeast are also composed of Smc1, Smc3, and Scc3, but the Scc1/Mcd1 subunit is replaced by the meiosis-specific variant Rec8 (Klein et al. 1999). The distribution of meiotic cohesins along chromosomes is similar to that of mitotic cohesion complexes (Glynn et al. 2004); however, an important difference exists between the ways in which mitotic and meiotic cohesins are removed from chromosomes. Whereas during mitosis cohesins are removed from chromosomes along their entire length at the metaphase–anaphase transition, cohesins are lost from meiotic chromosomes in a step-wise manner. At the metaphase I–anaphase I transition, cohesins are removed from chromosome arms but are maintained around centromeres until anaphase II. Loss of cohesins from chromosome arms is essential for the resolution of chiasmata that hold homologous chromosomes together on the metaphase I spindle (Buonomo et al. 2000). Maintenance of cohesins around centromeres beyond anaphase I and their loss at the metaphase II–anaphase II transition are required for the faithful segregation of sister chromatids during meiosis II.
We know of several factors responsible for maintaining cohesins around centromeres beyond meiosis I. The first factor implicated in this process was the Drosophila protein MEI-S332 (Kerrebrock et al. 1992, 1995; Tang et al. 1998). Subsequent studies in fission and budding yeast, metazoans as well as plants, identified homologs of MEI-S332, termed Sgo1 (Katis et al. 2004a; Kitajima et al. 2004; Marston et al. 2004; Rabitsch et al. 2004; Salic et al. 2004; McGuinness et al. 2005). In both fission and budding yeast, deletion of SGO1 results in premature loss of Rec8 from centromeric regions during anaphase I and a random meiosis II chromosome segregation pattern. The MEI-S332 proteins themselves localize to centromeric regions (Kerrebrock et al. 1995; Katis et al. 2004a; Kitajima et al. 2004; Marston et al. 2004; Rabitsch et al. 2004; Salic et al. 2004; McGuinness et al. 2005), which requires, in at least fission yeast and humans, the kinetochore and spindle checkpoint component Bub1 (Kitajima et al. 2004, 2005; Tang et al. 2004). Consistent with this role in regulating Sgo1 localization, cells lacking BUB1 lose centromeric cohesion prematurely (Bernard et al. 2001; Kitajima et al. 2004, 2005). Another factor required for protecting cohesins from removal around centromeres during meiosis I is the meiosis-specific protein Spo13 (Klein et al. 1999; Lee et al. 2002; Katis et al. 2004b; B.H. Lee et al. 2004). Spo13 is also necessary for the co-orientation of sister kinetochores, that is, the attaching of sister kinetochores to microtubules emanating from the same spindle pole in meiosis I (Katis et al. 2004b; B.H. Lee et al. 2004).
Immunolocalization studies of cohesins on anaphase I chromosomes show that cohesins are protected from removal during meiosis I at regions overlapping with centromeres (Klein et al. 1999; Watanabe and Nurse 1999; Lee et al. 2003). Where exactly on chromosomes cohesins are protected, however, is not known in any organism. In eukaryotes in which the core centromere is flanked by extensive repeated DNA elements that organize the pericentric heterochromatin, there is evidence to suggest that cohesins are protected from removal during meiosis I at these sites. For example, in Schizosaccharomyces pombe, proteins required to protect cohesins from being removed during meiosis I localize to the pericentric heterochromatic regions and appear excluded from the core centromere (Kitajima et al. 2004). Cytological studies in the fruit fly Drosophila melanogaster and mouse spermatocytes also suggest that cohesins are protected in regions that flank the core centromere (Kerrebrock et al. 1995; Moore et al. 1998; Blower and Karpen 2001; Lee et al. 2003). Budding yeast centromeres differ greatly from those of other eukaryotes. They are composed of a 120-base-pair (bp) conserved DNA sequence but lack flanking repeat elements. Whether protection of cohesins during meiosis I occurs only at the minimal centromere or whether it is maintained pericentrically in order to ensure functional cohesion between sister chromatids until anaphase II is not known.
Here we show that budding yeast Sgo1 localizes only to cohesin-associated regions (CARs) within a 50-kb region surrounding centromeres previously shown to exhibit enhanced cohesin association during mitosis (Weber et al. 2004). The association of Sgo1 with the core centromere and pericentric CARs requires the 120-bp core centromere as well as the spindle checkpoint protein Bub1 and the meiosis-specific factor Spo13. Binding of Sgo1 to pericentric CARs depends on cohesins themselves as well as the kinetochore components Iml3 and Chl4. Finally, we show that the CARs within the 50-kb pericentric region where Sgo1 associates are identical to the CARs where cohesins are protected from removal during meiosis I. Our studies define for the first time at the molecular level where on chromosomes cohesins are protected from removal during meiosis I and show that in budding yeast Spo13, the kinetochore components Bub1, Chl4, and Iml3 and cohesin itself function through Sgo1 to establish a chromatin domain where cohesin removal is prevented.
Results
Sgo1 localizes to cohesin-associated sites within a 50-kb region surrounding the centromere during meiosis I
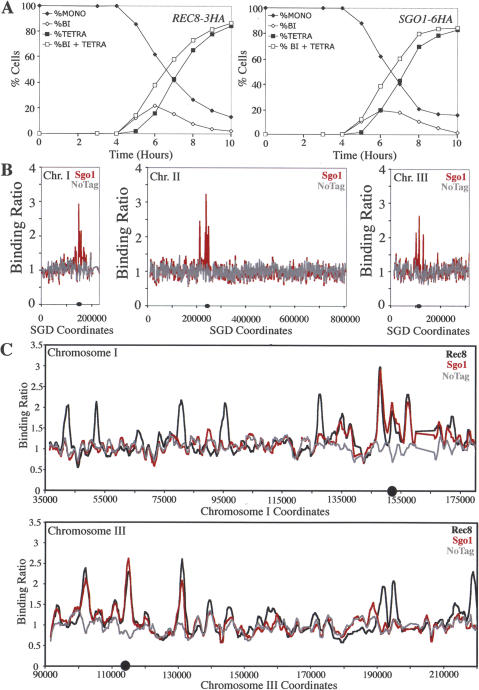
To obtain a molecular understanding of the chromosomal location where cohesins are protected from removal during meiosis I, we first compared the distribution of the cohesin protector Sgo1 with that of the cohesin subunit Rec8 using genome-wide location analysis. Shortly prior to the onset of the first meiotic division (5 h after transfer into sporulation medium) (Fig. 1A; see Materials and Methods), Rec8 was enriched at many regions along the arms of all 16 chromosomes and showed increased association with pericentric regions (Fig. 1C; Supplementary Fig. 1), which matched with previously published genome-wide mapping of cohesins during mitosis (Glynn et al. 2004; Lengronne et al. 2004; Weber et al. 2004).
Figure 1.
Sgo1 localizes to centromeric and pericentric cohesin-associated regions. (A) Wild-type diploid strains carrying REC8-3HA (A4758) and SGO1-6HA (A12282) fusions as well as a strain lacking them (A4962) were sporulated (see Materials and Methods). The percentage of mononucleated (closed diamonds), binucleated (open diamonds), and tetranucleated (closed squares), as well as the sum of binucleated and tetranucleated (open squares) was determined at the indicated time points for strains A4758 and A12282. Two-hundred cells were counted per time point. The data shown represent the average of two different cultures used for the genome-wide location analysis shown in B and C. (B) The binding ratios of immunoprecipitated Sgo1-6HA for chromosomes I, II, and III are shown. Values for a strain carrying the SGO1-6HA allele are shown in red, and those for a strain lacking tagged Sgo1 are shown in gray. The X-axis shows SGD coordinates for each chromosome. (C) Binding ratios for the REC8-3HA strain (black), the SGO1-6HA strain (red), and an untagged strain (No Tag, gray) around the centromere and arms of chromosomes I and III are shown. Shown in particular are the regions where the transition occurs from Sgo1 and Rec8 binding overlapping to a region where this is no longer the case. The gap to the right of the centromere of chromosome I represents a Ty element.
Using the same genome-wide location analysis, Sgo1 was found enriched around centromeres on all 16 chromosomes (Fig. 1B,C; Supplementary Fig. 2). Importantly, Sgo1 was not enriched at all sequences around centromeres. Instead, a comparison of the Sgo1 distribution with that of Rec8 revealed that the Sgo1- and Rec8-binding regions overlapped within a roughly 50-kb region surrounding the centromere but that this similarity in distribution was lost farther away from the centromere (Fig. 1C; Supplementary Figs. 3, 4). Our results indicate that Sgo1 localizes to the same sites as cohesins within a 50-kb domain surrounding the centromere.
Sgo1 associates with pericentric regions in mitotic cells
Although Sgo1 does not prevent cohesin removal during mitosis, the presence of the protein at pericentric regions until metaphase raised the possibility that Sgo1 affected the distribution of cohesins. However, we found that in cells lacking SGO1 that were arrested in metaphase by depleting cells of Cdc20, Scc1/Mcd1 association was not affected (Supplementary Fig. 5), indicating that in contrast to mammalian cells (McGuinness et al. 2005), Sgo1 does not affect cohesins during the mitotic divisions in budding yeast.
To determine whether differences in Sgo1 binding to chromosomes were responsible for the different behavior of Sgo1 in mitosis and meiosis, we compared the distribution of Sgo1 on chromosomes between mitotic and meiotic cells. Haploid yeast strains were arrested in metaphase using the microtubule depolymerizing drug nocodazole, and the Sgo1 distribution around the centromere of chromosome III was assessed by chromatin immunoprecipitation (ChIP) using primer sets based on our meiotic Sgo1 genome-wide location analysis (Supplementary Fig. 6A). Sgo1 localized to both centromeres and previously published regions of mitotic cohesin complex enrichment but failed to localize to negative control regions on the arm of chromosome III (Supplementary Fig. 6B,C). This distribution was qualitatively similar to the Sgo1 distribution observed in meiotic cells (Supplementary Fig. 6D). In diploid SK1 cells arrested in metaphase I by depleting Cdc20 (Lee and Amon 2003), Sgo1 was found at pericentric regions but not at arm regions. We note that cells are arrested in metaphase in these experiments and thus Sgo1 distribution could be affected. However, the meiotic distribution of Sgo1 in cells depleted for Cdc20 appeared similar to that obtained by location analysis in a synchronous meiosis (Fig. 1C; Supplementary Fig. 2), suggesting that changes in distribution due to arresting cells are minor, if they exist at all. Our results suggest that the distribution of Sgo1 at pericentric regions is qualitatively similar between metaphase I and nocodazole-treated mitotic cells, both conditions in which sister kinetochores are not under tension. Thus, it is likely that mechanisms other than Sgo1 distribution on chromosomes must be responsible for the different behavior of the protein during mitosis and meiosis I.
Sgo1 association with centromeric and pericentric regions depends on Bub1
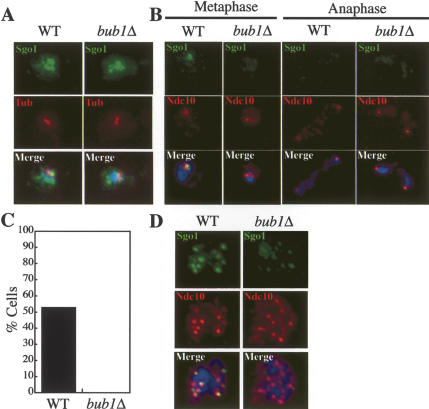
Next we examined which factors were necessary for Sgo1 to associate with centromeric and pericentric regions. The spindle checkpoint and kinetochore component Bub1 is required for the association of Sgo1 with chromosomes in fission yeast and mammalian cells (Kitajima et al. 2004, 2005; Tang et al. 2004). We found that Bub1 was also required for Sgo1 localization in budding yeast. Sgo1 accumulated in the nucleus in cells lacking BUB1, indicating that the protein was stable (Fig. 2A). The protein, however, failed to associate with kinetochores as judged by the lack of colocalization of Sgo1 with the kinetochore component Ndc10 on spread nuclei of mitotic and meiotic cells (Fig. 2B–D).
Figure 2.
BUB1 is required for Sgo1 localization during both mitosis and meiosis. (A–C) Exponentially growing wild-type cells (A11738) and cells deleted for BUB1 (A12634) carrying SGO1-9MYC and NDC10-6HA fusions were harvested to determine the localization of Sgo1-9Myc and Ndc10-6HA by whole-cell immunofluorescence (A) and on spread nuclei (B). Sgo1 is shown in green, tubulin (Tub) in red, Ndc10 in red, and DNA stained by DAPI (4′,6-diamidino-2-phenolindole) in blue. (C) The percentage of mononucleated cells from the experiment in B in which an Sgo1 signal colocalized with an Ndc10 signal. One-hundred mononucleated cells were counted for each strain. (D) Wild-type (A13179) and bub1Δ (A13177) cells carrying an SGO1-9MYC and NDC10-6HA fusion were induced to sporulate. After 6 h, samples were taken to determine the localization of Sgo1 (green) and Ndc10 (red) on spread nuclei. DNA stained by DAPI is shown in blue.
Spo13 is required for full association of Sgo1 with centromeric and pericentric regions
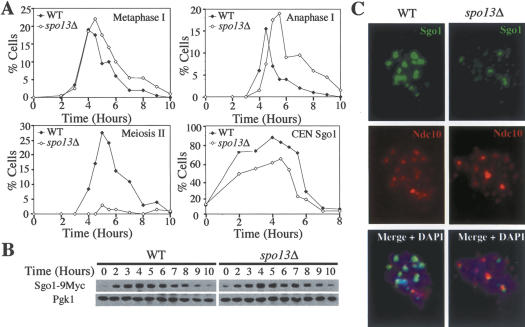
Spo13, which localizes to pericentric regions, is required to protect cohesins from removal during meiosis I (Klein et al. 1999; Katis et al. 2004b; B.H. Lee et al. 2004). Previous immunolocalization studies suggested that Sgo1 could associate with centromeric regions in the absence of SPO13, although it was noted that the Sgo1 signal was less intense (B.H. Lee et al. 2004). We confirmed that the Sgo1 signal on chromosome spreads was reduced in spo13Δ cells (Fig. 3A,C) and excluded the possibility that this decrease in Sgo1 association with centromeric regions was due to Sgo1 protein levels being lower in spo13Δ cells (Fig. 3B).
Figure 3.
SPO13 is required for full association of Sgo1 with chromosomes. (A–C) Wild-type (A10461, closed diamonds) and spo13Δ (A10755, open diamonds) cells carrying an SGO1-9MYC and NDC10-6HA fusion were induced to sporulate. The percentage of cells with metaphase I spindles (A, Metaphase I), anaphase I spindles (A, Anaphase I), and meiosis II spindles (A, Meiosis II); the percentage of cells with Sgo1 associated with centromeric regions (A, CEN Sgo1); and the amount of Sgo1 protein (B) were determined at the indicated time points. Pgk1 was used as a loading control in Western blots. (C) An example of Sgo1 localization on nuclear spreads of wild-type and spo13Δ cells. Sgo1 is shown in green, Ndc10 in red, and DNA in blue.
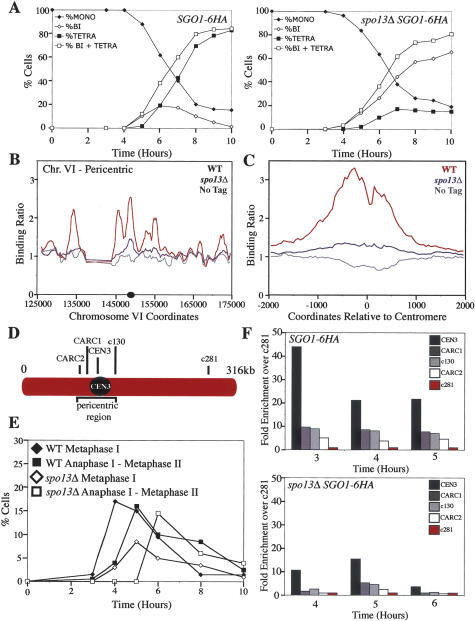
The decrease in Sgo1 localization to centromeric regions in spo13Δ cells was not only obvious on chromosome spreads but also when we analyzed Sgo1 binding along chromosomes using genome-wide location analysis. Wild-type cells and cells lacking SPO13 were induced to sporulate, and the distribution of Sgo1 along chromosomes was analyzed as cells entered the first meiotic division, 5 h after transfer of cells to meiosis-inducing conditions (Fig. 4A). Sgo1 binding at centromeric and pericentric regions was dramatically reduced in cells lacking SPO13 at all 16 chromosomes (Fig. 4B). The Sgo1-6HA signal was nevertheless above that detected in cells lacking a tagged version of Sgo1 (Fig. 4B), which was particularly obvious when the centromeric regions of all 16 chromosomes were analyzed together (Fig. 4C). We conclude that Spo13 is required for Sgo1 to associate with centromeric and pericentric regions and note that the low-level association of Sgo1 in spo13Δ mutants by genome-wide location analysis indicates that the cytological analysis likely overestimated that amount of Sgo1 associated with regions surrounding the centromere.
Figure 4.
SPO13 maintains Sgo1 association with chromosomes. (A–C) Wild-type (A12282; WT) and spo13Δ (A11967; spo13Δ) cells carrying an SGO1-6HA fusion, as well as a strain lacking the fusion (A4962; No Tag) were sporulated. (A) The percentage of mononucleated (closed diamonds), binucleated (open diamonds), and tetranucleated (closed squares), as well as the sum of binucleated and tetranucleated (open squares) was determined at the indicated time points for strains A12282 and A11967. The data shown represent the average of two different cultures used for the genome-wide location analysis experiments. (B) The binding ratios for Sgo1-6HA within a 50-kb region surrounding the centromere of chromosome VI 5 h after transfer into sporulation medium. (C) The binding ratios for Sgo1-6HA averaged across all 16 centromeres. The X-axis shows SGD coordinates relative to the centromere for each chromosome, taking into account centromere orientation. (D) Primer sets corresponding to CARs adjacent to the centromere of chromosome III (CEN3, CARC1, c130, CARC2) and a negative control region on the arm of chromosome III (c281). (E,F) Samples were taken for ChIP from wild-type (WT; A12282) and spo13Δ (A11967) cells. (E) Progression through meiosis as the percentage of wild-type metaphase I (closed diamonds), wild-type anaphase I–metaphase II (closed squares), spo13Δ metaphase I (open diamonds), and spo13Δ anaphase I–metaphase II (open squares) spindles. (F) The fold enrichment for sequences relative to a negative control sequence (c281) at the indicated time points as determined by semiquantitative PCR. Note that the strain deleted for SPO13 is delayed 1 h in entering meiosis.
Spo13 was not only necessary for the initial loading of Sgo1 onto centromeric regions but also appeared to be required for maintaining Sgo1 at centromeres. Sgo1 association with chromosomes diminished over time as judged by immunolocalization studies and ChIP analysis (Figs. 3A,C, 4D–F). This is reminiscent of the effects of Spo13 on the kinetochore-orientation factor Mam1. For this factor, too, it was found that Spo13 was required for its maintenance at kinetochores (Katis et al. 2004b; B.H. Lee et al. 2004). Our results indicate that Spo13 is essential for a wild-type level association of Sgo1 with both pericentric and centromeric regions. We furthermore note that the reduced localization, but not complete absence, of Sgo1 in spo13Δ cells could also explain the observation that not all Rec8 is lost prematurely in spo13Δ cells and that a small fraction of Rec8 persists around centromeres into anaphase I (Klein et al. 1999; Katis et al. 2004b; B.H. Lee et al. 2004).
Association of Sgo1 with pericentric sites requires cohesins and the kinetochore proteins Iml3 and Chl4
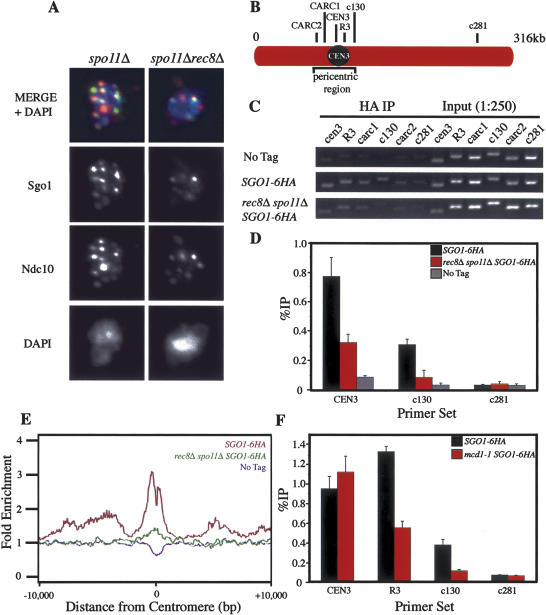
Which other factors are necessary for Sgo1 association with chromosomes? Owing to the observation that cohesin and Sgo1 physically interact in S. pombe (Kitajima et al. 2004), we considered the possibility that cohesins themselves were required for the association of Sgo1 with the core centromere and pericentric CARs. To test this, we first examined Sgo1 localization on chromosome spreads of meiotic cells lacking the cohesin subunit Rec8 that were also deleted for SPO11, which allows rec8Δ cells to progress beyond prophase I (Klein et al. 1999). Sgo1 localized to centromeric regions as judged by immunolocalization studies on chromosome spreads, although colocalization was poor and the signal was less intense than in a control strain (Fig. 5A). ChIP analysis revealed that Sgo1 association with the core centromere was reduced in rec8Δ spo11Δ cells (Fig. 5C,D). At a pericentric region the Sgo1 signal was even further reduced and only marginally above background (the signal seen at c281) (Fig. 5C,D). Similar results were obtained in a genome-wide location analysis, which was particularly apparent when all 16 chromosomes were analyzed together (Fig. 5E). Sgo1 was present at reduced levels at the core centromere and absent from pericentric regions. Cohesin was also required for Sgo1 to associate with pericentric regions during mitosis. Temperature-sensitive mcd1-1 cells were synchronized in G1 and then released into medium containing nocodazole at the restrictive temperature of 37°C. As in meiotic cells, Sgo1 associated with the core centromere but failed to efficiently associate with a pericentric CAR (c130) in the absence of functional cohesins (Fig. 5F). Our results suggest that Sgo1 associates with the core centromere in part in a cohesin-independent manner, as has been suggested in higher eukaryotes (Kitajima et al. 2004; J.Y. Lee et al. 2004), but cohesins appear critical for association of Sgo1 with pericentric CARs.
Figure 5.
The Sgo1 distribution is altered in rec8Δ and mcd1-1 cells. (A) spo11Δ (A10593) and rec8Δ spo11Δ (A11233) diploid strains carrying SGO1-9MYC and NDC10-6HA fusions were sporulated to examine Sgo1 localization by chromosome spreads. Sgo1 is shown in green, Ndc10 in red, and DNA in blue. (B–D) Wild-type (A12282) and spo11Δ rec8Δ (A13726) cells carrying the SGO1-6HA fusion were sporulated along with a strain lacking the tagged protein (A4962). (B) The location of primers used for PCR analysis. c281 is used as a negative control sequence. Primer set R3 amplifies a region ∼800 bp to the right of the core centromere. (C) PCR analysis of ChIP samples harvested 5 h after transfer into sporulation medium. (D) The percent immunoprecipitation (%IP) calculated as the percent of immunoprecipitated DNA signal returned in IP fractions. Immunoprecipitations were performed in triplicate from a single cell lysate. Error bars indicate the standard error of the mean. (E) The binding ratios for Sgo1-6HA averaged across all 16 centromeres in wild-type (A12282; red), spo11Δ rec8Δ (A13726; green), and a strain lacking the fusion (A4962; No Tag, blue) 5 h after transfer into sporulation medium. (F) Wild-type (A10652) and mcd1-1 (A13773) cells carrying the SGO1-6HA tag were arrested in G1 by addition of α-factor pheromone. Cells were released into fresh growth medium at 37°C in the presence of nocodazole. Samples were taken for ChIP after 2 h. Shown is the percent immunoprecipitation (%IP) calculated as the percent of immunoprecipitated DNA signal returned in IP fractions. Immunoprecipitations were performed in triplicate from a single cell lysate. Error bars indicate the standard error of the mean.
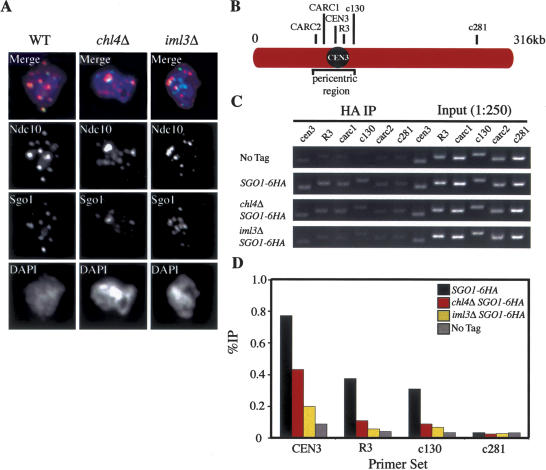
Factors required for Sgo1 localization could be proteins identified in a screen of the yeast knockout collection for genes required for maintaining cohesion around centromeres beyond anaphase I (Marston et al. 2004). Two such proteins are the kinetochore components Iml3 and Chl4. Cells carrying deletions in either gene lose cohesins around centromeres prematurely during meiosis I (Marston et al. 2004). Analysis of the localization of Sgo1 on chromosome spreads revealed that, like in cells lacking REC8, Sgo1 localized to centromeric regions poorly and many Sgo1 foci failed to colocalize with Ndc10 in the mutants (Fig. 6A). ChIP analysis revealed that Sgo1 association with the core centromere was reduced in iml3Δ and chl4Δ cells (Fig. 6C,D). At pericentric regions, the Sgo1 signal was only marginally above background (the signal seen at site c281) (Fig. 6C,D). Interestingly, iml3Δ cells exhibited a more severe defect than chl4Δ cells, matching with their reduced ability to protect cohesin beyond metaphase I (Marston et al. 2004). Our results indicate that cohesins, Iml3, and Chl4 are important for Sgo1 to associate with pericentric CARs, but the proteins are not essential for, although they contribute to, the association of Sgo1 with the core centromere.
Figure 6.
IML3 and CHL4 are required for Sgo1 to associate with pericentric CARs. (A) Wild-type (A10461), chl4Δ (A10629), and iml3Δ (A10628) diploid strains carrying SGO1-9MYC and NDC10-6HA fusions were sporulated to examine Sgo1 localization by chromosome spreads. Sgo1 is shown in green, Ndc10 in red, and DNA in blue. (B–D) Wild-type (A12282), chl4Δ (A13970), and iml3Δ (A13971) cells carrying the SGO1-6HA fusion were sporulated along with a strain lacking the tagged protein (A4962). (B) The location of primers used for ChIP analysis. c281 is used as a negative control sequence. Primer set R3 amplifies a region ∼800 bp to the right of the core centromere. (C) PCR analysis of ChIP samples harvested 4 h (A4962, A12282) or 5 h (A13970, A13971) after transfer into sporulation medium such that cells would be enriched for a population just prior to metaphase. Note that chl4Δ and iml3Δ cells are delayed 1 h in entering meiosis. (D) The percent immunoprecipitation (%IP) calculated as the percent of immunoprecipitated DNA signal returned in IP fractions. Each experiment was performed at least twice, and the average percent immunoprecipitation is shown.
The core centromere is sufficient to target Sgo1 to adjacent CARs
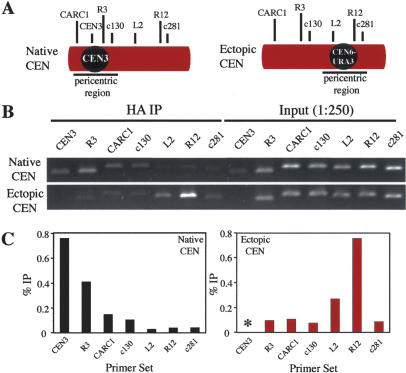
The requirement for the kinetochore components Iml3 and Chl4 in Sgo1 localization to pericentric sites raised the interesting possibility that the 120-bp centromere and proteins that associate with it function as a seed to establish a Sgo1 domain. To test this idea, we integrated a copy of the chromosome VI centromere at the right arm of chromosome III at the TRX3 locus and simultaneously deleted the native centromere of this chromosome. Introduction of CEN6 led to the association of Sgo1 with previously identified CARs (Weber et al. 2004) flanking the neo-centromere (Fig. 7B). For example, Sgo1 association is most highly enriched at the CAR amplified by the R12 primer set but remains low at the negative control region (c281) despite its location within the pericentromere (Fig. 7). Additionally, Sgo1 was no longer found at CARs flanking the region where the native CEN3 was deleted (Fig. 7B,C). These findings not only demonstrate that the core centromere is sufficient to establish an Sgo1-binding domain around itself but also show that the centromere directs Sgo1 specifically to adjacent CARs and not any specific DNA sequences flanking the native centromere.
Figure 7.
The 120-bp core centromere is sufficient to establish an Sgo1-binding domain around itself. A wild-type strain (A10652) and a strain carrying an ectopic centromere on the arm of chromosome III (A13806) and each carrying an SGO1-6HA fusion were arrested in G1 with α-factor pheromone. Cells were released into medium lacking the pheromone but containing the microtubule depolymerizing drug nocodazole (15 μg/mL). Samples were taken for ChIP 2 h after release. (A) Primer sets corresponding to CARs along chromosome III including adjacent to the native centromere (CEN3, R3, CARC1, c130), ectopic centromere (L2, R12), and a negative control region (c281). L2 is ∼2 kb away from CEN6-URA3 and R12 is ∼12 kb to the right. c281 is ∼20 kb to the right of the ectopic centromere. PCR reactions were performed using the immunoprecipitated DNA as well as input DNA for the wild-type strain and the ectopic centromere strain in B, and are quantified in C. An asterisk is shown in the place of CEN3 for the ectopic centromere strain as PCR amplification confirmed deletion of the locus.
Rec8 is protected from removal during meiosis I within the 50-kb Sgo1-binding region surrounding the centromere
The finding that Sgo1 localizes to CARs within a 50-kb domain surrounding the centromere suggests but does not prove that this is the chromosomal region where Rec8 is protected from removal during meiosis I. To determine where on chromosomes Rec8 persists until the onset of anaphase II, we compared the distribution of Rec8 on chromosomes between cell populations enriched for metaphase I cells, in which cohesins are present along the entire length of the chromosomes, and populations enriched for metaphase II cells, in which cohesins are present only around centromeres. This comparison has not been made before because progression through meiosis is too asynchronous to allow for the isolation of cell populations enriched in these particular cell cycle stages. To circumvent this limitation, we constructed strains that arrest in metaphase II by expressing a nondegradable version of the anaphase inhibitor Pds1 (Pds1dbΔ) (Cohen-Fix et al. 1996; Shonn et al. 2000) under the control of the meiosis II-specific SPS1 promoter (pSPS1-PDS1dbΔ) (see Materials and Methods). This construct drives PDS1dbΔ expression predominantly during meiosis II and acts in a dominant fashion to delay cells in metaphase II as judged by the analysis of Rec8 distribution. Ten hours after transfer into sporulation medium, 50% of the cohesin-containing cells exhibited cohesin localization at centromeric regions, and 25% of cells exhibited a metaphase II spindle morphology (data not shown) (note that this finding also demonstrates that degradation of Pds1 is required for the metaphase II–anaphase II transition). Thus, in a ChIP analysis that analyzes the distribution of cohesins in the pSPS1-PDS1dbΔ strains, we expect an enrichment of cohesins present around centromeres at later time points than at earlier time points when the majority of cells have not yet reached anaphase I.
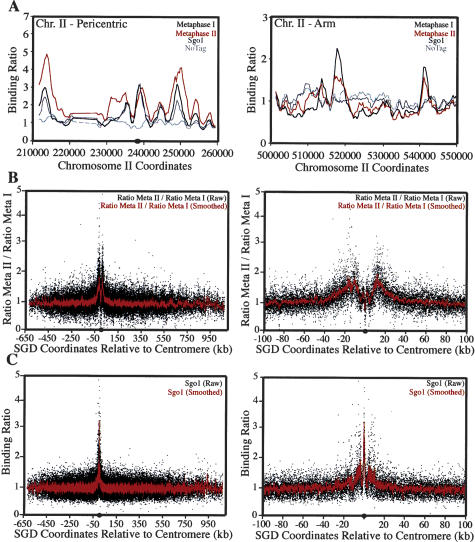
Analysis of the immunoprecipitated fractions using genome-wide location analysis (Fig. 8A,B) revealed a higher fold enrichment at centromeric and pericentric regions in PDS1dbΔ-expressing cells enriched for metaphase II cells (Fig. 8A–C). Five hours after transfer into sporulation medium when the majority of pSPS1-PDS1dbΔ cells were in a cell cycle stage prior to anaphase I, Rec8 was found to be distributed along the length of the chromosomes, with an average binding ratio of 3 at centromeric and pericentric regions and 2 at chromosome arm locations (Fig. 8). In contrast, 10 h after transfer into sporulation medium, when 50% of pSPS1-PDS1dbΔ cells were in cell cycle stages post-anaphase I, Rec8 showed binding ratios of 4–5 at centromeric and pericentric CARs but only 1.5 at chromosomal arm CARs (Fig. 8A). The pericentric enrichment of cohesins beyond anaphase I was particularly striking when all 16 chromosomes were analyzed together, thus creating an artificial “metachromosome” (Fig. 8B,C). This analysis indicates that cohesins are enriched in an ∼50-kb region surrounding each centromere (Fig. 8B). Sgo1 localized to similar regions of the chromosome (Fig. 8C), although in contrast to Rec8, its association with chromosomes decreased with distance from the centromere (Figs. 1C, 8C; Supplementary Figs. 3, 4). Very similar results were obtained using ChIP analysis of regions along chromosome III (Supplementary Fig. 7). Rec8 was enriched at centromeric and pericentric regions in PDS1dbΔ-expressing cells compared with Cdc20-depleted cells (Supplementary Fig. 7). We conclude that the 50-kb domain surrounding the centromere is not only the region where Sgo1 binds but also the region where cohesins are protected from removal during meiosis I.
Figure 8.
Cohesins are protected from removal during meiosis I within the 50-kb pericentromere. A strain carrying the pSPS1-PDS1dbΔ allele (A10008) was sporulated in duplicate. Samples were taken after 5 and 10 h for ChIP. (A) The binding ratios of immunoprecipitated Rec8-3HA for the pericentromere and an arm region of chromosome II. Binding ratios for the samples taken at 5 h are shown (black) as well as those taken at 10 h (red). Binding ratios for Sgo1 (dark gray) taken from the experiment in Figure 1 are also shown in addition to binding ratios for a strain lacking the tag (light gray). (B) The ratio of the binding ratio for REC8-3HA at 10 h (metaphase II-enriched cells) to the binding ratio at 5 h (metaphase I-enriched cells) for all 16 chromosomes with SGD coordinates relative to the centromere of each chromosome. Regions where the ratio of these binding ratios is high indicate portions of the genome that are more enriched for cohesins at 10 h. Raw data (Ratio Meta I/Ratio Meta II, black) are shown along with a smoothed line (Ratio Meta I/Ratio Meta II, red) created by averaging data over a moving 20-point window. A full “metachromosome” is shown (left) in addition to a version including only the 200 kb surrounding the centromere (right). (C) The binding ratio for SGO1-6HA for all 16 chromosomes with SGD coordinates relative to the centromere of each chromosome. The data are identical to that represented in Supplementary Figure 3. Raw data (black) are shown along with a smoothed line created by averaging data over a moving 10-point window. A full “metachromosome” is shown (left) in addition to a version including only the 200 kb surrounding the centromere (right).
Discussion
Inhibiting cohesin removal around centromeres during meiosis I is essential for accurate chromosome segregation during meiosis. The family of MEI-S332 proteins, of which S. cerevisiae Sgo1 is a member, is required for this process. Here we show that Sgo1 binds to CARs within a 50-kb region around centromeres that coincides with the chromosomal region where cohesins are protected from removal during meiosis I. Establishment of this Sgo1-binding domain requires the 120-bp core centromere, Bub1, and Spo13. Interestingly, whereas Bub1 and Spo13 are required for Sgo1 association with centromeric and pericentric regions, cohesins and the kinetochore proteins Iml3 and Chl4 are necessary for Sgo1 to associate with pericentric regions, but Sgo1 can load to some extent onto the core centromere in their absence, suggesting a multistep mechanism for Sgo1 to associate with chromosomes.
Defining centromeric cohesion at a molecular level
Cytological studies in many organisms have shown that sister chromatid cohesion is not removed at regions overlapping with centromeres during meiosis I (for review, see Miyazaki and Orr-Weaver 1994). Until now, however, we lacked a molecular understanding of the nature of this region in any organism. In eukaryotes other than budding yeast, the mapping of cohesins around centromeres using ChIP was not possible because the regions flanking the centromere are highly repetitive. Budding yeast pericentric regions are not repetitive, making this organism highly suitable for ChIP analysis. However, meiotic cell cycle progression is too asynchronous to obtain cell populations enriched for cells in which cohesins are solely present around centromeres. The generation of cells that express an anaphase inhibitor specifically during meiosis II enabled us to isolate cell populations enriched for cells in such cell cycle stages. Thus, we were able to determine that cohesins are protected from removal during meiosis I within an ∼50-kb region surrounding the centromere, which corresponds to the previously identified cohesin-rich domain in mitotic cells (Weber et al. 2004). Why is the region where cohesins are protected 50 kb long? During meiosis II, as during mitosis, sister kinetochores are under tension. The force exerted by microtubules can, at least during mitosis, separate sister chromatids in a domain of ∼20 kb around centromeres (He et al. 2000; Tanaka et al. 2000). Thus a 50-kb domain would be sufficiently large to resist the pulling force exerted by microtubules.
Our genome-wide location analysis of cohesins revealed the distribution of cohesins around centromeres to be dynamic as cells progress through meiosis. The peaks close to the centromere become broader in the cultures enriched for metaphase II cells (Fig. 8A; Supplementary Fig. 8), resembling more the distribution of cohesins during mitosis rather than that in metaphase I (Supplementary Fig. 8). This finding raises the interesting possibility that the cohesin distribution is being influenced by microtubule-dependent pulling forces exerted on sister kinetochores. During meiosis I, sister kinetochores are not under tension and cohesin peaks are less broad than in mitotic and meiosis II cells, when tension is exerted on sister kinetochores.
Sgo1 localizes to centromeric and pericentric regions in mitosis but does not protect cohesins
Sgo1 binding to chromosomes correlates directly with Rec8 binding, suggesting a direct role for Sgo1 in preventing cohesin removal. However, Sgo1 protects cohesins from removal around centromeres only during meiosis I but not during mitosis. Two scenarios or a combination thereof could be envisioned to explain this difference. Sgo1 could fail to associate with pericentric CARs and thus fail to prevent removal of cohesins from these sites. Alternatively, Sgo1 may not be present on chromosomes at the time when cohesin cleavage occurs. We observe little if any qualitative difference between the distribution of Sgo1 on mitotic metaphase and metaphase I chromosomes, suggesting that distribution differences do not account for the differential behavior of Sgo1 in mitosis and meiosis I. It is likely that the ability of Sgo1 to protect cohesins during meiosis I but not during mitosis requires its own protection from degradation by a ubiquitylation machinery defined by its ligase, the anaphase-promoting complex (APC) or Cyclosome (C), which targets proteins for degradation at the metaphase–anaphase transition (for review, see Harper et al. 2002).
The centromere directs Sgo1 localization at a distance
Our data show that Sgo1 association with chromosomes is not specific to the 50-kb region surrounding the centromere, but rather appears to be defined by proximity to the 120-bp core centromere. The core centromere is sufficient to direct Sgo1 to adjacent sequences, which is consistent with findings in Drosophila that MEI-S332 localization is determined by a functional centromere (Lopez et al. 2000). Remarkably, however, the centromere does not direct Sgo1 to any adjacent sequences but specifically to CARs. As the core centromere is also capable of establishing a cohesin-rich domain around itself (Weber et al. 2004) and given that Sgo1 associates with CARs, it is likely that Sgo1 associates directly or indirectly with pericentric cohesin complexes rather than binding to specific DNA sequences adjacent to the core centromere. Consistent with this idea is the finding that in fission yeast, Sgo1 is found in a complex with Rec8 (Kitajima et al. 2004). From our ChIP analyses it furthermore appears that Sgo1 levels decrease as the distance from the core centromere increases, raising the interesting possibility that Sgo1 “spreads,” perhaps in a cohesin-dependent manner, from the core centromere to pericentric regions.
The finding that the core centromere is capable of directing proteins such as cohesins and Sgo1 to adjacent sites could also provide an explanation for the observation that neo-centromeres are functional not only during mitosis and meiosis II but also during meiosis I. Several human neo-centromeres have been described that lack any obvious pericentric heterochromatin (Aagaard et al. 2000; Saffery et al. 2000; Amor and Choo 2002). The ability of the core centromere to establish a cohesin-rich and Sgo1-binding domain around itself would provide an explanation as to why these neo-centromeres are functional.
Association of Sgo1 with chromosomes
Our findings identified several proteins important for Sgo1 to associate with chromosomes. Bub1 and Spo13 are required for the association of Sgo1 with the core centromere and pericentric regions. Rec8, Chl4, and Iml3 are necessary for Sgo1 to associate with pericentric sites but less so for Sgo1 binding to centromeric sites.
The requirement for Bub1 in Sgo1 localization appears to be conserved across species (Kitajima et al. 2004, 2005; Tang et al. 2004). Whether Bub1 binds to centromeric and pericentric sites to localize Sgo1 or whether the protein functions as an Sgo1 “receptor” at the kinetochore and, together with other kinetochore proteins such as Iml3 and Chl4, directs Sgo1 to adjacent CARs is not yet known. Similarly, whether Bub1's role in localizing Sgo1 is connected to the protein's function in the spindle assembly checkpoint remains to be determined.
Spo13 is also important for Sgo1 to associate with both centromeric and pericentric sites. However, in contrast to Bub1, the role of Spo13 in regulating Sgo1 and cohesin removal appears more complex. Chromosome-wide location analysis of Spo13 in budding yeast has shown the protein to predominantly localize to pericentric regions during meiosis I (Katis et al. 2004b), putting Spo13 in the right place at the right time to affect Sgo1 localization. Genetic interactions between SPO13 and components of the APC/C-dependent ubiquitylation machinery suggest that Spo13's function in regulating Sgo1 is to protect the protein from degradation during meiosis I (Katis et al. 2004b). This idea is consistent with our observation that in spo13Δ cells, Sgo1 association with centromeric and pericentric regions diminishes over time. Spo13, however, not only regulates Sgo1 localization to prevent premature loss of Sgo1 from chromosomes but also functions to regulate chromosome segregation in an Sgo1-independent manner. Overexpression of SPO13 in mitotic cells causes a cell cycle arrest in metaphase (Lee et al. 2002; Shonn et al. 2002). This arrest is independent of SGO1 (B.H. Lee et al. 2004), indicating that Spo13 not only functions upstream of Sgo1 but also in parallel to regulate the metaphase–anaphase transition.
In cells lacking IML3, CHL4, or REC8, Sgo1 association with chromosomes is partially disrupted, consistent with the observation that random meiosis II segregation of chromosome V is seen in only a fraction of iml3Δ and chl4Δ cells (Marston et al. 2004). The association of Sgo1 with the core centromere is reduced in cells lacking IML3, CHL4, or REC8. The effects of deleting these genes on Sgo1 at pericentric regions was more dramatic. Sgo1 levels were reduced to almost background levels (and background levels in the case of the rec8Δ) at pericentric sites. These results suggest that Sgo1 binding to the core centromere is only in part dependent on these factors but is completely dependent at pericentric CARs. It is possible that cohesin, Iml3, and Chl4 decrease Sgo1 loading onto chromosomes overall. We favor the idea that the partial dependence of Sgo1 localization on IML3, CHL4, and REC8 to the core centromere reflects two modes of association of Sgo1 with this genomic region. Sgo1 binds to CARs near the core centromere in a REC8-, IML3-, and CHL4-dependent manner and in addition to the core centromere in a REC8-, IML3-, and CHL4-independent manner. At pericentric sites, Sgo1 binding is solely dependent on cohesins and ILM3 and CHL4. How these factors collaborate to regulate Sgo1 localization is not known. It is possible that Iml3 and Chl4 promote the association of Sgo1 with cohesins at pericentric CARs. It is also possible that Iml3 and Chl4 affect cohesins, thereby preventing Sgo1 from associating efficiently with chromosomes.
In S. pombe and Drosophila, association of Sgo1 with chromosomes is cohesin-independent (Kitajima et al. 2004; J.Y. Lee et al. 2004). In contrast, in maize Sgo1 association with regions surrounding the centromere requires Rec8 (Hamant et al. 2005). Our studies of factors regulating Sgo1 localization suggest that in S. cerevisae, Sgo1 localization to chromosomes is both cohesin-dependent and cohesin-independent, which reflects two steps in the assembly of Sgo1 onto chromosomes. These two steps could reflect different modes of association of Sgo1 with the core centromere and pericentric CARs. Alternatively, it is possible that the core centromere and kinetochore proteins function as a seed for Sgo1 association with chromosomes. From there, the protein spreads to pericentric CARs, which is mediated by the kinetochore proteins Iml3 and Chl4 and cohesins and perhaps involves sliding of cohesins. Consistent with this model is the finding that the 120-bp core centromere is sufficient to direct Sgo1 to a 50-kb domain around itself. It is tempting to speculate that the core centromere functions as an epigenetic organizer of chromosome segregation. It establishes—in an epigenetic manner—a chromosome domain around itself that is essential for the accurate segregation of the chromosome. On a sequence level, the organization of the S. cerevisiae centromere differs dramatically from that of other eukaryotes, in that budding yeast lacks extensive repeated heterochromatic DNA elements (Bernard et al. 2001; Kitajima et al. 2004, 2005; Tang et al. 2004). Our studies suggest that on a functional level, S. cerevisiae centromeres may not be that different from those of other eukaryotes after all. Budding yeast centromeres are also surrounded by a large chromatin domain that binds proteins essential for accurate chromosome segregation.
Materials and methods
Strains and plasmids
The strains used in this study are described in Supplementary Table 1 and were derivatives of SK1 unless otherwise noted. The pCLB2-CDC20 fusion is described in Lee and Amon (2003). The pSPS1-PDS1dbΔ construct was generated by cloning the SPS1 promoter upstream of PDS1 lacking the destruction box (PDS1dbΔ) (Cohen-Fix et al. 1996; Shonn et al. 2000). The construct was integrated at the PDS1 locus while maintaining an intact copy of wild-type PDS1. bub1Δ::KanMX6 was created by a one-step PCR-based gene replacement method (Longtine et al. 1998). sgo1Δ::KanMX6, SGO1-9MYC, and SGO1-6HA were described in Marston et al. (2004). NDC10-6HA and rec8Δ::KanMX4 were described in Toth et al. (2000). REC8-3HA, spo13::hisG, and spo11::URA3 were described in Klein et al. (1999). The strain carrying an ectopic centromere was created by a dual transformation that included the integration of CEN6-URA3 at the TRX3 locus on the arm of chromosome III (Weber et al. 2004) and deletion of the endogenous CEN3 in a strain carrying the SGO1-6HA fusion.
Sporulation conditions
Cells were grown to saturation in YPD (YEP + 2% glucose) for 24 h, diluted into YPA (YEP + 2% KAc) at OD600 = 0.3, and grown overnight. Cells were then washed with water and resuspended in SPO medium (0.3% KAc at pH 7.0) at OD600 = 1.9 at 30°C to induce sporulation.
Genome-wide location analysis
Genome-wide location analyses were performed in duplicate as described in Pokholok et al. (2005). The yeast array (Agilent) contains >41,000 probes designed against the entire yeast genome. In total, the probes cover ∼12 Mb of the yeast genome (or 85%). Most of the missing regions (represented by flat lines in the graphs) are telomeric or other highly repetitive regions. For genomic regions that are covered, there is a probe approximately every 266 bp. Binding ratios represent the ratio of signal between differentially labeled immunoprecipitated and input fractions and were normalized such that the median binding ratio for each data set equals unity. For the Sgo1 location analysis in Figures 1 and 3, input DNA fractions from the wild-type (WT) and spo13Δ samples were mixed and split prior to labeling to allow for a normalization to compare IP signals. For the Rec8 location analysis in Figure 8, input DNA fractions from the 5- and 10-h samples were mixed and split prior to labeling to allow for a normalization to compare IP signals. All data, except the data shown in Figure 8, are shown after smoothing by calculating a moving average across five data points. Complete data sets for all experiments are available upon request.
The raw data for the genome-wide location analysis shown in Figures 1, 4, and 8, and Supplementary Figures 1, 2, 3, 4, and 8 are included as an Excel file named “Raw Data” in the Supplemental Material. The data presented therein represent median-normalized binding rations after error analysis performed on duplicate experiments as described in Pokholok et al. (2005). Strain numbers and chromosomal coordinates are indicated.
To calculate the percent enrichment of Rec8 at pericentric regions in cell populations enriched for metaphase II cells (pSPS1-PDS1dbΔ cells at the 10-h time point), we first divided each binding ratio of Rec8 at 10 h by the binding ratio at 5 h to obtain a ratio of the binding ratios. This number quantifies the enrichment for a given feature in metaphase II-enriched cells. We then compared this ratio of the binding ratios at pericentric regions (50 kb around each centromere) and at arm regions (the rest of the chromosome). By dividing the value at pericentromeres and arms, we obtained an average value for the fold enrichment at pericentromeres at 10 h. The average percent increase in enrichment for all chromosomes was 46.3% ± 6.75%.
ChIP
ChIPs were performed as described in B.H. Lee et al. (2004). An internal control product was amplified using a primer set that amplified an ∼500-bp region of the nonreplicative vector pRS306 (Sikorski and Hieter 1989). Sequences of primers are shown in Supplementary Table 2.
Whole-cell immunofluorescence
Indirect in situ immunofluorescence was carried out as described in Visintin et al. (1998). Rat anti-tubulin antibodies (Oxford Biotechnology) and anti-rat FITC antibodies (Jackson Immunoresearch) were used at a 1:100 dilution. Sgo1-9Myc was detected using a mouse anti-Myc antibody (BabCO) at a 1:2000 dilution and an anti-mouse Cy3 secondary antibody (Jackson Immunoresearch) at a 1:2000 dilution.
Immunolocalization analysis on chromosome spreads.
Chromosomes were spread as described in Nairz and Klein (1997). Sgo1-9Myc was detected using rabbit anti-Myc antibodies (Gramsch) at a 1:150 dilution and anti-rabbit FITC antibodies (Jackson Immunoresearch) at a 1:300 dilution. Ndc10-6HA was detected using a mouse anti-HA antibody (BabCO) at a 1:200 dilution and an anti-mouse Cy3 antibody at a 1:300 dilution. Quantification of Sgo1 at centromeres was performed as described in B.H. Lee et al. (2004).
Western blot analysis
Cells were harvested, incubated in 5% trichloroacetic acid (TCA), and lysed as described in Moll et al. (1991). Immunoblots were performed as described in Cohen-Fix et al. (1996). Sgo1-9Myc was detected using a mouse anti-Myc antibody (BabCO) at a 1:1000 dilution. Pgk1 was detected using a mouse anti-PGK1 antibody (Molecular Probes) at a 1:5000 dilution. The secondary antibody used was a goat anti-mouse antibody conjugated to horseradish peroxidase (HRP; Jackson Immunoresearch) at a 1:5000 dilution.
Supplementary Material
Acknowledgments
We are grateful to Daniel Wells for making strain A13806. We thank D. Koshland and Hong-Guo Yu for advice on ChIP and communication of results prior to publication, and Terry Orr-Weaver and members of the Amon laboratory for their critical reading of the manuscript. This research was supported by NIH Grants GM62207 (to A.A.) and GM6213 (to P.M.). A.A. is also an investigator of the Howard Hughes Medical Institute.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1373005.
Supplemental material is available at http://www.genesdev.org.
References
- Aagaard L., Schmid, M., Warburton, P., and Jenuwein, T. 2000. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 113: 817-829. [DOI] [PubMed] [Google Scholar]
- Amor D.J. and Choo, K.H. 2002. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71: 695-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure, J.F., and Javerzat, J.P. 2001. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat. Cell Biol. 3: 522-526. [DOI] [PubMed] [Google Scholar]
- Blower M.D. and Karpen, G.H. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo S.B., Clyne, R.K., Fuchs, J., Loidl, J., Uhlmann, F., and Nasmyth, K. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387-398. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters, J.M., Kirschner, M.W., and Koshland, D. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 10: 3081-3093. [DOI] [PubMed] [Google Scholar]
- Glynn E.F., Megee, P.C., Yu, H.G., Mistrot, C., Unal, E., Koshland, D.E., DeRisi, J.L., and Gerton, J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Golubovskaya, I., Meeley, R., Fiume, E., Timofejeva, L., Schleiffer, A., Nasmyth, K., and Cande, W.Z. 2005. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 15: 948-954. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Burton, J.L., and Solomon, M.J. 2002. The anaphase-promoting complex: It's not just for mitosis any more. Genes & Dev. 16: 2179-2206. [DOI] [PubMed] [Google Scholar]
- He X., Asthana, S., and Sorger, P.K. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101: 763-775. [DOI] [PubMed] [Google Scholar]
- Katis V.L., Galova, M., Rabitsch, K.P., Gregan, J., and Nasmyth, K. 2004a. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 14: 560-572. [DOI] [PubMed] [Google Scholar]
- Katis V.L., Matos, J., Mori, S., Shirahige, K., Zachariae, W., and Nasmyth, K. 2004b. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr. Biol. 14: 2183-2196. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W., Miyazaki, W.Y., Birnby, D., and Orr-Weaver, T.L. 1992. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics 130: 827-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W., Moore, D.P., Wu, J.S., and Orr-Weaver, T.L. 1995. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 83: 247-256. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima, S.A., and Watanabe, Y. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510-517. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Hauf, S., Ohsugi, M., Yamamoto, T., and Watanabe, Y. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15: 353-359. [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91-103. [DOI] [PubMed] [Google Scholar]
- Lee B.H. and Amon, A. 2003. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300: 482-486. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Amon, A., and Prinz, S. 2002. Spo13 regulates cohesin cleavage. Genes & Dev. 16: 1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Iwai, T., Yokota, T., and Yamashita, M. 2003. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J. Cell Sci. 116: 2781-2790. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Kiburz, B.M., and Amon, A. 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr. Biol. 14: 2168-2182. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Dej, K.J., Lopez, J.M., and Orr-Weaver, T.L. 2004. Control of centromere localization of the MEI-S332 cohesion protection protein. Curr. Biol. 14: 1277-1283. [DOI] [PubMed] [Google Scholar]
- Lengronne A., Katou, Y., Mori, S., Yokobayashi, S., Kelly, G.P., Itoh, T., Watanabe, Y., Shirahige, K., and Uhlmann, F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- Lopez J.M., Karpen, G.H., and Orr-Weaver, T.L. 2000. Sister-chromatid cohesion via MEI-S332 and kinetochore assembly are separable functions of the Drosophila centromere. Curr. Biol. 10: 997-1000. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Tham, W.H., Shah, H., and Amon, A. 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303: 1367-1370. [DOI] [PubMed] [Google Scholar]
- McGuinness B.E., Hirota, T., Kudo, N.R., Peters, J.M., and Nasmyth, K. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki W.Y. and Orr-Weaver, T.L. 1994. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 28: 167-187. [DOI] [PubMed] [Google Scholar]
- Moll T., Tebb, G., Surana, U., Robitsch, H., and Nasmyth, K. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66: 743-758. [DOI] [PubMed] [Google Scholar]
- Moore D.P., Page, A.W., Tang, T.T., Kerrebrock, A.W., and Orr-Weaver, T.L. 1998. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J. Cell Biol. 140: 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz K. and Klein, F. 1997. mre11S—A yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes & Dev. 11: 2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35: 673-745. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison, C.T., Levine, S., Cole, M., Hannett, N.M., Lee, T.I., Bell, G.W., Walker, K., Rolfe, P.A., Herbolsheimer, E., et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517-527. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P., Gregan, J., Schleiffer, A., Javerzat, J.P., Eisenhaber, F., and Nasmyth, K. 2004. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 14: 287-301. [DOI] [PubMed] [Google Scholar]
- Saffery R., Irvine, D.V., Griffiths, B., Kalitsis, P., Wordeman, L., and Choo, K.H. 2000. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum. Mol. Genet. 9: 175-185. [DOI] [PubMed] [Google Scholar]
- Salic A., Waters, J.C., and Mitchison, T.J. 2004. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 118: 567-578. [DOI] [PubMed] [Google Scholar]
- Shonn M.A., McCarroll, R., and Murray, A.W. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289: 300-303. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes & Dev. 16: 1659-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter, P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs, J., Loidl, J., and Nasmyth, K. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2: 492-499. [DOI] [PubMed] [Google Scholar]
- Tang T.T., Bickel, S.E., Young, L.M., and Orr-Weaver, T.L. 1998. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes & Dev. 12: 3843-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Sun, Y., Harley, S.E., Zou, H., and Yu, H. 2004. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. 101: 18012-18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A., Rabitsch, K.P., Galova, M., Schleiffer, A., Buonomo, S.B., and Nasmyth, K. 2000. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis I. Cell 103: 1155-1168. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig, K., Hwang, E.S., Prinz, S., Tyers, M., and Amon, A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2: 709-718. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. and Nurse, P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461-464. [DOI] [PubMed] [Google Scholar]
- Weber S.A., Gerton, J.L., Polancic, J.E., DeRisi, J.L., Koshland, D., and Megee, P.C. 2004. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2: E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.